882755436ae423dbc85e1af32aeadcfa.ppt
- Количество слайдов: 87

Linking Multiple Ontologies: The OBO Foundry Approach Chris Mungall NIAID Cell Ontology Workshop May 2008

Outline • Introduction to ontologies – The OBO perspective – Case study in the Gene Ontology • • The OBO Foundry: goals and principles The OBO relation ontology Organization of ontologies in OBO Modularity – An example from CL • Linking CL to the OBO Foundry
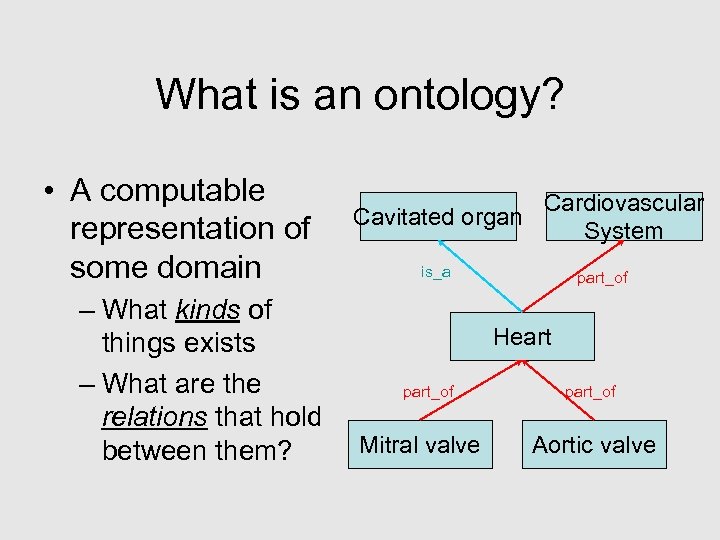
What is an ontology? • A computable representation of some domain – What kinds of things exists – What are the relations that hold between them? Cardiovascular Cavitated organ System is_a part_of Heart part_of Mitral valve part_of Aortic valve

Aspects of an ontology • Identifiers – Uniquely identify a class / term • E. g. CL: 0000037 is ID for the term “hematopoietic stem cell” – Identifier metadata • Terminological aspects – Names and synonyms/alternate labels • CL: 0000037 has “hemopoietic progenitor cell” as a related synonym and “hemopoietic stem cell” as exact synonym • Logical aspects – Relations – Definitions Provenance

Some ontologies and their uses • The Gene Ontology – Annotation of gene products – Analyzing high-throughput datasets • Anatomical ontologies (including CL) – – Experimental metadata Image annotation Indicating location of gene expression Creating Phenotypic descriptions • Others – NLP – Annotating information models
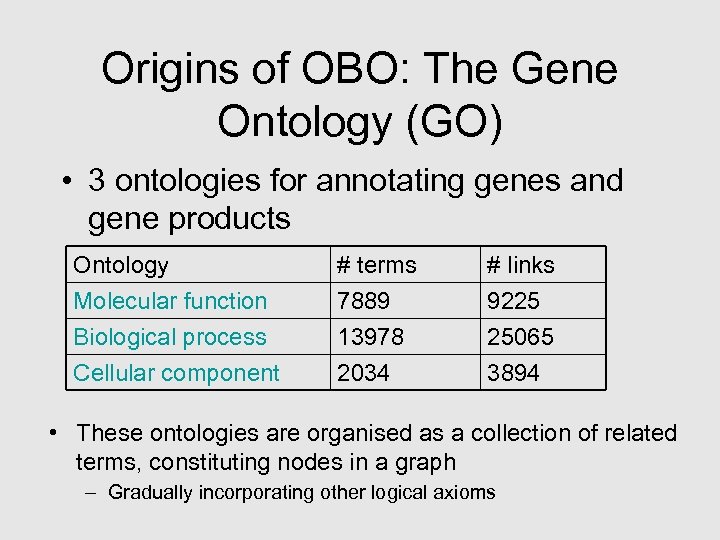
Origins of OBO: The Gene Ontology (GO) • 3 ontologies for annotating genes and gene products Ontology Molecular function Biological process Cellular component # terms 7889 13978 2034 # links 9225 25065 3894 • These ontologies are organised as a collection of related terms, constituting nodes in a graph – Gradually incorporating other logical axioms

Annotation and GO • GO Annotations: – Associations between genes and GO terms, with evidence – Met 17 : “methionine metabolism” GO: 0006555 • 222, 000 genes and gene products have high quality annotations to GO terms – 3. 4 m including automated predictions – 66, 000 publications curated • Variety of analysis tools – http: //www. geneontology. org/GO. tools. shtml#micro

GO and high-throughput biology: Over-representation of GO terms for gene sets GO: : Term. Finder Sherlock et al

GO and the need for OBO • GO terms implicitly reference kinds of entities outwith the scope of GO chemical cell anatomy OBO was born from the need to create source quality – – • Methionine biosynthesis Neural crest cell migration Cardiac muscle morphogenesis Regulation of vascular permeability ontologies for GO term ‘cross-products’ – Define composite classes in terms of simpler ones

The Open Biomedical Ontologies (OBO) Foundry • A collection of orthogonal reference ontologies in the biological/biomedical domain • The OBO Foundry: Each is committed to an agreed upon set of principles governing best practices in ontology development

Some OBO ontologies Gene Ontology • SO - sequence features Ch. EBI - chemical entities • Model organism anatomy OBI - investigations – ZFA PATO, MP - phenotypes – Fly_anat CL - cells – Dicty_anat ENVO - environment and – Mouse_anat habitat –… • DO - Human diseases • CARO - common anatomy • OBO Relation Ontology • FMA - human anatomy • • •

OBO Foundry: criteria, v 1 • Open • Well-defined exchange format E. g. OBO or OWL • • • Uses identifiers according to OBO ID policy Ontology Life-cycle / versioning Has clearly specified and delineated content Has unambiguous definitions Uses or extends relations in the OBO Relation Ontology Well documented Has a plurality of users (and a mail list & issue tracker) Developed collaboratively Orthogonal, modular http: //obofoundry. org/

OBO Relation Ontology • Edges can link nodes… – Within ontologies – Across ontologies • The precise meaning of the relation is important – Relations have formal definitions – Rules for composing relations together – http: //obofoundry. org/ro/
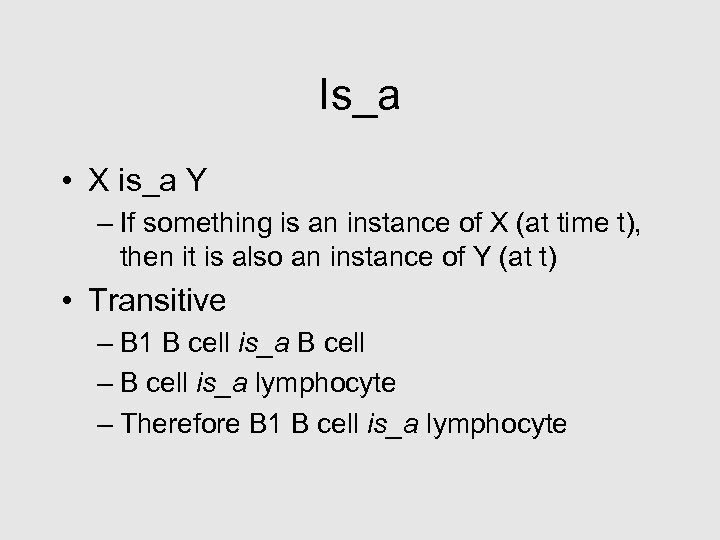
Is_a • X is_a Y – If something is an instance of X (at time t), then it is also an instance of Y (at t) • Transitive – B 1 B cell is_a B cell – B cell is_a lymphocyte – Therefore B 1 B cell is_a lymphocyte

Part_of • Instance level part_of relation is primitive • Between classes: – X part_of Y : • • Every instance of X is part_of some instance of Y Paneth cell part_of intestine : YES Nucleus part_of Cell : YES Neuron part_of brain : NO – (there are some neurons that are part of others parts of the nervous system) • Transitive – X part_of Y, Y part_of Z • Therefore, X part_of Z

Has_part • Instance level inverse of part_of • X has_part Y – Every X has some Y as part – Cell has_part nucleus : NO – Nucleate erythrocyte has_part nucleus : YES
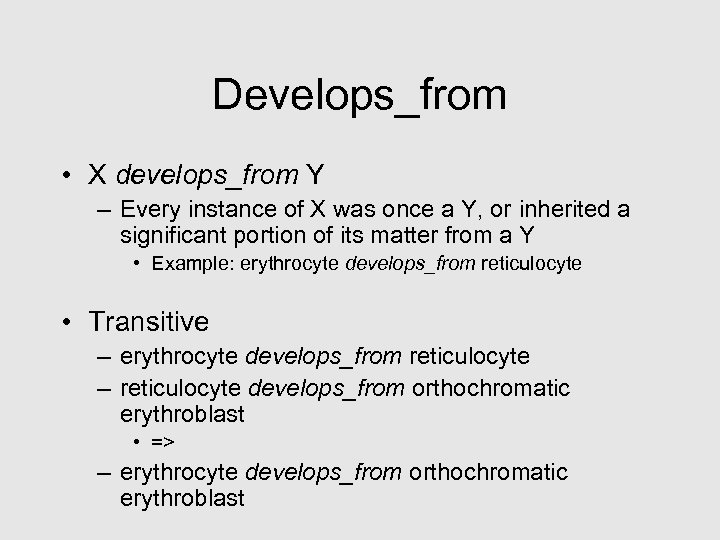
Develops_from • X develops_from Y – Every instance of X was once a Y, or inherited a significant portion of its matter from a Y • Example: erythrocyte develops_from reticulocyte • Transitive – erythrocyte develops_from reticulocyte – reticulocyte develops_from orthochromatic erythroblast • => – erythrocyte develops_from orthochromatic erythroblast
Transformation and derivation • Develops_from relation can be refined into two cases: – Transformation_of • X transformation_of Y : – Any instance of X was previously an instance of Y – Example: erythrocyte transformation_of reticulocyte – Derives_from • X derives_from Y : – Holds between distinct instances where Y inherits matter from X • Most OBO ontologies just use the develops_from relation
Other relations • Inherence – Between a quality and an object – E. g. between a specific shape and a cell • Participation – Between a process and an object – E. g. between a B cell and an immune process
Definitions state necessary and sufficient conditions • Links in the ontology graph state necessary conditions for a class • E. g. erythroid progenitor cell develops_from megakaryocyte erythroid progenitor – These characteristics may not be unique • A definition should state necessary and sufficient conditions for a class – The characteristics must be unique to the defined class • E. g. “progenitor cell that is committed to the erythroid lineage” • Definition should be precise and (as far as possible) translated / translatable to logical
Genus differentia definitions • Of the form – An X is a G that D – G should be in the same ontology – D is discriminating characteristics that differentiate (in the classification sense) Xs from other Gs. • Relations to terms in an ontology (the same ontology or a different one) • Example: – A B cell is a lymphocyte that expresses an immunoglubulin complex

Orthogonality of ontologies • No two ontologies should represent the same kind of entity – E. g. “B-cell” should only be represented in one ontology – Related entities should be coordinated across ontologies • GO: “B-cell differentiation” • Exceptions: – The term “cell” connects GO Cellular Component (cell parts) and CL (cells) • Advantages: – Reduces redundancy and work – Easier to make the union consistent
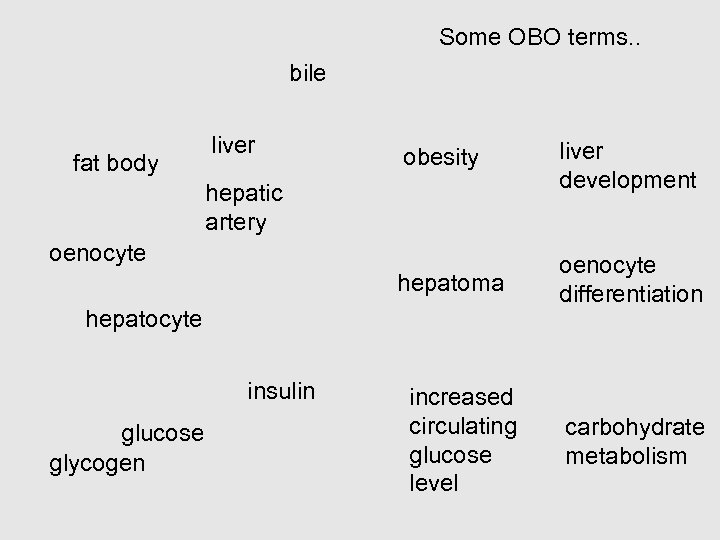
Some OBO terms. . bile liver obesity liver development hepatoma fat body oenocyte differentiation hepatic artery oenocyte hepatocyte insulin glucose glycogen increased circulating glucose level carbohydrate metabolism
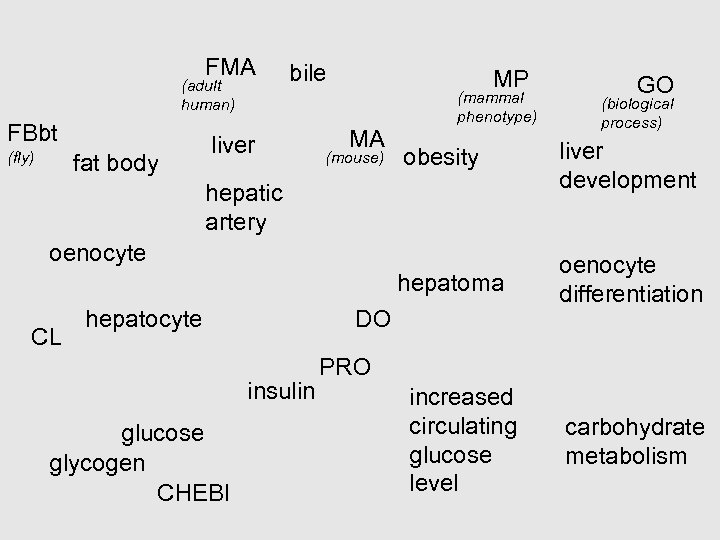
FMA (adult human) FBbt bile liver MP MA (mammal phenotype) GO (biological process) fat body (mouse) obesity liver development hepatoma (fly) oenocyte differentiation hepatic artery oenocyte CL hepatocyte DO insulin glucose glycogen CHEBI PRO increased circulating glucose level carbohydrate metabolism
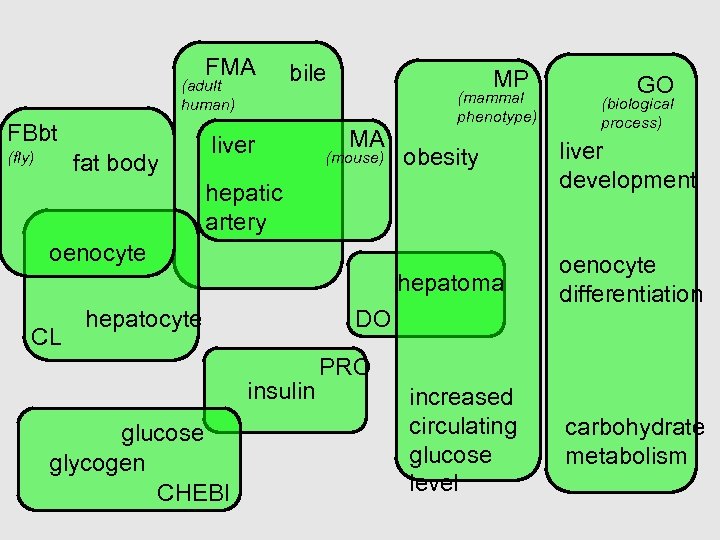
FMA (adult human) FBbt bile liver MP MA (mammal phenotype) GO (biological process) fat body (mouse) obesity liver development hepatoma (fly) oenocyte differentiation hepatic artery oenocyte CL hepatocyte DO insulin glucose glycogen CHEBI PRO increased circulating glucose level carbohydrate metabolism
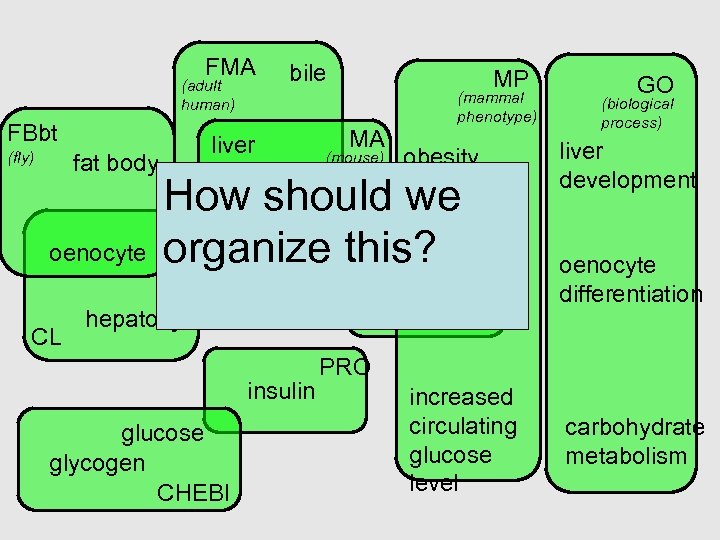
FMA (adult human) FBbt bile liver MP MA (mammal phenotype) GO (biological process) fat body oenocyte CL (mouse) obesity liver development hepatoma (fly) oenocyte differentiation hepatic How should we artery organize this? hepatocyte DO insulin glucose glycogen CHEBI PRO increased circulating glucose level carbohydrate metabolism

Top-level organisation (BFO: Basic Formal Ontology) • General categories – 3 D things (continuants) • Independent – Cells, organs, molecules • Dependent – Shapes, sizes, concentrations, … – 4 D things (processes) • Processes • Useful organisational principle for OBO • is_a and part_of should not cross top level categories • Levels of granularity (scale) – – – Population Organism Organ Cell Molecule • part_of relations can cross levels
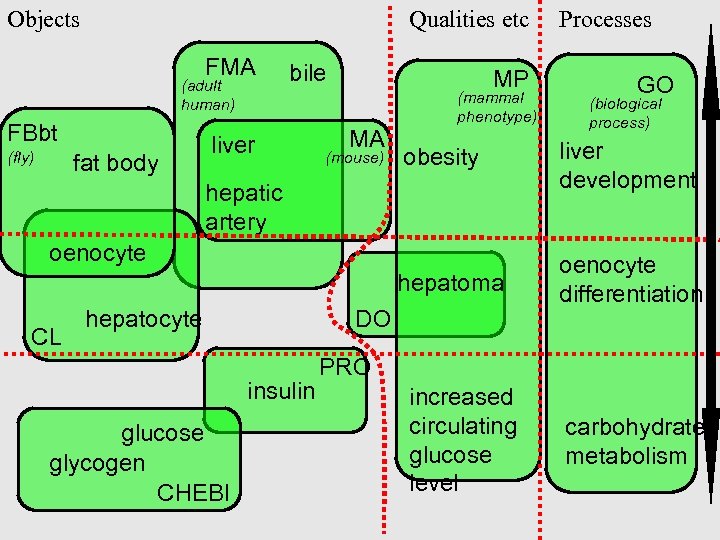
Objects Qualities etc FMA (adult human) FBbt bile liver MP MA (mammal phenotype) Processes GO (biological process) fat body (mouse) obesity liver development hepatoma (fly) oenocyte differentiation hepatic artery oenocyte CL hepatocyte DO insulin glucose glycogen CHEBI PRO increased circulating glucose level carbohydrate metabolism


The OBO Foundry can help with modular ontology design • Biology is complex – So our ontologies will be complex – Multiple purposes – Multiple means of classifying • Separate out different aspects – Modular approach – Avoid multiple inheritance (>1 is_a parent) • Don’t over-use is_a • Don’t cross aspects with is_a • Make complex descriptions from simpler parts – Polyhierarchies arise from composition
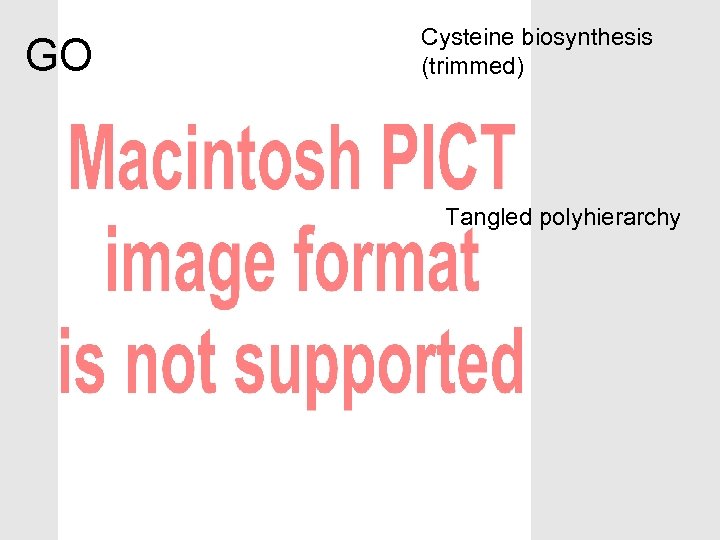
GO Cysteine biosynthesis (trimmed) Tangled polyhierarchy
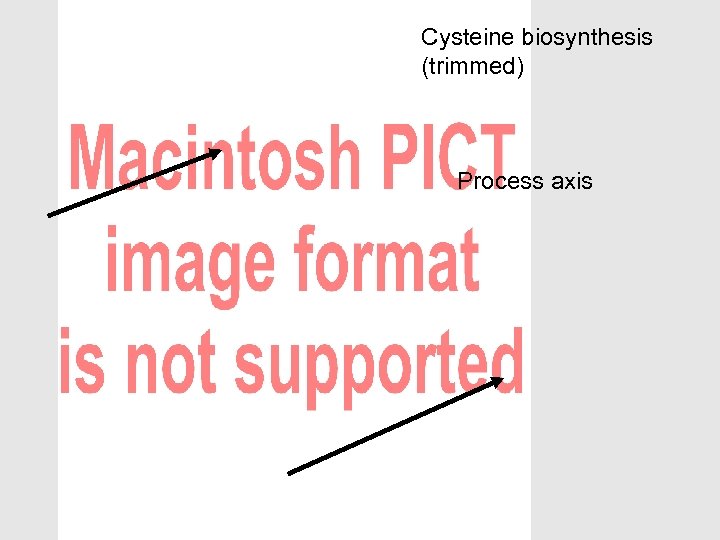
Cysteine biosynthesis (trimmed) Process axis
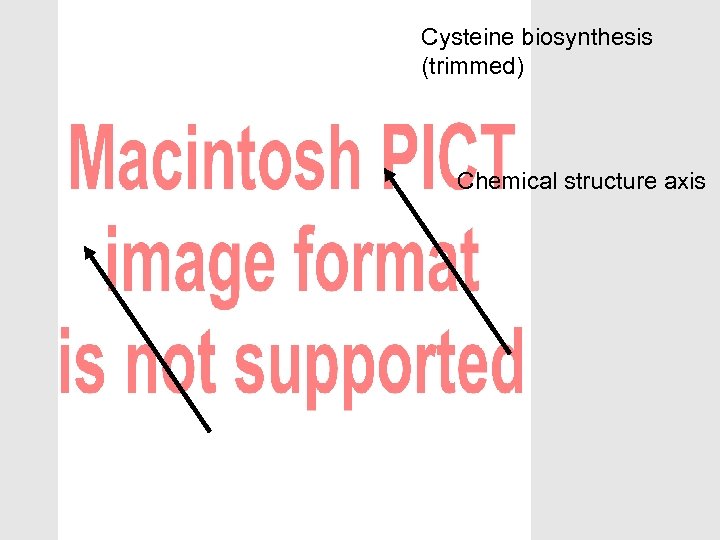
Cysteine biosynthesis (trimmed) Chemical structure axis
Cysteine biosynthesis (trimmed) Ch. EBI (trimmed)
Cysteine biosynthesis (trimmed) Ch. EBI (trimmed)
Cysteine biosynthesis (trimmed) Ch. EBI (trimmed)
Cysteine biosynthesis (trimmed) We can do more than simply link terms: Cross-products (aka logical definitions, Computable genusdifferentia definitions) Ch. EBI (trimmed)
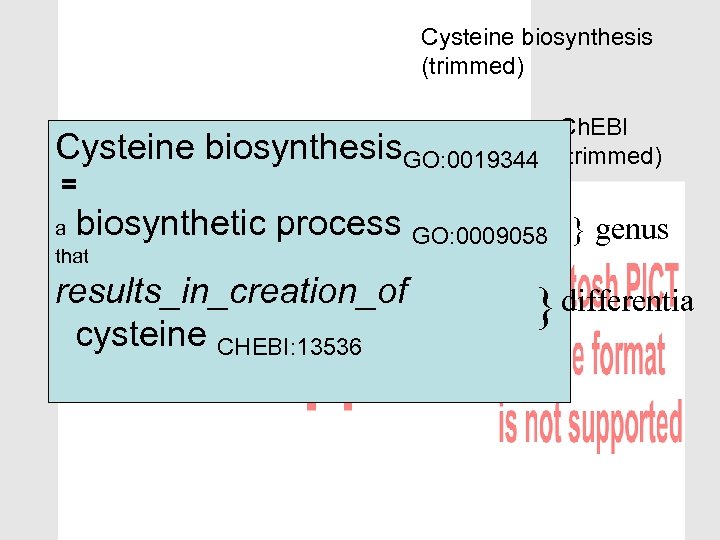
Cysteine biosynthesis (trimmed) Cysteine = Ch. EBI biosynthesis. GO: 0019344 (trimmed) biosynthetic process GO: 0009058 } genus a that results_in_creation_of cysteine CHEBI: 13536 } differentia

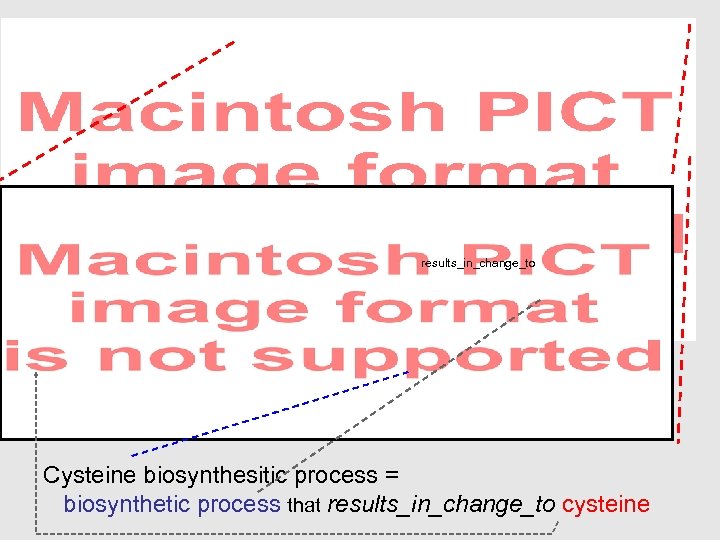
results_in_change_to Cysteine biosynthesitic process = biosynthetic process that results_in_change_to cysteine
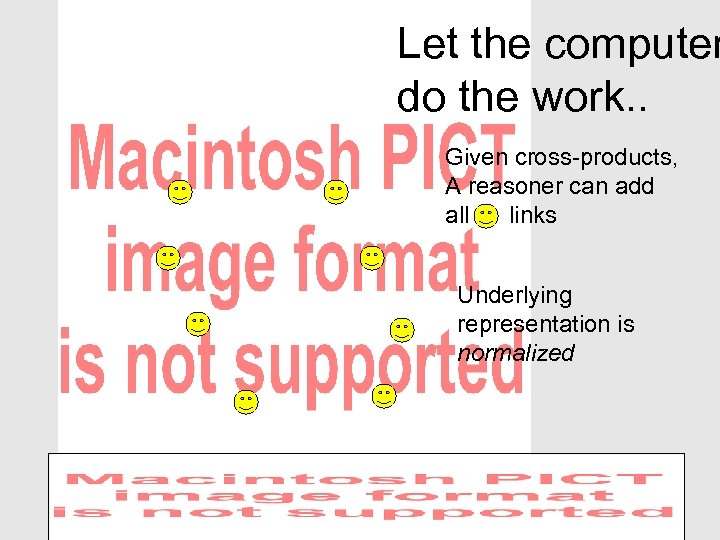
Let the computer do the work. . Given cross-products, A reasoner can add all links Underlying representation is normalized
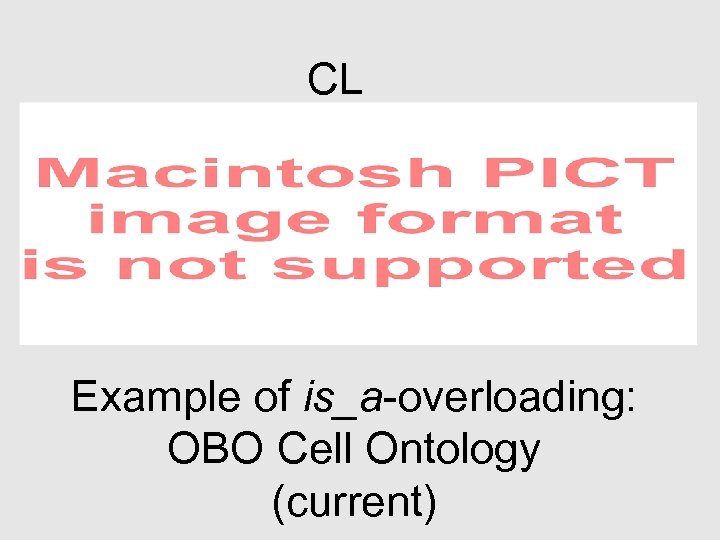
CL Example of is_a-overloading: OBO Cell Ontology (current)

CL X • Try not to assert too many is_a parents

CL GO X Has function • Reuse existing ontologies • Non-is_a relation ?

How CL can use other OBO ontologies • GO Cellular component – Mononuclear phagocyte – B cell (expresses immunoglubulin complex) • GO Biological process – Photosynthetic cell • PATO Qualities – Spiny neuron • CHEBI Chemical entities – X secreting cell • Anatomy Ontologies – CNS neuron Molecular function, PRO - CD 4 positive cell
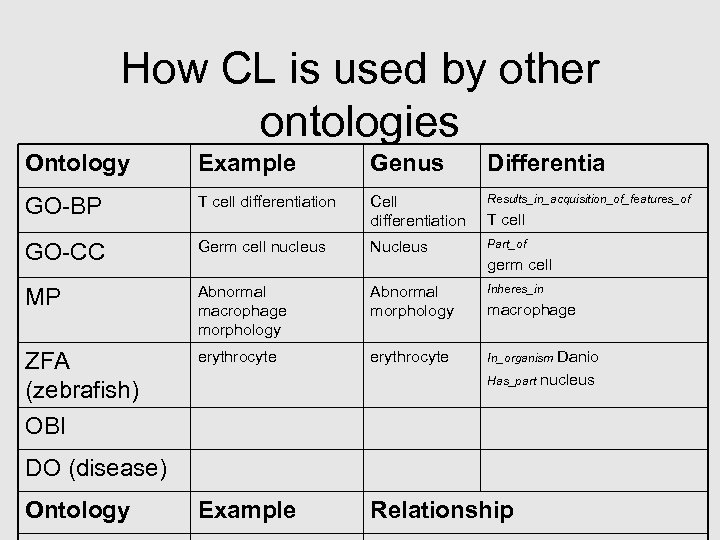
How CL is used by other ontologies Ontology Example Genus Differentia GO-BP T cell differentiation Cell differentiation Results_in_acquisition_of_features_of GO-CC Germ cell nucleus Nucleus Part_of MP Abnormal macrophage morphology Abnormal morphology Inheres_in ZFA (zebrafish) OBI erythrocyte In_organism Example Relationship T cell germ cell macrophage Danio Has_part nucleus DO (disease) Ontology
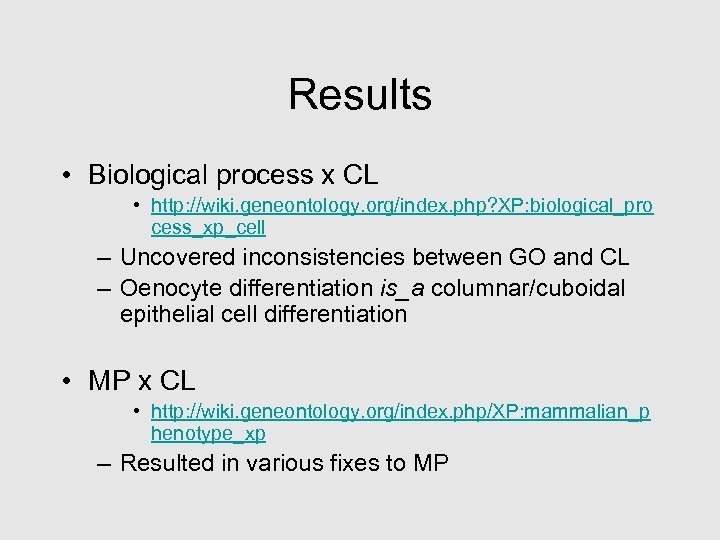
Results • Biological process x CL • http: //wiki. geneontology. org/index. php? XP: biological_pro cess_xp_cell – Uncovered inconsistencies between GO and CL – Oenocyte differentiation is_a columnar/cuboidal epithelial cell differentiation • MP x CL • http: //wiki. geneontology. org/index. php/XP: mammalian_p henotype_xp – Resulted in various fixes to MP

OBD: Ontology Annotation Database

Summary • The cell ontology is a representation of the types of cell that exist • The OBO Foundry provides – Principles – A framework for connecting ontologies • There are many points of coordination between CL and other OBO ontologies • CL could benefit from the gradual introduction of a modular approach

The Gene Ontology; and beyond • Curation of genes and gene products – Molecular function – Biological process – Cellular component Multiple databases using the same ontology GO
The Gene Ontology; and beyond • Curation of genes and gene products – Molecular function – Biological process – Cellular component • What about curation of other data types? – Expression, transcriptomics – Genetics, phenotypes and disease – Many others. . • OBO – Open Bio-Ontologies – Arose partly in response to requirements outside scope of GO GO
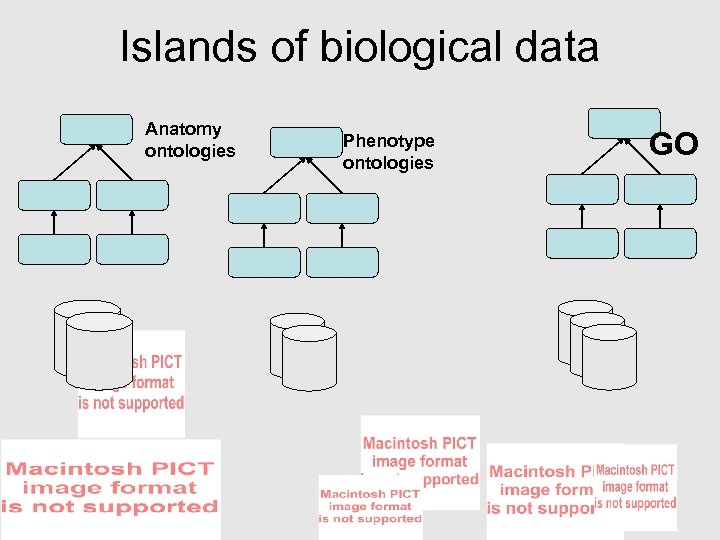
Islands of biological data Anatomy ontologies Phenotype ontologies GO
Connecting the islands
Connecting the islands

Amino acid cross-products in GO: Bada et al : GO to Ch. EBI http: //www. berkeleybop. org/obol

http: //www. berkeleybop. org/obol

• GO approach is retrospective – Text based approaches to ‘decompose’ terms • Obol • Bada/Hunter – Born of necessity • OBO did not exist when GO started – Hard work • New ontologies should take the prospective approach – Separate out aspects from the outset – No heuristic parsing necessary

Prospective approach: Sequence Ontology Separate hierarchies created from the outset - cross-products made from the beginning

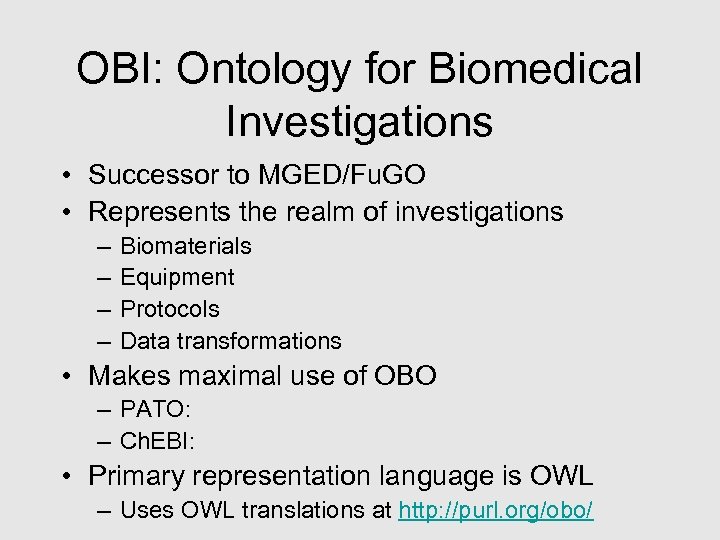
OBI: Ontology for Biomedical Investigations • Successor to MGED/Fu. GO • Represents the realm of investigations – – Biomaterials Equipment Protocols Data transformations • Makes maximal use of OBO – PATO: – Ch. EBI: • Primary representation language is OWL – Uses OWL translations at http: //purl. org/obo/
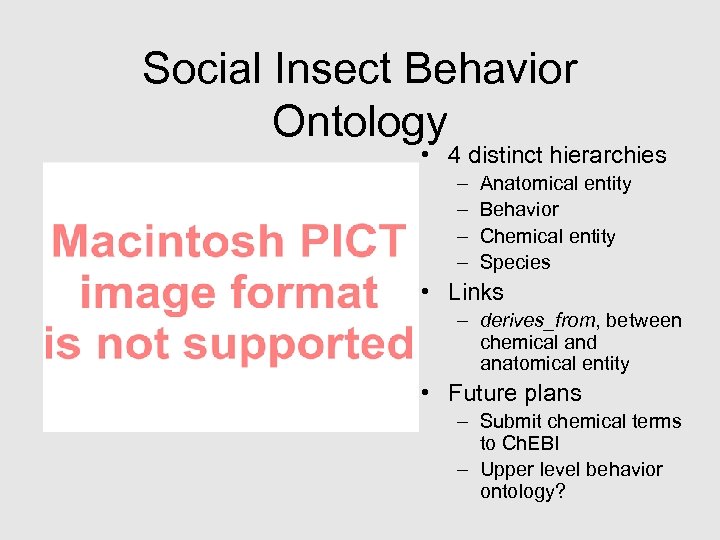
Social Insect Behavior Ontology • 4 distinct hierarchies – – Anatomical entity Behavior Chemical entity Species • Links – derives_from, between chemical and anatomical entity • Future plans – Submit chemical terms to Ch. EBI – Upper level behavior ontology?

Anatomy • GO is relevant for all kingdoms of life • Development of anatomical ontologies has been less coordinated – Cell & subcellular: one ontology applicable to all – Gross Anatomy: multiple ontologies • Vertebrate: – – – MA + EMAP: Mouse FMA: Human (adult) EHDA: Human ZFA: Zebrafish TAO: teleost anatomy XAO: Xenopus • Invertebrate: –FBbt: Drosophila anatomy –Tick anatomy –Mosquito anatomy

Anatomy: Ongoing work • CARO – Upper level shared anatomical ontology – Very general terms • Teleost anatomy ontology – Broader than zebrafish anatomy ontology – Will include homology links • Linking cells to gross anatomical entity poster – Purkinje cell part_of cerebellum – Spans ontologies (CL + ss. AO) • BIRNLex • Stages and development talk

Using multiple ontologies: Pre vs post composition • Complex descriptions (aka cross-products) can be composed from 2 or more terms – By ontology editors (pre) – By curators (post) • Example: – Liver hyperplasia • Precomposed phenotype ontology – MP: 0005141 “liver hyperplasia” increased size of liver due to increased hepatocyte cell number • Post-composition at time of genotype curation – PATO: 0000644 “hyperplastic” – MA: 0000358 “liver” • Which strategy to choose?

• Either strategy can be used • Or mixed and matched – Caveat: • Pre-composed terms must have computable definitions (cross-products) • Currently created retrospectively • Current progress : – MP (Mammalian Phenotype): • 4136/5760 xp defs, partially vetted • Caveat: species-specificity – Worm. Phenotype: • 350/1569 xp defs – Plant. Trait: • 340/765 xp defs, partially vetted

Other ontologies • Envo + GAZ – Environmental ontology and gazetteer – Habitats: • Host (anatomy) • Geographical features (eg hydrothermal vents) – Qualities, chemical entities • BIRNLex • Protein Ontology – Links to/from GO • Complexes • Functions of ancestral proteins

Envo-based annotation in Phenote
Technical consequences of modular approach • Dependencies – Technical issues • Dependence on network? • Formats - converters – Social & management issues – Change and versioning • http: //www. bioontologies. org/ • Managing dependencies • http: //obofoundry. org/wiki/index. php/Mappings – Stable URLs for downloading ontologies in obo or owl http: //purl. org/obo/ – OBO Identifier policy
Conclusions • Be modular – Distinct hierarchies – Avoid is_a overloading – Link to existing ontologies • Rewards – Standards – Increases value of curated data – Reduces duplication of effort and maximises curation effort – Ontologies are long term infrastructure • It’s worth getting them right

Learning more • http: //www. bioontology. org – National Center for Biomedical Ontology – Browse and search OBO – Coming soon: inter-ontology links • http: //obofoundry. org – Principles and recommendations – Participation • Mailing lists • Trackers

Restructuring Cell. obo

OBO Cell Ontology • Current version – Overloading of is_a hierarchy – Difficult to maintain – Leads to “true path” violations • Refactoring – Replace is links with has_function – Keep main axis structure-based (but not religiously so)

• For every term immediately under cell-by-function, we made a new function term • • • • • propagation of genome to circulate to secrete to metabolise to contract Electrical absorption Barrier Motility Structural to accumulate stuff signaling (mitogenic) to die Defense Transport to photosynthesize to support Valve to fix nitrogen • Also create grouping terms
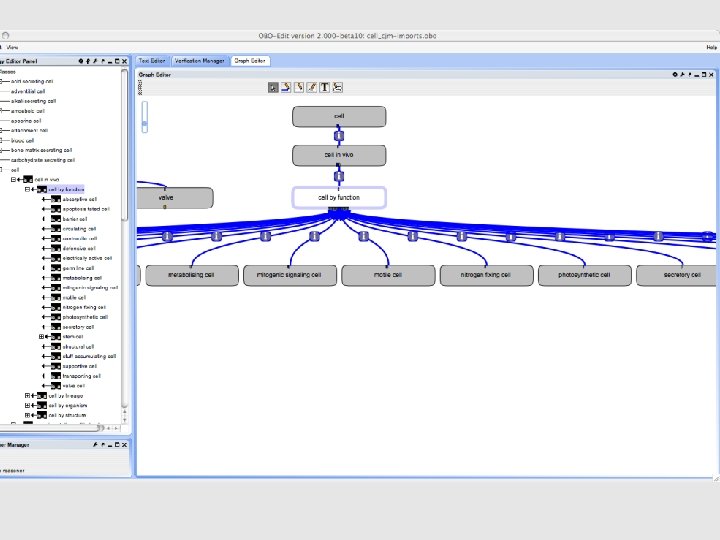
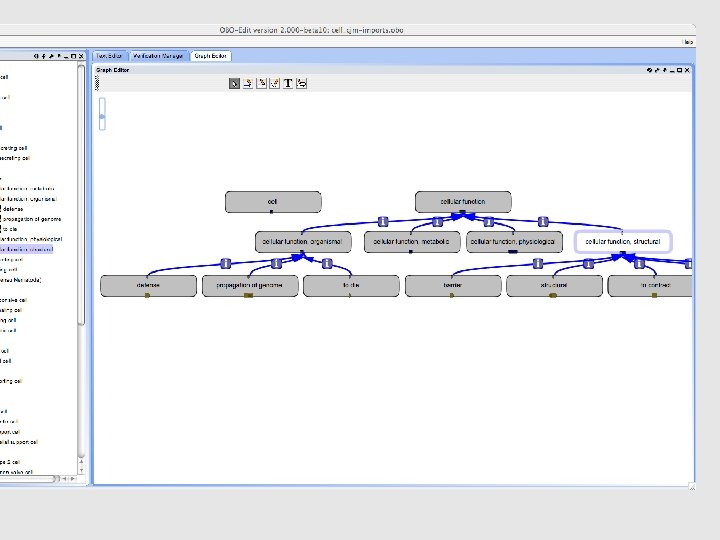

• Replaced is_a links to cell-by-function terms with has_function links to corresponding function terms
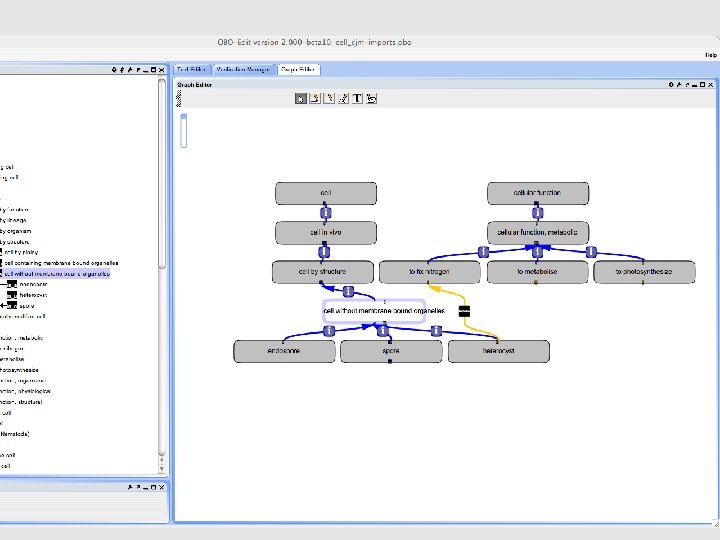

• What do we do about the old cell-by-function terms? • We can eliminate them. . • OR we can support them, but infer the ‘tangled DAG’ • Requires xp defs: – Nitrogen fixing cell = cell THAT has_function nitrogen-fixing
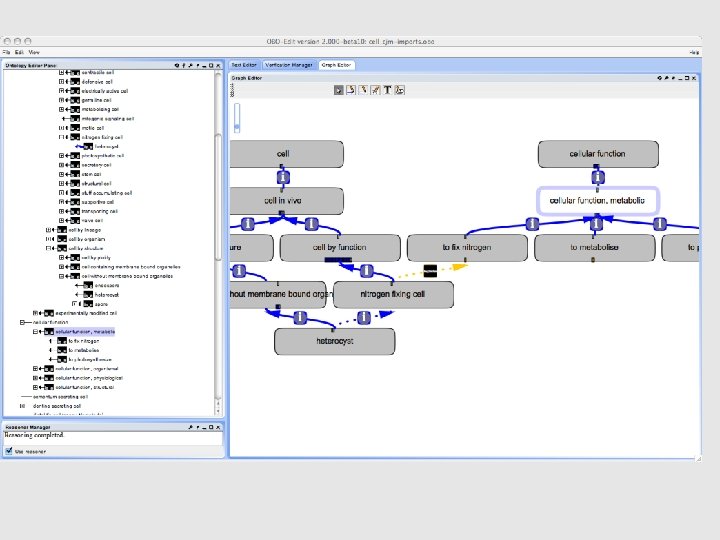

• Future work / ongoing issues: • Redundancy between cell functions & GO biological process? • Cell-by-lineage
Synchronizing ss. AOs and CL • Fly_anat, zfa, plant_anat all represent cell types – Part_of links from cells to gross anatomy • E. g. purkinje_cell part_of cerebellum • Methodology – – Xrefs from ss. AOs to CL IDs Treat as ss subtypes Use reasoner to stay in sync http: //www. bioontology. org/wiki/index. php/CL: Aligning_speci es-specific_anatomy_ontologies_with_CL – Examples: • http: //www. berkeleybop. org/obol/#fly_anatomy_xp_cell-obol
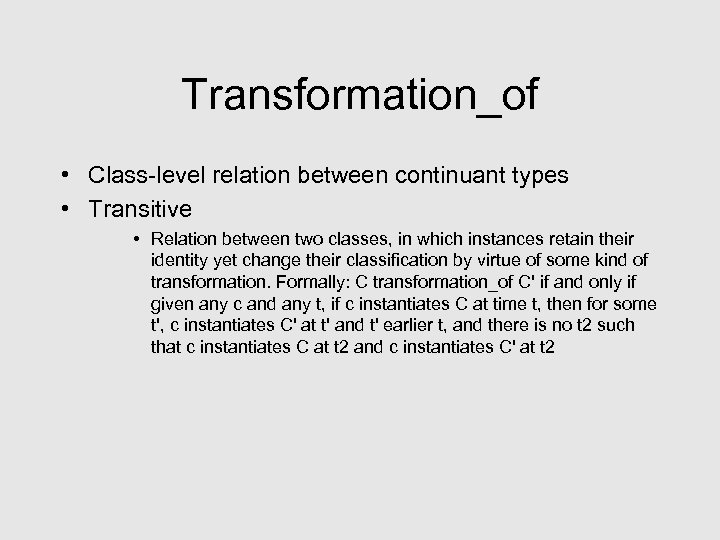
Transformation_of • Class-level relation between continuant types • Transitive • Relation between two classes, in which instances retain their identity yet change their classification by virtue of some kind of transformation. Formally: C transformation_of C' if and only if given any c and any t, if c instantiates C at time t, then for some t', c instantiates C' at t' and t' earlier t, and there is no t 2 such that c instantiates C at t 2 and c instantiates C' at t 2


Derives_from • Holds between continuants • transitive • Derivation on the instance level (*derives_from*) holds between distinct material continuants when one succeeds the other across a temporal divide in such a way that at least a biologically significant portion of the matter of the earlier continuant is inherited by the later • We say that one class C derives_from class C' if instances of C are connected to instances of C' via some chain of instancelevel derivation relations. • Examples: – osteocyte derives_from osteoblast


882755436ae423dbc85e1af32aeadcfa.ppt