e0aaace1bd06029918c2e114b54d23aa.ppt
- Количество слайдов: 36
Magnetic Tweezer System Development Probing mechanical properties across multiple scales Jason Sherfey Senior BME, Vanderbilt University Advisor: Dr. Franz Baudenbacher

• Purpose of device: To make quantifying cell-cell adhesion quick and easy. (i. e. , finding mechanical properties) • Specific structures to quantify: 1. cell-cell linkage 2. adhesion protein linker system 3. cytoskeleton

Motivation: - Cell-cell adhesion is essential to establishing and maintaining cell and tissue morphology and in cellular migration - Specific issue - alterations in cell morphology and migration are essential to tumor growth and metastasis - Idea. Quantify cell-cell adhesion Better understand cellular morphology and migration Improve diagnostics and treatments for cancer Principle Components of the Design Process: 1. System development 2. Model testing (E-cadherin system in p 120 KO vs WT mdck) for error analysis and concept testing & validation
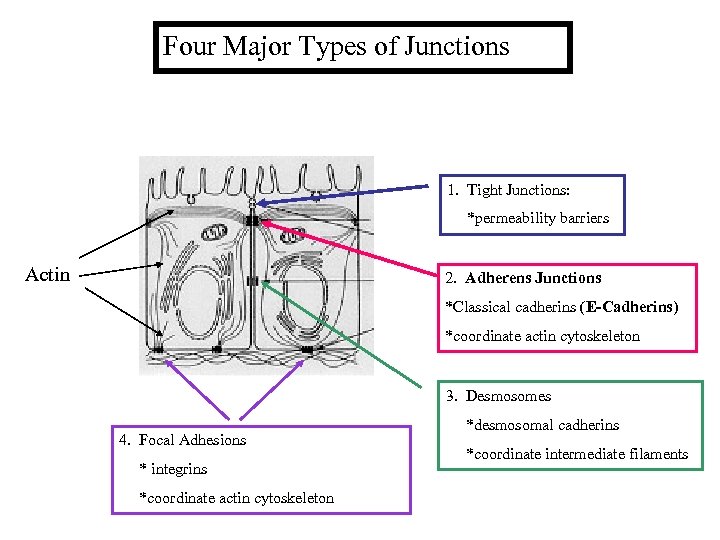
Four Major Types of Junctions 1. Tight Junctions: *permeability barriers Actin 2. Adherens Junctions *Classical cadherins (E-Cadherins) *coordinate actin cytoskeleton 3. Desmosomes 4. Focal Adhesions * integrins *coordinate actin cytoskeleton *desmosomal cadherins *coordinate intermediate filaments
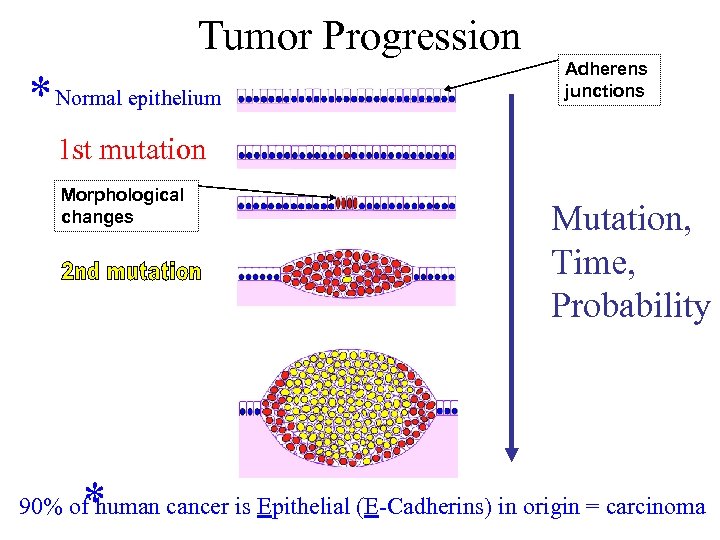
Tumor Progression * Normal epithelium Adherens junctions 1 st mutation Morphological changes * Mutation, Time, Probability 90% of human cancer is Epithelial (E-Cadherins) in origin = carcinoma
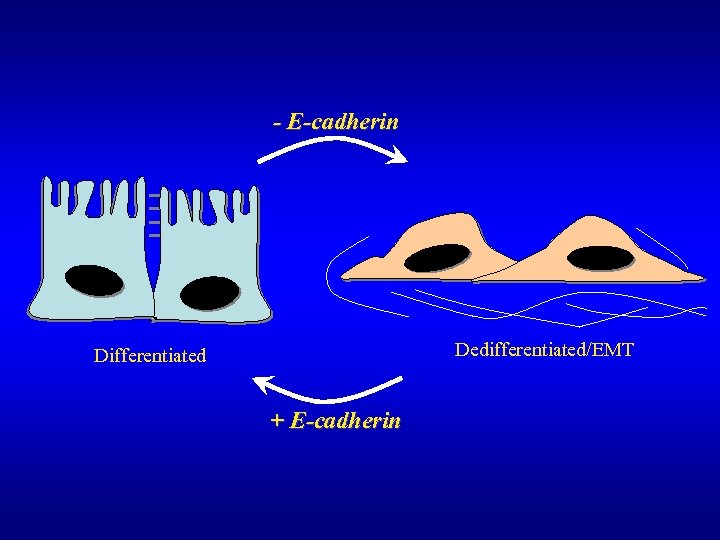
- E-cadherin Dedifferentiated/EMT Differentiated + E-cadherin
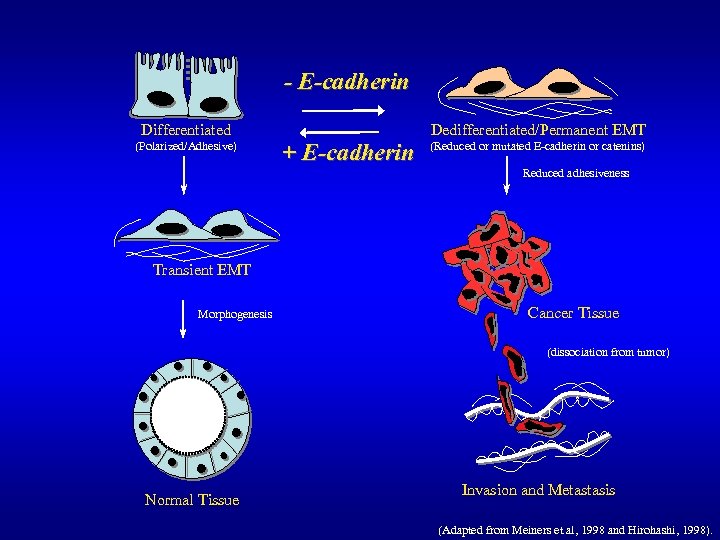
- E-cadherin Differentiated (Polarized/Adhesive) Dedifferentiated/Permanent EMT + E-cadherin (Reduced or mutated E-cadherin or catenins) Reduced adhesiveness Transient EMT Morphogenesis Cancer Tissue (dissociation from tumor) Normal Tissue Invasion and Metastasis (Adapted from Meiners et al, 1998 and Hirohashi, 1998).

What is the best way to objectively measure changes in cell-cell adhesion relevant to metastasis? Is it sufficient to directly quantify E-cadherin activity?
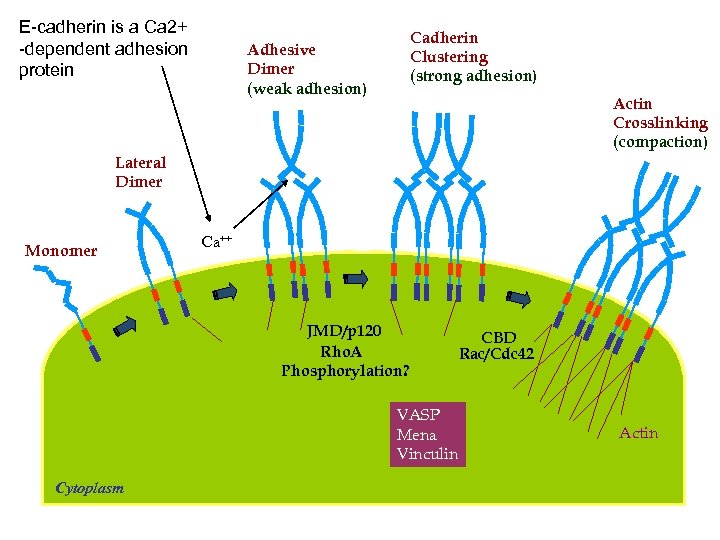
E-cadherin is a Ca 2+ -dependent adhesion protein Cadherin Clustering (strong adhesion) Adhesive Dimer (weak adhesion) Actin Crosslinking (compaction) Lateral Dimer Monomer Ca++ JMD/p 120 Rho. A Phosphorylation? VASP Mena Vinculin Cytoplasm CBD Rac/Cdc 42 Actin
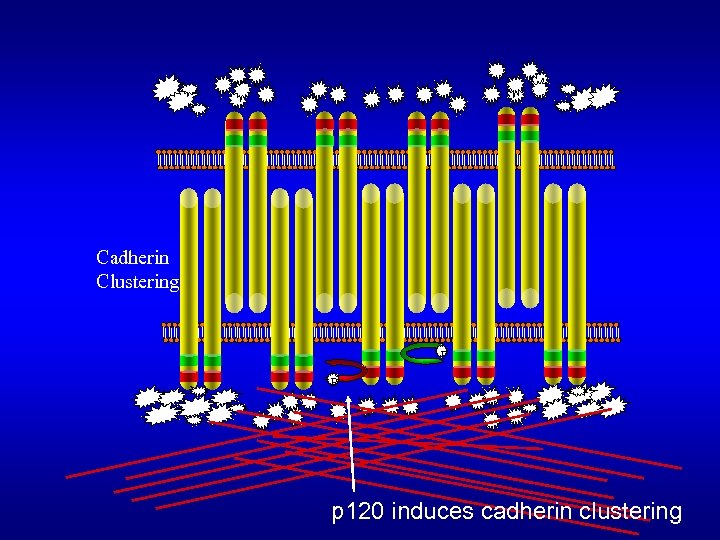
Cadherin Clustering P P p 120 induces cadherin clustering

1. The amount of E-cadherin is directly relevant to adhesive strength (all things being equal). 2. The amount of E-cadherin does not necessarily reflect adhesive activity. (eg, Rac, rho experiments)

Where does p 120 fit in to all of this?
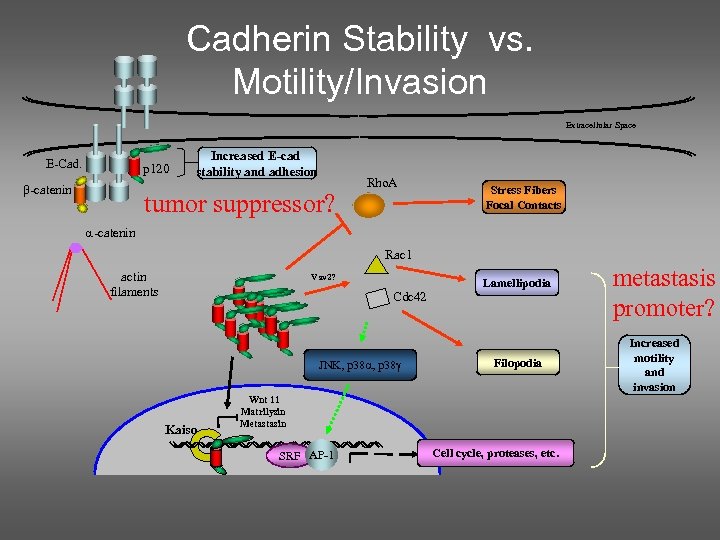
Cadherin Stability vs. Motility/Invasion Extracellular Space E-Cad. p 120 -catenin Increased E-cad stability and adhesion tumor suppressor? Rho. A Stress Fibers Focal Contacts -catenin Rac 1 actin filaments Vav 2? Cdc 42 JNK, p 38 Kaiso Lamellipodia Filopodia Wnt 11 Matrilysin Metastasin SRF AP-1 Cell cycle, proteases, etc. metastasis promoter? Increased motility and invasion
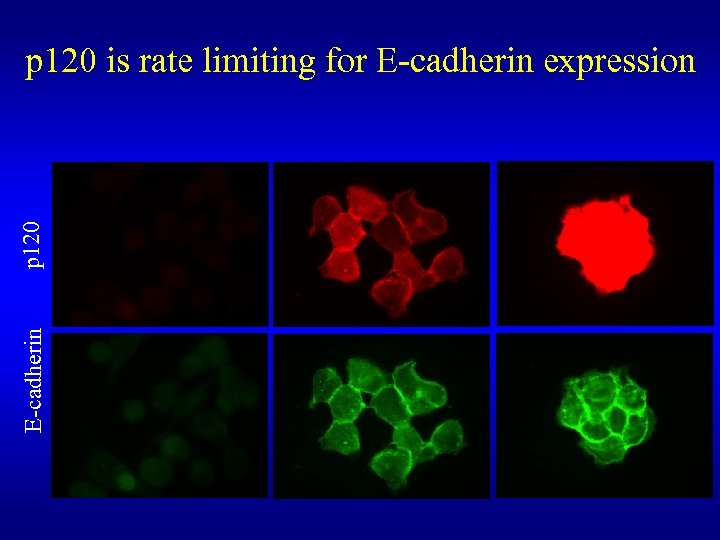
E-cadherin p 120 is rate limiting for E-cadherin expression
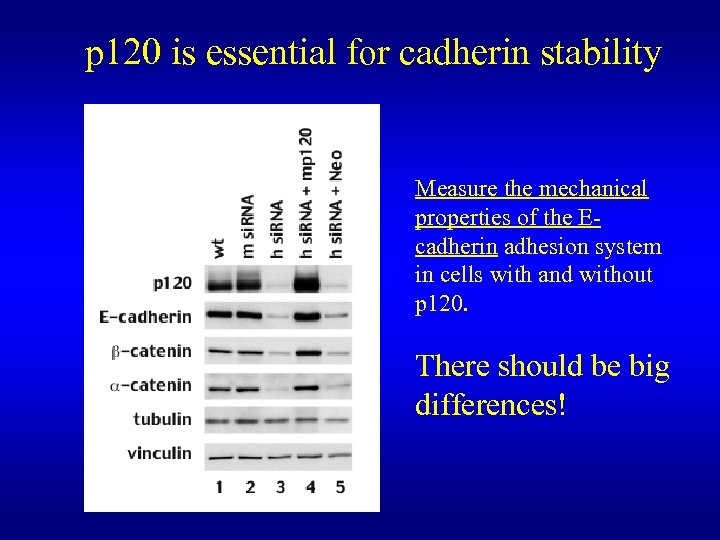
p 120 is essential for cadherin stability Measure the mechanical properties of the Ecadherin adhesion system in cells with and without p 120. There should be big differences!

Target Systems 1. E-cadherin activity: linker system mechanics 2. (magnetic tweezer) 2. Mechanics of underlying actin and the homophilic 3. E-cadherin binding junction (fluorescent beads, inversed microgrippers? )
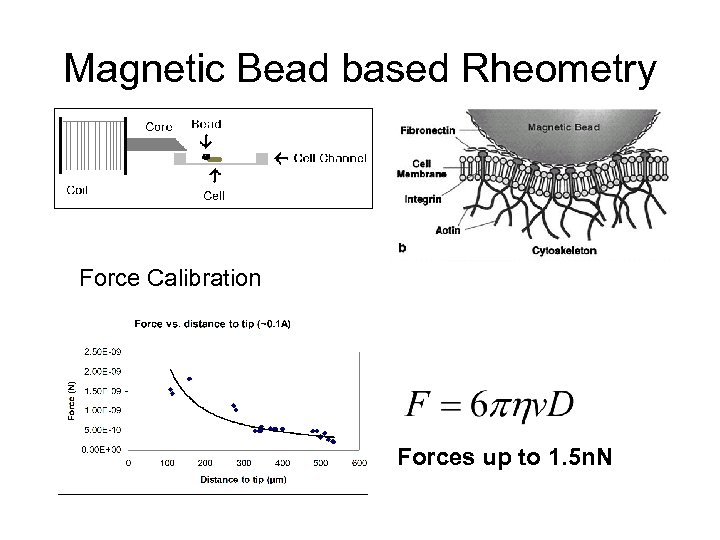
Magnetic Bead based Rheometry Force Calibration Forces up to 1. 5 n. N
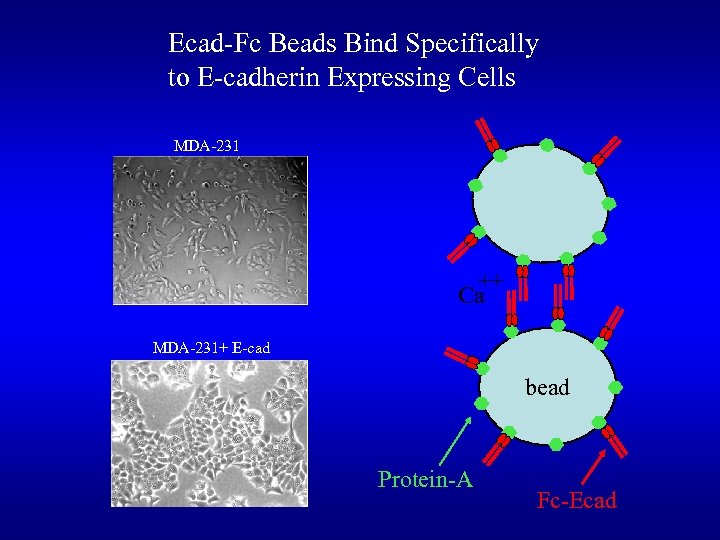
Ecad-Fc Beads Bind Specifically to E-cadherin Expressing Cells MDA-231 ++ Ca MDA-231+ E-cad bead Protein-A Fc-Ecad

Accomplishments • Implemented particle tracking software • Fabricated magnetic tweezer • Protocol to quantify the elastic properties of E-cadherins using magnetic bead based microrheology – Validation of the linker system
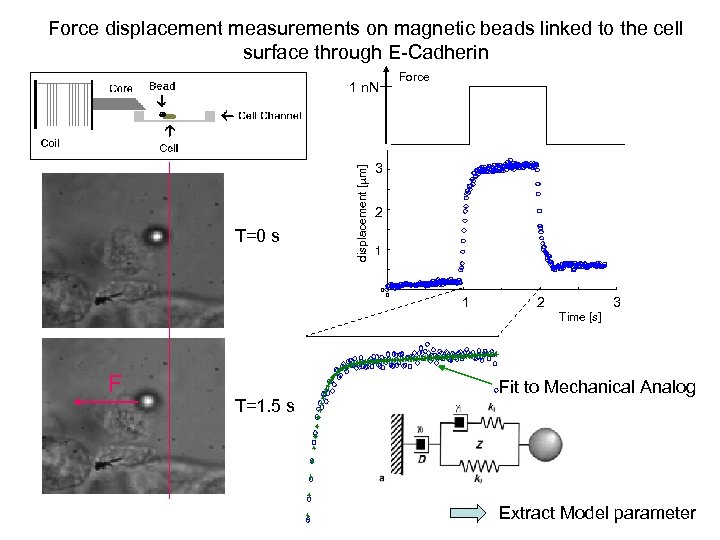
Force displacement measurements on magnetic beads linked to the cell surface through E-Cadherin Force T=0 s displacement [mm] 1 n. N 3 2 1 0 0 1 2 3 Time [s] F T=1. 5 s Fit to Mechanical Analog Extract Model parameter
Force-induced Displacement
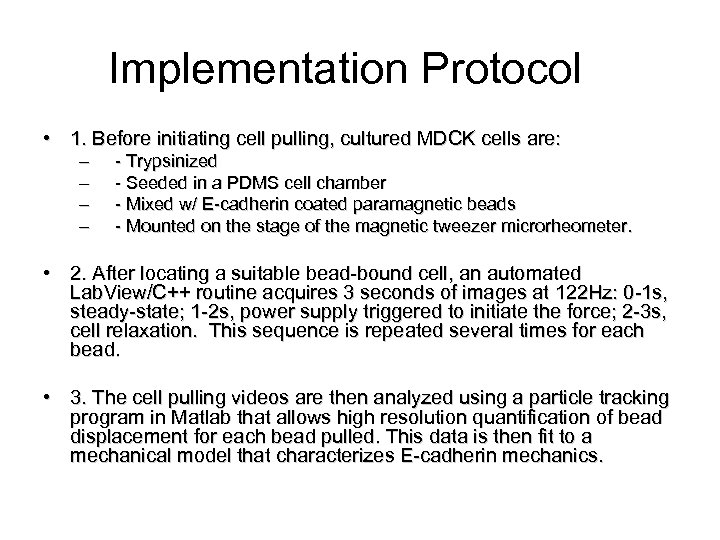
Implementation Protocol • 1. Before initiating cell pulling, cultured MDCK cells are: – – - Trypsinized - Seeded in a PDMS cell chamber - Mixed w/ E-cadherin coated paramagnetic beads - Mounted on the stage of the magnetic tweezer microrheometer. • 2. After locating a suitable bead-bound cell, an automated Lab. View/C++ routine acquires 3 seconds of images at 122 Hz: 0 -1 s, steady-state; 1 -2 s, power supply triggered to initiate the force; 2 -3 s, cell relaxation. This sequence is repeated several times for each bead. • 3. The cell pulling videos are then analyzed using a particle tracking program in Matlab that allows high resolution quantification of bead displacement for each bead pulled. This data is then fit to a mechanical model that characterizes E-cadherin mechanics.
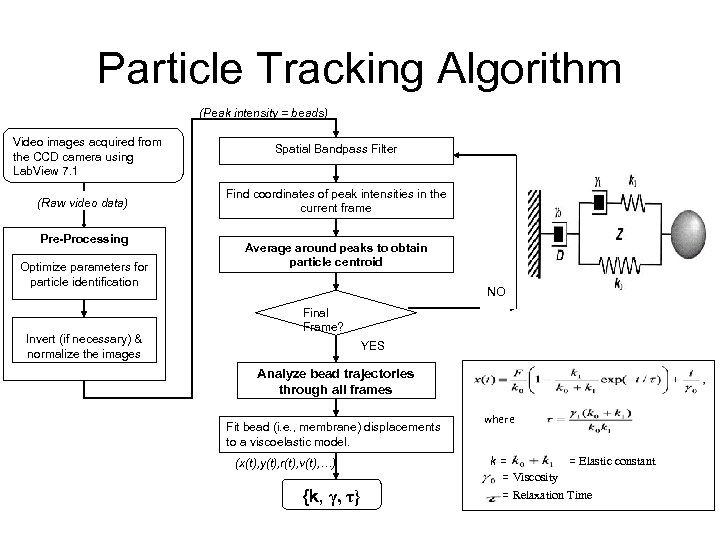
Particle Tracking Algorithm (Peak intensity = beads) Video images acquired from the CCD camera using Lab. View 7. 1 (Raw video data) Pre-Processing Optimize parameters for particle identification Invert (if necessary) & normalize the images Spatial Bandpass Filter Find coordinates of peak intensities in the current frame Average around peaks to obtain particle centroid NO Final Frame? YES Analyze bead trajectories through all frames Fit bead (i. e. , membrane) displacements to a viscoelastic model. (x(t), y(t), r(t), v(t), …) {k, γ, τ} where k= = Elastic constant = Viscosity = Relaxation Time
![Analysis of viscoeleastic response curves based on three observables [1] Number of different cells Analysis of viscoeleastic response curves based on three observables [1] Number of different cells](https://present5.com/presentation/e0aaace1bd06029918c2e114b54d23aa/image-24.jpg)
Analysis of viscoeleastic response curves based on three observables [1] Number of different cells = 7 Viscosity (Pa s m) 0. 01 0. 008 0. 006 0. 004 0. 002 0 0 2 4 6 8 10 12 14 16 18 20 [1] Local Measurements of Viscoelastic Parameters of Adherent Cell Surfaces by Magnetic Bead Microrheometry Andreas R. Bausch et. al. , Biophysical Journal Volume 75 October 1998 2038– 2049
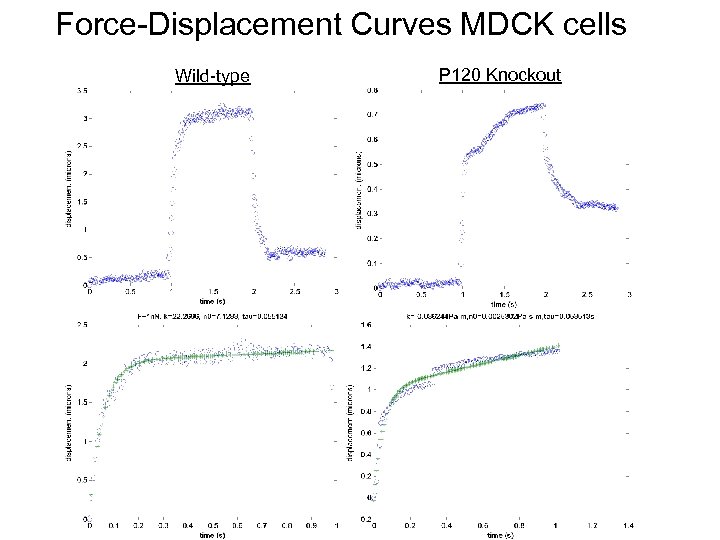
Force-Displacement Curves MDCK cells Wild-type P 120 Knockout
What next? How to quantify mechanical properties at different scales?
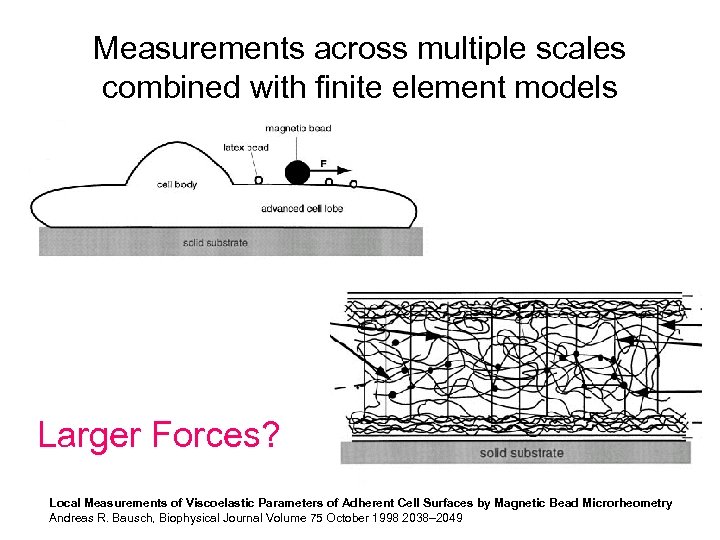
Measurements across multiple scales combined with finite element models Larger Forces? Local Measurements of Viscoelastic Parameters of Adherent Cell Surfaces by Magnetic Bead Microrheometry Andreas R. Bausch, Biophysical Journal Volume 75 October 1998 2038– 2049

Intracellular and membrane heterogeneity?
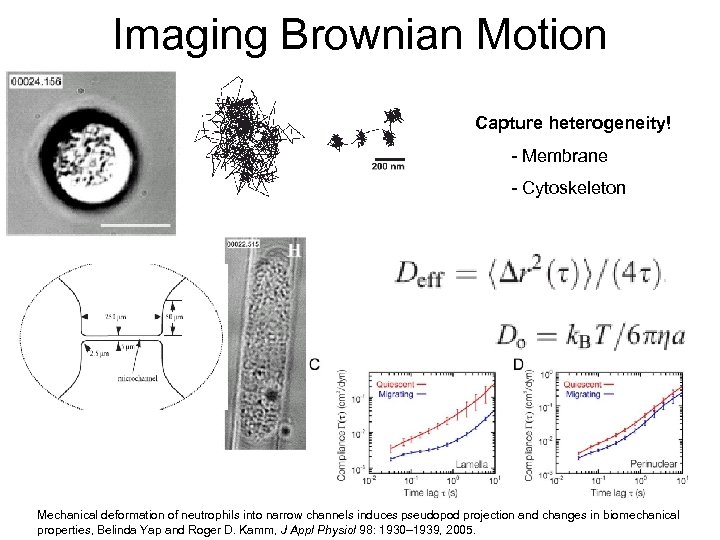
Imaging Brownian Motion Capture heterogeneity! - Membrane - Cytoskeleton Mechanical deformation of neutrophils into narrow channels induces pseudopod projection and changes in biomechanical properties, Belinda Yap and Roger D. Kamm, J Appl Physiol 98: 1930– 1939, 2005.

Larger forces (>100 n. N) for increased spatial scales?
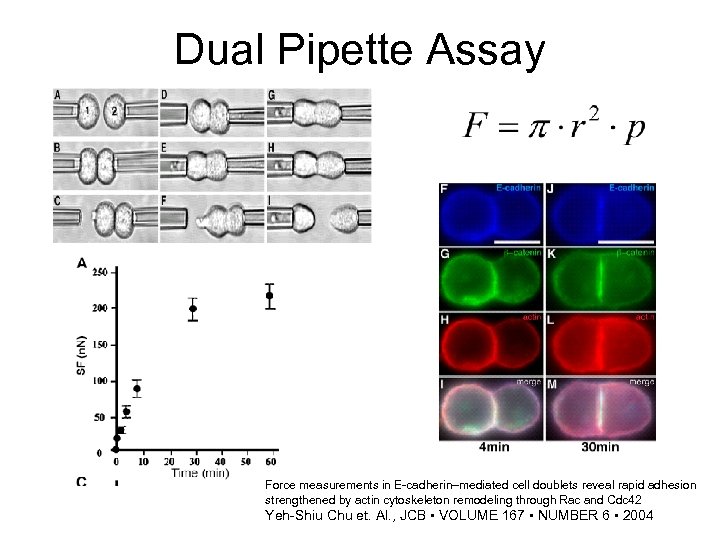
Dual Pipette Assay Force measurements in E-cadherin–mediated cell doublets reveal rapid adhesion strengthened by actin cytoskeleton remodeling through Rac and Cdc 42 Yeh-Shiu Chu et. Al. , JCB • VOLUME 167 • NUMBER 6 • 2004
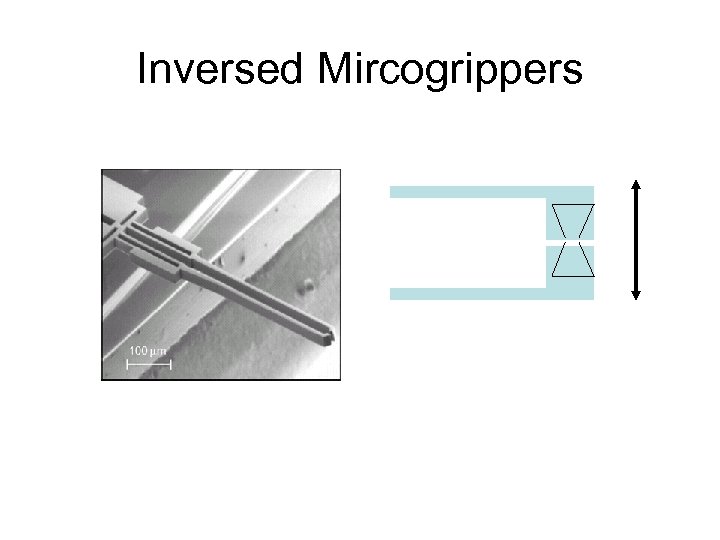
Inversed Mircogrippers
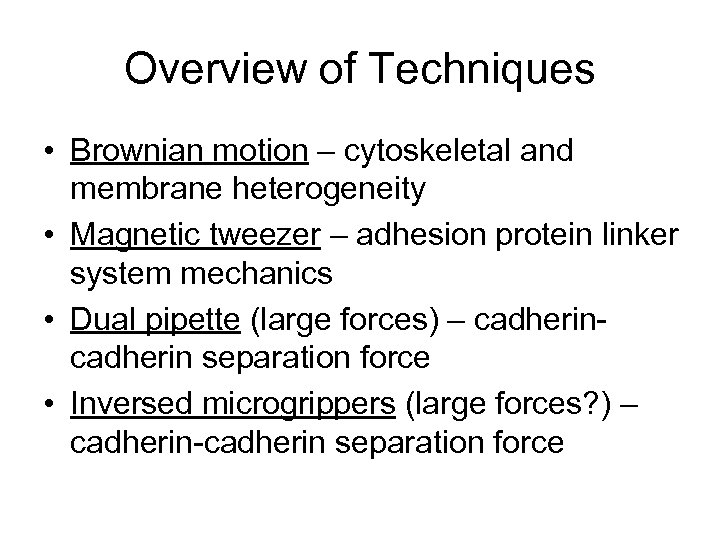
Overview of Techniques • Brownian motion – cytoskeletal and membrane heterogeneity • Magnetic tweezer – adhesion protein linker system mechanics • Dual pipette (large forces) – cadherin separation force • Inversed microgrippers (large forces? ) – cadherin-cadherin separation force
Single device for multi-scale measurements of cell-cell adhesion Now that the tracking and analysis software works and bead-based microrheology has been validated, how can a device be designed that characterizes the mechanical properties of the adhesion system of any cell line over multiple spatial and temporal scales?
Signaling from Cadherins E-cadherin adhesion molecules, receptors, etc. Cadherin Activated Signals Cadherin Dependent Signals
Integration and Miniaturization! Fluorescent microbeads or quantum dots? - imaging brownian motion - tracking force-induced changes in heterogeneities On chip electrical components (e. g. , CMOS)? - signal conditioning - increase signal-to-noise ratio High density GMR sensor array substrate with multiplexing? - removes need for expensive microscope & CCD - directly senses XY-position Microfabricated electromagnets? - removes need for expensive micromanipulators Microfluidics with cell traps? - fixed cell positions (don’t have to search for good cells) Micropatterning? …etc…
e0aaace1bd06029918c2e114b54d23aa.ppt