ebe81edde0f7243daa3a3a4363250e1e.ppt
- Количество слайдов: 29

Machine-learning in building bioinformatics databases for infectious diseases ASEAN-China International Bioinformatics Workshop 2008 17 Apr 2008 Victor Tong Institute for Infocomm Research A*STAR, Singapore
Overview n Definitions and background n Architectures of existing immunological databases n Machine-learning for biological databases n Conclusion
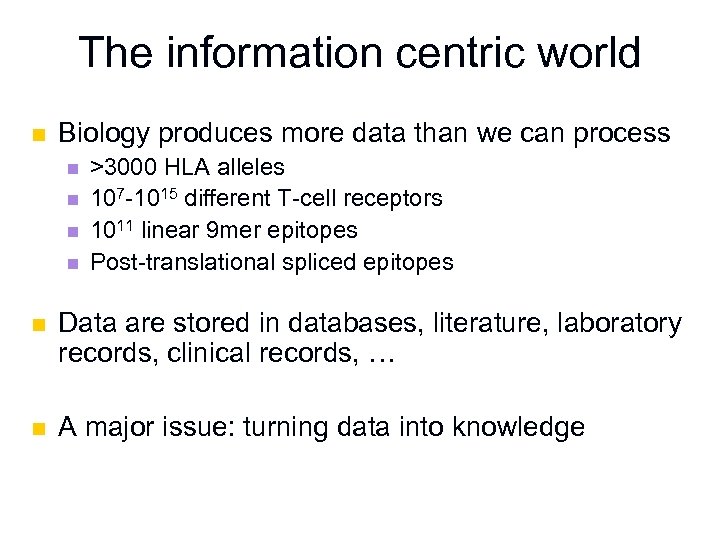
The information centric world n Biology produces more data than we can process n n >3000 HLA alleles 107 -1015 different T-cell receptors 1011 linear 9 mer epitopes Post-translational spliced epitopes n Data are stored in databases, literature, laboratory records, clinical records, … n A major issue: turning data into knowledge

Use of bioinformatics n Impractical to do manual curation n Large amounts of data that are difficult to interpret n n ≥ 16 million Pub. Med abstracts ~80 K immunology related references Protein-protein interaction extraction from text Bioinformatics: systematic construction and updating of databases

Ad hoc bioinformatics Biological system Computational analysis Biological interpretation
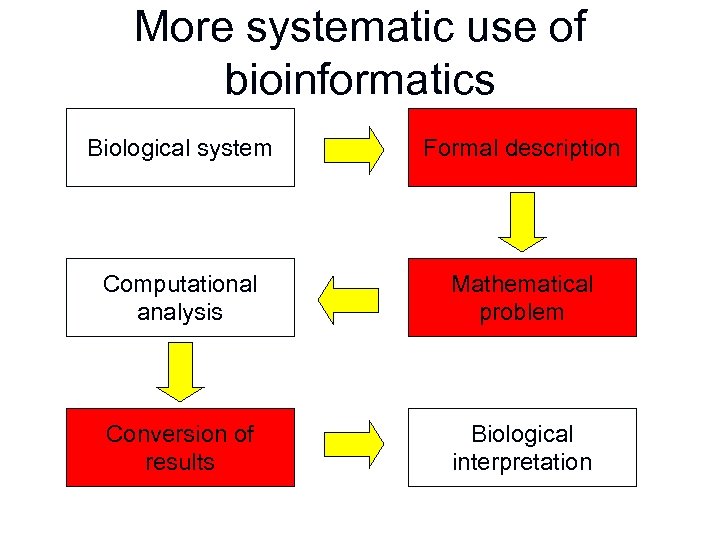
More systematic use of bioinformatics Biological system Formal description Computational analysis Mathematical problem Conversion of results Biological interpretation

Knowledge discovery from databases is the process of automated extraction of useful information or knowledge from individual or multiple databases
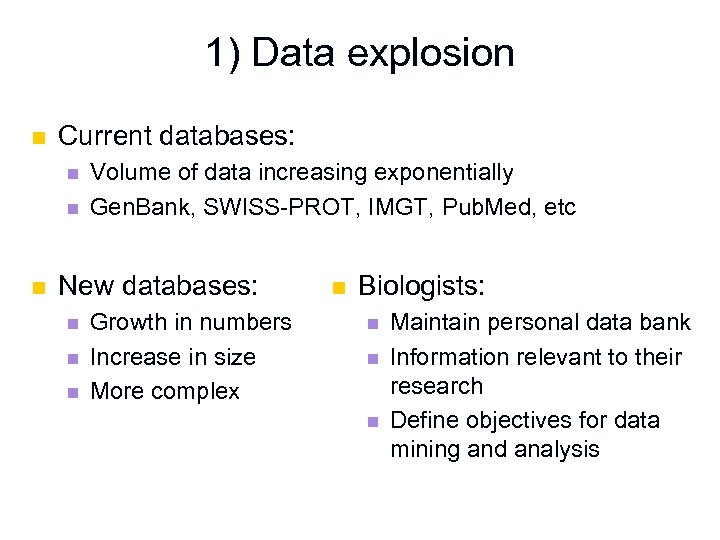
1) Data explosion n Current databases: n n n Volume of data increasing exponentially Gen. Bank, SWISS-PROT, IMGT, Pub. Med, etc New databases: n n n Growth in numbers Increase in size More complex n Biologists: n n n Maintain personal data bank Information relevant to their research Define objectives for data mining and analysis

2) Data quality n Nature of biological data: n n n Fuzzy and complex Varying interpretations Problems with raw data: n n n Inconsistent Inaccurate Redundant Irrelevant Incomplete Incorrect n Data cleaning: n n n Limit on the percentage error that can be tolerated in the data Prevent propagation of errors to our databases Prevent depreciation of data quality

3) Database creation and maintenance n Software tools and programming efforts: n n n Data collection Constructing databases Integrating data mining tools Updating the databases Nature of the databases: n n Short lifespan Hard to maintain
4) Data integration n Disparities in data sources: n n n Data structures Data formats Views Search mechanisms Location
Overview n Definitions and background n Architectures of existing immunological databases n Machine-learning for biological databases n Conclusion
Web-resources for immune epitope information n Immune Epitope Database and Analysis Resource (IEDB) Contains B-cell epitopes, T-cell epitopes, MHC ligands for humans, nonhuman primates, rodents, and other animal species. URL: http: //www. immuneepitope. org n The international Im. Muno. Gene. Tics information system (IMGT) Specializes in Ig, T-cell receptors, MHC, Ig superfamily, MHC superfamily, and related proteins of the immune system of human and other vertebrate species URL: http: //imgt. cines. fr/ n SYFPEITHI Contains ~3, 500 T-cell epitopes, MHC ligands and peptide motifs for humans and rodents URL: http: //www. syfpeithi. de/
Web-resources for immune epitope information n MHCBN Contains T-cell epitopes, TAP ligands, MHC binding peptides and MHC non-binding peptides for humans and rodents URL: http: //www. imtech. res. in/raghava/mhcbn/ n MPID-T Contains 3 D structural information of 187 T-cell receptors, MHCs and interacting epitopes for humans and rodents, spanning 40 alleles URL: http: //surya. bic. nus. edu. sg/mpidt/ n Anti. Jen/Jen. Pep Contains T-cell epitopes, MHC ligands, TAP ligands and B-cell epitopes. URL: http: //www. jenner. ac. uk/antijen/
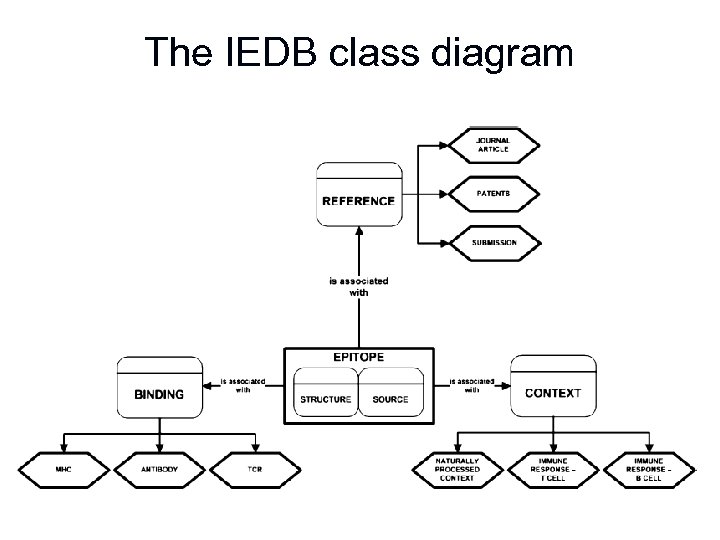
The IEDB class diagram

Relationships between an epitope & contexts


Overview n Definitions and background n Architectures of existing immunological databases n Machine-learning for biological databases n Conclusion
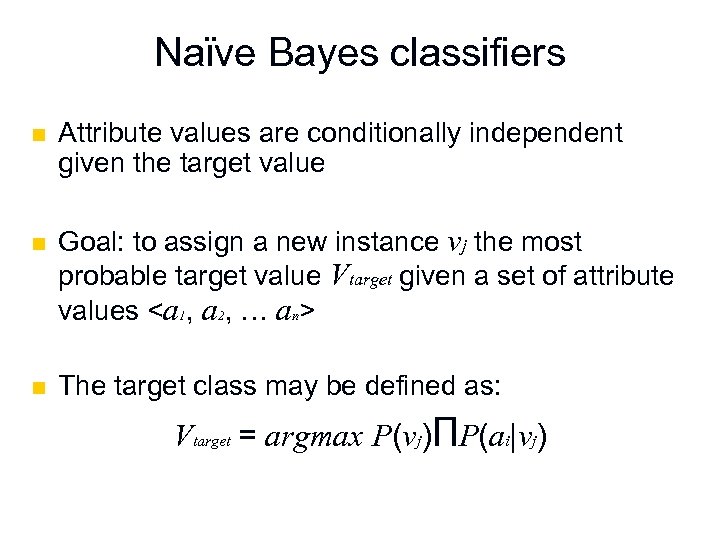
Naϊve Bayes classifiers n n n Attribute values are conditionally independent given the target value Goal: to assign a new instance vj the most probable target value Vtarget given a set of attribute values <a 1, a 2, … an> The target class may be defined as: Vtarget = argmax P(vj)ΠP(ai|vj)

Comparison of popular text classification algorithms n Dataset n n n AROC n n Wang et al. , BMC Bioinformatics 2007, 8: 269 20, 910 Pub. Med abstracts 181, 299 unique words NBC: 0. 838 ANN: 0. 831 SVM: 0. 825 DT: 0. 809

Feature selection (FS) n Data source n n n Pub. Med abstracts Medical Subject Headings (Me. SH) - National Library of Medicine's controlled vocabulary used for indexing articles, for cataloging books and other holdings Publication title Author(s) etc

Feature selection (FS) n Algorithms n n Document frequency (DF) – ranks features based on the number of abstracts they appear in Information gain (IG) – measures the number of bits of information obtained for category prediction based on their occurrence in a document IG(u) = -∑ P(ci) log P(ci) + P(u) ∑ P(ci|u) log P(ci|u) + P(t) ∑ P(ci|ū) log P(ci|ū) where u is the feature of interest, ci (i = 1, …, m) denotes the set of categories the documents belong to
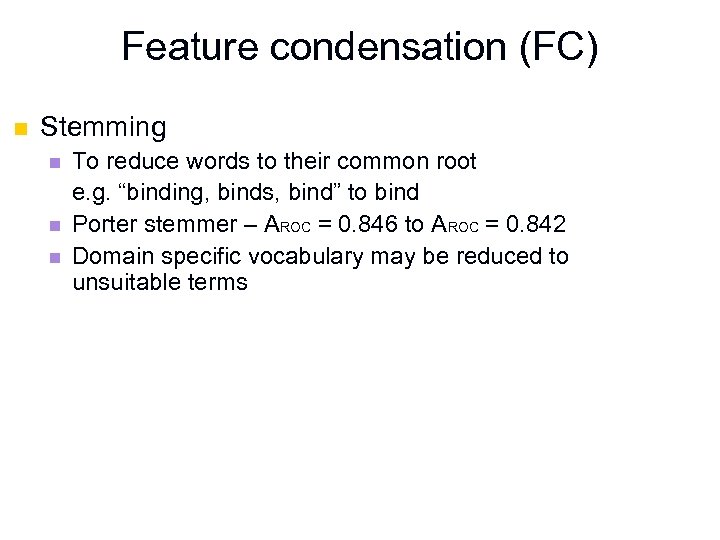
Feature condensation (FC) n Stemming n n n To reduce words to their common root e. g. “binding, binds, bind” to bind Porter stemmer – AROC = 0. 846 to AROC = 0. 842 Domain specific vocabulary may be reduced to unsuitable terms

Feature extraction (FE) n Rules to capture immune related expressions and group them together n n n Reduction of feature space (i. e. no. of unique words) Enrichment of information content Better performance?

Feature extraction (FE) n Examples: n n n Sequence length – identify sequence length and replace with “~range<50~” or “~range>50~” if sequences to be mapped stretches 50 amino acids MHC alleles – identify MHC alleles and replace with “~mhc_allele~” Protein sequences – identify sequences as a) exclusively containing characters representing the 20 aa, b) in upper case, length > threshold, and replace with “~sequence~”
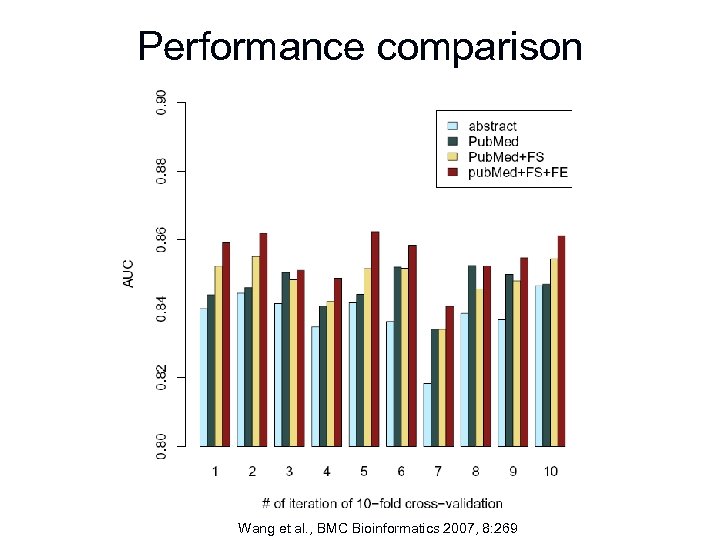
Performance comparison Wang et al. , BMC Bioinformatics 2007, 8: 269
Overview n Definitions and background n Architectures of existing immunological databases n Machine-learning for biological databases n Conclusion

Conclusion n Machine-learning algorithms enable systematic approach to database construction and facilitates scientific discovery n It must be performed with due care and must be scientifically and technically sound
ebe81edde0f7243daa3a3a4363250e1e.ppt