768b73832dd6f8567c14553cc4699c72.ppt
- Количество слайдов: 23
M. S. Thesis Defense Protein Structure Database for Structural Genomics Group Jessica Lau December 13, 2004
• Bioinformatics is • • • At the Northeast Structural Genomics Consortium, database management systems play a large role in its daily operation • • Analysis of biological data: gene expression, DNA sequence, protein sequence. Data mining and management of biological information through database systems. Data collection and mining of experimental results Track target progress – status milestones Exchange information with rest of the world My thesis presents work in database management systems at the NESG. • • • Part 1: Zeba. View Part 2: Worm Structure Gallery Part 3: Prototype of NESG Structure Gallery
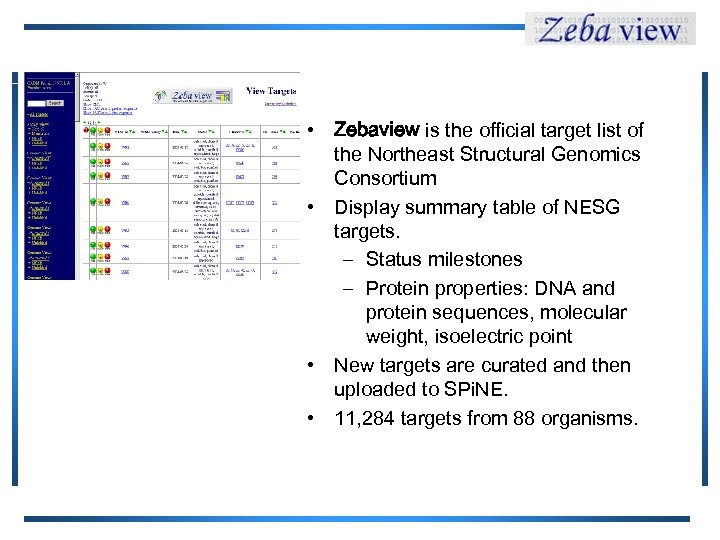
• Zebaview is the official target list of the Northeast Structural Genomics Consortium • Display summary table of NESG targets. – Status milestones – Protein properties: DNA and protein sequences, molecular weight, isoelectric point • New targets are curated and then uploaded to SPi. NE. • 11, 284 targets from 88 organisms.
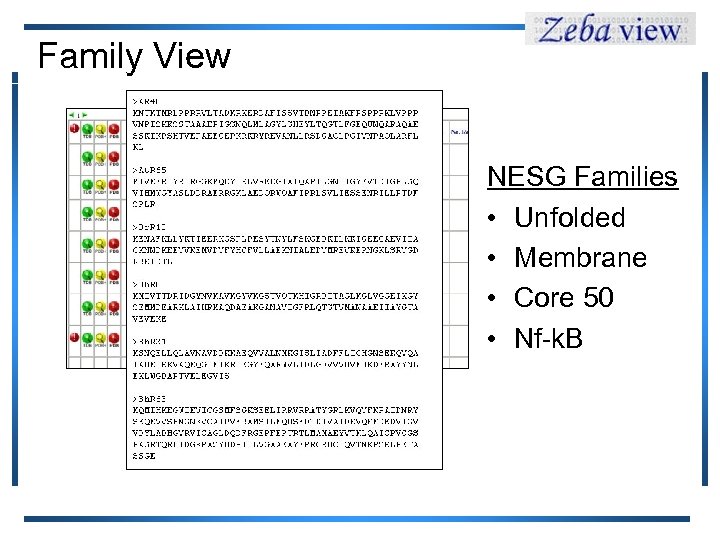
Family View NESG Families • Unfolded • Membrane • Core 50 • Nf-k. B
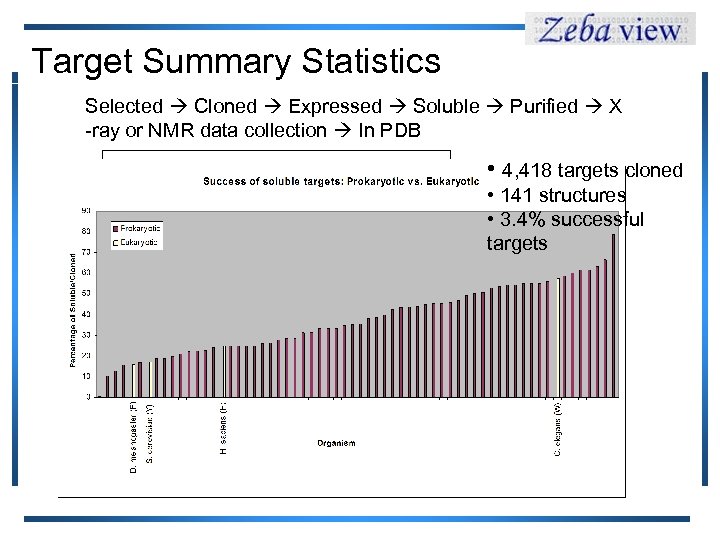
Target Summary Statistics Selected Cloned Expressed Soluble Purified X -ray or NMR data collection In PDB • 4, 418 targets cloned • 141 structures • 3. 4% successful targets
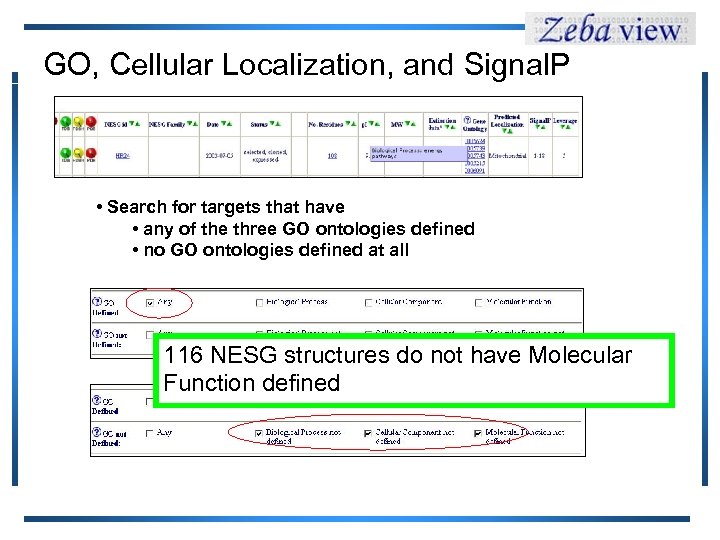
GO, Cellular Localization, and Signal. P • Search for targets that have • any of the three GO ontologies defined • no GO ontologies defined at all 116 NESG structures do not have Molecular Function defined
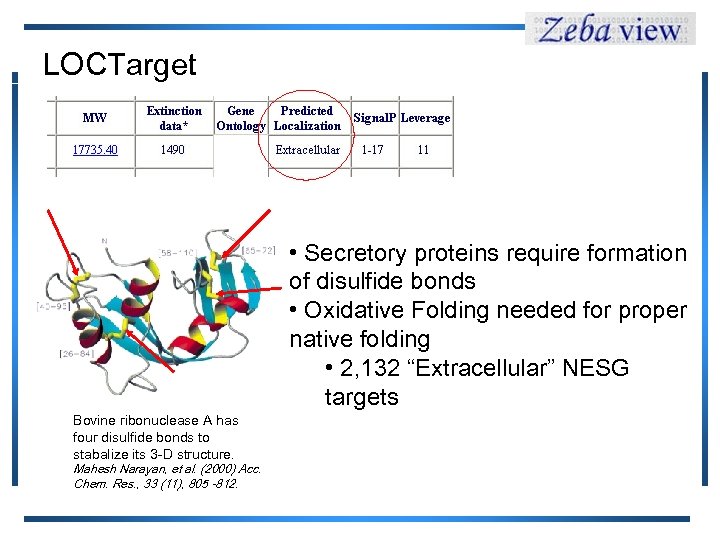
LOCTarget Bovine ribonuclease A has four disulfide bonds to stabalize its 3 -D structure. Mahesh Narayan, et al. (2000) Acc. Chem. Res. , 33 (11), 805 -812. • Secretory proteins require formation of disulfide bonds • Oxidative Folding needed for proper native folding • 2, 132 “Extracellular” NESG targets
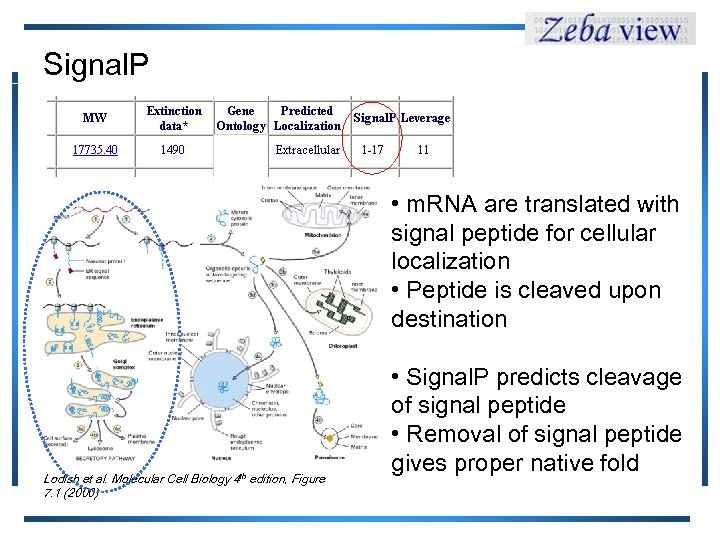
Signal. P • m. RNA are translated with signal peptide for cellular localization • Peptide is cleaved upon destination Lodish et al. Molecular Cell Biology 4 th edition, Figure 7. 1 (2000) • Signal. P predicts cleavage of signal peptide • Removal of signal peptide gives proper native fold
Part 2 – Worm Structure Gallery
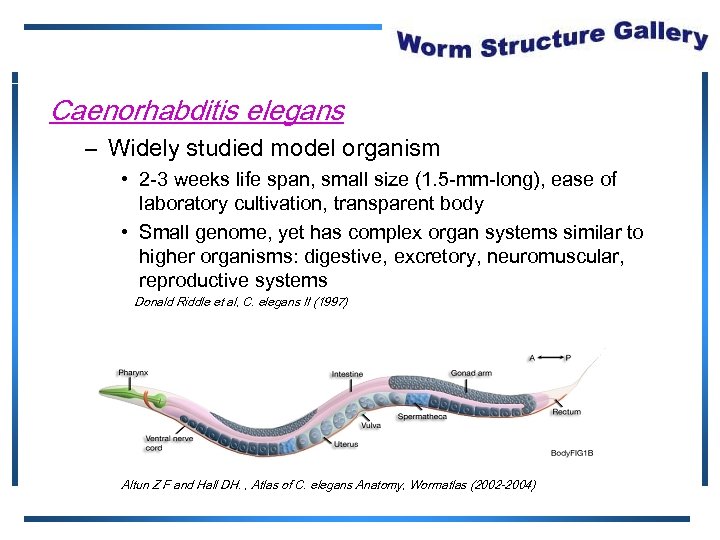
Caenorhabditis elegans – Widely studied model organism • 2 -3 weeks life span, small size (1. 5 -mm-long), ease of laboratory cultivation, transparent body • Small genome, yet has complex organ systems similar to higher organisms: digestive, excretory, neuromuscular, reproductive systems Donald Riddle et al, C. elegans II (1997) Altun Z F and Hall DH. , Atlas of C. elegans Anatomy, Wormatlas (2002 -2004)
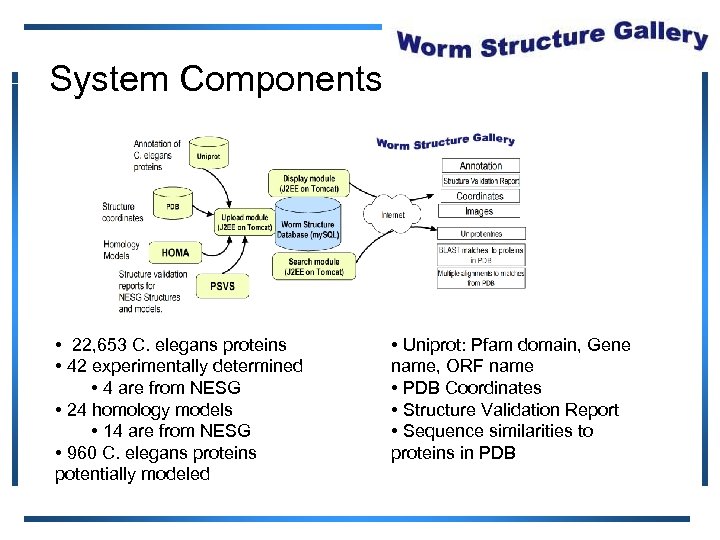
System Components • 22, 653 C. elegans proteins • 42 experimentally determined • 4 are from NESG • 24 homology models • 14 are from NESG • 960 C. elegans proteins potentially modeled • Uniprot: Pfam domain, Gene name, ORF name • PDB Coordinates • Structure Validation Report • Sequence similarities to proteins in PDB
Protein Structure Validation Software • Suite of quality validation software – PROCHECK • Quality of experimental data • Distribution of φ, ψ angles in Ramachandran plot – Mol. Probity Clashscore • Number of H atom clashes per 1, 000 atoms • With respect to a set of scores from 129 high resolution X-ray crystal structures • < 500 residues, of resolution <= 1. 80 Å, R-factor <= 0. 25 and R-free <= 0. 28; Bahattacharya, A et al. to be published
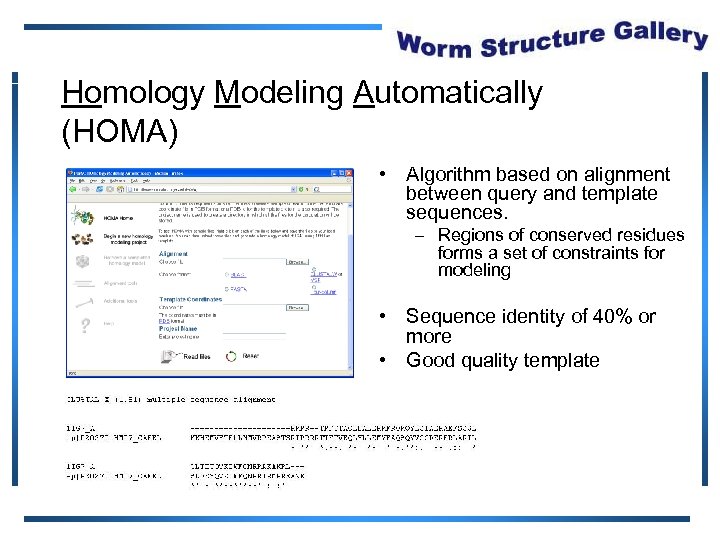
Homology Modeling Automatically (HOMA) • Algorithm based on alignment between query and template sequences. – Regions of conserved residues forms a set of constraints for modeling • Sequence identity of 40% or more • Good quality template
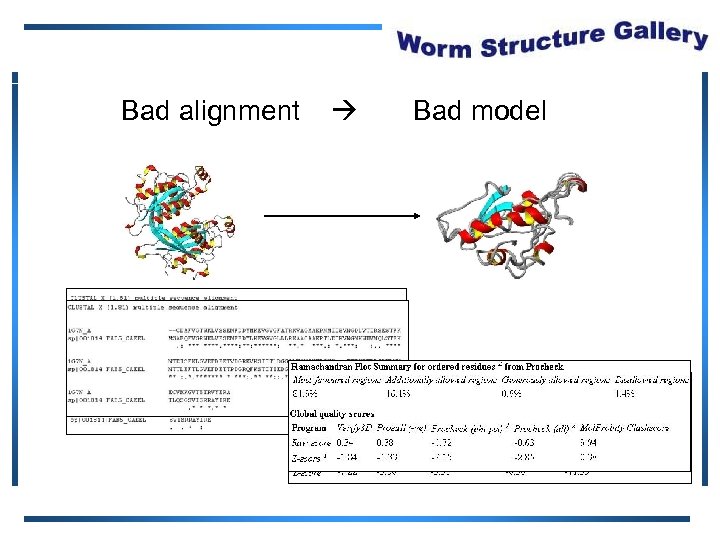
Bad alignment Bad model
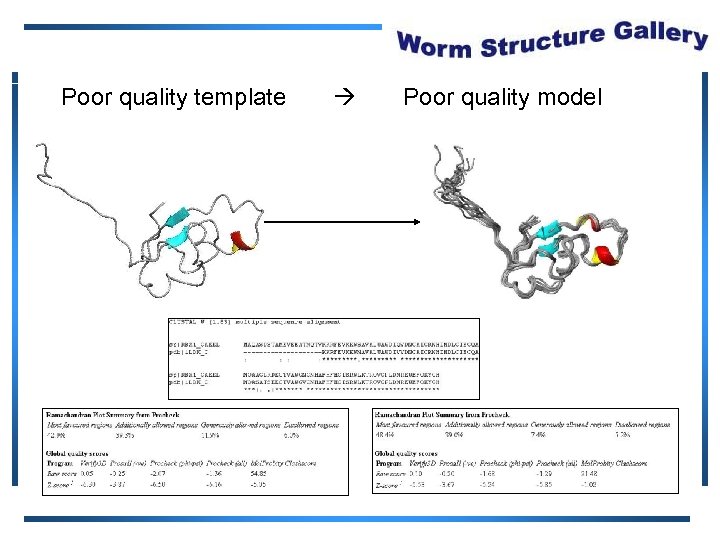
Poor quality template Poor quality model
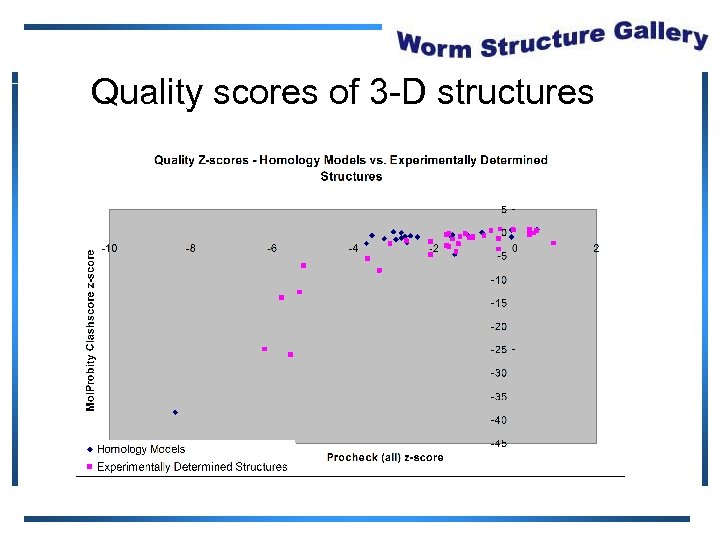
Quality scores of 3 -D structures
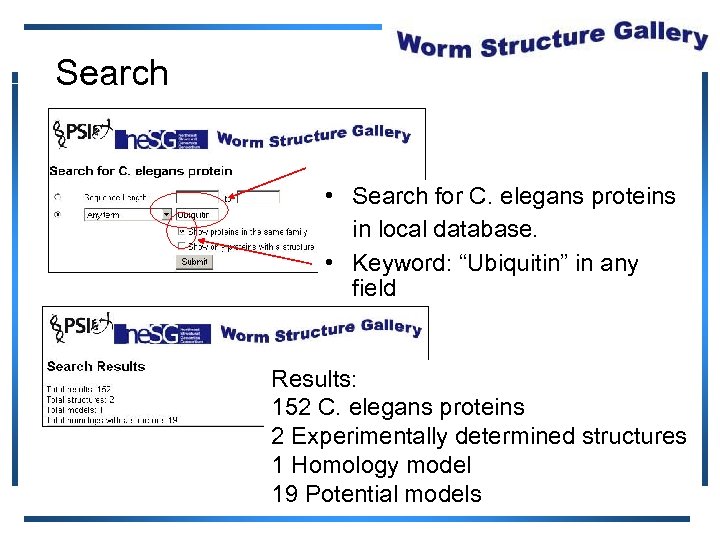
Search • Search for C. elegans proteins in local database. • Keyword: “Ubiquitin” in any field Results: 152 C. elegans proteins 72 C. elegans proteins 2 Experimentally determined structures 1 Homology model 19 11 Potential models
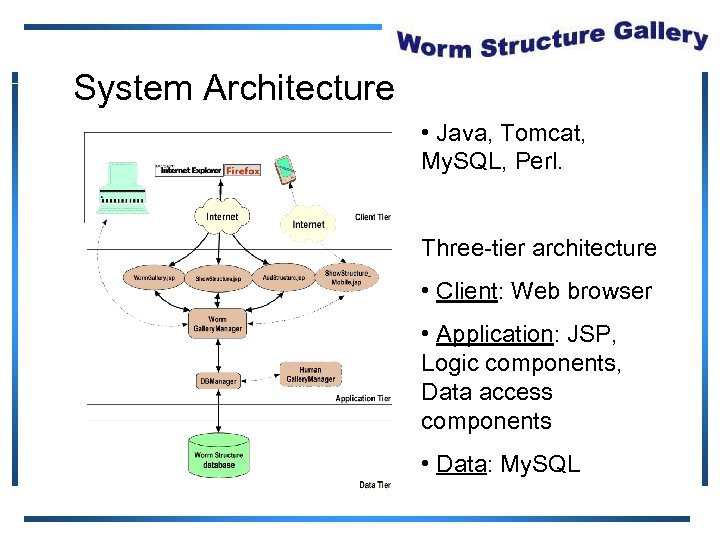
System Architecture • Java, Tomcat, My. SQL, Perl. Three-tier architecture • Client: Web browser • Application: JSP, Logic components, Data access components • Data: My. SQL
Part 3 – NESG Structure Gallery
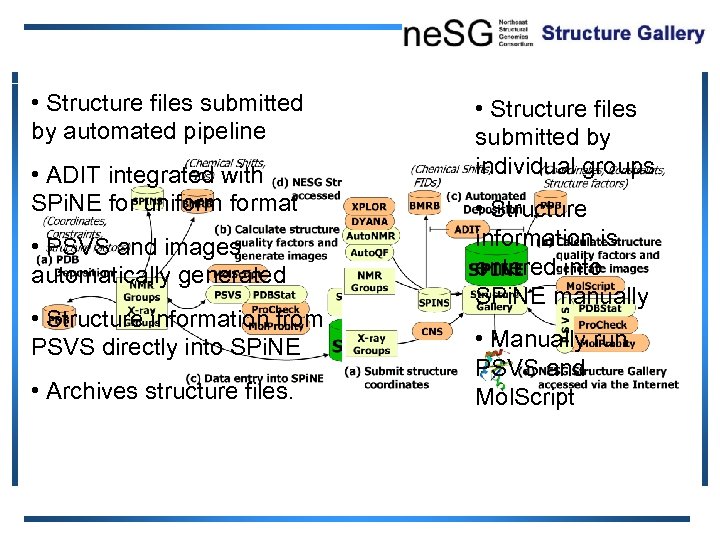
• Structure files submitted by automated pipeline • ADIT integrated with SPi. NE for uniformat • PSVS and images automatically generated • Structure information from PSVS directly into SPi. NE • Archives structure files. • Structure files submitted by individual groups • Structure information is entered into SPi. NE manually • Manually run PSVS and Mol. Script
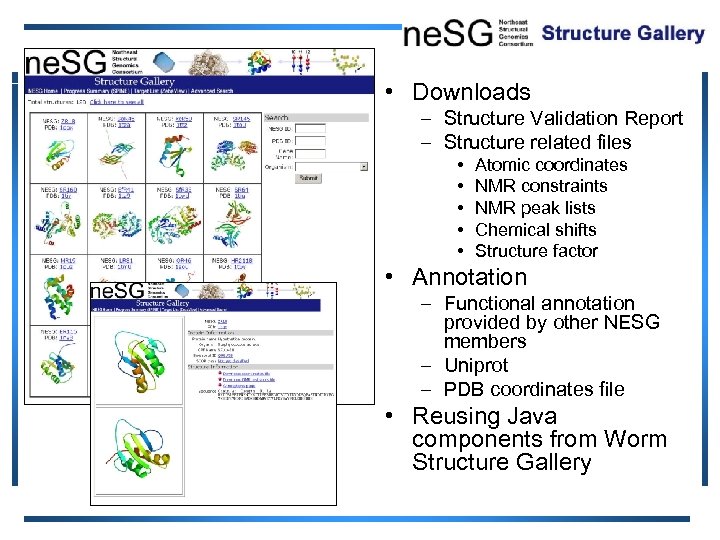
• Downloads – Structure Validation Report – Structure related files • • • Atomic coordinates NMR constraints NMR peak lists Chemical shifts Structure factor • Annotation – Functional annotation provided by other NESG members – Uniprot – PDB coordinates file • Reusing Java components from Worm Structure Gallery
– Enhance Zeba. View performance to handle increased load and functionalities – Integrate annotation from other protein and structure databases. – Make modules available for other java-based applications within structural genomics. – Develop a gallery for other organisms: yeast, fruit fly, human – Continue specifications for the new NESG Structure Gallery

Advisor: Dr. Gaetano Montelione Thanks to everyone at the Protein NMR lab and NESG! Aneerban Bhattacharya John Everett All the scientists who solved the structures!
768b73832dd6f8567c14553cc4699c72.ppt