b9e2fdcc50d5274eb5238cf303b74d14.ppt
- Количество слайдов: 39
 Manual De Novo Peptide MS/MS Interpretation For Evaluating Database Search Results Karl R. Clauser Broad Institute of MIT and Harvard Cold Spring Harbor Proteomics Course July, 2010 Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 16 PM 1
Manual De Novo Peptide MS/MS Interpretation For Evaluating Database Search Results Karl R. Clauser Broad Institute of MIT and Harvard Cold Spring Harbor Proteomics Course July, 2010 Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 16 PM 1
 Outline • AA properties • Fragmentation pathways and ion types • b/y pairs • Fragment charge from mass defect • Non-mobile proton • Neutral loss ion types • Phosphosite ambiguity • Sample handling chemistry artifacts • Isobaric co-eluters • Mass tolerance units and isobaric AA’s • Other Tutorials • Dominant ions • AA adjacencies • Positions Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 16 PM 2
Outline • AA properties • Fragmentation pathways and ion types • b/y pairs • Fragment charge from mass defect • Non-mobile proton • Neutral loss ion types • Phosphosite ambiguity • Sample handling chemistry artifacts • Isobaric co-eluters • Mass tolerance units and isobaric AA’s • Other Tutorials • Dominant ions • AA adjacencies • Positions Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 16 PM 2
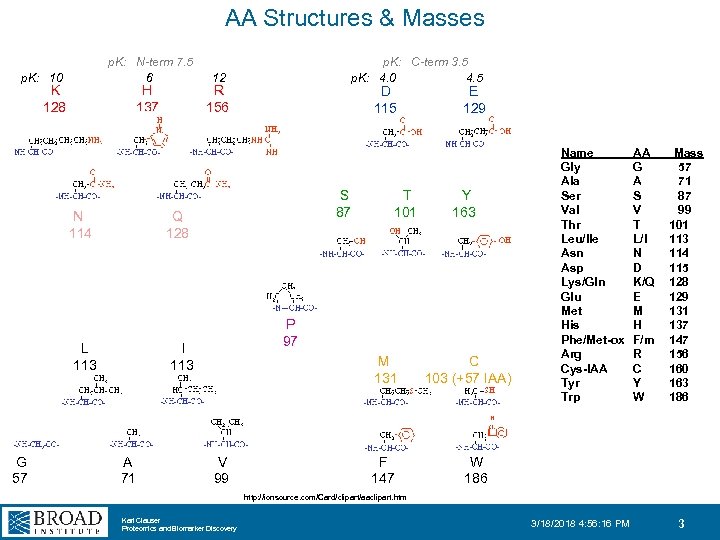 AA Structures & Masses p. K: 10 p. K: N-term 7. 5 6 p. K: C-term 3. 5 p. K: 4. 0 4. 5 12 K H R 128 137 156 D E 115 129 S T Y 87 101 163 N Q 114 128 L I 113 G A V 57 71 99 P 97 M C 131 103 (+57 IAA) Name Gly Ala Ser Val Thr Leu/Ile Asn Asp Lys/Gln Glu Met His Phe/Met-ox Arg Cys-IAA Tyr Trp AA G A S V T L/I N D K/Q E M H F/m R C Y W Mass 57 71 87 99 101 113 114 115 128 129 131 137 147 156 160 163 186 F W 147 186 http: //ionsource. com/Card/clipart/aaclipart. htm Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 16 PM 3
AA Structures & Masses p. K: 10 p. K: N-term 7. 5 6 p. K: C-term 3. 5 p. K: 4. 0 4. 5 12 K H R 128 137 156 D E 115 129 S T Y 87 101 163 N Q 114 128 L I 113 G A V 57 71 99 P 97 M C 131 103 (+57 IAA) Name Gly Ala Ser Val Thr Leu/Ile Asn Asp Lys/Gln Glu Met His Phe/Met-ox Arg Cys-IAA Tyr Trp AA G A S V T L/I N D K/Q E M H F/m R C Y W Mass 57 71 87 99 101 113 114 115 128 129 131 137 147 156 160 163 186 F W 147 186 http: //ionsource. com/Card/clipart/aaclipart. htm Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 16 PM 3
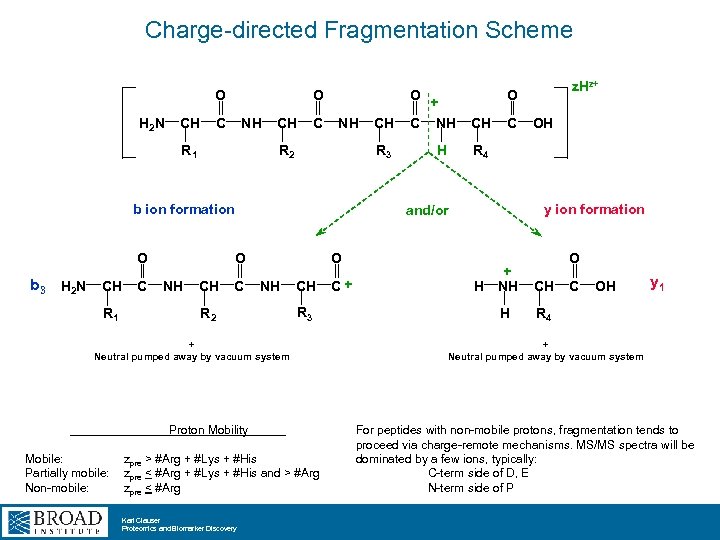 Charge-directed Fragmentation Scheme O H 2 N CH O C NH R 1 CH C NH R 2 R 3 b ion formation O O R 2 R 3 Proton Mobility zpre > #Arg + #Lys + #His zpre < #Arg + #Lys + #His and > #Arg zpre < #Arg Karl Clauser Proteomics and Biomarker Discovery CH C OH R 4 y ion formation O + Neutral pumped away by vacuum system Mobile: Partially mobile: Non-mobile: H z. Hz+ O and/or b 3 H 2 N CH C NH CH C + R 1 CH O + C NH O + NH CH H H R 4 C OH y 1 + Neutral pumped away by vacuum system For peptides with non-mobile protons, fragmentation tends to proceed via charge-remote mechanisms. MS/MS spectra will be dominated by a few ions, typically: C-term side of D, E N-term side of P
Charge-directed Fragmentation Scheme O H 2 N CH O C NH R 1 CH C NH R 2 R 3 b ion formation O O R 2 R 3 Proton Mobility zpre > #Arg + #Lys + #His zpre < #Arg + #Lys + #His and > #Arg zpre < #Arg Karl Clauser Proteomics and Biomarker Discovery CH C OH R 4 y ion formation O + Neutral pumped away by vacuum system Mobile: Partially mobile: Non-mobile: H z. Hz+ O and/or b 3 H 2 N CH C NH CH C + R 1 CH O + C NH O + NH CH H H R 4 C OH y 1 + Neutral pumped away by vacuum system For peptides with non-mobile protons, fragmentation tends to proceed via charge-remote mechanisms. MS/MS spectra will be dominated by a few ions, typically: C-term side of D, E N-term side of P
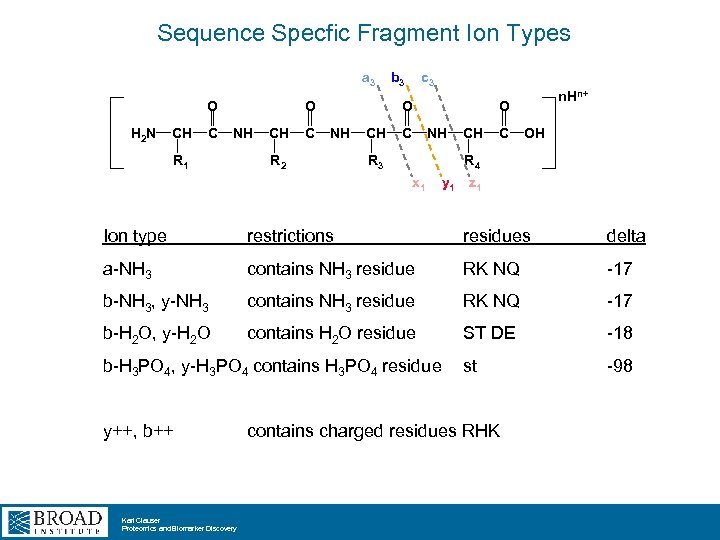 Sequence Specfic Fragment Ion Types a 3 O H 2 N CH C O NH R 1 CH C b 3 c 3 O NH R 2 CH C n. Hn+ O NH R 3 CH C OH R 4 x 1 y 1 z 1 Ion type restrictions residues delta a-NH 3 contains NH 3 residue RK NQ -17 b-NH 3, y-NH 3 contains NH 3 residue RK NQ -17 b-H 2 O, y-H 2 O contains H 2 O residue ST DE -18 st -98 b-H 3 PO 4, y-H 3 PO 4 contains H 3 PO 4 residue y++, b++ Karl Clauser Proteomics and Biomarker Discovery contains charged residues RHK
Sequence Specfic Fragment Ion Types a 3 O H 2 N CH C O NH R 1 CH C b 3 c 3 O NH R 2 CH C n. Hn+ O NH R 3 CH C OH R 4 x 1 y 1 z 1 Ion type restrictions residues delta a-NH 3 contains NH 3 residue RK NQ -17 b-NH 3, y-NH 3 contains NH 3 residue RK NQ -17 b-H 2 O, y-H 2 O contains H 2 O residue ST DE -18 st -98 b-H 3 PO 4, y-H 3 PO 4 contains H 3 PO 4 residue y++, b++ Karl Clauser Proteomics and Biomarker Discovery contains charged residues RHK
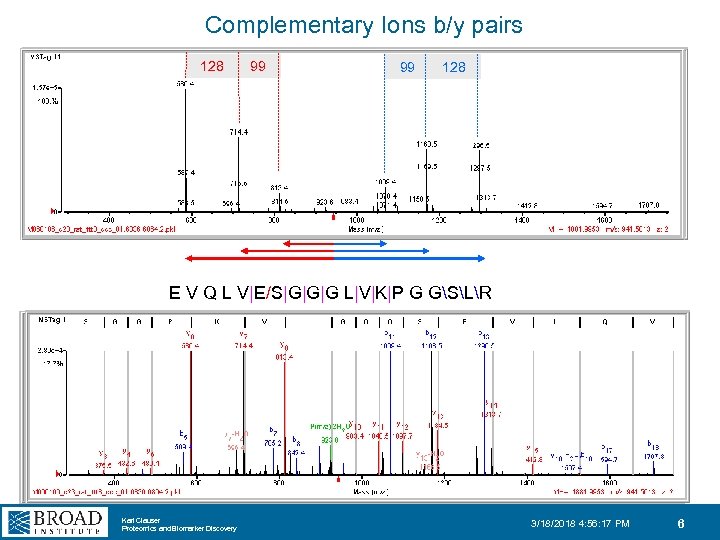 Complementary Ions b/y pairs 128 99 99 128 E V Q L V|E/S|G|G|G L|V|K|P G GSLR Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 17 PM 6
Complementary Ions b/y pairs 128 99 99 128 E V Q L V|E/S|G|G|G L|V|K|P G GSLR Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 17 PM 6
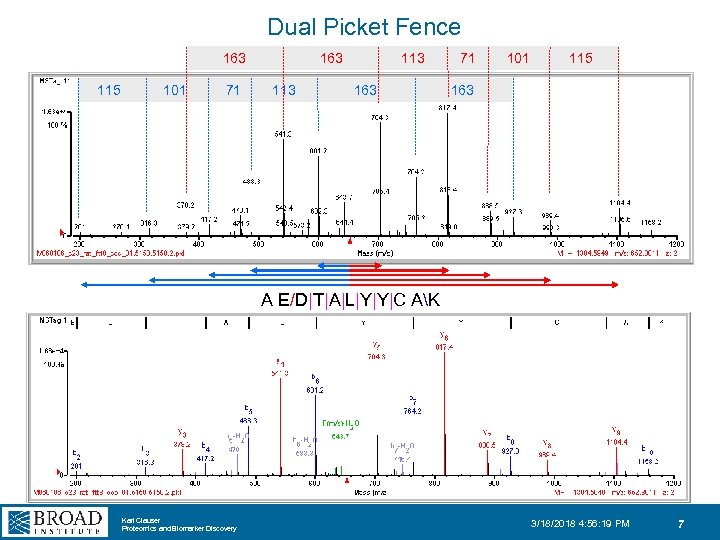 Dual Picket Fence 163 113 71 101 115 101 71 113 163 A E/D|T|A|L|Y|Y|C AK Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 19 PM 7
Dual Picket Fence 163 113 71 101 115 101 71 113 163 A E/D|T|A|L|Y|Y|C AK Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 19 PM 7
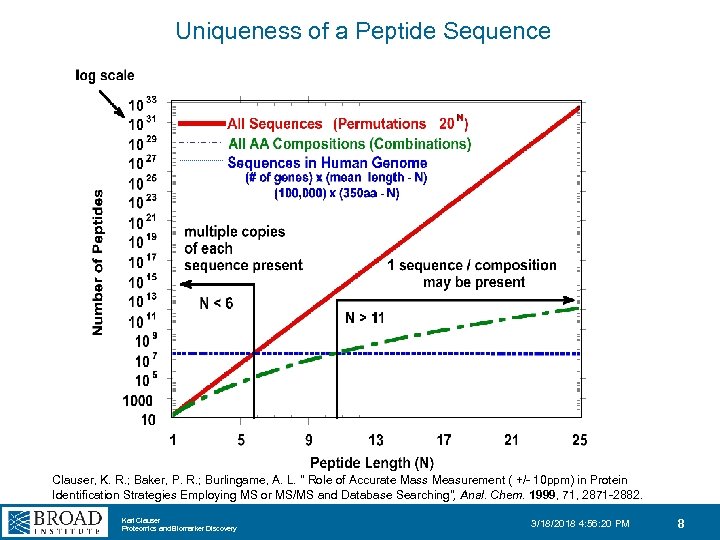 Uniqueness of a Peptide Sequence Clauser, K. R. ; Baker, P. R. ; Burlingame, A. L. " Role of Accurate Mass Measurement ( +/- 10 ppm) in Protein Identification Strategies Employing MS or MS/MS and Database Searching", Anal. Chem. 1999, 71, 2871 -2882. Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 20 PM 8
Uniqueness of a Peptide Sequence Clauser, K. R. ; Baker, P. R. ; Burlingame, A. L. " Role of Accurate Mass Measurement ( +/- 10 ppm) in Protein Identification Strategies Employing MS or MS/MS and Database Searching", Anal. Chem. 1999, 71, 2871 -2882. Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 20 PM 8
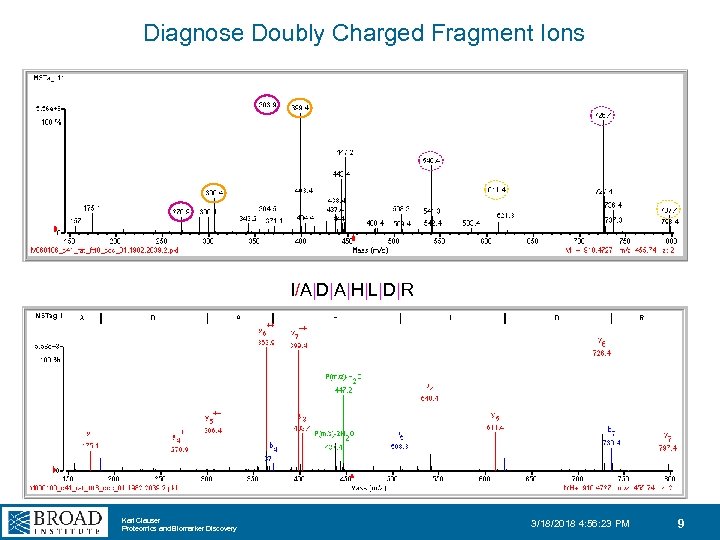 Diagnose Doubly Charged Fragment Ions I/A|D|A|H|L|D|R Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 23 PM 9
Diagnose Doubly Charged Fragment Ions I/A|D|A|H|L|D|R Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 23 PM 9
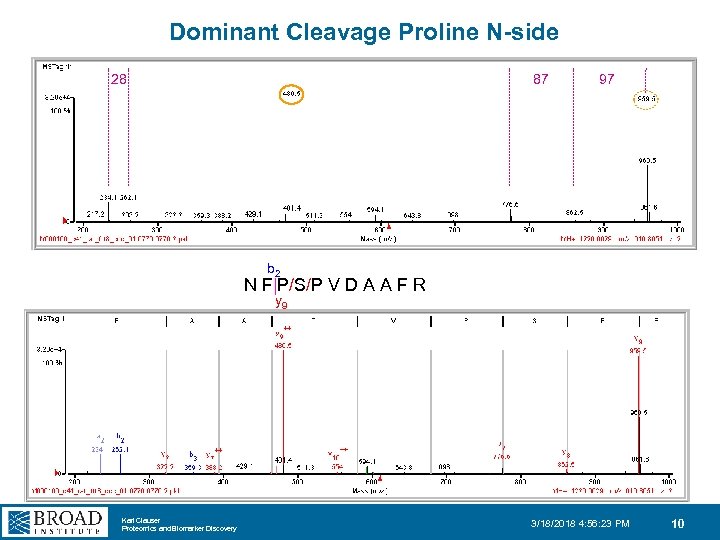 Dominant Cleavage Proline N-side 28 87 97 b 2 N F|P/S/P V D A A F R y 9 Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 23 PM 10
Dominant Cleavage Proline N-side 28 87 97 b 2 N F|P/S/P V D A A F R y 9 Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 23 PM 10
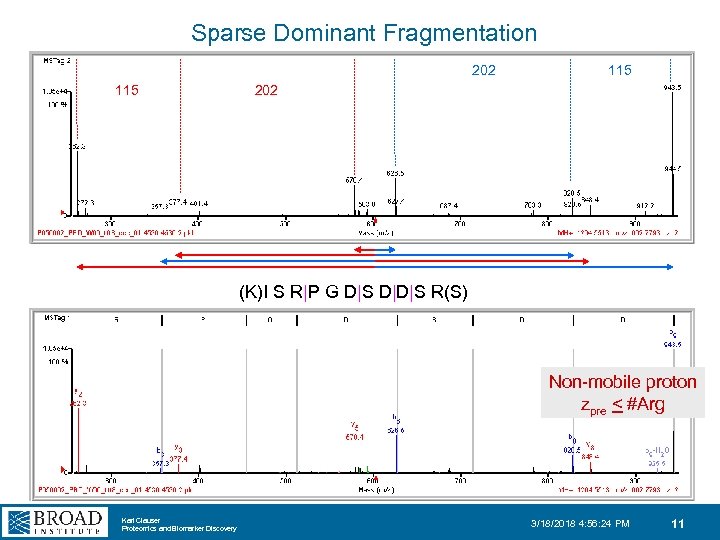 Sparse Dominant Fragmentation 202 115 202 (K)I S R|P G D|S D|D|S R(S) Non-mobile proton zpre < #Arg Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 24 PM 11
Sparse Dominant Fragmentation 202 115 202 (K)I S R|P G D|S D|D|S R(S) Non-mobile proton zpre < #Arg Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 24 PM 11
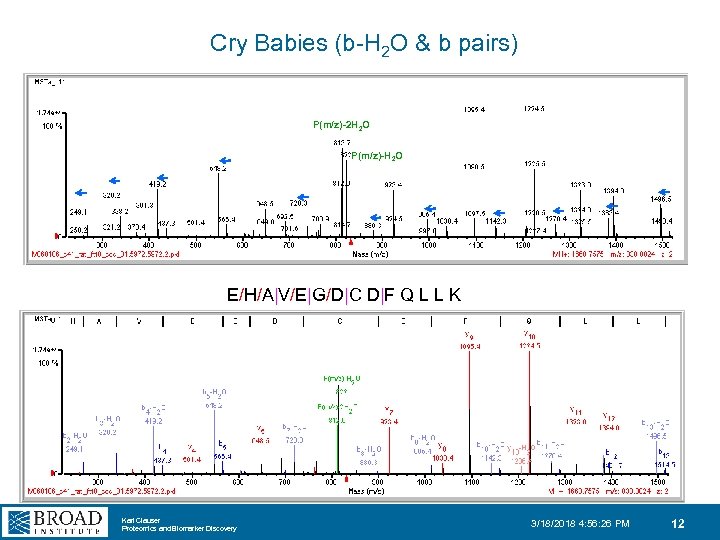 Cry Babies (b-H 2 O & b pairs) P(m/z)-2 H 2 O P(m/z)-H 2 O E/H/A|V/E|G/D|C D|F Q L L K Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 26 PM 12
Cry Babies (b-H 2 O & b pairs) P(m/z)-2 H 2 O P(m/z)-H 2 O E/H/A|V/E|G/D|C D|F Q L L K Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 26 PM 12
 Interpreting MS/MS Spectra is Fun!! Andrea Kaitlin Aidan Jack Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 26 PM 13
Interpreting MS/MS Spectra is Fun!! Andrea Kaitlin Aidan Jack Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 26 PM 13
 Interpreting MS/MS Spectra is Fun!! Andrea Kaitlin Aidan Jack Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 28 PM 14
Interpreting MS/MS Spectra is Fun!! Andrea Kaitlin Aidan Jack Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 28 PM 14
 Source of Incorrect MS/MS Interpretations Major Database Peptide not in database. Mutation. MS/MS not from a peptide. Unanticipated Protein Chemistry Chemical modification, post-translational modification. Enzyme/Ion Source Non-specific cleavage. In-source fragmentation yields MS 3. Minor Algorithm Fragment ion types of instrument not accounted for. Peak Detection. Instrument Resolution Wrong parent charge. Wrong fragment charge. User Competence Wrong parameters selected. Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 29 PM 15
Source of Incorrect MS/MS Interpretations Major Database Peptide not in database. Mutation. MS/MS not from a peptide. Unanticipated Protein Chemistry Chemical modification, post-translational modification. Enzyme/Ion Source Non-specific cleavage. In-source fragmentation yields MS 3. Minor Algorithm Fragment ion types of instrument not accounted for. Peak Detection. Instrument Resolution Wrong parent charge. Wrong fragment charge. User Competence Wrong parameters selected. Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 29 PM 15
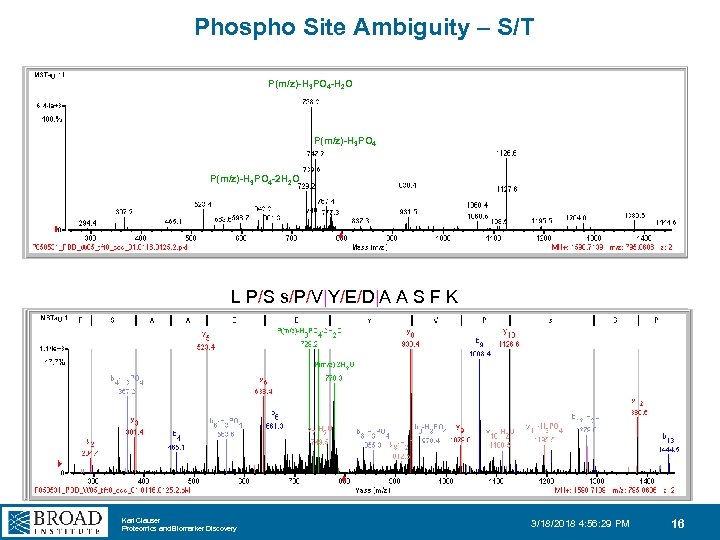 Phospho Site Ambiguity – S/T P(m/z)-H 3 PO 4 -H 2 O P(m/z)-H 3 PO 4 -2 H 2 O L P/S s/P/V|Y/E/D|A A S F K Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 29 PM 16
Phospho Site Ambiguity – S/T P(m/z)-H 3 PO 4 -H 2 O P(m/z)-H 3 PO 4 -2 H 2 O L P/S s/P/V|Y/E/D|A A S F K Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 29 PM 16
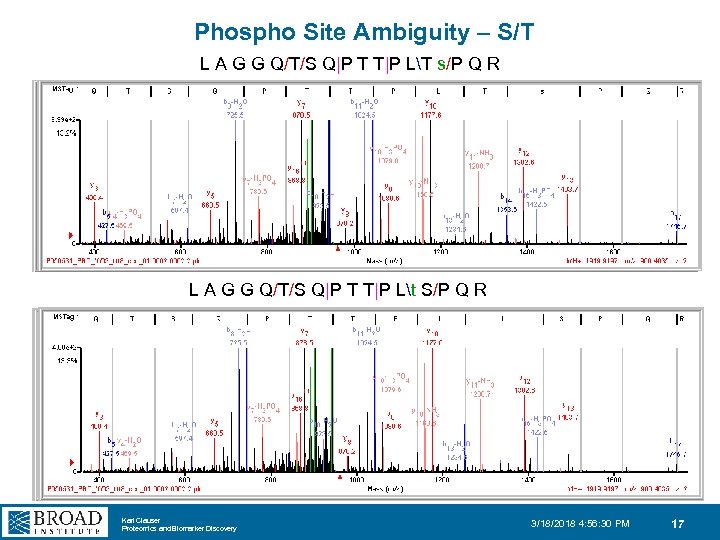 Phospho Site Ambiguity – S/T L A G G Q/T/S Q|P T T|P LT s/P Q R L A G G Q/T/S Q|P T T|P Lt S/P Q R Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 30 PM 17
Phospho Site Ambiguity – S/T L A G G Q/T/S Q|P T T|P LT s/P Q R L A G G Q/T/S Q|P T T|P Lt S/P Q R Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 30 PM 17
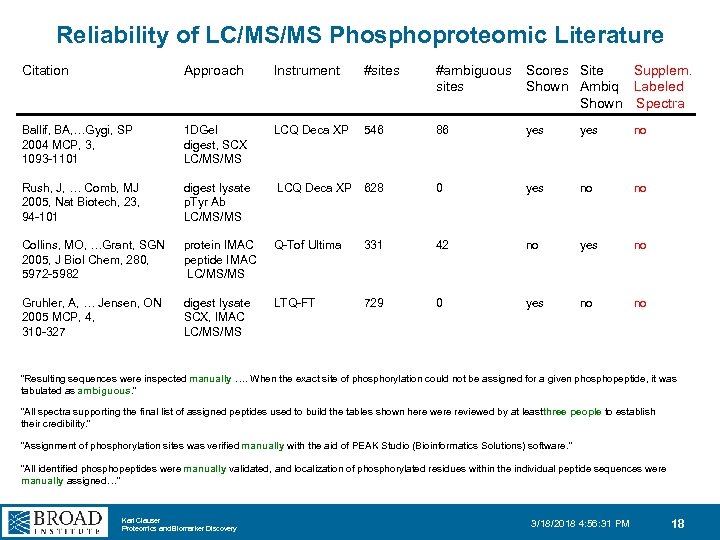 Reliability of LC/MS/MS Phosphoproteomic Literature Citation Approach Instrument #sites #ambiguous Scores Site Supplem. sites Shown Ambiq Labeled Shown Spectra Ballif, BA, …Gygi, SP 2004 MCP, 3, 1093 -1101 1 DGel digest, SCX LC/MS/MS LCQ Deca XP 546 86 yes no Rush, J, … Comb, MJ 2005, Nat Biotech, 23, 94 -101 digest lysate p. Tyr Ab LC/MS/MS LCQ Deca XP 628 0 yes no no Collins, MO, …Grant, SGN 2005, J Biol Chem, 280, 5972 -5982 protein IMAC peptide IMAC LC/MS/MS Q-Tof Ultima 331 42 no yes no Gruhler, A, … Jensen, ON 2005 MCP, 4, 310 -327 digest lysate SCX, IMAC LC/MS/MS LTQ-FT 729 0 yes no no “Resulting sequences were inspected manually …. When the exact site of phosphorylation could not be assigned for a given phosphopeptide, it was tabulated as ambiguous. ” “All spectra supporting the final list of assigned peptides used to build the tables shown here were reviewed by at least hree people to establish t their credibility. ” “Assignment of phosphorylation sites was verified manually with the aid of PEAK Studio (Bioinformatics Solutions) software. ” “All identified phosphopeptides were manually validated, and localization of phosphorylated residues within the individual peptide sequences were manually assigned…” Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 31 PM 18
Reliability of LC/MS/MS Phosphoproteomic Literature Citation Approach Instrument #sites #ambiguous Scores Site Supplem. sites Shown Ambiq Labeled Shown Spectra Ballif, BA, …Gygi, SP 2004 MCP, 3, 1093 -1101 1 DGel digest, SCX LC/MS/MS LCQ Deca XP 546 86 yes no Rush, J, … Comb, MJ 2005, Nat Biotech, 23, 94 -101 digest lysate p. Tyr Ab LC/MS/MS LCQ Deca XP 628 0 yes no no Collins, MO, …Grant, SGN 2005, J Biol Chem, 280, 5972 -5982 protein IMAC peptide IMAC LC/MS/MS Q-Tof Ultima 331 42 no yes no Gruhler, A, … Jensen, ON 2005 MCP, 4, 310 -327 digest lysate SCX, IMAC LC/MS/MS LTQ-FT 729 0 yes no no “Resulting sequences were inspected manually …. When the exact site of phosphorylation could not be assigned for a given phosphopeptide, it was tabulated as ambiguous. ” “All spectra supporting the final list of assigned peptides used to build the tables shown here were reviewed by at least hree people to establish t their credibility. ” “Assignment of phosphorylation sites was verified manually with the aid of PEAK Studio (Bioinformatics Solutions) software. ” “All identified phosphopeptides were manually validated, and localization of phosphorylated residues within the individual peptide sequences were manually assigned…” Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 31 PM 18
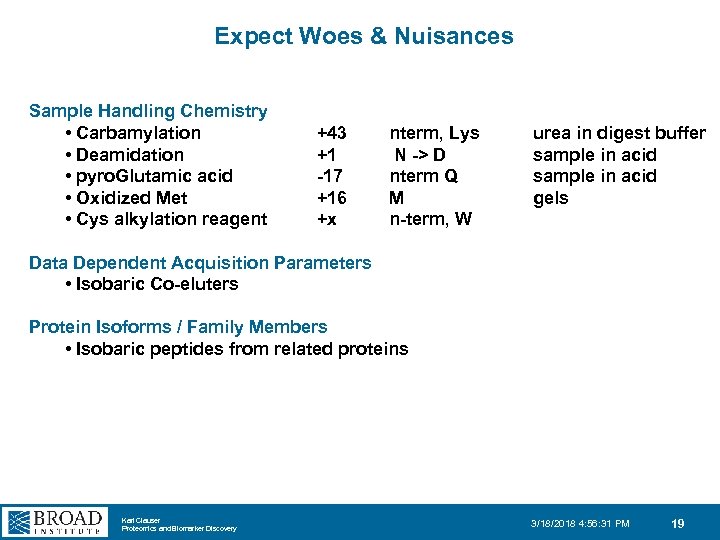 Expect Woes & Nuisances Sample Handling Chemistry • Carbamylation • Deamidation • pyro. Glutamic acid • Oxidized Met • Cys alkylation reagent +43 +1 -17 +16 +x nterm, Lys N -> D nterm Q M n-term, W urea in digest buffer sample in acid gels Data Dependent Acquisition Parameters • Isobaric Co-eluters Protein Isoforms / Family Members • Isobaric peptides from related proteins Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 31 PM 19
Expect Woes & Nuisances Sample Handling Chemistry • Carbamylation • Deamidation • pyro. Glutamic acid • Oxidized Met • Cys alkylation reagent +43 +1 -17 +16 +x nterm, Lys N -> D nterm Q M n-term, W urea in digest buffer sample in acid gels Data Dependent Acquisition Parameters • Isobaric Co-eluters Protein Isoforms / Family Members • Isobaric peptides from related proteins Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 31 PM 19
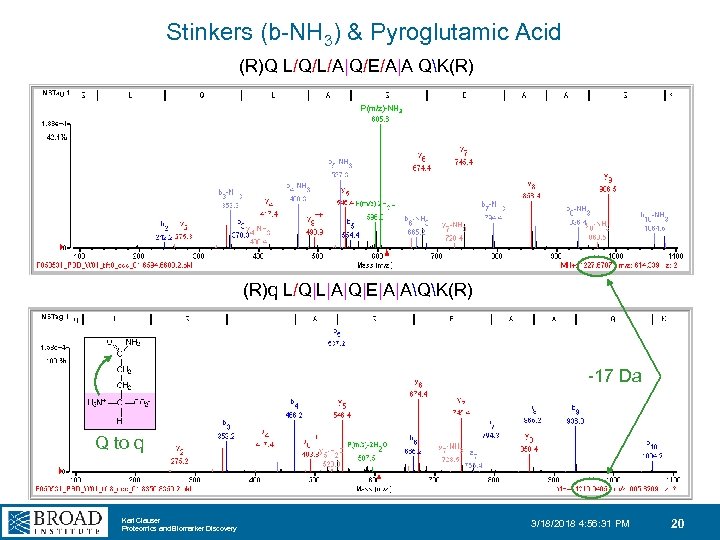 Stinkers (b-NH 3) & Pyroglutamic Acid (R)Q L/Q/L/A|Q/E/A|A QK(R) P(m/z)-NH 3 (R)q L/Q|L|A|Q|E|A|AQK(R) -17 Da Q to q Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 31 PM 20
Stinkers (b-NH 3) & Pyroglutamic Acid (R)Q L/Q/L/A|Q/E/A|A QK(R) P(m/z)-NH 3 (R)q L/Q|L|A|Q|E|A|AQK(R) -17 Da Q to q Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 31 PM 20
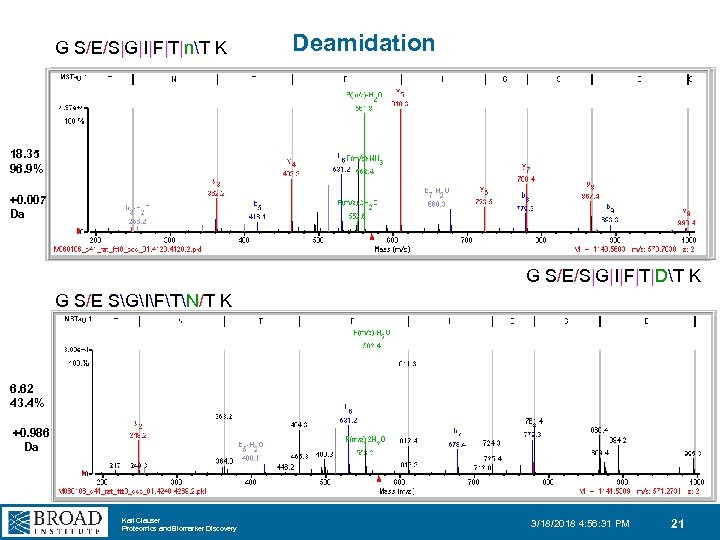 G S/E/S|G|I|F|T|nT K Deamidation 18. 35 96. 9% +0. 007 Da G S/E/S|G|I|F|T|DT K G S/E SGIFTN/T K 6. 62 43. 4% +0. 986 Da Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 31 PM 21
G S/E/S|G|I|F|T|nT K Deamidation 18. 35 96. 9% +0. 007 Da G S/E/S|G|I|F|T|DT K G S/E SGIFTN/T K 6. 62 43. 4% +0. 986 Da Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 31 PM 21
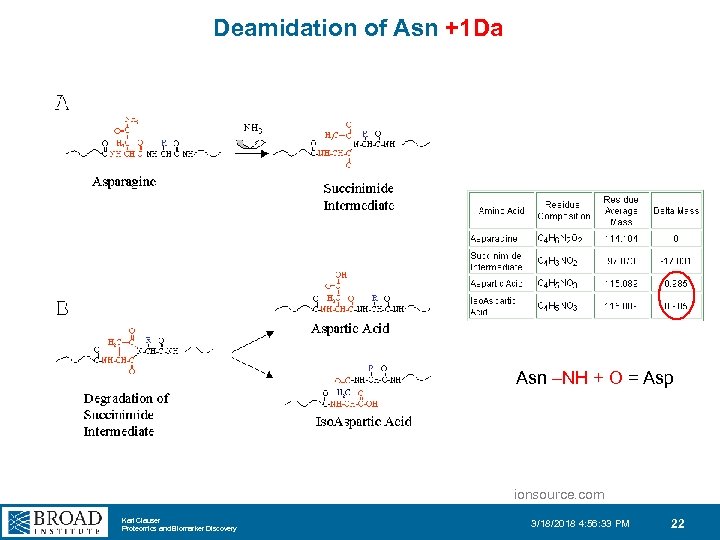 Deamidation of Asn +1 Da Asn –NH + O = Asp ionsource. com Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 33 PM 22
Deamidation of Asn +1 Da Asn –NH + O = Asp ionsource. com Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 33 PM 22
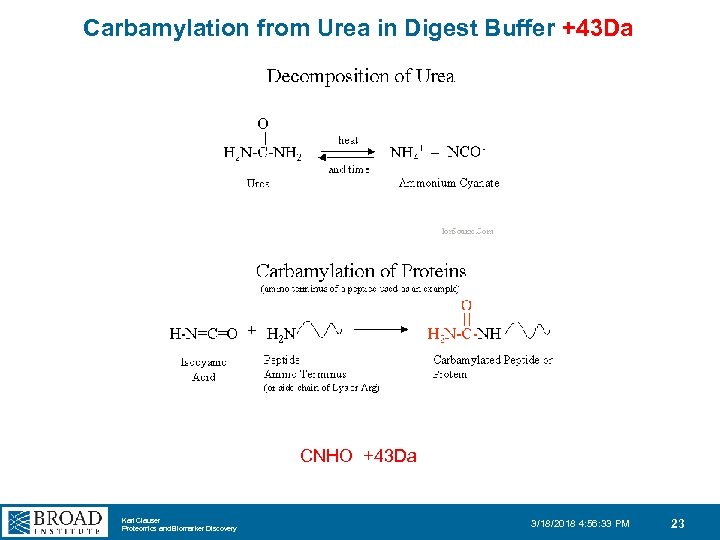 Carbamylation from Urea in Digest Buffer +43 Da CNHO +43 Da Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 33 PM 23
Carbamylation from Urea in Digest Buffer +43 Da CNHO +43 Da Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 33 PM 23
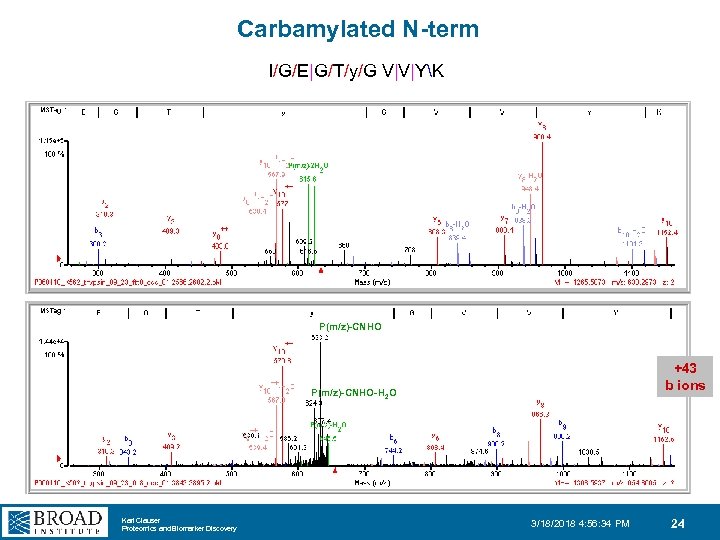 Carbamylated N-term I/G/E|G/T/y/G V|V|YK P(m/z)-CNHO +43 b ions P(m/z)-CNHO-H 2 O Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 34 PM 24
Carbamylated N-term I/G/E|G/T/y/G V|V|YK P(m/z)-CNHO +43 b ions P(m/z)-CNHO-H 2 O Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 34 PM 24
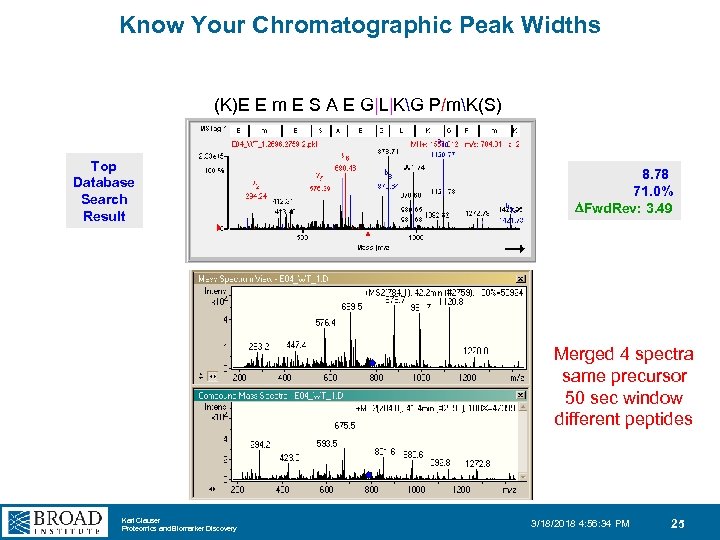 Know Your Chromatographic Peak Widths (K)E E m E S A E G|L|KG P/mK(S) Top Database Search Result 8. 78 71. 0% DFwd. Rev: 3. 49 Merged 4 spectra same precursor 50 sec window different peptides Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 34 PM 25
Know Your Chromatographic Peak Widths (K)E E m E S A E G|L|KG P/mK(S) Top Database Search Result 8. 78 71. 0% DFwd. Rev: 3. 49 Merged 4 spectra same precursor 50 sec window different peptides Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 34 PM 25
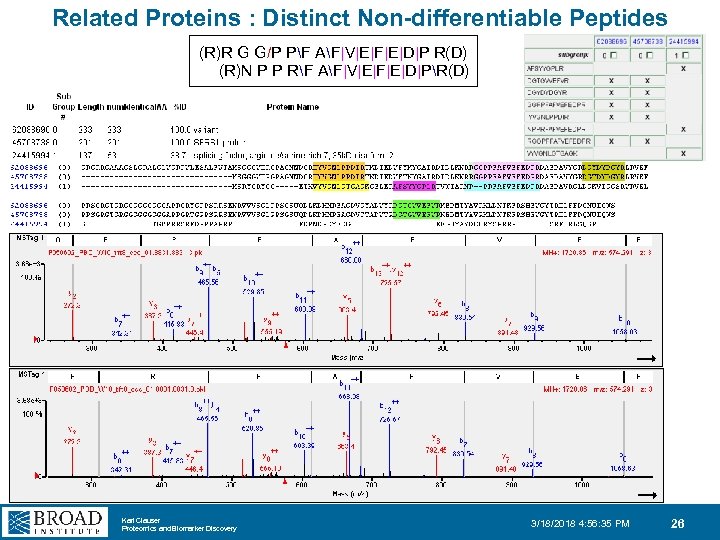 Related Proteins : Distinct Non-differentiable Peptides (R)R G G/P PF AF|V|E|F|E|D|P R(D) (R)N P P RF AF|V|E|F|E|D|PR(D) Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 35 PM 26
Related Proteins : Distinct Non-differentiable Peptides (R)R G G/P PF AF|V|E|F|E|D|P R(D) (R)N P P RF AF|V|E|F|E|D|PR(D) Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 35 PM 26
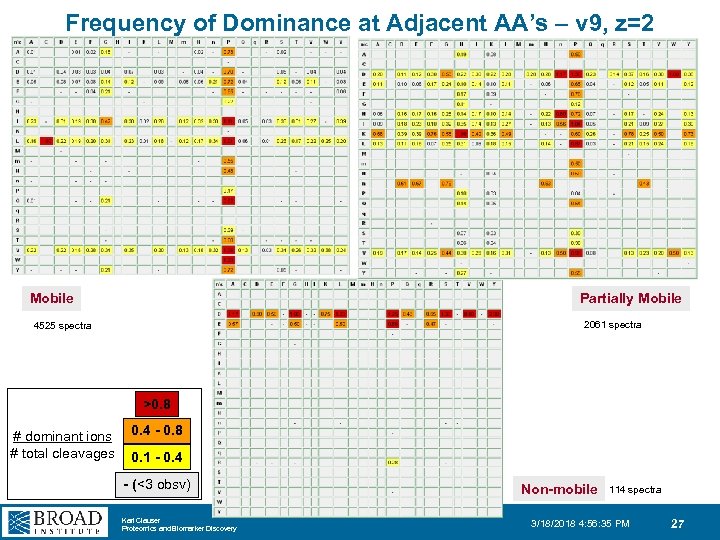 Frequency of Dominance at Adjacent AA’s – v 9, z=2 Mobile Partially Mobile 2061 spectra 4525 spectra >0. 8 # dominant ions # total cleavages 0. 4 - 0. 8 0. 1 - 0. 4 - (<3 obsv) Karl Clauser Proteomics and Biomarker Discovery Non-mobile 114 spectra 3/18/2018 4: 56: 35 PM 27
Frequency of Dominance at Adjacent AA’s – v 9, z=2 Mobile Partially Mobile 2061 spectra 4525 spectra >0. 8 # dominant ions # total cleavages 0. 4 - 0. 8 0. 1 - 0. 4 - (<3 obsv) Karl Clauser Proteomics and Biomarker Discovery Non-mobile 114 spectra 3/18/2018 4: 56: 35 PM 27
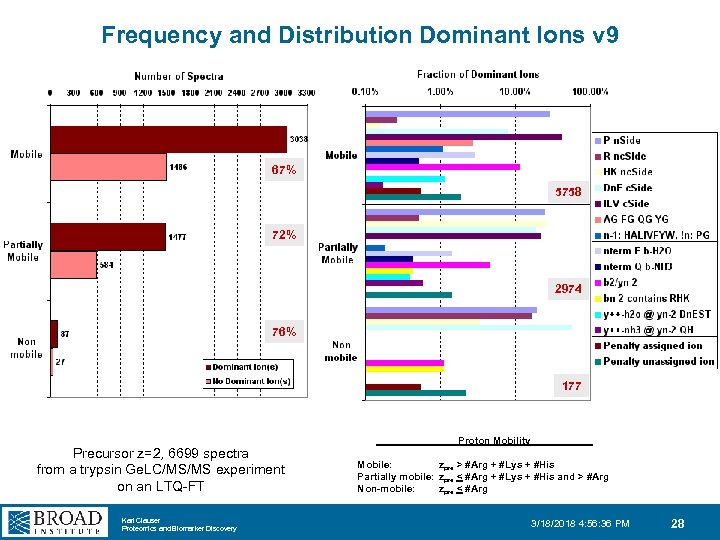 Frequency and Distribution Dominant Ions v 9 67% 5758 72% 2974 76% 177 Precursor z=2, 6699 spectra from a trypsin Ge. LC/MS/MS experiment on an LTQ-FT Karl Clauser Proteomics and Biomarker Discovery Proton Mobility Mobile: zpre > #Arg + #Lys + #His Partially mobile: zpre < #Arg + #Lys + #His and > #Arg Non-mobile: zpre < #Arg 3/18/2018 4: 56: 36 PM 28
Frequency and Distribution Dominant Ions v 9 67% 5758 72% 2974 76% 177 Precursor z=2, 6699 spectra from a trypsin Ge. LC/MS/MS experiment on an LTQ-FT Karl Clauser Proteomics and Biomarker Discovery Proton Mobility Mobile: zpre > #Arg + #Lys + #His Partially mobile: zpre < #Arg + #Lys + #His and > #Arg Non-mobile: zpre < #Arg 3/18/2018 4: 56: 36 PM 28
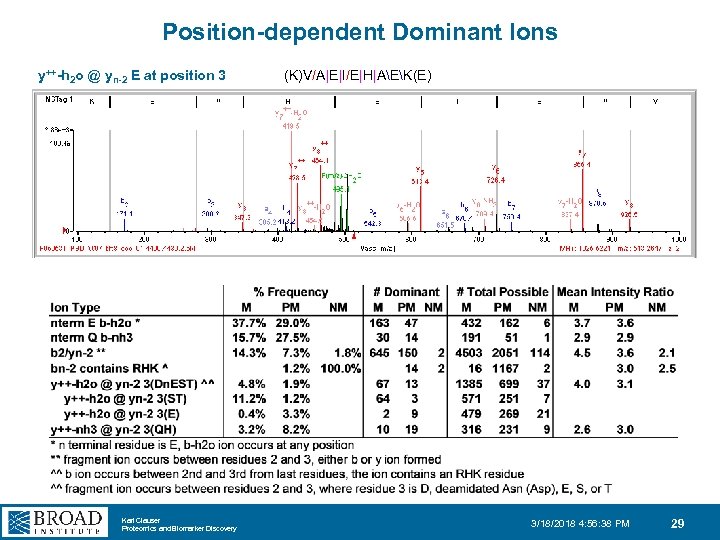 Position-dependent Dominant Ions y++-h 2 o @ yn-2 E at position 3 Karl Clauser Proteomics and Biomarker Discovery (K)V/A|E|I/E|H|AEK(E) 3/18/2018 4: 56: 38 PM 29
Position-dependent Dominant Ions y++-h 2 o @ yn-2 E at position 3 Karl Clauser Proteomics and Biomarker Discovery (K)V/A|E|I/E|H|AEK(E) 3/18/2018 4: 56: 38 PM 29
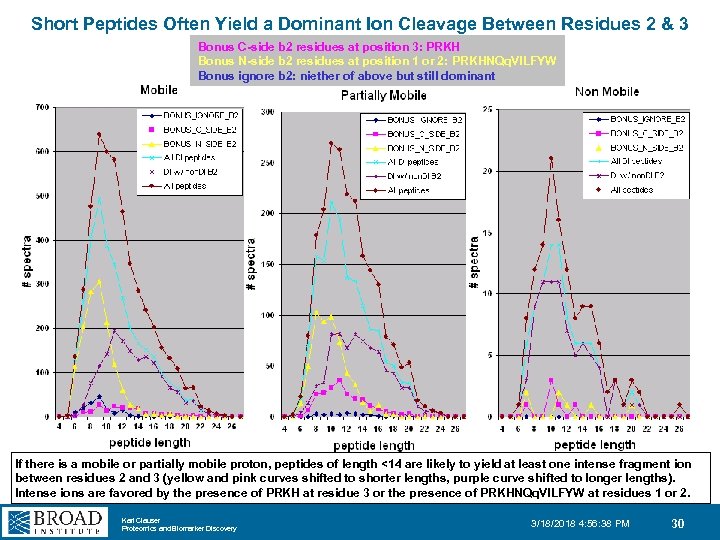 Short Peptides Often Yield a Dominant Ion Cleavage Between Residues 2 & 3 Bonus C-side b 2 residues at position 3: PRKH Bonus N-side b 2 residues at position 1 or 2: PRKHNQq. VILFYW Bonus ignore b 2: niether of above but still dominant If there is a mobile or partially mobile proton, peptides of length <14 are likely to yield at least one intense fragment ion between residues 2 and 3 (yellow and pink curves shifted to shorter lengths, purple curve shifted to longer lengths). Intense ions are favored by the presence of PRKH at residue 3 or the presence of PRKHNQq. VILFYW at residues 1 or 2. Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 38 PM 30
Short Peptides Often Yield a Dominant Ion Cleavage Between Residues 2 & 3 Bonus C-side b 2 residues at position 3: PRKH Bonus N-side b 2 residues at position 1 or 2: PRKHNQq. VILFYW Bonus ignore b 2: niether of above but still dominant If there is a mobile or partially mobile proton, peptides of length <14 are likely to yield at least one intense fragment ion between residues 2 and 3 (yellow and pink curves shifted to shorter lengths, purple curve shifted to longer lengths). Intense ions are favored by the presence of PRKH at residue 3 or the presence of PRKHNQq. VILFYW at residues 1 or 2. Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 38 PM 30
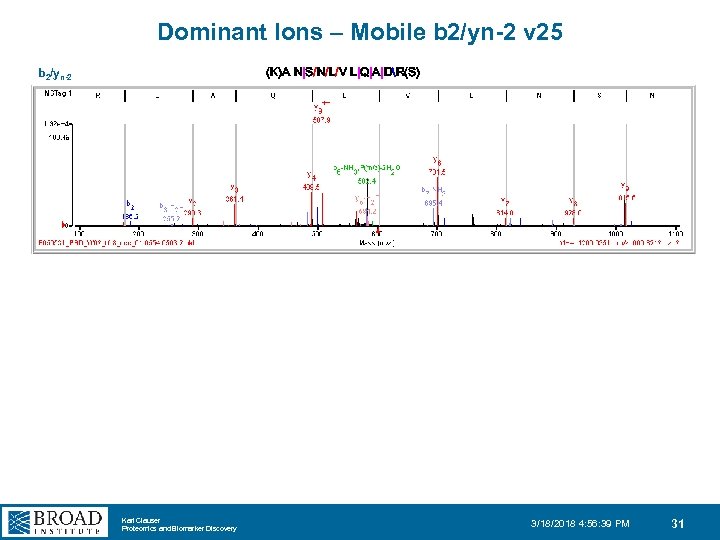 Dominant Ions – Mobile b 2/yn-2 v 25 (K)A N|S/N/L/V L|Q|A|DR(S) b 2/yn-2 Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 39 PM 31
Dominant Ions – Mobile b 2/yn-2 v 25 (K)A N|S/N/L/V L|Q|A|DR(S) b 2/yn-2 Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 39 PM 31
 Physiochemical Complications to Computational Interpretation • Incomplete Fragmentation • Inconsistent intensity of fragment ion types Instrument type dependent Amino acid dependent • Isobaric AA’s • I = L (C 6 H 11 N 1 O) • K = Q (C 6 H 12 N 2 O, C 5 H 8 N 2 O 2) • Isobaric AA combinations • GG=N (C 4 H 6 N 2 O 2 , C 4 H 6 N 2 O 2) • GA=K=Q (C 5 H 8 N 2 O 2, C 6 H 12 N 2 O, C 5 H 8 N 2 O 2) • W=DA=VS (C 11 H 11 N 2 O, C 7 H 10 N 2 O 4, C 8 H 14 N 2 O 3) • Parent charge uncertainty • Fragment charge uncertainty • Chemical or post-translational modifications Karl Clauser Proteomics and Biomarker Discovery
Physiochemical Complications to Computational Interpretation • Incomplete Fragmentation • Inconsistent intensity of fragment ion types Instrument type dependent Amino acid dependent • Isobaric AA’s • I = L (C 6 H 11 N 1 O) • K = Q (C 6 H 12 N 2 O, C 5 H 8 N 2 O 2) • Isobaric AA combinations • GG=N (C 4 H 6 N 2 O 2 , C 4 H 6 N 2 O 2) • GA=K=Q (C 5 H 8 N 2 O 2, C 6 H 12 N 2 O, C 5 H 8 N 2 O 2) • W=DA=VS (C 11 H 11 N 2 O, C 7 H 10 N 2 O 4, C 8 H 14 N 2 O 3) • Parent charge uncertainty • Fragment charge uncertainty • Chemical or post-translational modifications Karl Clauser Proteomics and Biomarker Discovery
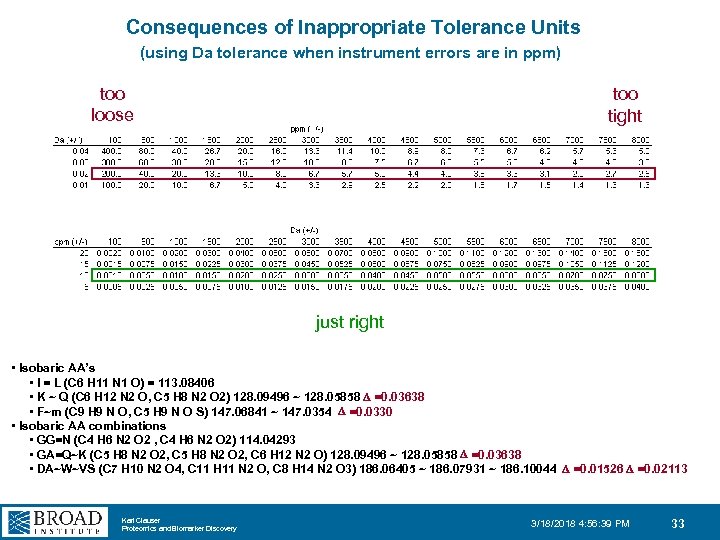 Consequences of Inappropriate Tolerance Units (using Da tolerance when instrument errors are in ppm) too loose too tight just right • Isobaric AA’s • I = L (C 6 H 11 N 1 O) = 113. 08406 • K ~ Q (C 6 H 12 N 2 O, C 5 H 8 N 2 O 2) 128. 09496 ~ 128. 05858 D =0. 03638 • F~m (C 9 H 9 N O, C 5 H 9 N O S) 147. 06841 ~ 147. 0354 D =0. 0330 • Isobaric AA combinations • GG=N (C 4 H 6 N 2 O 2 , C 4 H 6 N 2 O 2) 114. 04293 • GA=Q~K (C 5 H 8 N 2 O 2, C 6 H 12 N 2 O) 128. 09496 ~ 128. 05858 D =0. 03638 • DA~W~VS (C 7 H 10 N 2 O 4, C 11 H 11 N 2 O, C 8 H 14 N 2 O 3) 186. 06405 ~ 186. 07931 ~ 186. 10044 D =0. 01526 D =0. 02113 Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 39 PM 33
Consequences of Inappropriate Tolerance Units (using Da tolerance when instrument errors are in ppm) too loose too tight just right • Isobaric AA’s • I = L (C 6 H 11 N 1 O) = 113. 08406 • K ~ Q (C 6 H 12 N 2 O, C 5 H 8 N 2 O 2) 128. 09496 ~ 128. 05858 D =0. 03638 • F~m (C 9 H 9 N O, C 5 H 9 N O S) 147. 06841 ~ 147. 0354 D =0. 0330 • Isobaric AA combinations • GG=N (C 4 H 6 N 2 O 2 , C 4 H 6 N 2 O 2) 114. 04293 • GA=Q~K (C 5 H 8 N 2 O 2, C 6 H 12 N 2 O) 128. 09496 ~ 128. 05858 D =0. 03638 • DA~W~VS (C 7 H 10 N 2 O 4, C 11 H 11 N 2 O, C 8 H 14 N 2 O 3) 186. 06405 ~ 186. 07931 ~ 186. 10044 D =0. 01526 D =0. 02113 Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 39 PM 33
 Additional Resources Google: “de novo sequencing tutorial” Don Hunt and Jeff Shabanowitz - manual http: //www. ionsource. com/tutorial/De. Novo. TOC. htm Rich Johnson - manual http: //www. abrf. org/Research. Groups/Mass. Spectrometry/EPosters/ms 97 quiz/Sequencing. Tutorial. html PEAKS - automated http: //www. bioinformaticssolutions. com/products/peaks/support/tutorials/PEAKS_De_Novo. html Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 40 PM 34
Additional Resources Google: “de novo sequencing tutorial” Don Hunt and Jeff Shabanowitz - manual http: //www. ionsource. com/tutorial/De. Novo. TOC. htm Rich Johnson - manual http: //www. abrf. org/Research. Groups/Mass. Spectrometry/EPosters/ms 97 quiz/Sequencing. Tutorial. html PEAKS - automated http: //www. bioinformaticssolutions. com/products/peaks/support/tutorials/PEAKS_De_Novo. html Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 40 PM 34
 Acknowledgements Broad Institute MIT Steve Carr Terri Addona Jinyan Du Phillip Mertins Michael Yaffe Majbrit Hjerrld Drew Lowery Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 40 PM 35
Acknowledgements Broad Institute MIT Steve Carr Terri Addona Jinyan Du Phillip Mertins Michael Yaffe Majbrit Hjerrld Drew Lowery Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 40 PM 35
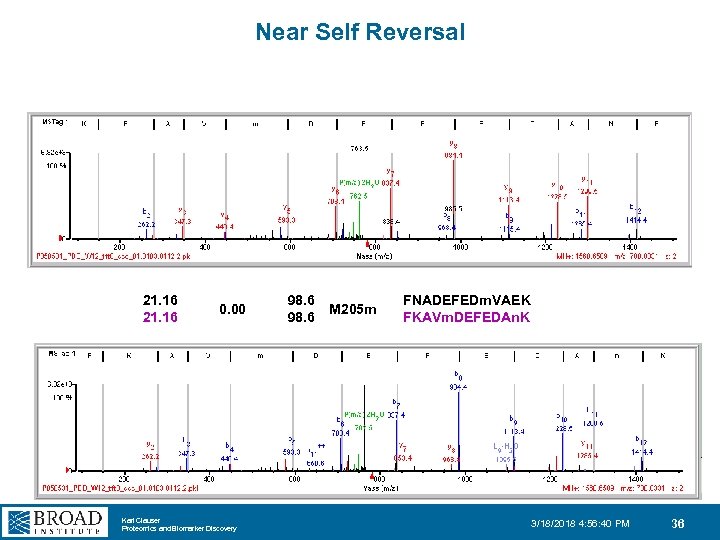 Near Self Reversal 21. 16 0. 00 Karl Clauser Proteomics and Biomarker Discovery 98. 6 M 205 m FNADEFEDm. VAEK FKAVm. DEFEDAn. K 3/18/2018 4: 56: 40 PM 36
Near Self Reversal 21. 16 0. 00 Karl Clauser Proteomics and Biomarker Discovery 98. 6 M 205 m FNADEFEDm. VAEK FKAVm. DEFEDAn. K 3/18/2018 4: 56: 40 PM 36
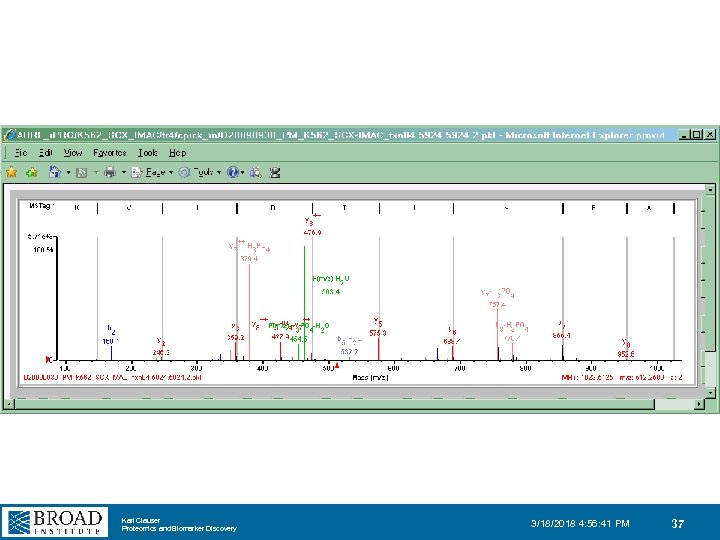 Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 41 PM 37
Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 41 PM 37
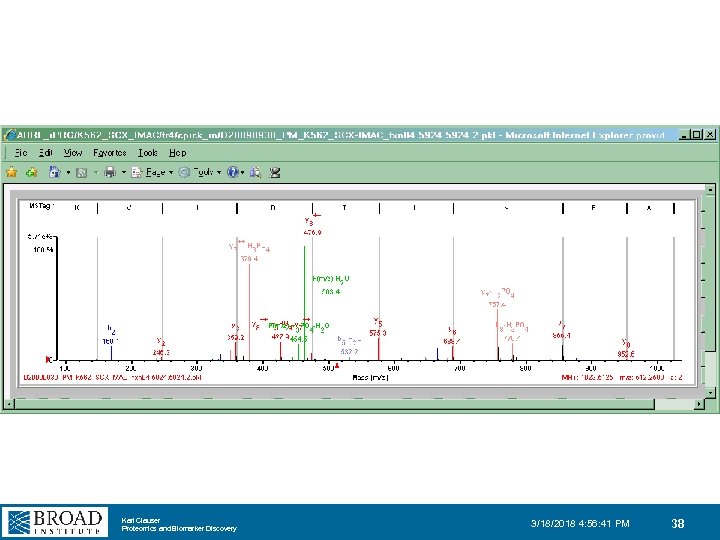 Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 41 PM 38
Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 41 PM 38
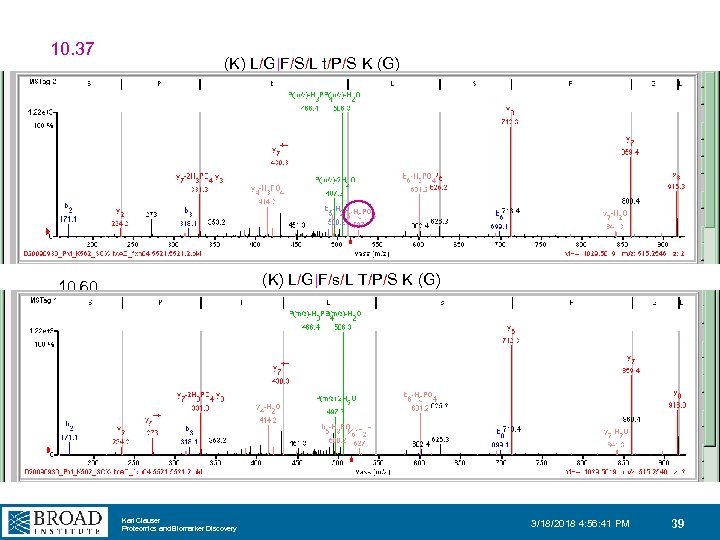 10. 37 (K) L/G|F/S/L t/P/S K (G) (K) L/G|F/s/L T/P/S K (G) 10. 60 Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 41 PM 39
10. 37 (K) L/G|F/S/L t/P/S K (G) (K) L/G|F/s/L T/P/S K (G) 10. 60 Karl Clauser Proteomics and Biomarker Discovery 3/18/2018 4: 56: 41 PM 39


