625c3faceccbdb5c2fdc98452694e1cb.ppt
- Количество слайдов: 36
 Lengths, Energies and Time Scales in Photosynthesis. Implications for Artificial Systems. Dror Noy Plant Sciences Dept. Weizmann Institute of Science Rehovot, Israel
Lengths, Energies and Time Scales in Photosynthesis. Implications for Artificial Systems. Dror Noy Plant Sciences Dept. Weizmann Institute of Science Rehovot, Israel
 How does Nature exploits fundamental physical principles in the construction of biological energy conversion systems?
How does Nature exploits fundamental physical principles in the construction of biological energy conversion systems?
 How can we implement the Natural strategies in manmade energy conversion systems?
How can we implement the Natural strategies in manmade energy conversion systems?
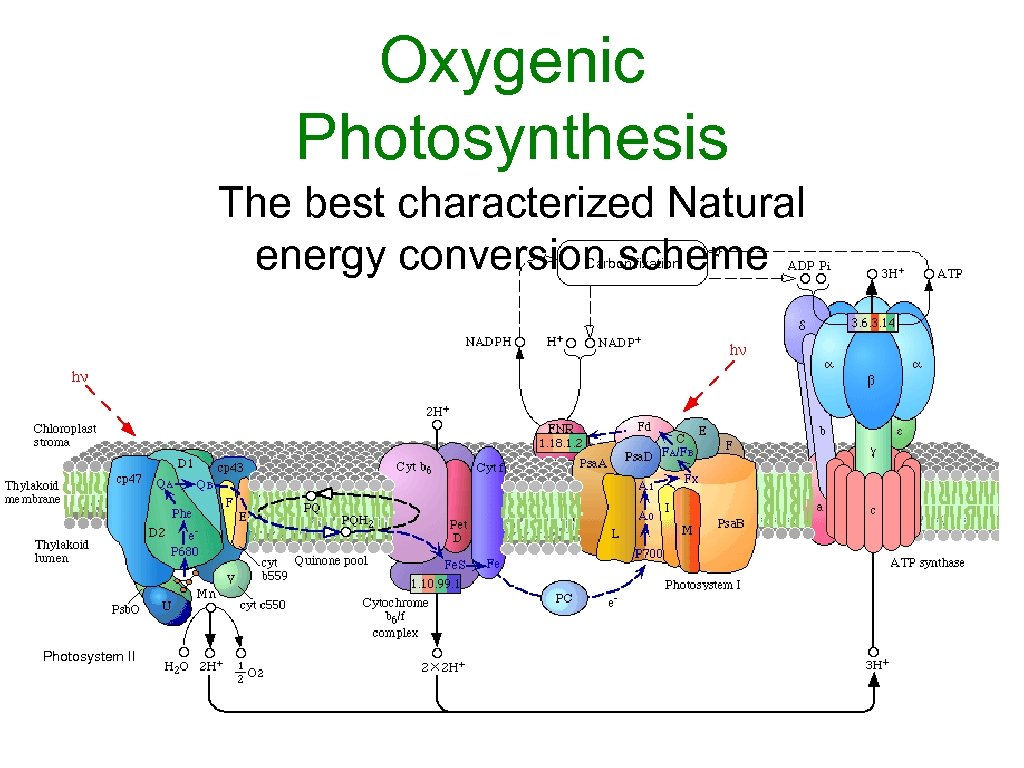 Oxygenic Photosynthesis The best characterized Natural energy conversion scheme Carbon fixation Photosystem II
Oxygenic Photosynthesis The best characterized Natural energy conversion scheme Carbon fixation Photosystem II
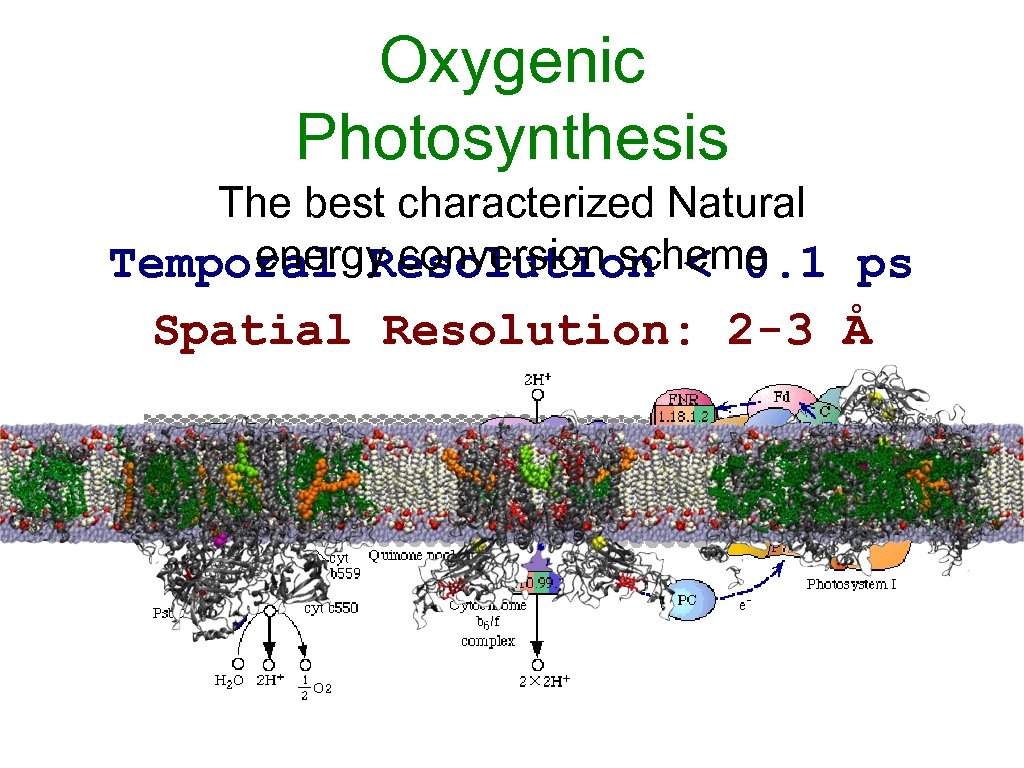 Oxygenic Photosynthesis The best characterized Natural energy conversion scheme Temporal Resolution < 0. 1 ps Spatial Resolution: 2 -3 Å
Oxygenic Photosynthesis The best characterized Natural energy conversion scheme Temporal Resolution < 0. 1 ps Spatial Resolution: 2 -3 Å
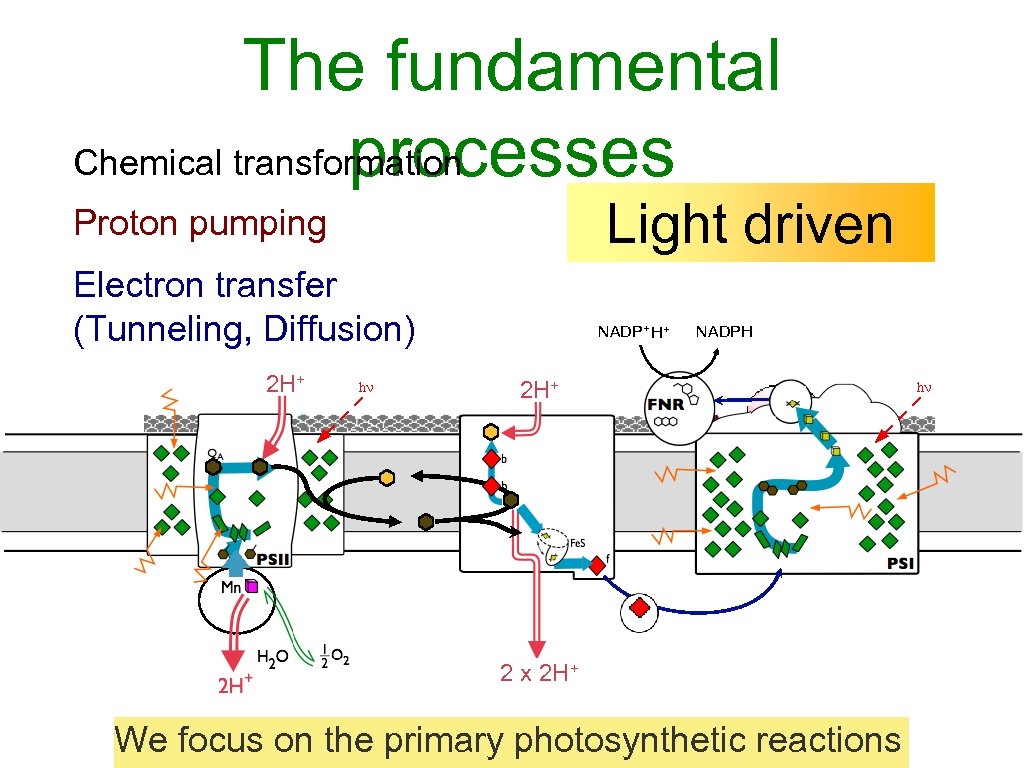 The fundamental Chemical transformation processes Light driven Proton pumping Electron transfer (Tunneling, Diffusion) 2 H+ NADP+ H+ NADPH 2 H+ 2 x 2 H+ We focus on the primary photosynthetic reactions
The fundamental Chemical transformation processes Light driven Proton pumping Electron transfer (Tunneling, Diffusion) 2 H+ NADP+ H+ NADPH 2 H+ 2 x 2 H+ We focus on the primary photosynthetic reactions
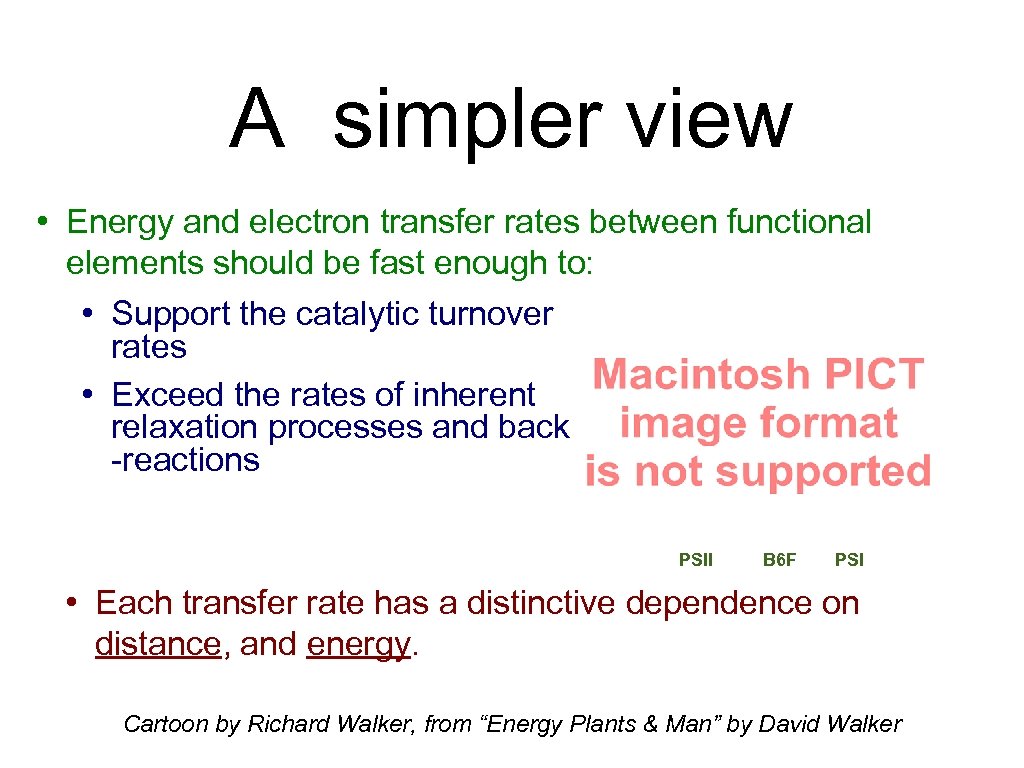 A simpler view • Energy and electron transfer rates between functional elements should be fast enough to: • Support the catalytic turnover rates • Exceed the rates of inherent relaxation processes and back -reactions PSII B 6 F PSI • Each transfer rate has a distinctive dependence on distance, and energy. Cartoon by Richard Walker, from “Energy Plants & Man” by David Walker
A simpler view • Energy and electron transfer rates between functional elements should be fast enough to: • Support the catalytic turnover rates • Exceed the rates of inherent relaxation processes and back -reactions PSII B 6 F PSI • Each transfer rate has a distinctive dependence on distance, and energy. Cartoon by Richard Walker, from “Energy Plants & Man” by David Walker
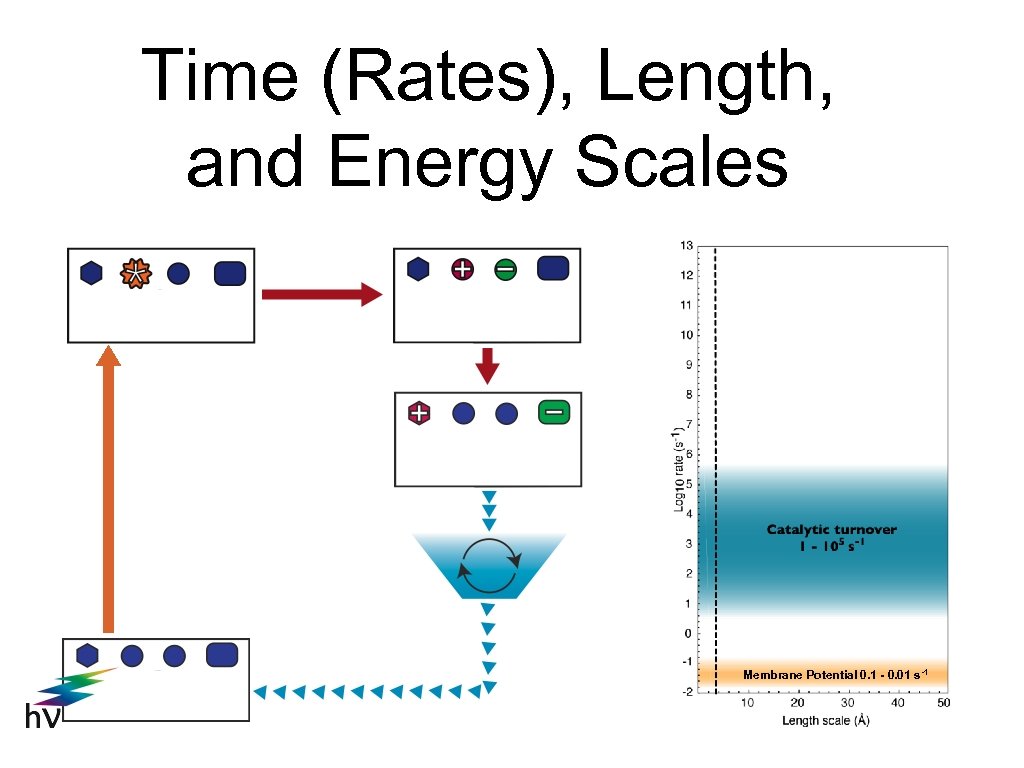 Time (Rates), Length, and Energy Scales Membrane Potential 0. 1 - 0. 01 s-1
Time (Rates), Length, and Energy Scales Membrane Potential 0. 1 - 0. 01 s-1
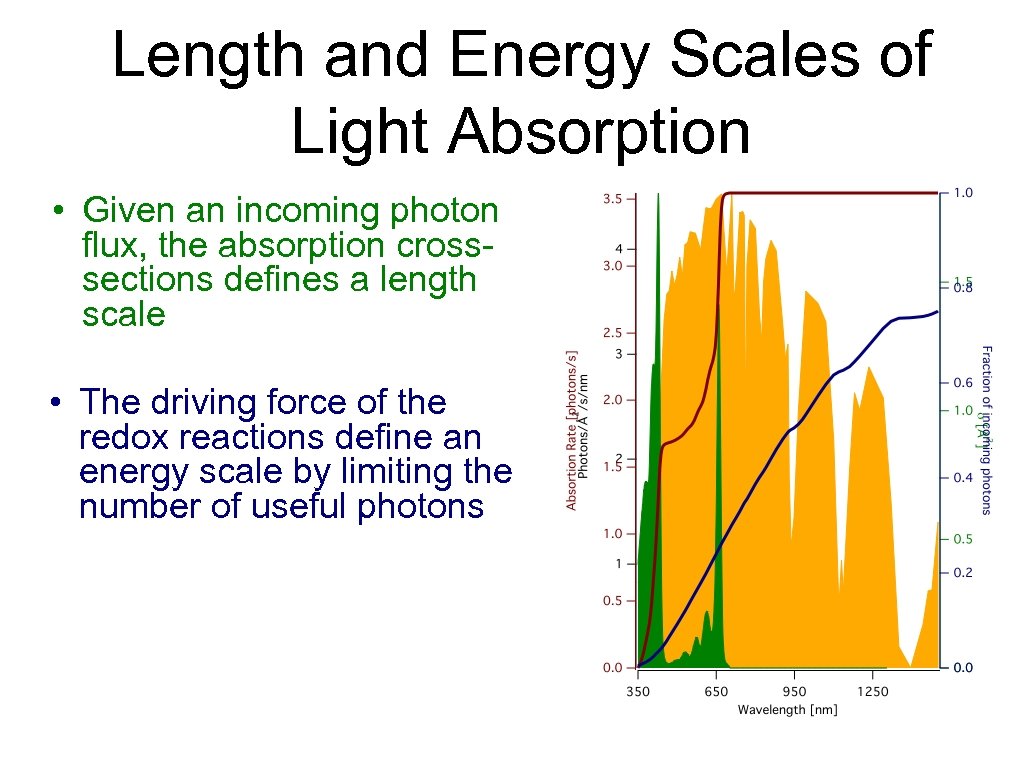 Length and Energy Scales of Light Absorption • Given an incoming photon flux, the absorption crosssections defines a length scale • The driving force of the redox reactions define an energy scale by limiting the number of useful photons
Length and Energy Scales of Light Absorption • Given an incoming photon flux, the absorption crosssections defines a length scale • The driving force of the redox reactions define an energy scale by limiting the number of useful photons
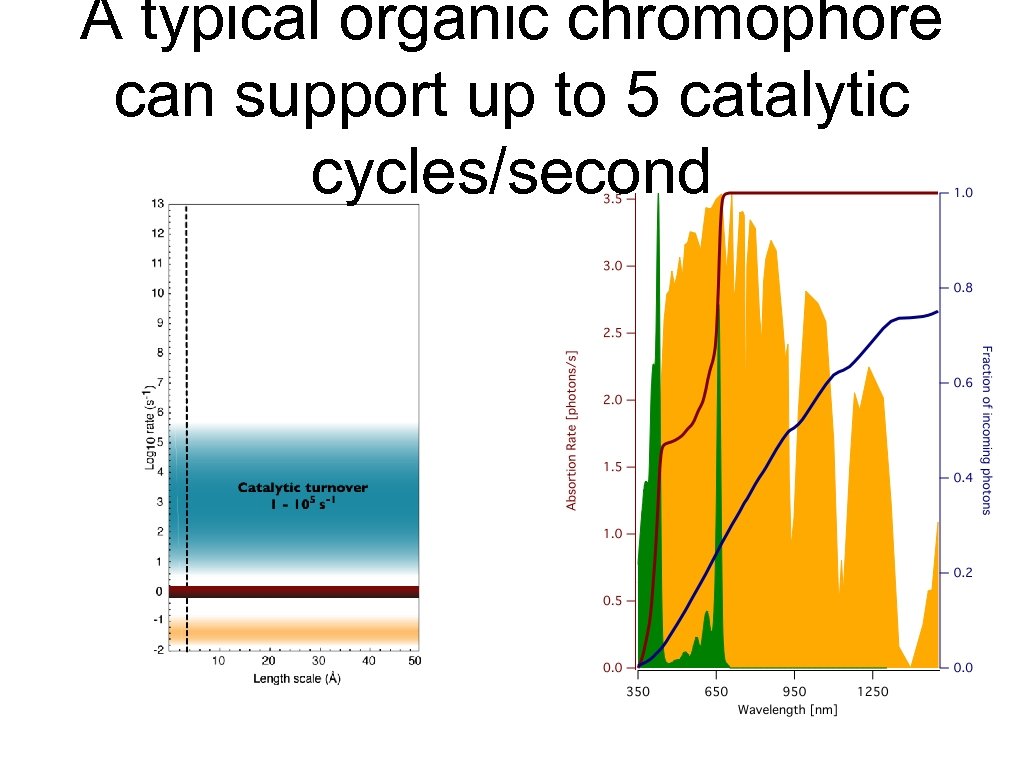 A typical organic chromophore can support up to 5 catalytic cycles/second
A typical organic chromophore can support up to 5 catalytic cycles/second
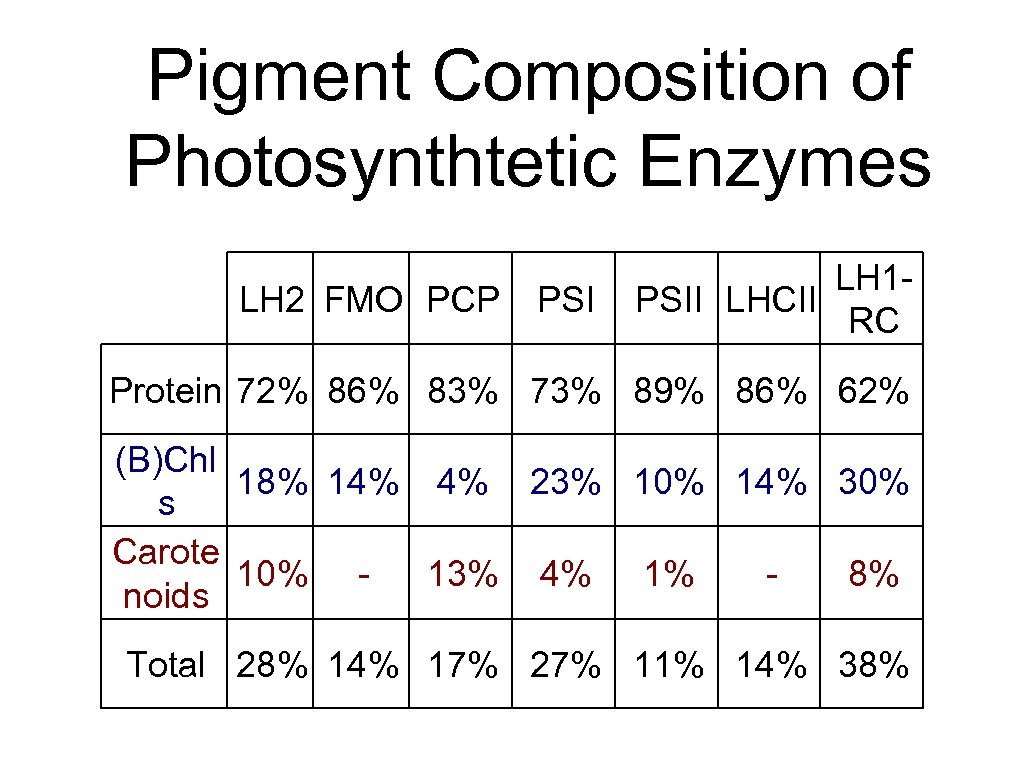 Pigment Composition of Photosynthtetic Enzymes LH 2 FMO PCP PSI LH 1 PSII LHCII RC Protein 72% 86% 83% 73% 89% 86% 62% (B)Chl 18% 14% 4% 23% 10% 14% 30% s Carote 10% 13% 4% 1% 8% noids Total 28% 14% 17% 27% 11% 14% 38%
Pigment Composition of Photosynthtetic Enzymes LH 2 FMO PCP PSI LH 1 PSII LHCII RC Protein 72% 86% 83% 73% 89% 86% 62% (B)Chl 18% 14% 4% 23% 10% 14% 30% s Carote 10% 13% 4% 1% 8% noids Total 28% 14% 17% 27% 11% 14% 38%
 Different distance and energy dependence for electron and energy transfer Electron tunneling Energy transfer Electron Transfer ∝ 10^(-0. 6 r-3. 1⋅(ΔG+λ)2/λ) Energy Transfer ∝ (ro/r)6 Membrane Potential 0. 1 - 0. 01 s-1
Different distance and energy dependence for electron and energy transfer Electron tunneling Energy transfer Electron Transfer ∝ 10^(-0. 6 r-3. 1⋅(ΔG+λ)2/λ) Energy Transfer ∝ (ro/r)6 Membrane Potential 0. 1 - 0. 01 s-1
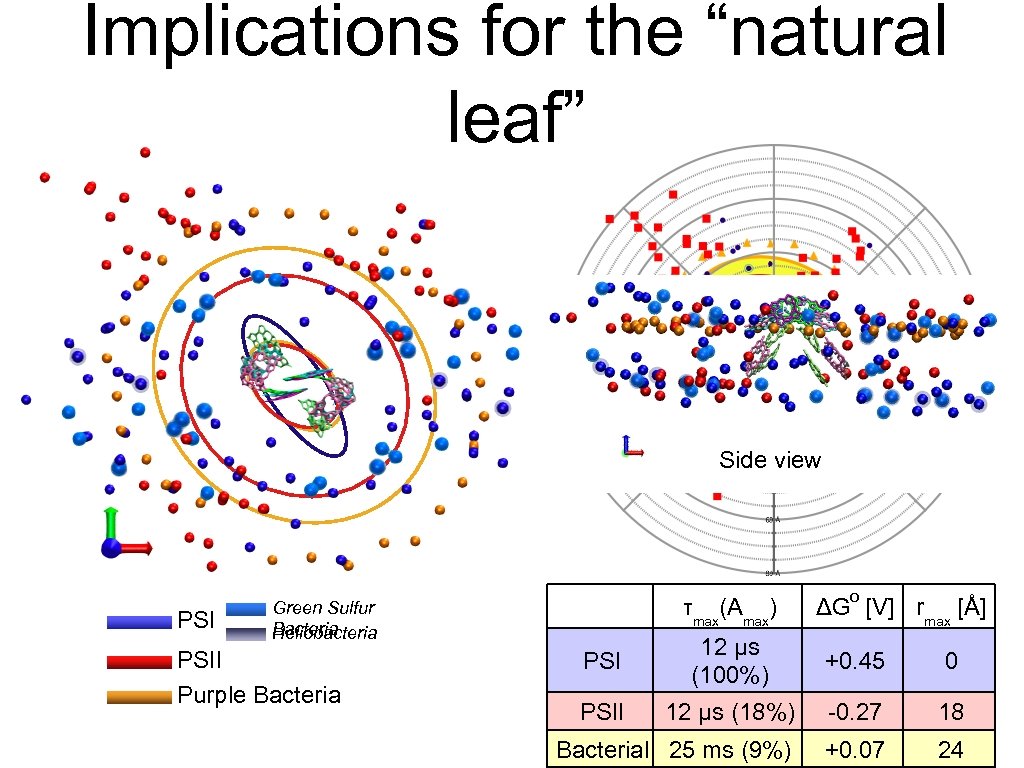 Implications for the “natural leaf” Side view PSI τmax(Amax) Green Sulfur Bacteria Heliobacteria PSII Purple Bacteria o ΔG [V] rmax [Å] PSI 12 μs (100%) +0. 45 0 PSII 12 μs (18%) -0. 27 18 Bacterial 25 ms (9%) +0. 07 24
Implications for the “natural leaf” Side view PSI τmax(Amax) Green Sulfur Bacteria Heliobacteria PSII Purple Bacteria o ΔG [V] rmax [Å] PSI 12 μs (100%) +0. 45 0 PSII 12 μs (18%) -0. 27 18 Bacterial 25 ms (9%) +0. 07 24
 Conclusions The basic physics of the transfer processes allow for a large degree of tolerance In photosystems, natural selection favors robust design with the predominant parameter being control over cofactor distances
Conclusions The basic physics of the transfer processes allow for a large degree of tolerance In photosystems, natural selection favors robust design with the predominant parameter being control over cofactor distances
 However. . .
However. . .
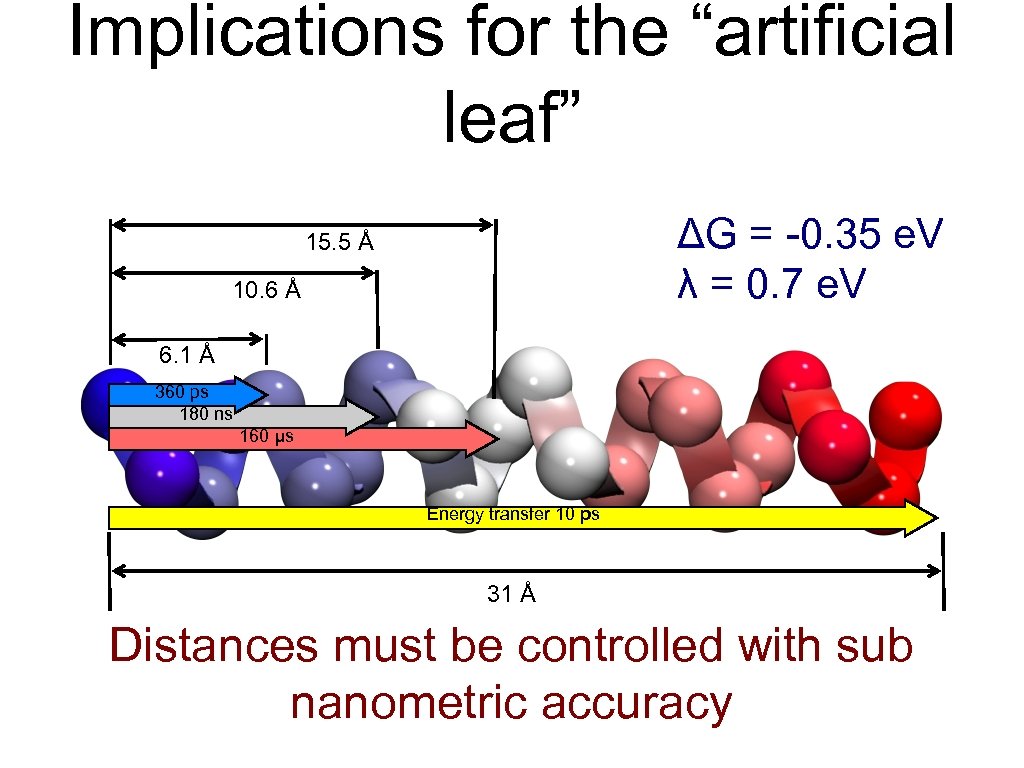 Implications for the “artificial leaf” ΔG = -0. 35 e. V λ = 0. 7 e. V 15. 5 Å 10. 6 Å 6. 1 Å 360 ps 180 ns 160 μs Energy transfer 10 ps 31 Å Distances must be controlled with sub nanometric accuracy
Implications for the “artificial leaf” ΔG = -0. 35 e. V λ = 0. 7 e. V 15. 5 Å 10. 6 Å 6. 1 Å 360 ps 180 ns 160 μs Energy transfer 10 ps 31 Å Distances must be controlled with sub nanometric accuracy
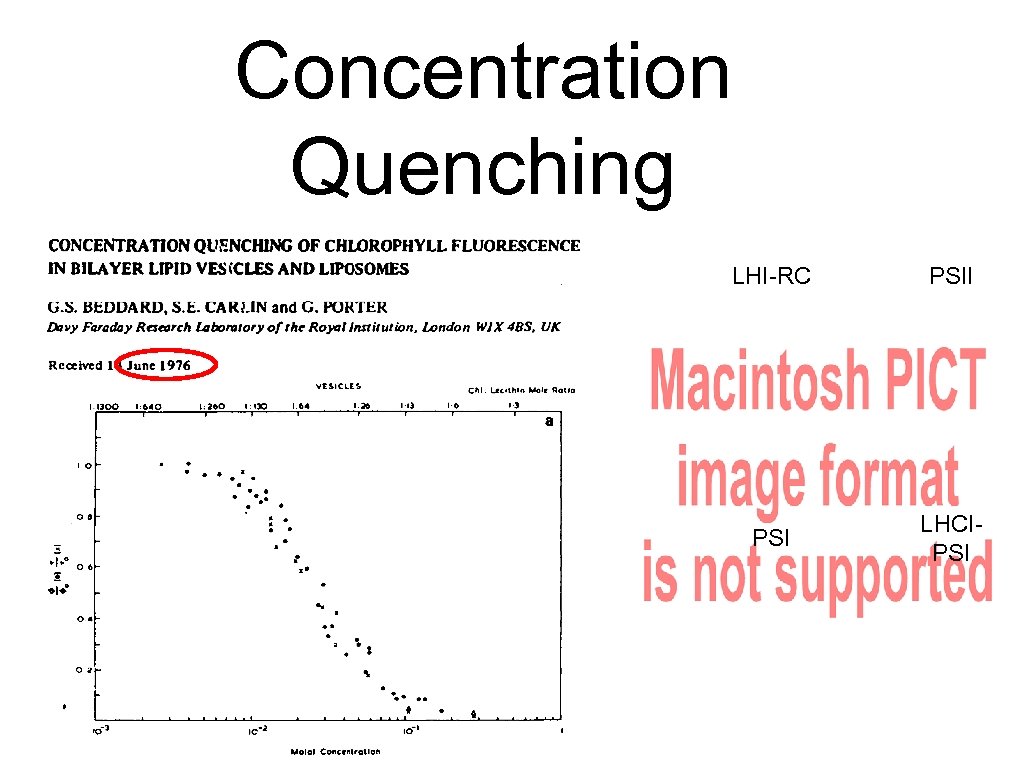 Concentration Quenching LHI-RC PSII PSI LHCIPSI
Concentration Quenching LHI-RC PSII PSI LHCIPSI
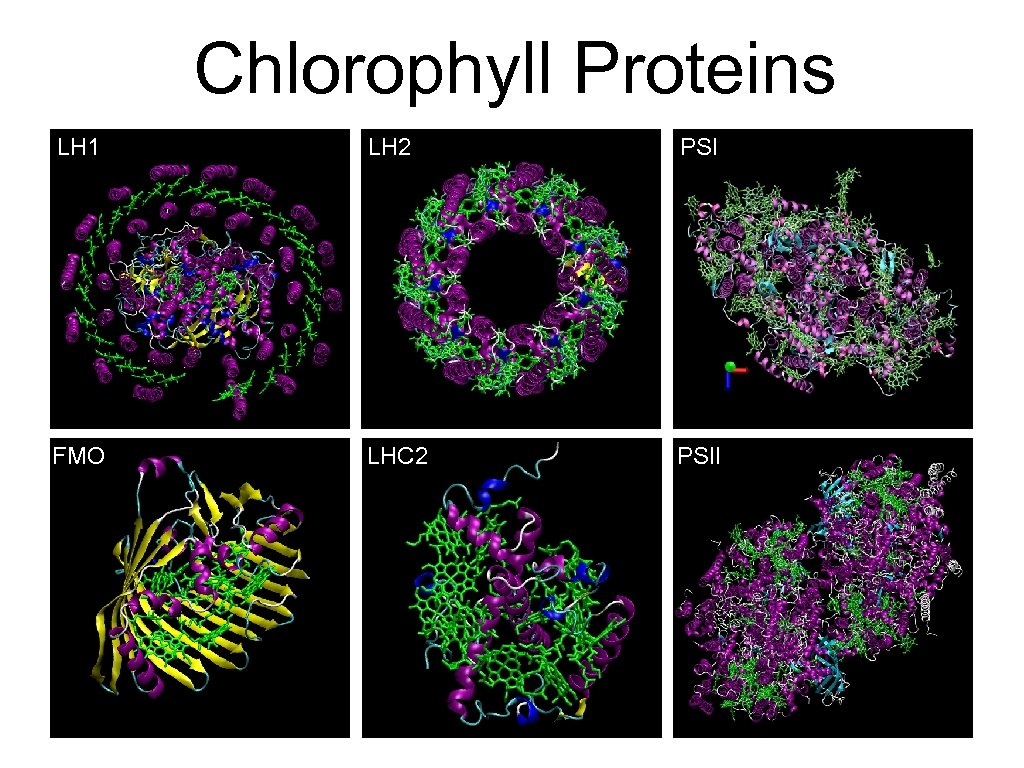 Chlorophyll Proteins LH 1 LH 2 PSI FMO LHC 2 PSII
Chlorophyll Proteins LH 1 LH 2 PSI FMO LHC 2 PSII
 Non-natural Systems • Rudimentary • Iterative • High structures design resolution structural information, only a bonus
Non-natural Systems • Rudimentary • Iterative • High structures design resolution structural information, only a bonus
 De Novo Designed Protein Building Blocks for Energy and Electron Transfer Relays
De Novo Designed Protein Building Blocks for Energy and Electron Transfer Relays
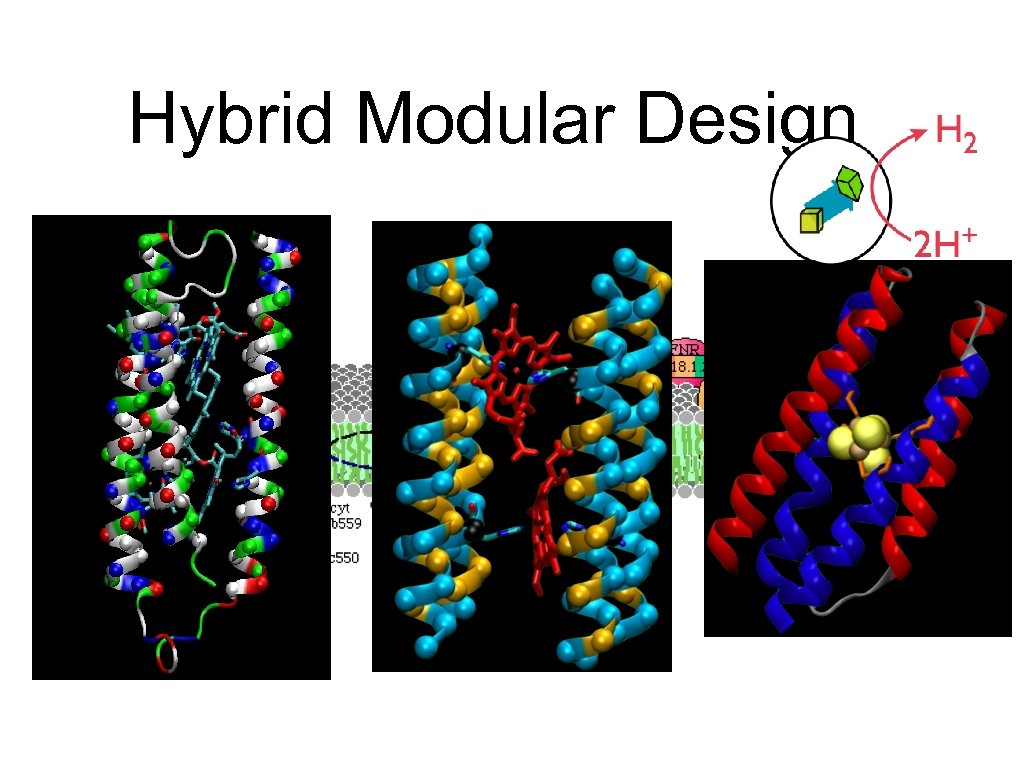 Hybrid Modular Design
Hybrid Modular Design
 De Novo Design of a Non. Natural Fold for an Iron. Sulfur Protein
De Novo Design of a Non. Natural Fold for an Iron. Sulfur Protein
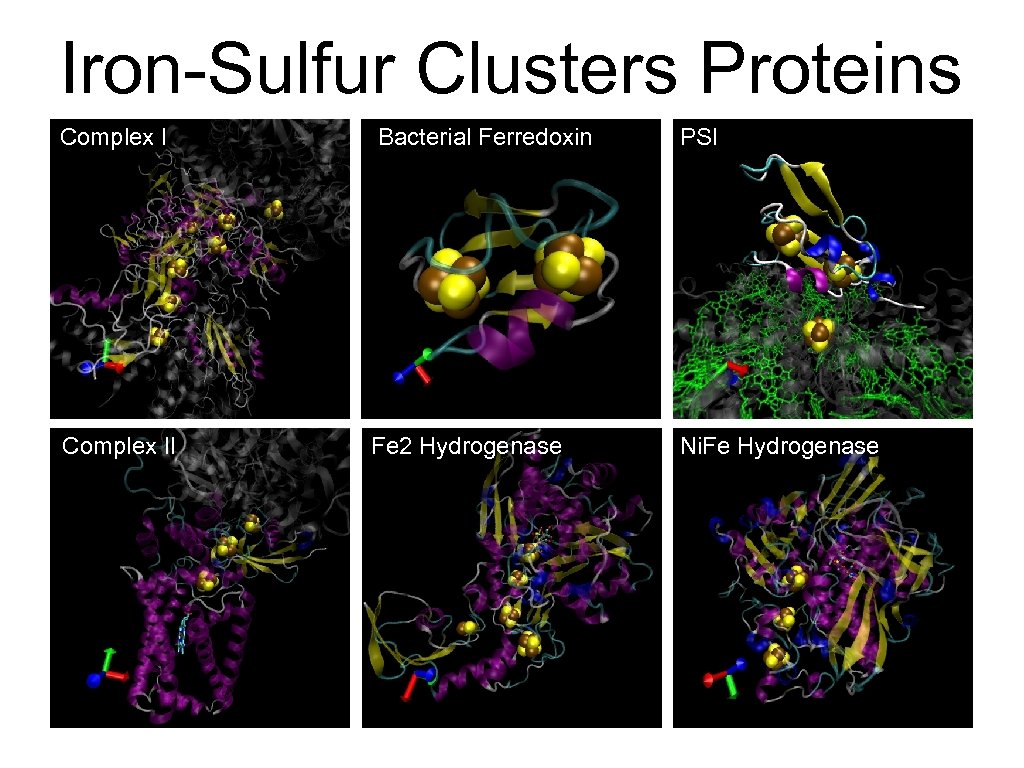 Iron-Sulfur Clusters Proteins Complex I Bacterial Ferredoxin PSI Complex II Fe 2 Hydrogenase Ni. Fe Hydrogenase
Iron-Sulfur Clusters Proteins Complex I Bacterial Ferredoxin PSI Complex II Fe 2 Hydrogenase Ni. Fe Hydrogenase
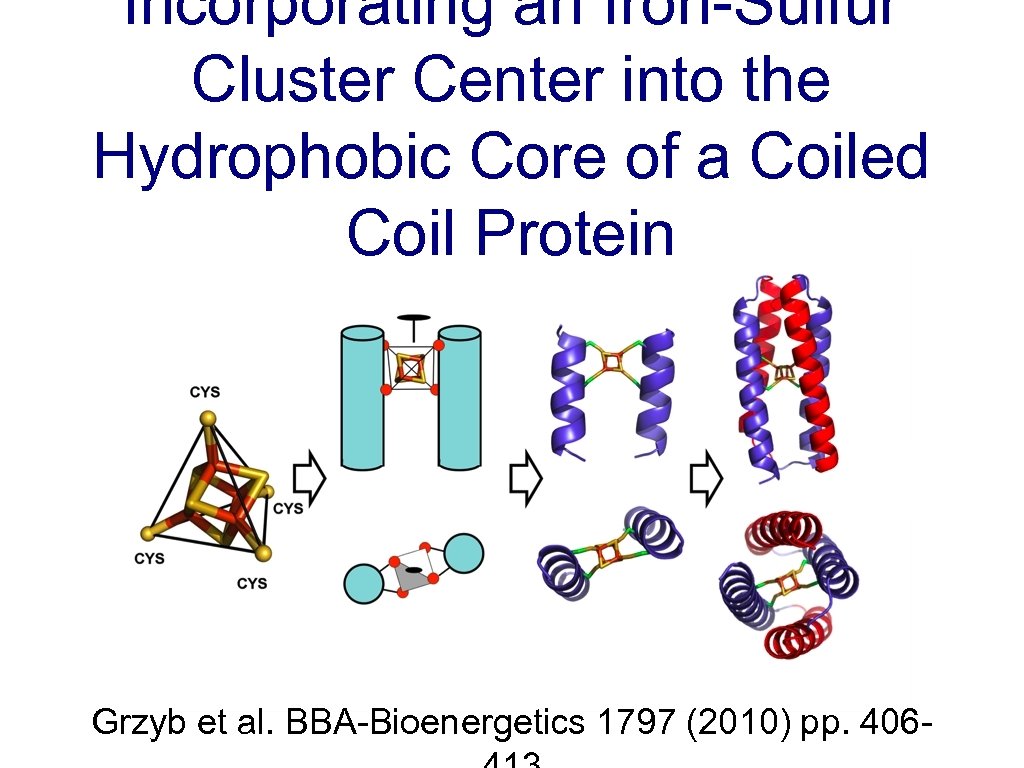 Incorporating an Iron-Sulfur Cluster Center into the Hydrophobic Core of a Coiled Coil Protein Grzyb et al. BBA-Bioenergetics 1797 (2010) pp. 406 -
Incorporating an Iron-Sulfur Cluster Center into the Hydrophobic Core of a Coiled Coil Protein Grzyb et al. BBA-Bioenergetics 1797 (2010) pp. 406 -
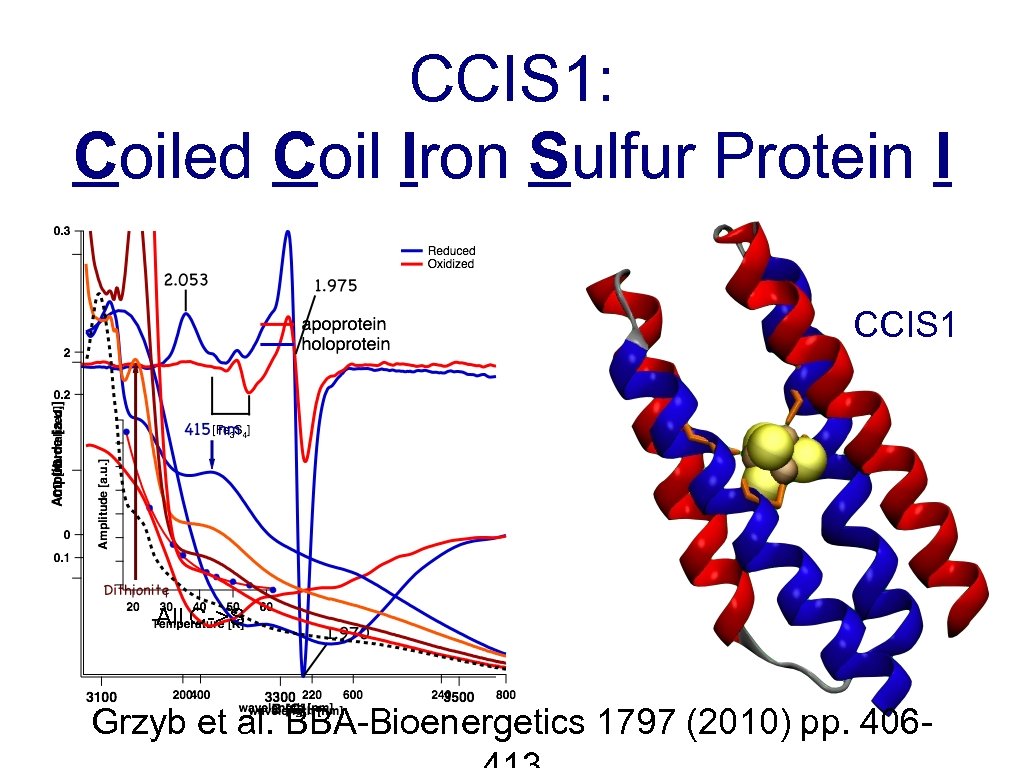 CCIS 1: Coiled Coil Iron Sulfur Protein I CCIS 1 All C->S Grzyb et al. BBA-Bioenergetics 1797 (2010) pp. 406 -
CCIS 1: Coiled Coil Iron Sulfur Protein I CCIS 1 All C->S Grzyb et al. BBA-Bioenergetics 1797 (2010) pp. 406 -
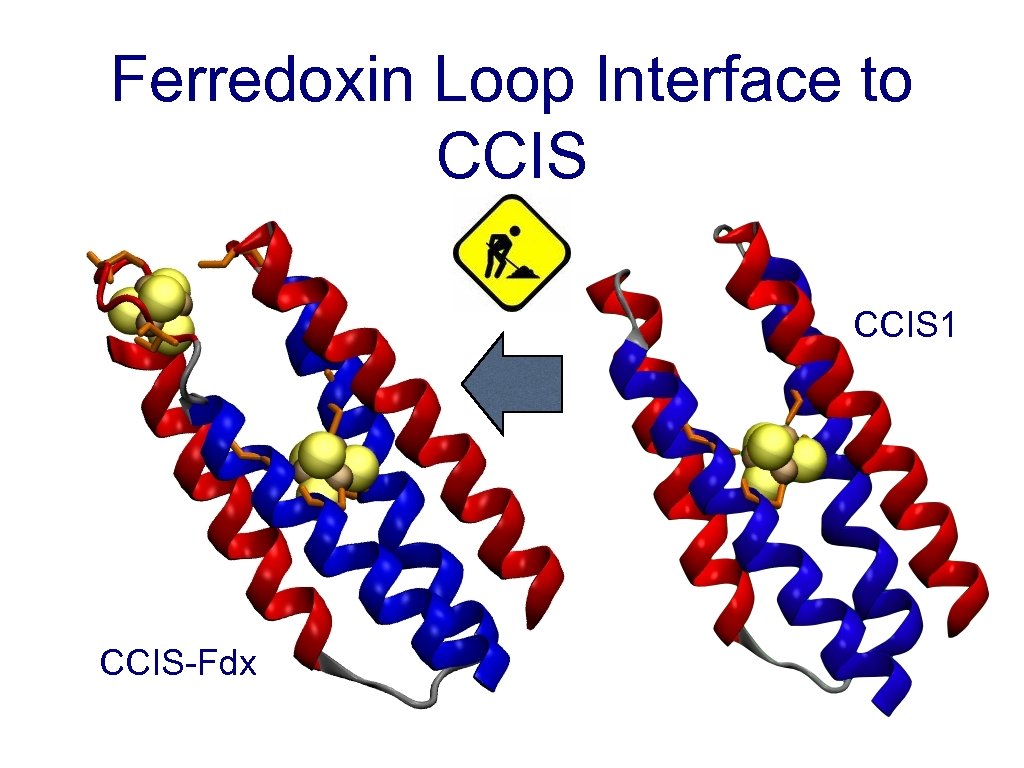 Ferredoxin Loop Interface to CCIS 1 CCIS-Fdx
Ferredoxin Loop Interface to CCIS 1 CCIS-Fdx
 De Novo Design of a Water Soluble Analog of Transmembranal Chlorophyll Proteins
De Novo Design of a Water Soluble Analog of Transmembranal Chlorophyll Proteins
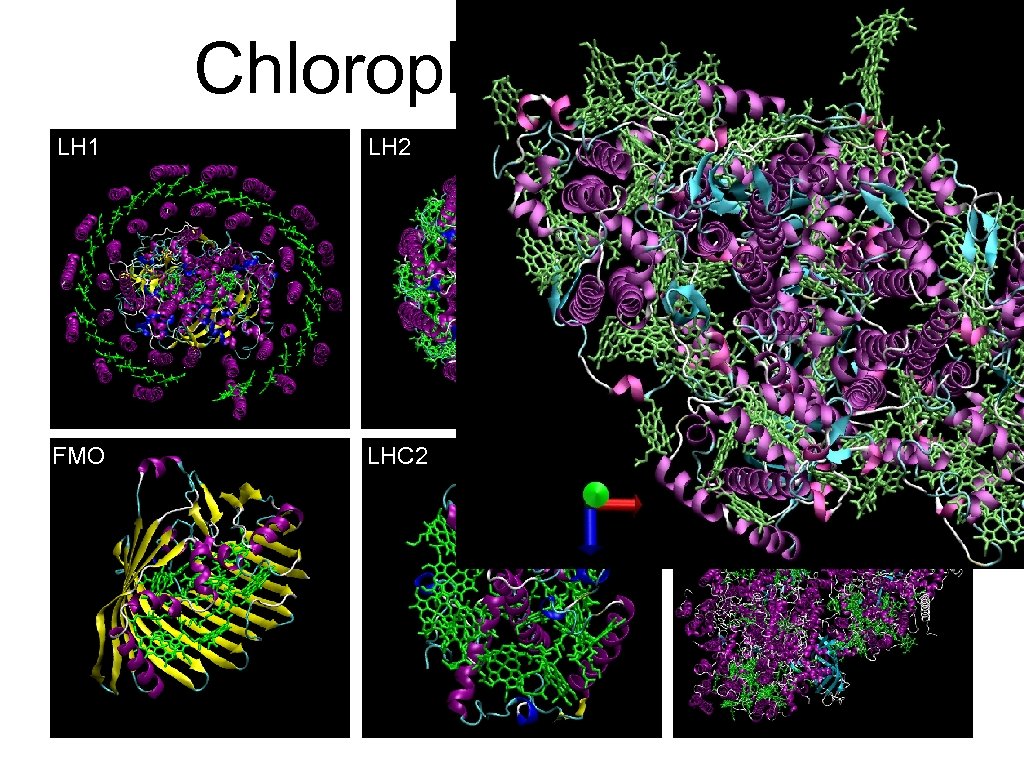 Chlorophyll Proteins LH 1 LH 2 PSI FMO LHC 2 PSII
Chlorophyll Proteins LH 1 LH 2 PSI FMO LHC 2 PSII
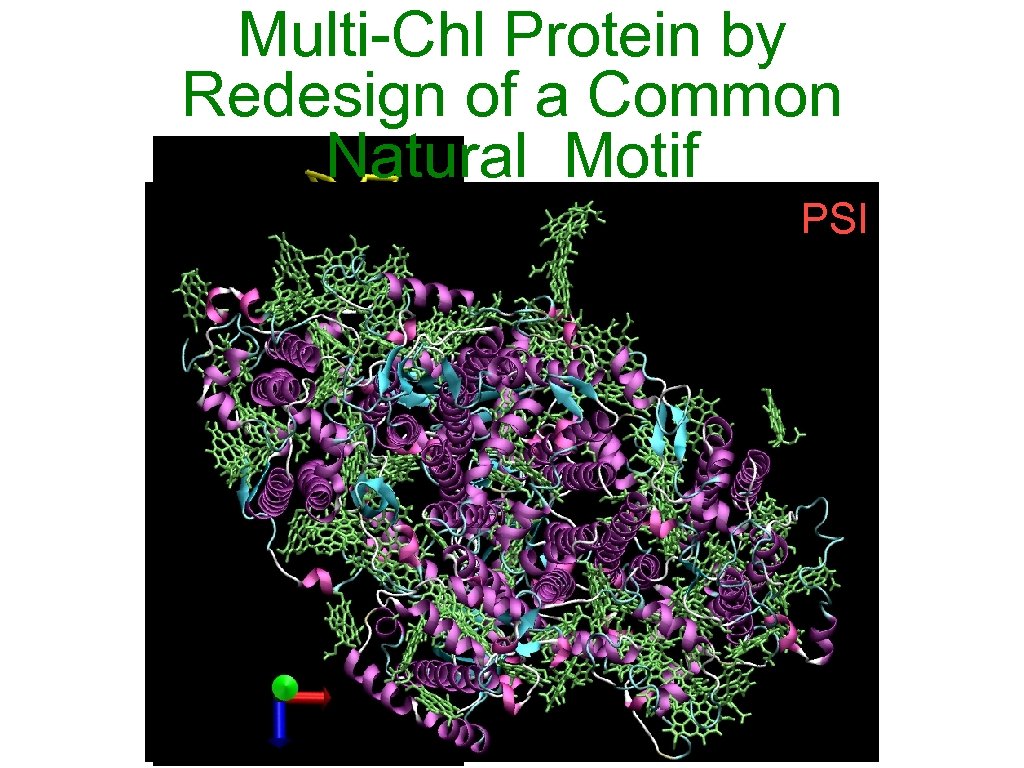 Multi-Chl Protein by Redesign of a Common Natural Motif PSII
Multi-Chl Protein by Redesign of a Common Natural Motif PSII
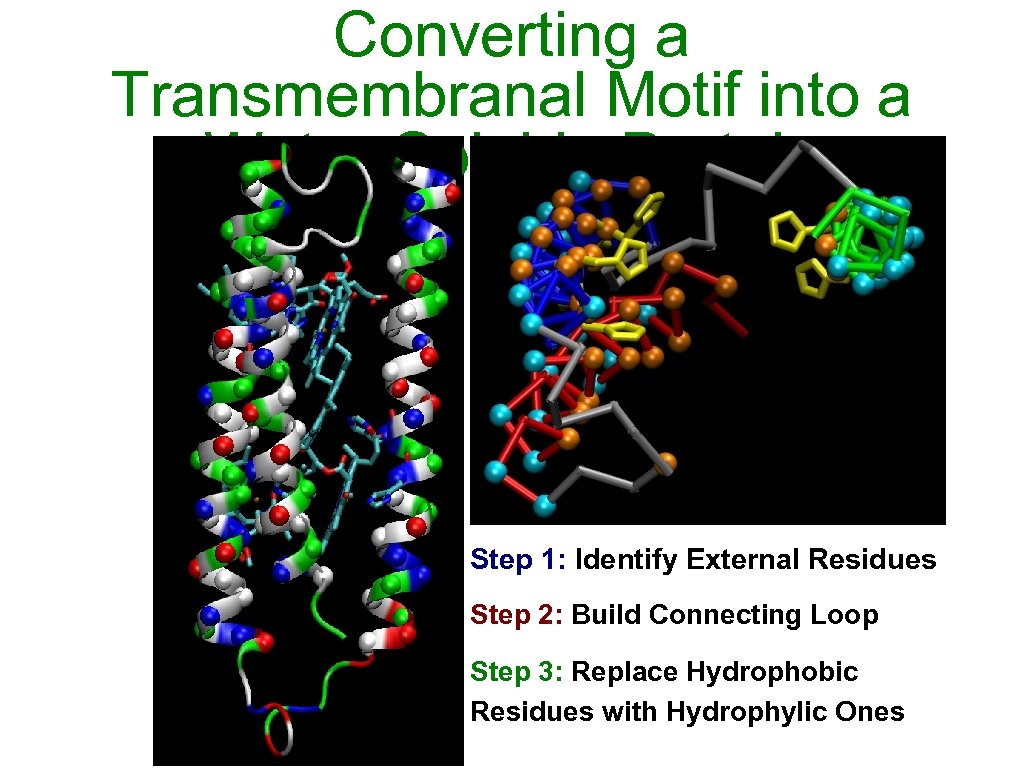 Converting a Transmembranal Motif into a Water-Soluble Protein Step 1: Identify External Residues Step 2: Build Connecting Loop Step 3: Replace Hydrophobic Residues with Hydrophylic Ones
Converting a Transmembranal Motif into a Water-Soluble Protein Step 1: Identify External Residues Step 2: Build Connecting Loop Step 3: Replace Hydrophobic Residues with Hydrophylic Ones
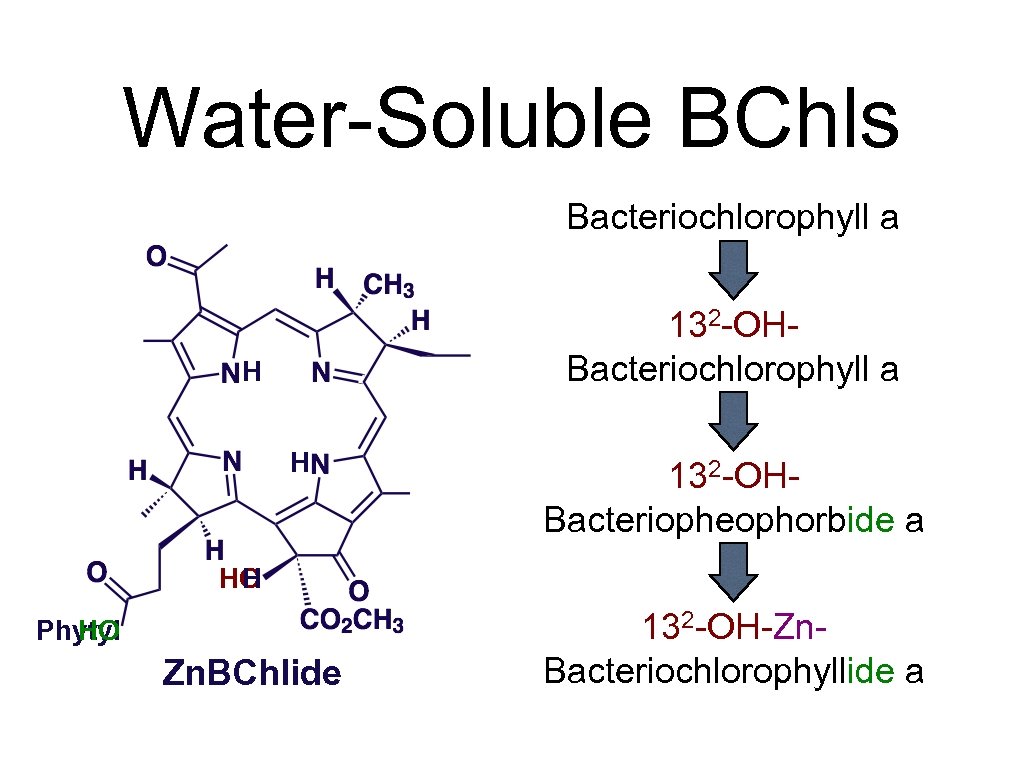 Water-Soluble BChls Bacteriochlorophyll a H Mg Zn H 132 -OHBacteriochlorophyll a 132 -OHBacteriopheophorbide a HO H Phytyl HO Zn. BChlide 132 -OH-Zn. Bacteriochlorophyllide a
Water-Soluble BChls Bacteriochlorophyll a H Mg Zn H 132 -OHBacteriochlorophyll a 132 -OHBacteriopheophorbide a HO H Phytyl HO Zn. BChlide 132 -OH-Zn. Bacteriochlorophyllide a
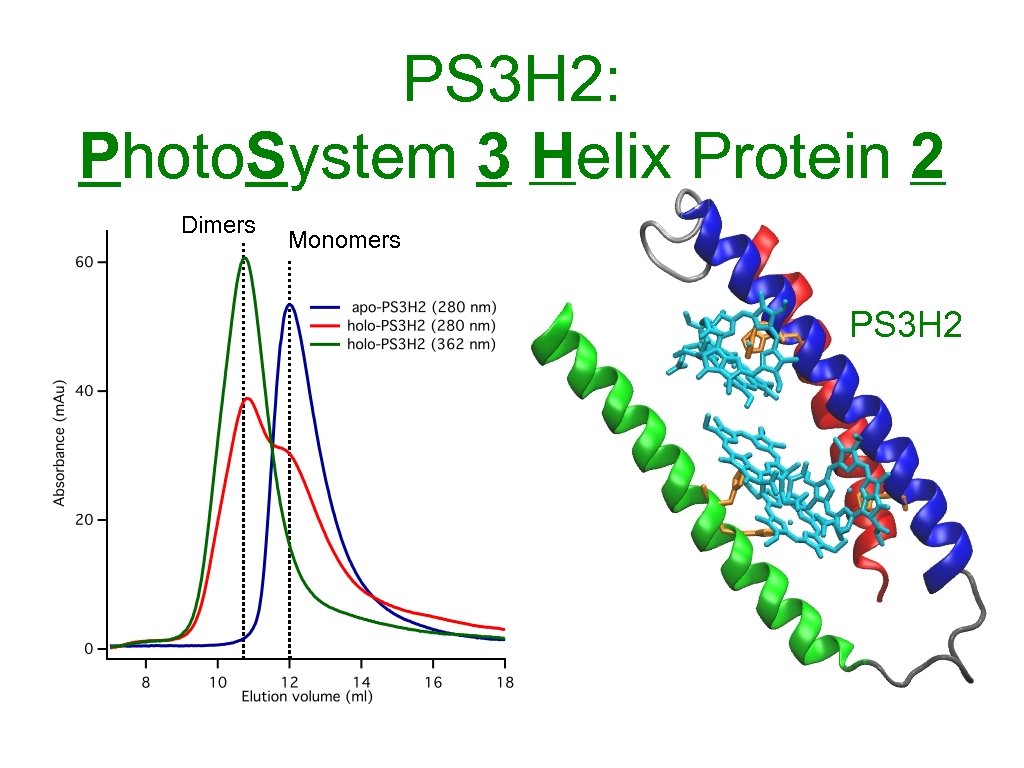 PS 3 H 2: Photo. System 3 Helix Protein 2 Dimers Monomers PS 3 H 2
PS 3 H 2: Photo. System 3 Helix Protein 2 Dimers Monomers PS 3 H 2
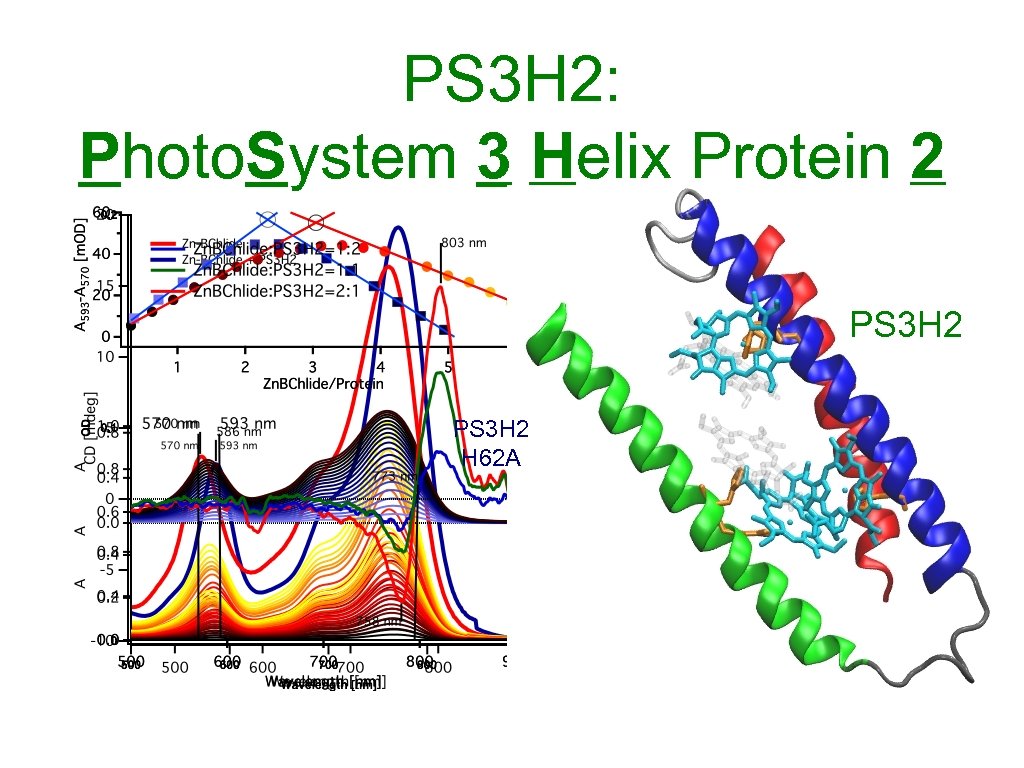 PS 3 H 2: Photo. System 3 Helix Protein 2 PS 3 H 2 H 62 A
PS 3 H 2: Photo. System 3 Helix Protein 2 PS 3 H 2 H 62 A
 Conclusions Two examples of designing de novo protein cofactor complexes were presented: • An iron-sulfur cluster with a non-natural fold • A multi-Chl binding protein that is a water-soluble analog of a highly conserved transmembranal Chlbinding motif These examples demonstrate: • The viability of protein de novo design for making novel functional proteins • The effectivity of the iterative design approach in identifying and correcting design flaws
Conclusions Two examples of designing de novo protein cofactor complexes were presented: • An iron-sulfur cluster with a non-natural fold • A multi-Chl binding protein that is a water-soluble analog of a highly conserved transmembranal Chlbinding motif These examples demonstrate: • The viability of protein de novo design for making novel functional proteins • The effectivity of the iterative design approach in identifying and correcting design flaws
 Conclusions By focusing on simple and robust energy and electron transfer relays we can achieve functional variability by “mixing and matching” a few unique catalytic centers Protein de novo design is a useful way of constructing the relays that will provide building blocks for energy conversion systems
Conclusions By focusing on simple and robust energy and electron transfer relays we can achieve functional variability by “mixing and matching” a few unique catalytic centers Protein de novo design is a useful way of constructing the relays that will provide building blocks for energy conversion systems
 Acknowledgments Noy Group • Ilit Cohen-Ofri • Joanna Grzyb • Jebasingh Tennyson • Iris Margalit Wolfgang Lubitz • Maurice van Gastel Vik Nanda Zx ab Ron Koder Collaboration Avigdor Scherz Zx • Alex Brandis ab • Oksana Shlyk-Kerner Noam Adir, Technion Lev Weiner, Daniella Goldfarb Israel Proteomics Center • Shira Albeck, Yoav Peleg, Tamar Unger Les Dutton • Chris Moser • Funding $Human Frontiers Science Program Organization
Acknowledgments Noy Group • Ilit Cohen-Ofri • Joanna Grzyb • Jebasingh Tennyson • Iris Margalit Wolfgang Lubitz • Maurice van Gastel Vik Nanda Zx ab Ron Koder Collaboration Avigdor Scherz Zx • Alex Brandis ab • Oksana Shlyk-Kerner Noam Adir, Technion Lev Weiner, Daniella Goldfarb Israel Proteomics Center • Shira Albeck, Yoav Peleg, Tamar Unger Les Dutton • Chris Moser • Funding $Human Frontiers Science Program Organization


