114208280b4a6dc3fa48b2f53a92873d.ppt
- Количество слайдов: 62
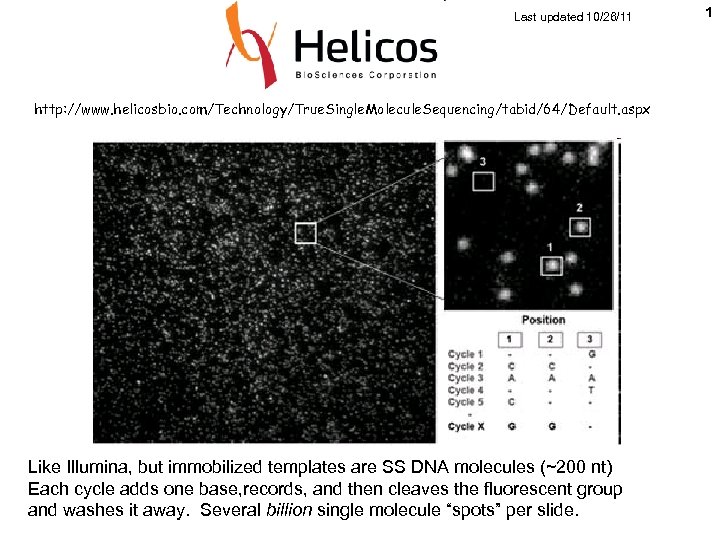
Last updated 10/26/11 http: //www. helicosbio. com/Technology/True. Single. Molecule. Sequencing/tabid/64/Default. aspx Like Illumina, but immobilized templates are SS DNA molecules (~200 nt) Each cycle adds one base, records, and then cleaves the fluorescent group and washes it away. Several billion single molecule “spots” per slide. 1
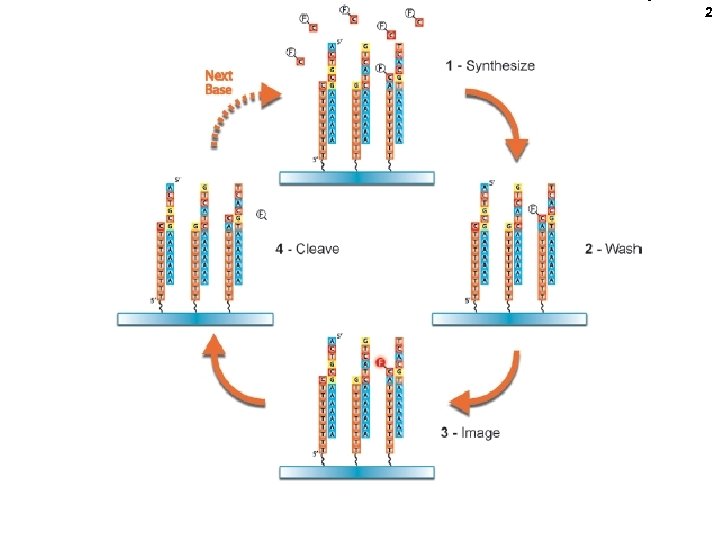
2
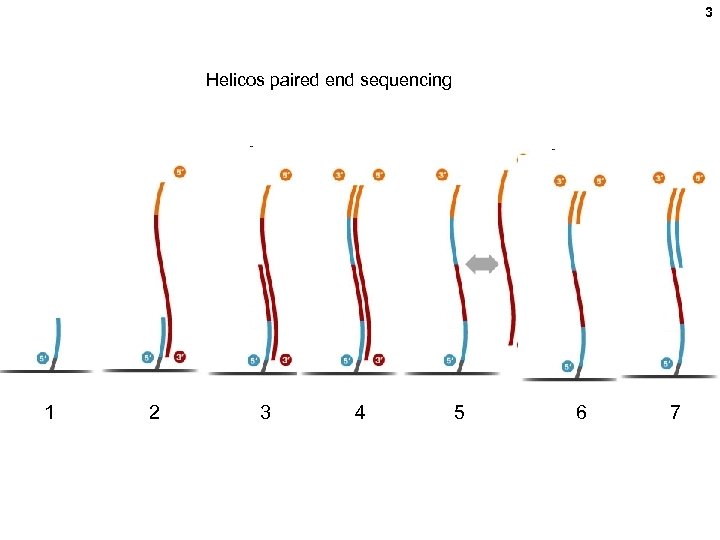
3 Helicos paired end sequencing 1 2 3 4 5 6 7
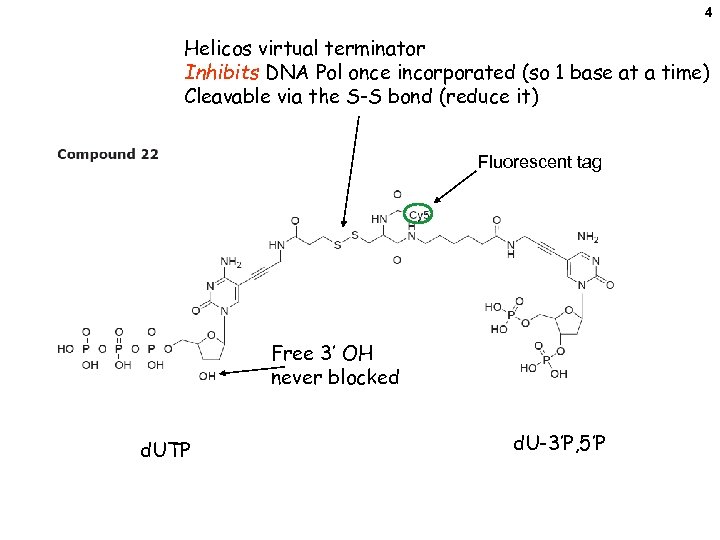
4 Helicos virtual terminator Inhibits DNA Pol once incorporated (so 1 base at a time) Cleavable via the S-S bond (reduce it) Fluorescent tag Free 3’ OH never blocked d. UTP d. U-3’P, 5’P
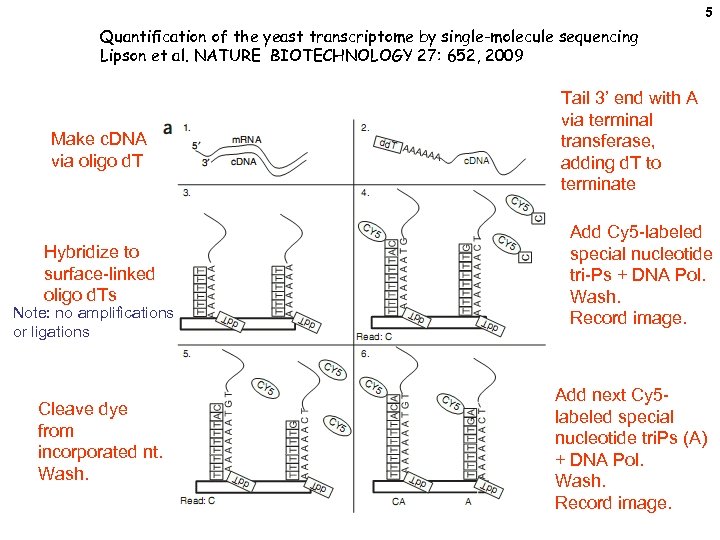
5 Quantification of the yeast transcriptome by single-molecule sequencing Lipson et al. NATURE BIOTECHNOLOGY 27: 652, 2009 Make c. DNA via oligo d. T Hybridize to surface-linked oligo d. Ts Note: no amplifications or ligations Cleave dye from incorporated nt. Wash. Tail 3’ end with A via terminal transferase, adding d. T to terminate Add Cy 5 -labeled special nucleotide tri-Ps + DNA Pol. Wash. Record image. Add next Cy 5 labeled special nucleotide tri. Ps (A) + DNA Pol. Wash. Record image.
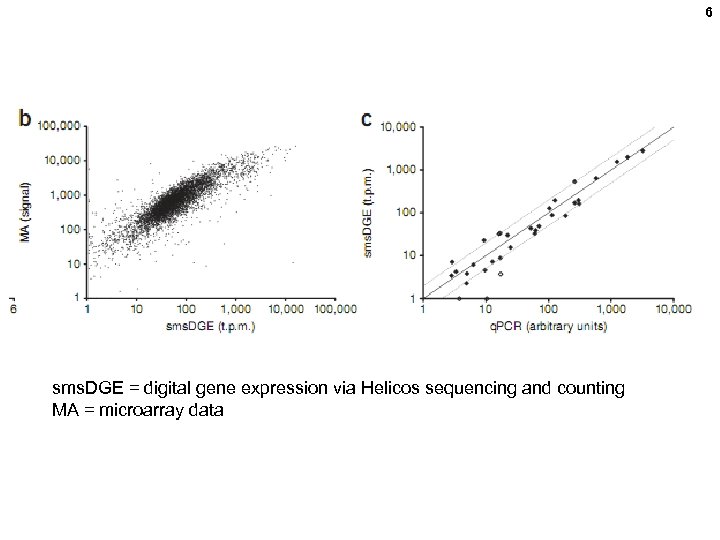
6 sms. DGE = digital gene expression via Helicos sequencing and counting MA = microarray data
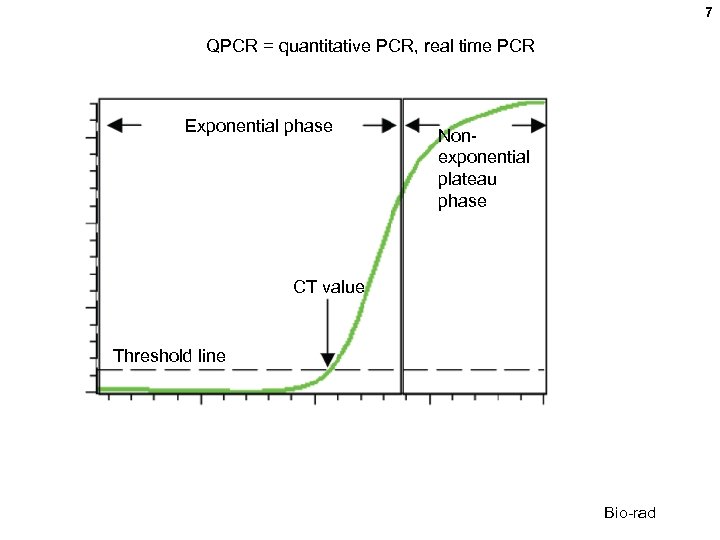
7 QPCR = quantitative PCR, real time PCR Exponential phase Nonexponential plateau phase CT value Threshold line Bio-rad
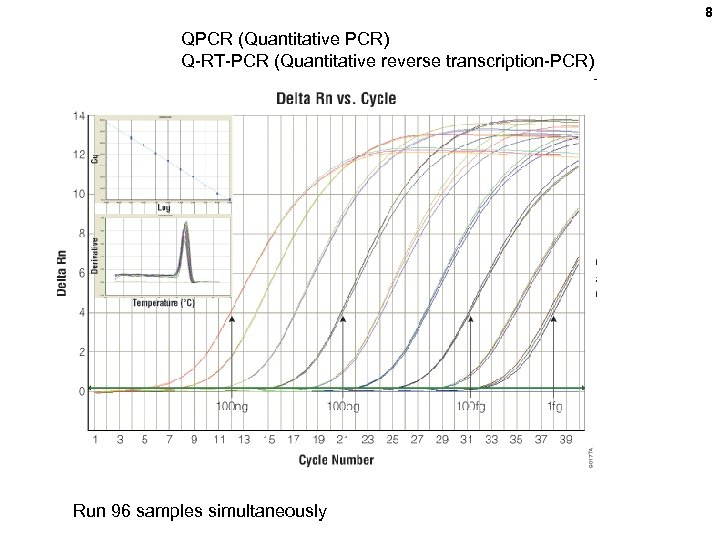
8 QPCR (Quantitative PCR) Q-RT-PCR (Quantitative reverse transcription-PCR) Run 96 samples simultaneously
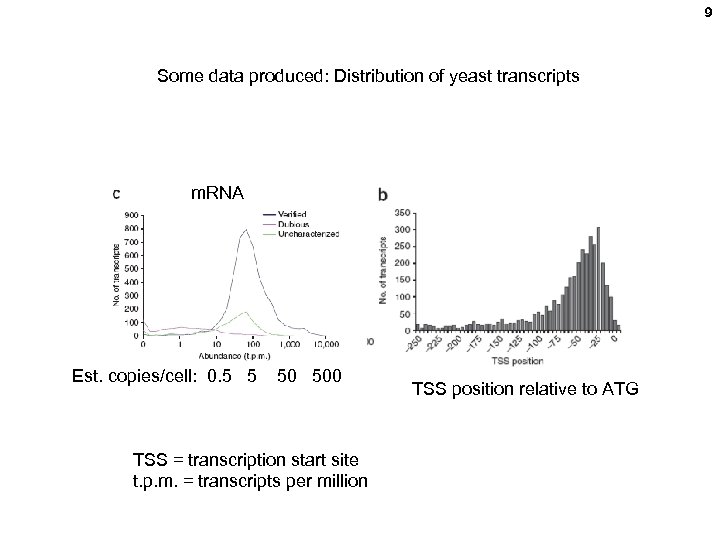
9 Some data produced: Distribution of yeast transcripts m. RNA Est. copies/cell: 0. 5 5 50 500 TSS = transcription start site t. p. m. = transcripts per million TSS position relative to ATG
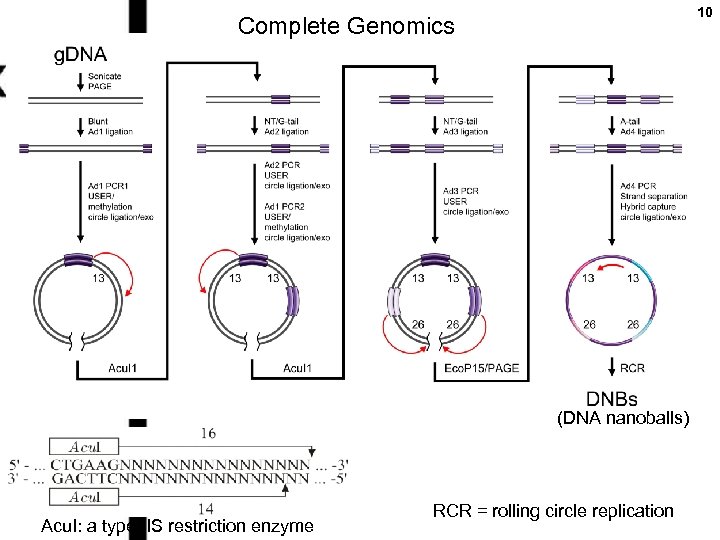
10 Complete Genomics (DNA nanoballs) Acu. I: a type IIS restriction enzyme RCR = rolling circle replication
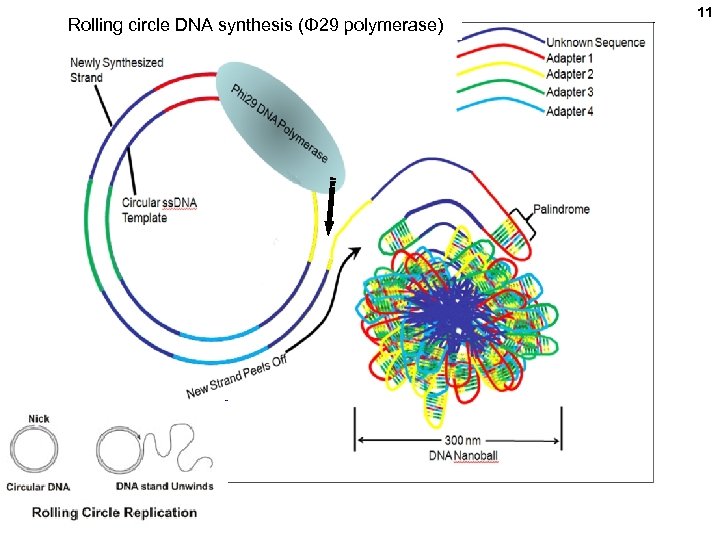
Rolling circle DNA synthesis (Φ 29 polymerase) 11
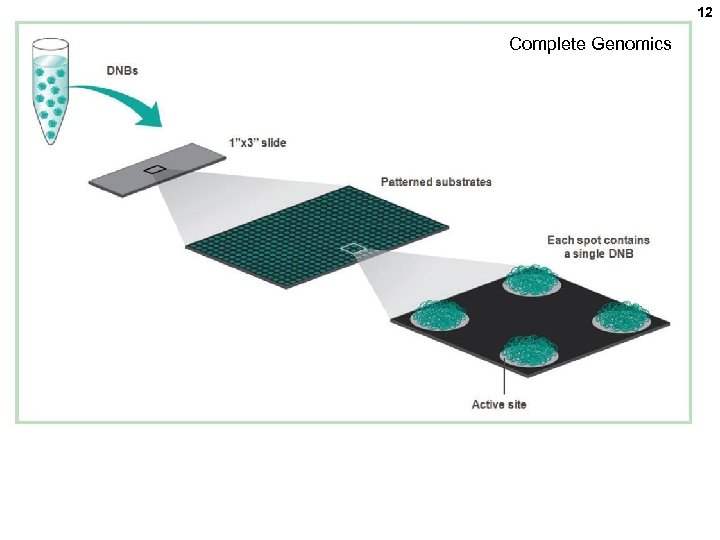
12 Complete Genomics
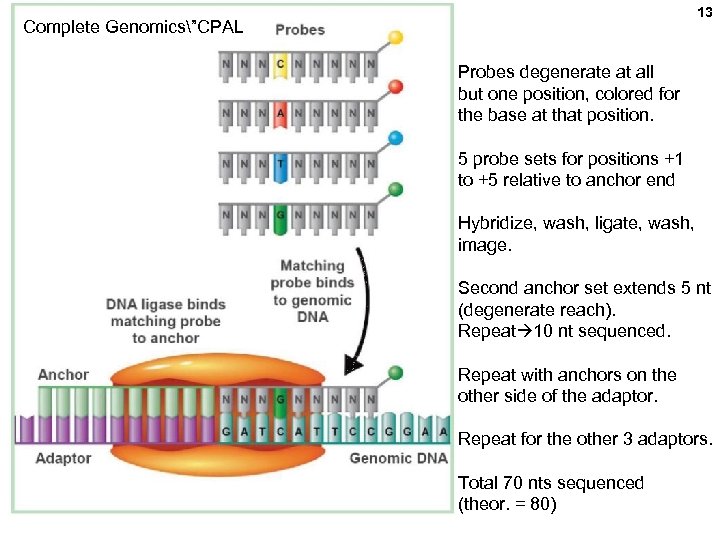
13 Complete Genomics”CPAL Probes degenerate at all but one position, colored for the base at that position. 5 probe sets for positions +1 to +5 relative to anchor end Hybridize, wash, ligate, wash, image. Second anchor set extends 5 nt (degenerate reach). Repeat 10 nt sequenced. Repeat with anchors on the other side of the adaptor. Repeat for the other 3 adaptors. Total 70 nts sequenced (theor. = 80)

14 Complete Genomics Est. 1 billion spots (reads) per slide Lower cost 200 human genomes sequenced Business plan: sell sequencing service, not machines
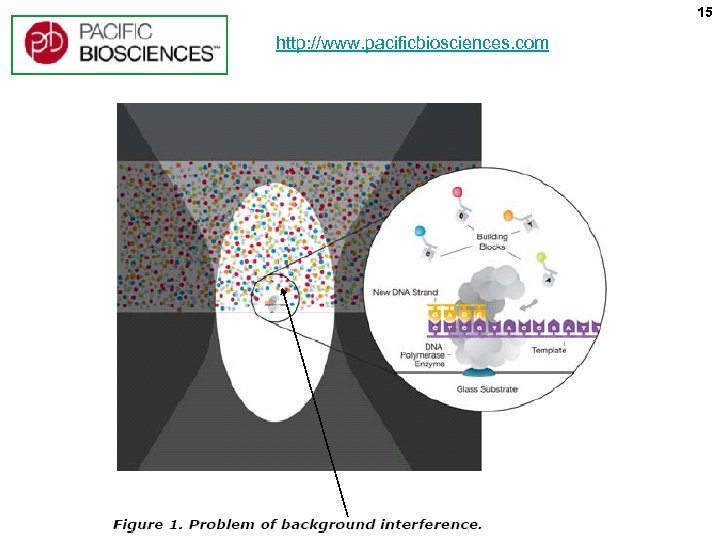
15 http: //www. pacificbiosciences. com
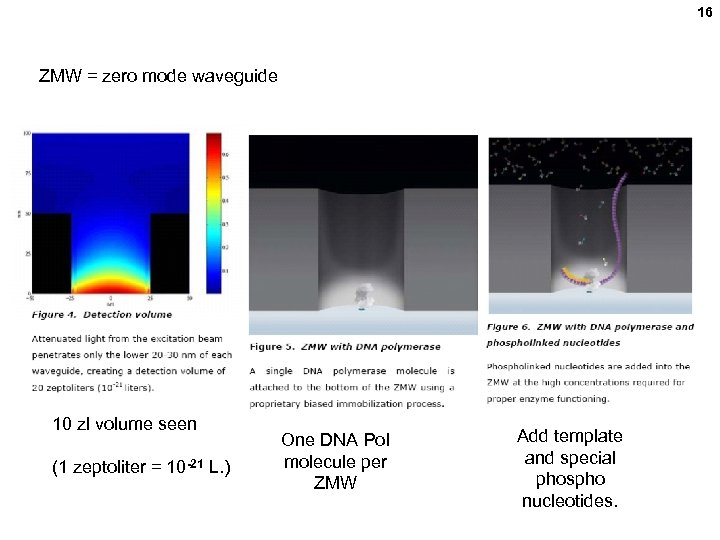
16 ZMW = zero mode waveguide 10 zl volume seen (1 zeptoliter = 10 -21 L. ) One DNA Pol molecule per ZMW Add template and special phospho nucleotides.
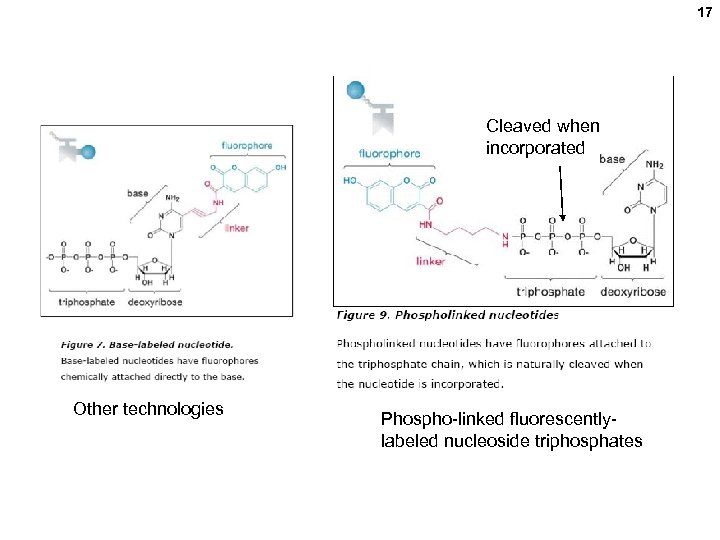
17 Cleaved when incorporated Other technologies Phospho-linked fluorescentlylabeled nucleoside triphosphates
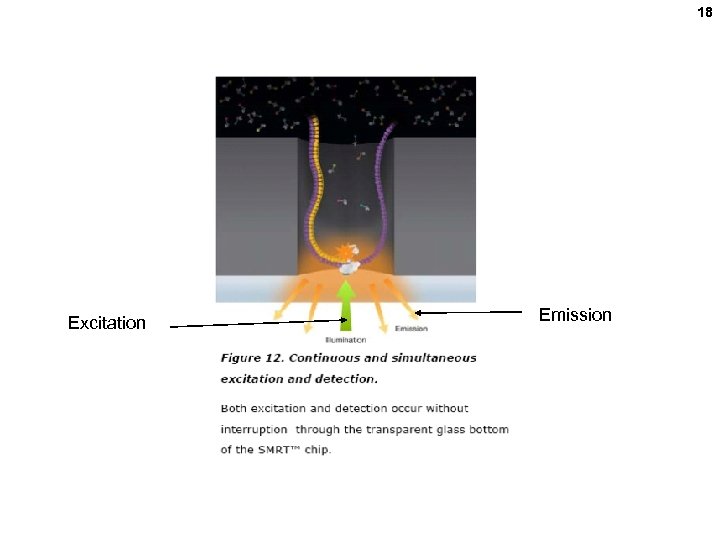
18 Excitation Emission
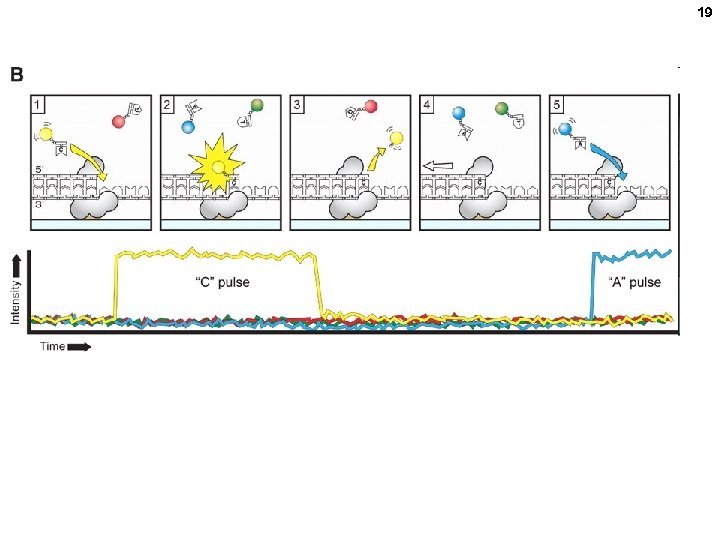
19
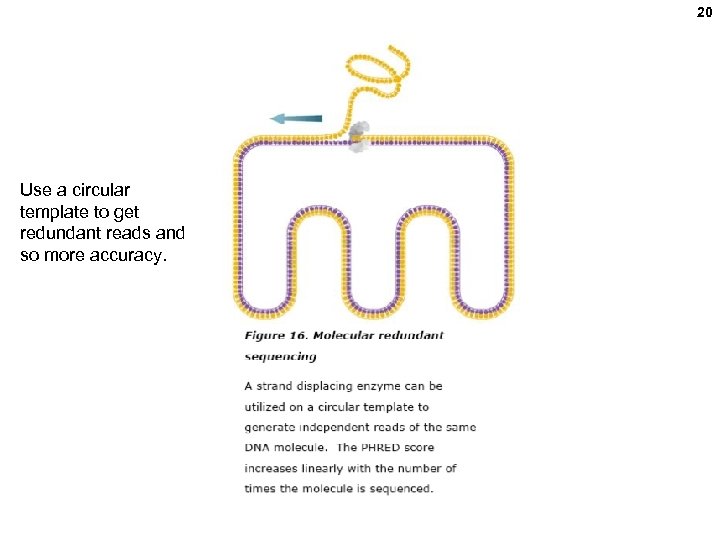
20 Use a circular template to get redundant reads and so more accuracy.

21 Pacific Biosciences • 50, 000 ZMWs (Aug. , 2011), and density may climb • Long reads (e. g. , full molecule analysis for splicing isoform) • Direct RNA sequecning possible. • DNA methylation detectable
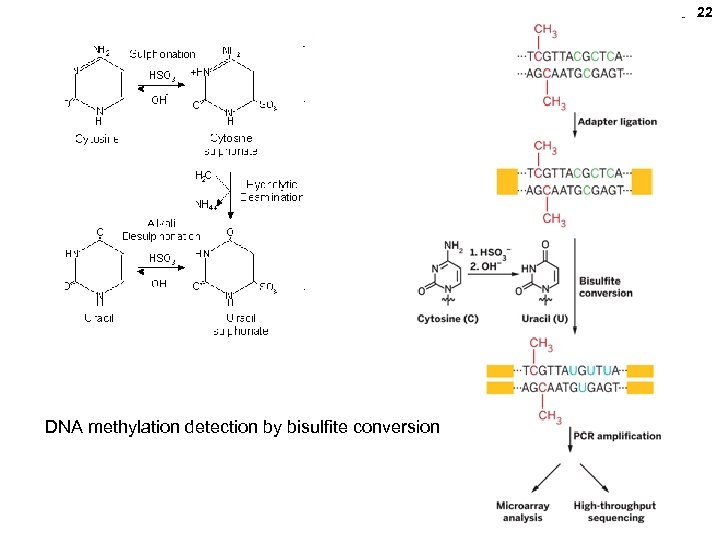
22 DNA methylation detection by bisulfite conversion
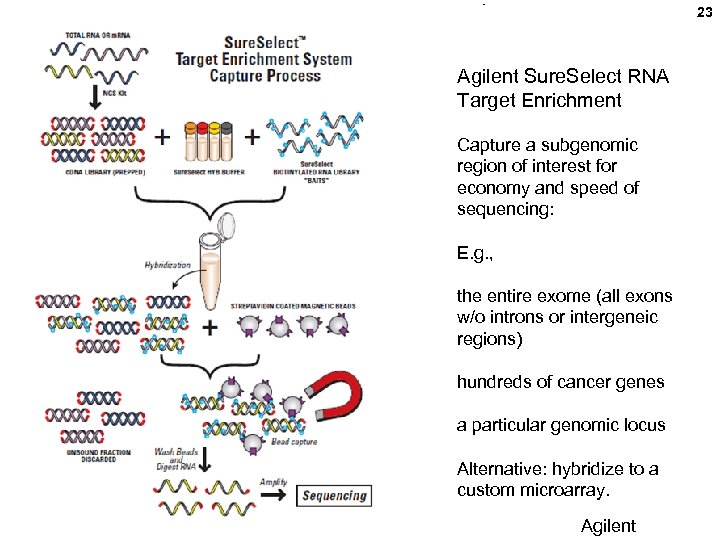
23 Agilent Sure. Select RNA Target Enrichment Capture a subgenomic region of interest for economy and speed of sequencing: E. g. , the entire exome (all exons w/o introns or intergeneic regions) hundreds of cancer genes a particular genomic locus Alternative: hybridize to a custom microarray. Agilent
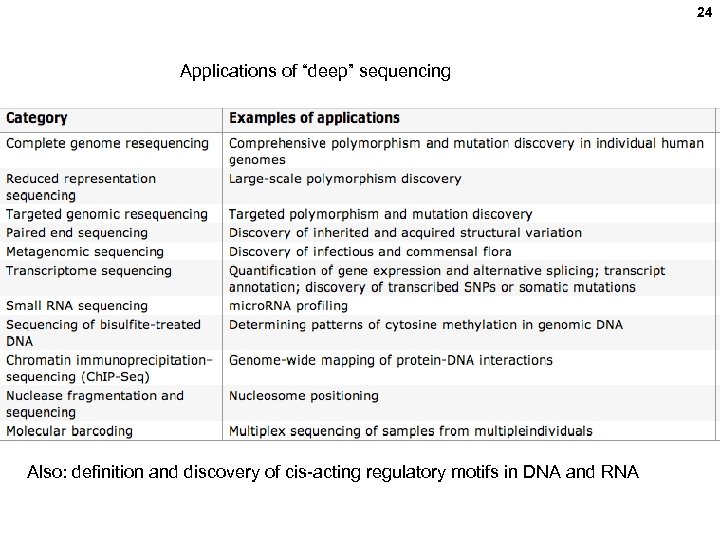
24 Applications of “deep” sequencing Also: definition and discovery of cis-acting regulatory motifs in DNA and RNA
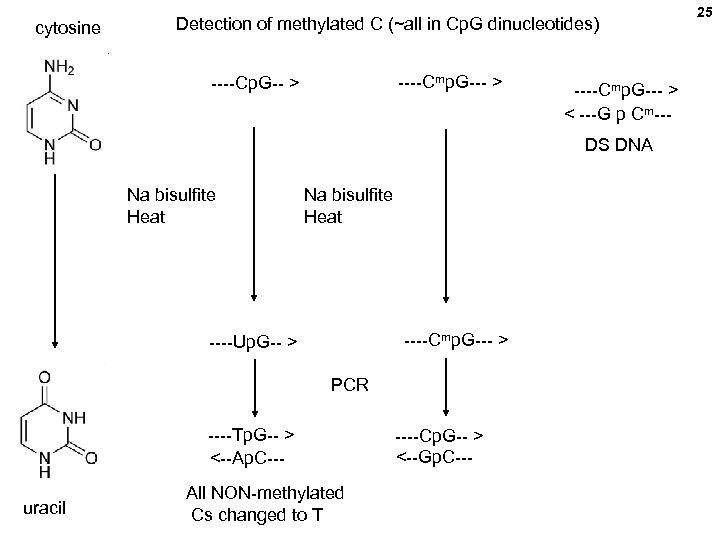
cytosine Detection of methylated C (~all in Cp. G dinucleotides) ----Cmp. G--- > ----Cp. G-- > ----Cmp. G--- > < ---G p Cm--DS DNA Na bisulfite Heat ----Cmp. G--- > ----Up. G-- > PCR ----Tp. G-- > <--Ap. C--uracil All NON-methylated Cs changed to T ----Cp. G-- > <--Gp. C--- 25

26 DEEP SEQUENCING (Next generation sequencing, High throughput sequencing, Massively parallel sequencing) applications: Human genome re-sequencing (mutations, SNPs, haplotypes, disease associations, personalized medicine) Tumor genome sequencing Microbial flora sequencing (microbiome) Metagenomic sequencing (without cell culturing) RNA sequencing (RNAseq; gene expression levels, mi. RNAs, lnc. RNAs, splicing isoforms) Chromatin structure (Ch. IP-seq; histone modifications, nucleosome positioning) Epigenetic modifications (DNA Cp. G methylation and hydroxymethylation) Transcription kinetics (GROseq; nascent RNA, pulse labeled RNA) High throughput genetics (QUEPASA; cis-acting regulatory motif discovery) Drug discovery (bar-coded organic molecule libraries)
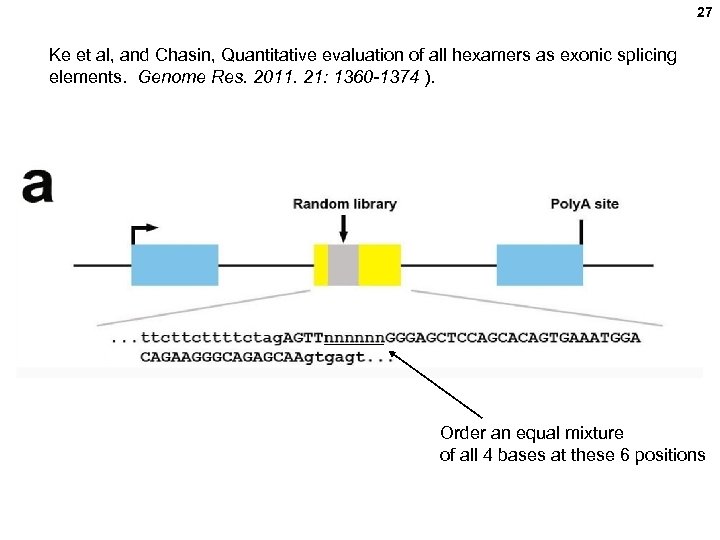
27 Ke et al, and Chasin, Quantitative evaluation of all hexamers as exonic splicing elements. Genome Res. 2011. 21: 1360 -1374 ). Order an equal mixture of all 4 bases at these 6 positions
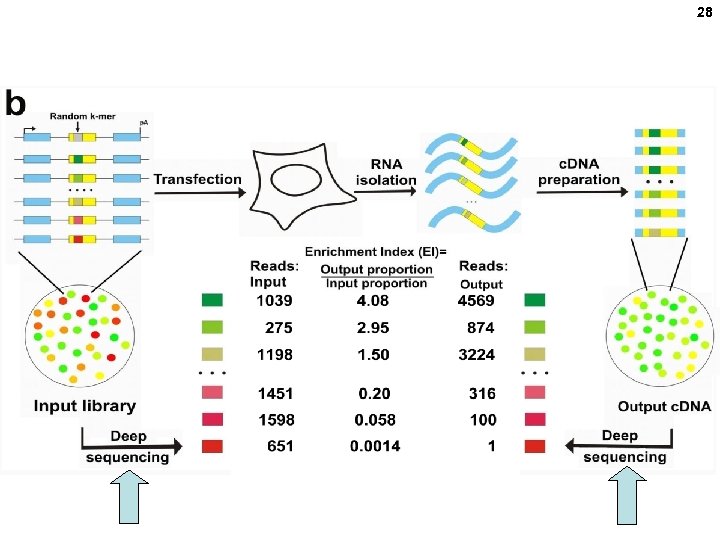
28
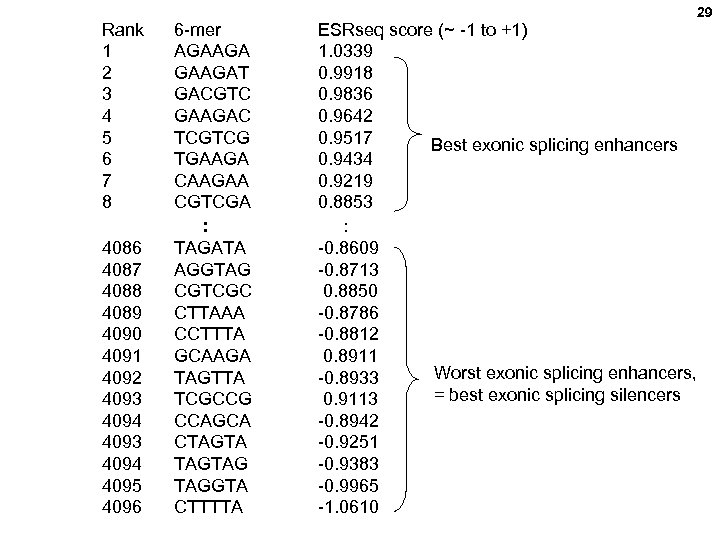
Rank 1 2 3 4 5 6 7 8 6 -mer AGAAGAT GACGTC GAAGAC TCGTCG TGAAGA CAAGAA CGTCGA : 4086 4087 4088 4089 4090 4091 4092 4093 4094 4095 4096 TAGATA AGGTAG CGTCGC CTTAAA CCTTTA GCAAGA TAGTTA TCGCCG CCAGCA CTAGTAG TAGGTA CTTTTA ESRseq score (~ -1 to +1) 1. 0339 0. 9918 0. 9836 0. 9642 0. 9517 Best exonic splicing enhancers 0. 9434 0. 9219 0. 8853 : -0. 8609 -0. 8713 0. 8850 -0. 8786 -0. 8812 0. 8911 Worst exonic splicing enhancers, -0. 8933 = best exonic splicing silencers 0. 9113 -0. 8942 -0. 9251 -0. 9383 -0. 9965 -1. 0610 29
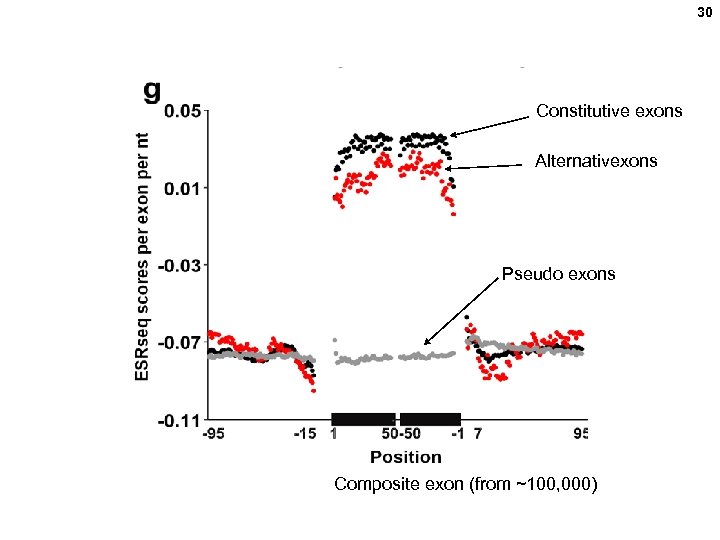
30 Constitutive exons Alternativexons Pseudo exons Composite exon (from ~100, 000)
![31 31 Sequence of 36 Quality code CGCACTGTGCTGGAGCTCCCGGGGTTAACTCTAGAA ab. U^Vaa`aaaa]a. Wa. TNZ`aa`Q][TE[Ua. P_U] TACACTGTGCTGGAGCTCCCAACGGCAACTCTAGAA 31 31 Sequence of 36 Quality code CGCACTGTGCTGGAGCTCCCGGGGTTAACTCTAGAA ab. U^Vaa`aaaa]a. Wa. TNZ`aa`Q][TE[Ua. P_U] TACACTGTGCTGGAGCTCCCAACGGCAACTCTAGAA](https://present5.com/presentation/114208280b4a6dc3fa48b2f53a92873d/image-31.jpg)
31 31 Sequence of 36 Quality code CGCACTGTGCTGGAGCTCCCGGGGTTAACTCTAGAA ab. U^Vaa`aaaa]a. Wa. TNZ`aa`Q][TE[Ua. P_U] TACACTGTGCTGGAGCTCCCAACGGCAACTCTAGAA a`P^Wa`[`Wa^`X_X_XWVa^NSP]_]S^X_TX^ CGCACTGTGCTGGAGCTCCCATGGAGAACTCTAGAA a. Ta`^b``baaaa^aab^Ya. TQLOHIa`^a``TX]] TACACTGTGCTGGAGCTCCCAAACTCTAGAA I_`aaaaaaa_a_^[KZIGIGZ`U`^P^^` CGCACTGTGCTGGAGCTCCCAATAGTAACTTTAGAA a. Y_abb[Tabaaa`a`b. Z[HXXIZa_`_LGMS[` TATACTGTGCTGGAGCTCCCGACGTAAACTCTAGAA aba]^aa_a]`aa]_]`XWSMFGGIPX[P]X`V_Y^ TACACTGTGCTGGAGCTCCCTGGTAAAACTCTAGAA a_^a^aa`a. Yaaa_a. Y`Y_^[I]VY`]V]RW]VV TACACTGTGCTGGAGCTCCCAATAAAAACTCTAGAA XZababa`a. Zaaaaa. YXX`baa``\Ta. Uaa. W` Variable region Constant regions (peculiar to our expt. ) 2 nt barcode (TA or CG) Experiment: 1 1 1 2 2 1+2 2 2 1 2
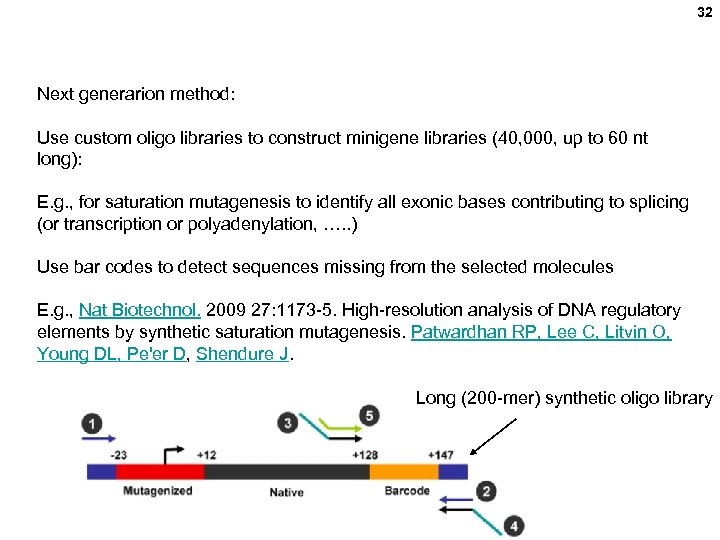
32 Next generarion method: Use custom oligo libraries to construct minigene libraries (40, 000, up to 60 nt long): E. g. , for saturation mutagenesis to identify all exonic bases contributing to splicing (or transcription or polyadenylation, …. . ) Use bar codes to detect sequences missing from the selected molecules E. g. , Nat Biotechnol. 2009 27: 1173 -5. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Patwardhan RP, Lee C, Litvin O, Young DL, Pe'er D, Shendure J. Long (200 -mer) synthetic oligo library

OUTLINE OF NEXT LECTURE TOPICS Expression and manipulation of transgenes in the laboratory • In vitro mutagenesis to isolate variants of your protein/gene with desirable properties – – • To study the protein: Express your transgene – – – • • • Single base mutations Deletions Overlap extension PCR Cassette mutagenesis Usually in E. coli, for speed, economy Expression in eukaryotic hosts Drive it with a promoter/enhancer Purify it via a protein tag Cleave it to get the pure protein Explore protein-protein interaction Co-immunoprecipitation (co-IP) from extracts 2 -hybrid formation surface plasmon resonance FRET (Fluorescence resonance energy transfer) Complementation readout 33 33
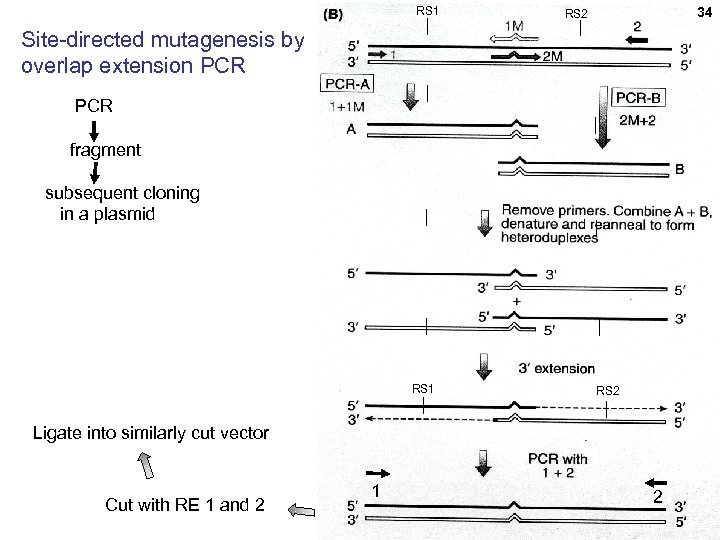
RS 1 34 34 RS 2 Site-directed mutagenesis by overlap extension PCR fragment subsequent cloning in a plasmid RS 1 RS 2 Ligate into similarly cut vector Cut with RE 1 and 2 1 2
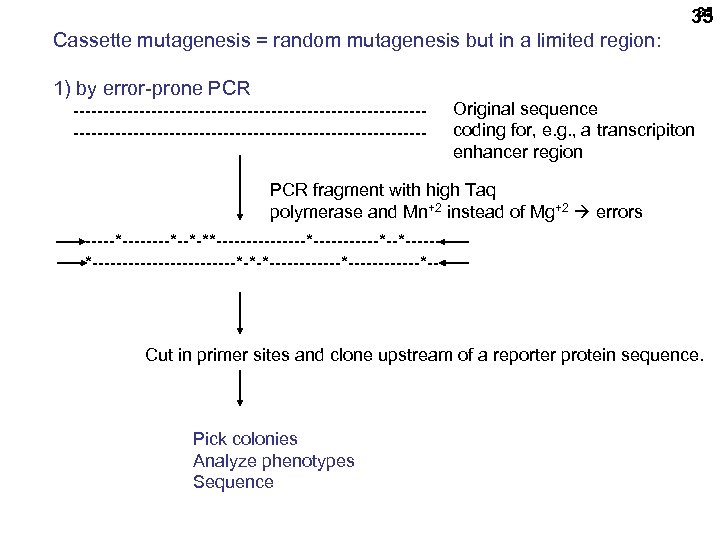
35 35 Cassette mutagenesis = random mutagenesis but in a limited region: 1) by error-prone PCR ----------------------------------------------------------- Original sequence coding for, e. g. , a transcripiton enhancer region PCR fragment with high Taq polymerase and Mn+2 instead of Mg+2 errors ------*--*-**--------*------*--*--------------*-*-*------------*-- Cut in primer sites and clone upstream of a reporter protein sequence. Pick colonies Analyze phenotypes Sequence
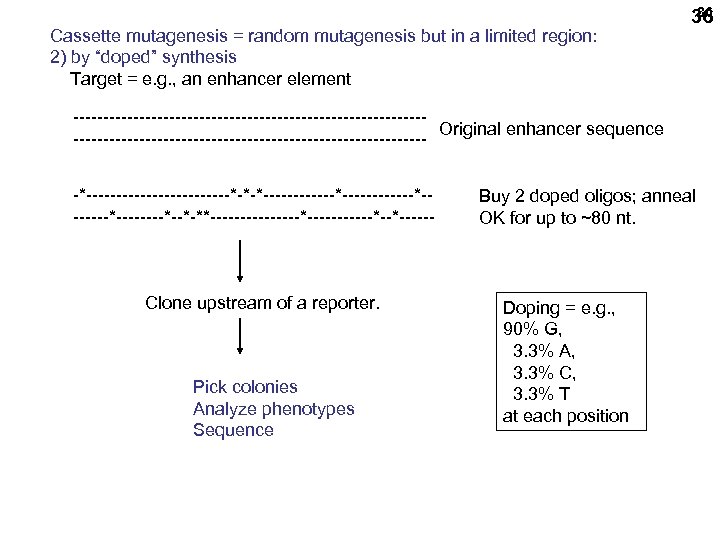
Cassette mutagenesis = random mutagenesis but in a limited region: 2) by “doped” synthesis Target = e. g. , an enhancer element 36 36 -----------------------------Original enhancer sequence ------------------------------*------------*-*-*------------*--------*--*-**--------*------*--*------ Clone upstream of a reporter. Pick colonies Analyze phenotypes Sequence Buy 2 doped oligos; anneal OK for up to ~80 nt. Doping = e. g. , 90% G, 3. 3% A, 3. 3% C, 3. 3% T at each position

37 Got this far

38 38 E. coli as a host • PROs: Easy, flexible, high tech, fast, cheap; but problems • • • CONs Folding (can misfold) Sorting -> can form inclusion bodies Purification -- endotoxins Modification -- not done (glycosylation, phosphorylation, etc. ) • • • Modifications: Glycoproteins Acylation: acetylation, myristoylation Methylation (arg, lys) Phosphorylation (ser, thr, tyr) Sulfation (tyr) Prenylation (farnesyl, geranyl on cys) Vitamin C-Dependent Modifications (hydroxylation of proline and lysine) Vitamin K-Dependent Modifications (gamma carboxylation of glu) Selenoproteins (seleno-cys t. RNA at UGA stop)

39 39 Some alternative hosts • • Yeasts (Saccharomyces , Pichia) Insect cells with baculovirus vectors Mammalian cells in culture (later) Whole organisms (mice, goats, corn) (not discussed) • In vitro (cell-free), for analysis only (good for radiolabeled proteins)
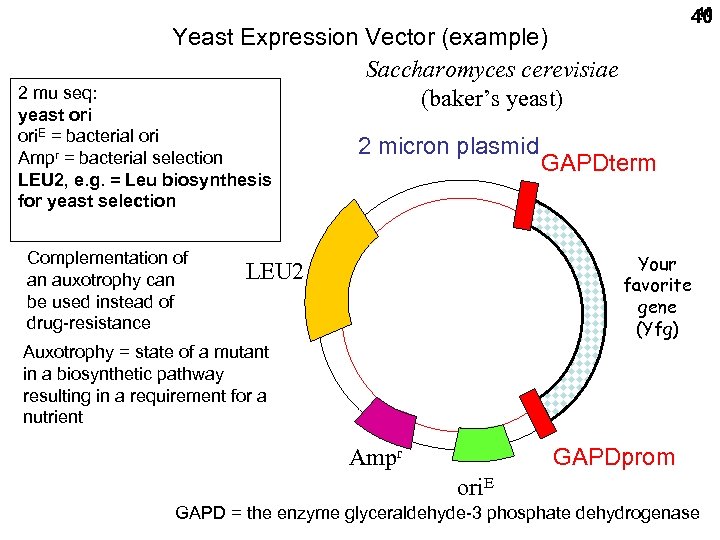
40 40 Yeast Expression Vector (example) Saccharomyces cerevisiae (baker’s yeast) 2 mu seq: yeast ori. E = bacterial ori Ampr = bacterial selection LEU 2, e. g. = Leu biosynthesis for yeast selection Complementation of an auxotrophy can be used instead of drug-resistance 2 micron plasmid GAPDterm Your favorite gene (Yfg) LEU 2 Auxotrophy = state of a mutant in a biosynthetic pathway resulting in a requirement for a nutrient GAPDprom Ampr ori. E GAPD = the enzyme glyceraldehyde-3 phosphate dehydrogenase
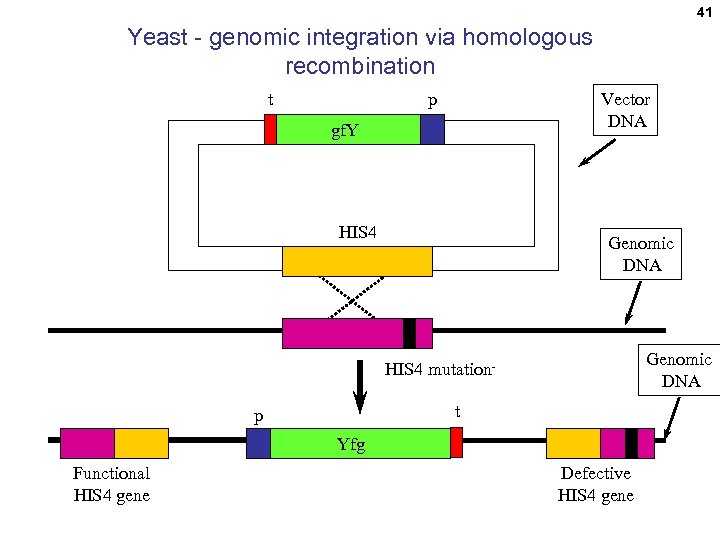
41 Yeast - genomic integration via homologous recombination t Vector DNA p gf. Y HIS 4 Genomic DNA HIS 4 mutationt p Yfg Functional HIS 4 gene Defective HIS 4 gene
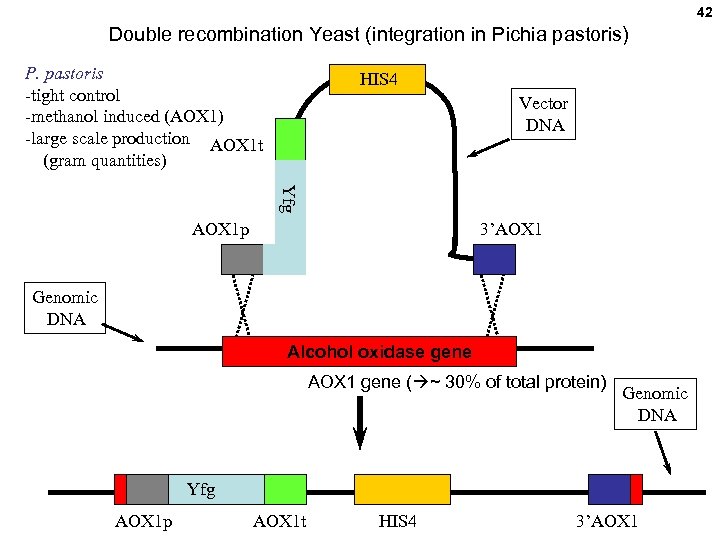
42 Double recombination Yeast (integration in Pichia pastoris) P. pastoris -tight control -methanol induced (AOX 1) -large scale production AOX 1 t (gram quantities) HIS 4 Vector DNA Yfg AOX 1 p 3’AOX 1 Genomic DNA Alcohol oxidase gene AOX 1 gene ( ~ 30% of total protein) Genomic DNA Yfg AOX 1 p AOX 1 t HIS 4 3’AOX 1
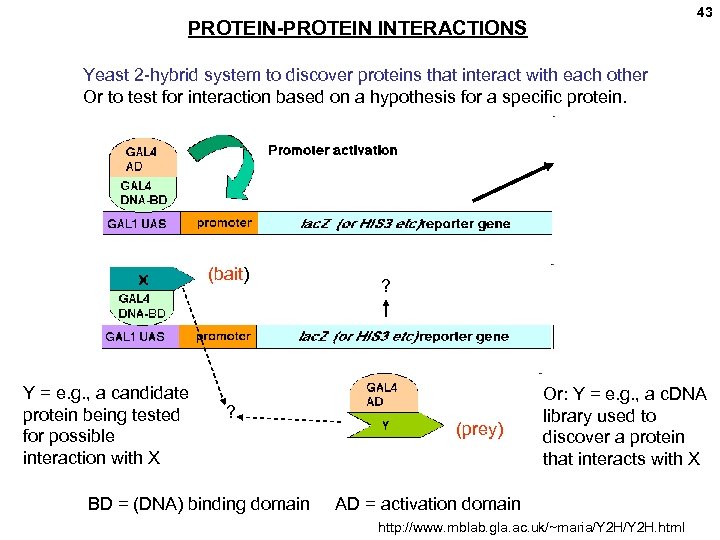
43 PROTEIN-PROTEIN INTERACTIONS Yeast 2 -hybrid system to discover proteins that interact with each other Or to test for interaction based on a hypothesis for a specific protein. (bait) Y = e. g. , a candidate protein being tested for possible interaction with X ? BD = (DNA) binding domain ? (prey) Or: Y = e. g. , a c. DNA library used to discover a protein that interacts with X AD = activation domain http: //www. mblab. gla. ac. uk/~maria/Y 2 H. html
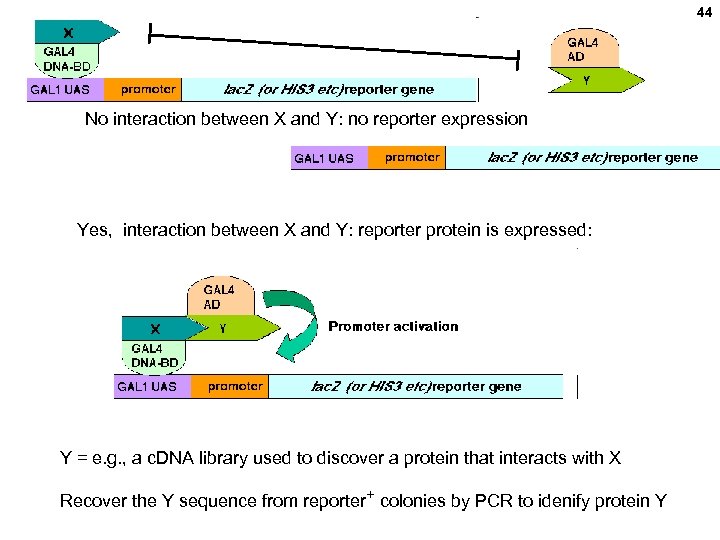
44 No interaction between X and Y: no reporter expression Yes, interaction between X and Y: reporter protein is expressed: Y = e. g. , a c. DNA library used to discover a protein that interacts with X Recover the Y sequence from reporter+ colonies by PCR to idenify protein Y
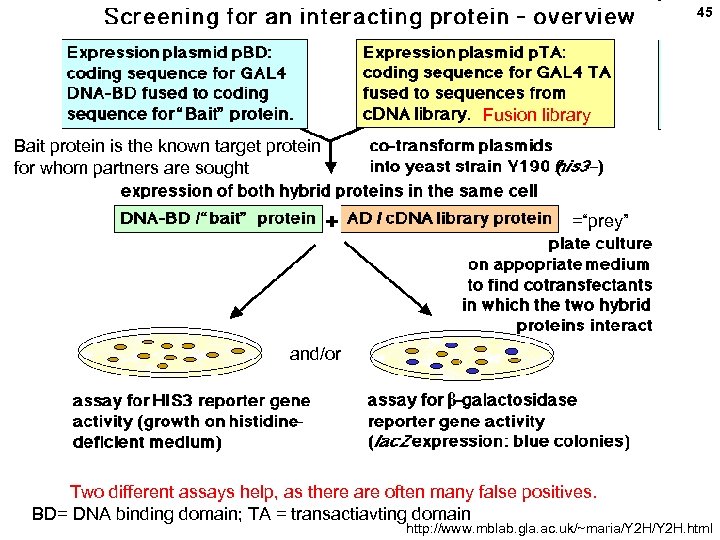
45 Fusion library Bait protein is the known target protein for whom partners are sought =“prey” and/or Two different assays help, as there are often many false positives. BD= DNA binding domain; TA = transactiavting domain http: //www. mblab. gla. ac. uk/~maria/Y 2 H. html
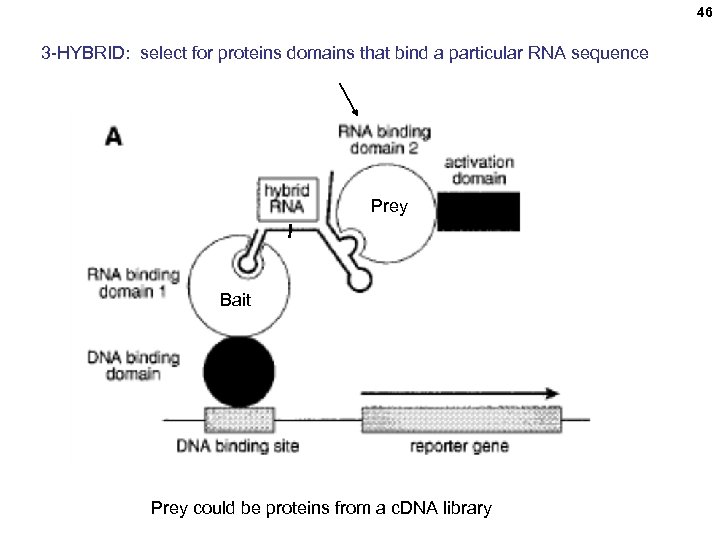
46 3 -HYBRID: select for proteins domains that bind a particular RNA sequence Prey Bait Prey could be proteins from a c. DNA library
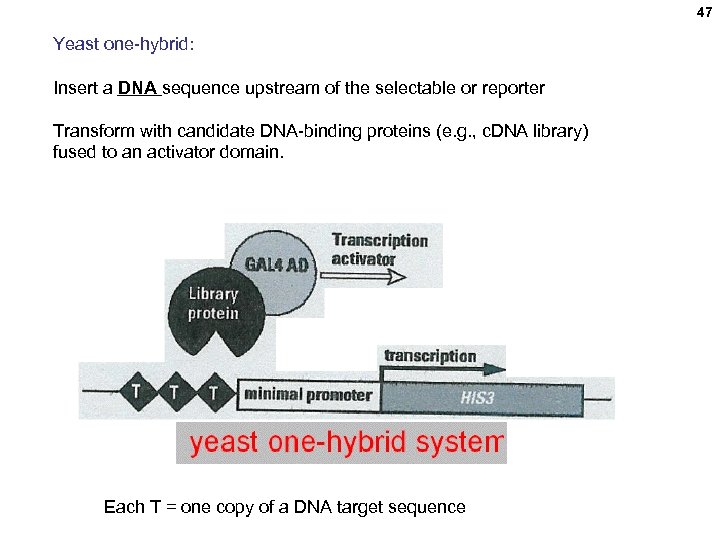
47 Yeast one-hybrid: Insert a DNA sequence upstream of the selectable or reporter Transform with candidate DNA-binding proteins (e. g. , c. DNA library) fused to an activator domain. Each T = one copy of a DNA target sequence
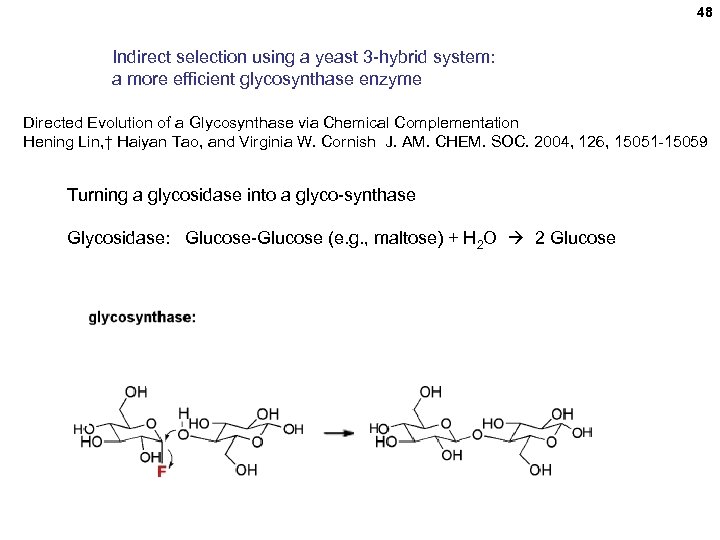
48 Indirect selection using a yeast 3 -hybrid system: a more efficient glycosynthase enzyme Directed Evolution of a Glycosynthase via Chemical Complementation Hening Lin, † Haiyan Tao, and Virginia W. Cornish J. AM. CHEM. SOC. 2004, 126, 15051 -15059 Turning a glycosidase into a glyco-synthase Glycosidase: Glucose-Glucose (e. g. , maltose) + H 2 O 2 Glucose
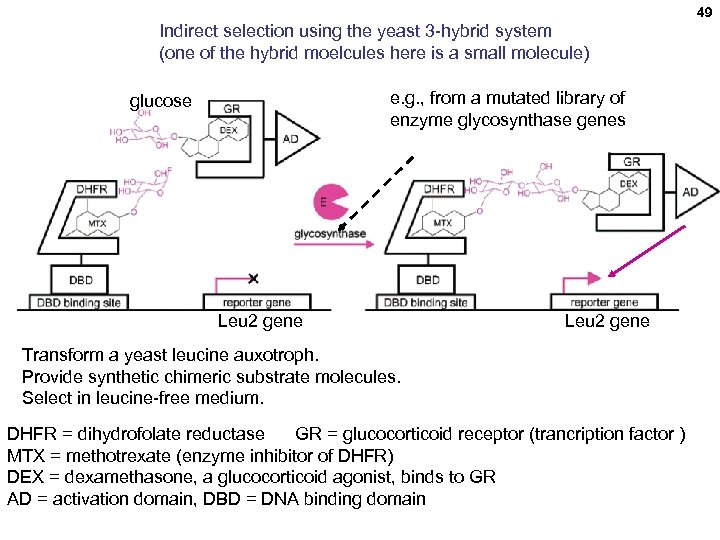
49 Indirect selection using the yeast 3 -hybrid system (one of the hybrid moelcules here is a small molecule) e. g. , from a mutated library of enzyme glycosynthase genes glucose Leu 2 gene Transform a yeast leucine auxotroph. Provide synthetic chimeric substrate molecules. Select in leucine-free medium. DHFR = dihydrofolate reductase GR = glucocorticoid receptor (trancription factor ) MTX = methotrexate (enzyme inhibitor of DHFR) DEX = dexamethasone, a glucocorticoid agonist, binds to GR AD = activation domain, DBD = DNA binding domain
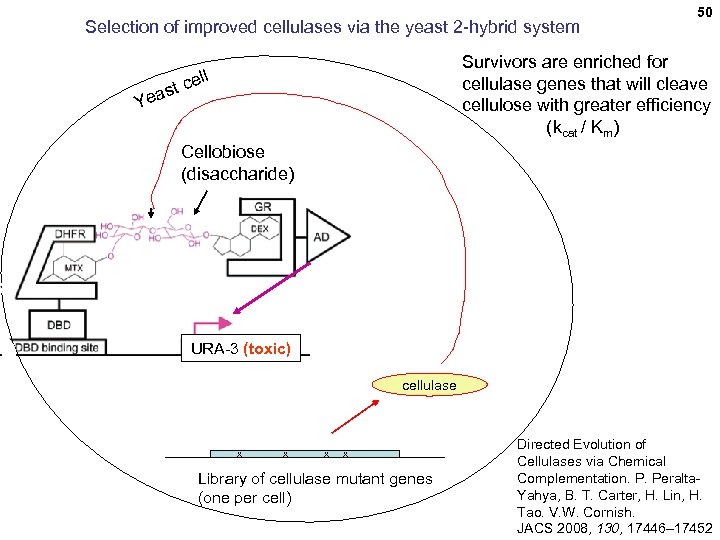
Selection of improved cellulases via the yeast 2 -hybrid system 50 Survivors are enriched for cellulase genes that will cleave cellulose with greater efficiency (kcat / Km) ll e st c Yea Cellobiose (disaccharide) URA-3 (toxic) cellulase x x Library of cellulase mutant genes (one per cell) Directed Evolution of Cellulases via Chemical Complementation. P. Peralta. Yahya, B. T. Carter, H. Lin, H. Tao. V. W. Cornish. JACS 2008, 130, 17446– 17452
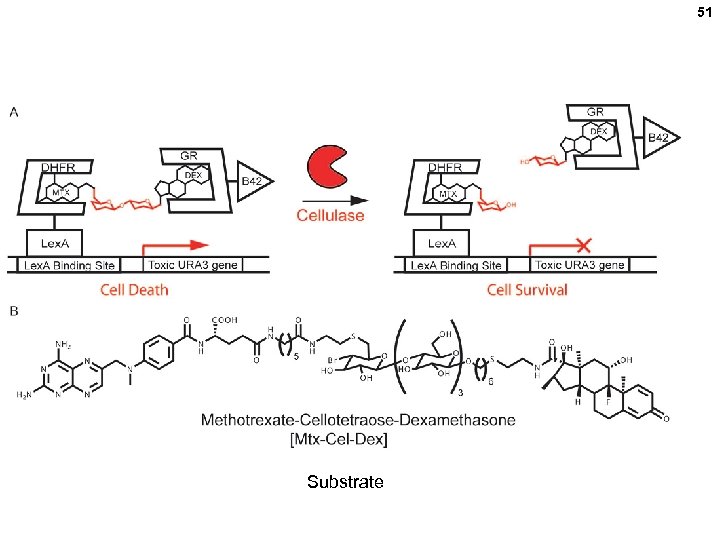
51 Substrate
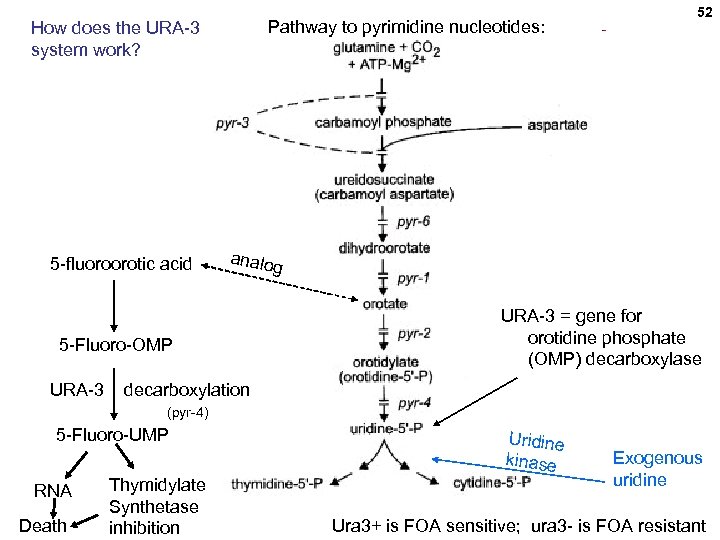
Pathway to pyrimidine nucleotides: How does the URA-3 system work? 5 -fluoroorotic acid analo 5 -Fluoro-OMP URA-3 52 g URA-3 = gene for orotidine phosphate (OMP) decarboxylase decarboxylation (pyr-4) 5 -Fluoro-UMP RNA Death Thymidylate Synthetase inhibition Uridine kinase Exogenous uridine Ura 3+ is FOA sensitive; ura 3 - is FOA resistant
53 Measuring protein-protein interactions in vitro X=one protein Y= another protein Pull-downs: Binding between defined purified proteins, at least one being purified. Tag each protein differently. Examples: His 6 -X + HA-Y; Bind to nickel ion column, elute (his), Western with HA Ab GST-X + HA-Y; Bind to glutathione ion column, elute (glutathione), Western with HA Ab His 6 -X + 35 S-Y (made in vitro); Bind Ni column, elute (his), gel + autoradiography. No antibody needed. (HA = influenza virus flu hemagglutinin) glutathione = Gamma-glutamyl-cysteinyl-glycine.
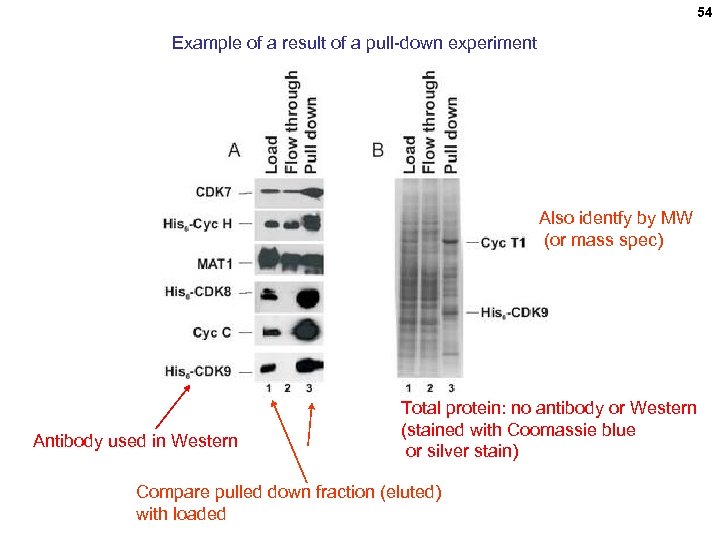
54 Example of a result of a pull-down experiment Also identfy by MW (or mass spec) Antibody used in Western Total protein: no antibody or Western (stained with Coomassie blue or silver stain) Compare pulled down fraction (eluted) with loaded
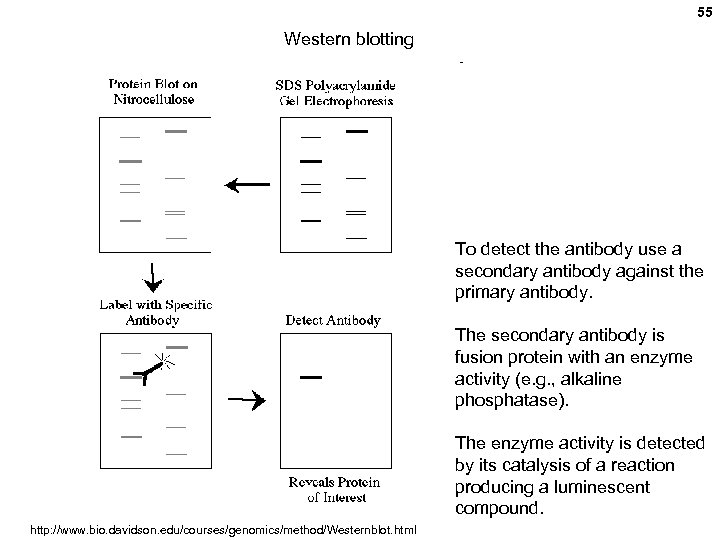
55 Western blotting To detect the antibody use a secondary antibody against the primary antibody. The secondary antibody is fusion protein with an enzyme activity (e. g. , alkaline phosphatase). The enzyme activity is detected by its catalysis of a reaction producing a luminescent compound. http: //www. bio. davidson. edu/courses/genomics/method/Westernblot. html
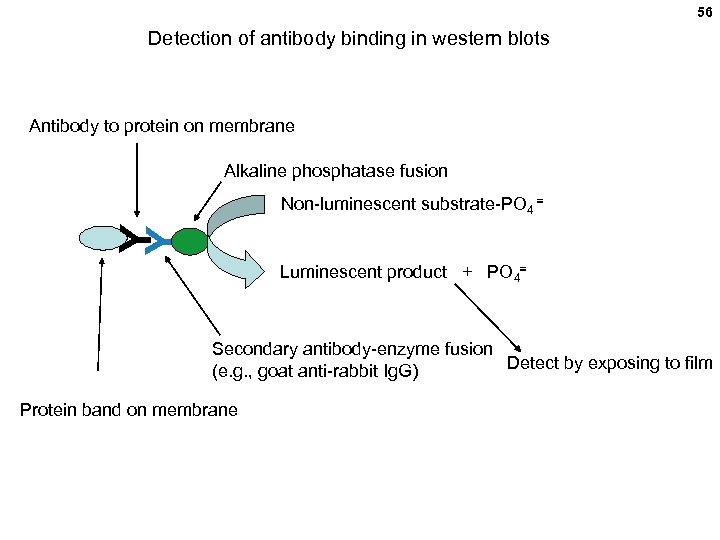
56 Detection of antibody binding in western blots Antibody to protein on membrane Alkaline phosphatase fusion Y Y Non-luminescent substrate-PO 4 = Luminescent product + PO 4= Secondary antibody-enzyme fusion Detect by exposing to film (e. g. , goat anti-rabbit Ig. G) Protein band on membrane
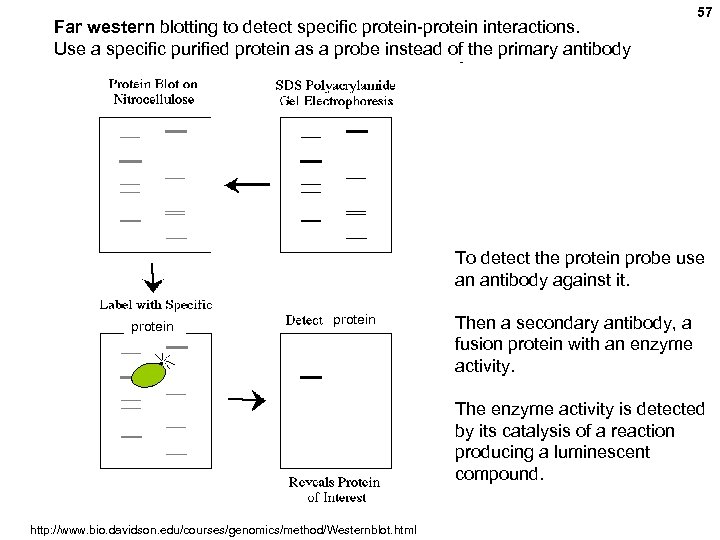
Far western blotting to detect specific protein-protein interactions. Use a specific purified protein as a probe instead of the primary antibody 57 To detect the protein probe use an antibody against it. protein Then a secondary antibody, a fusion protein with an enzyme activity. The enzyme activity is detected by its catalysis of a reaction producing a luminescent compound. http: //www. bio. davidson. edu/courses/genomics/method/Westernblot. html
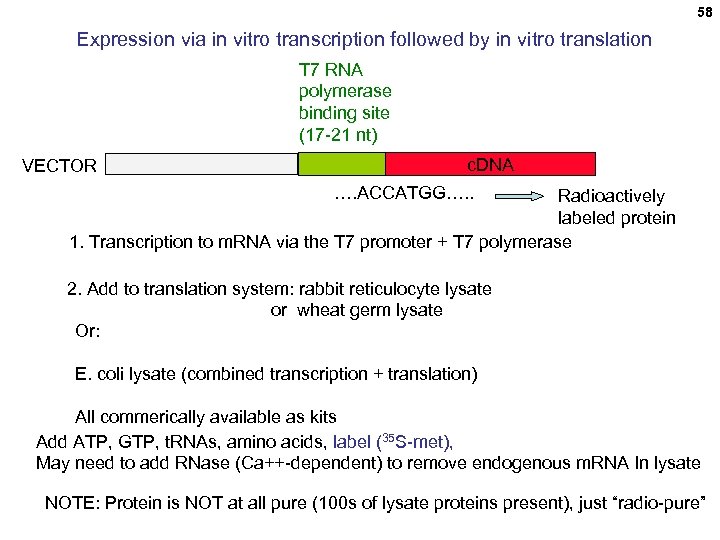
58 Expression via in vitro transcription followed by in vitro translation T 7 RNA polymerase binding site (17 -21 nt) VECTOR c. DNA …. ACCATGG…. . Radioactively labeled protein 1. Transcription to m. RNA via the T 7 promoter + T 7 polymerase 2. Add to translation system: rabbit reticulocyte lysate or wheat germ lysate Or: E. coli lysate (combined transcription + translation) All commerically available as kits Add ATP, GTP, t. RNAs, amino acids, label (35 S-met), May need to add RNase (Ca++-dependent) to remove endogenous m. RNA In lysate NOTE: Protein is NOT at all pure (100 s of lysate proteins present), just “radio-pure”
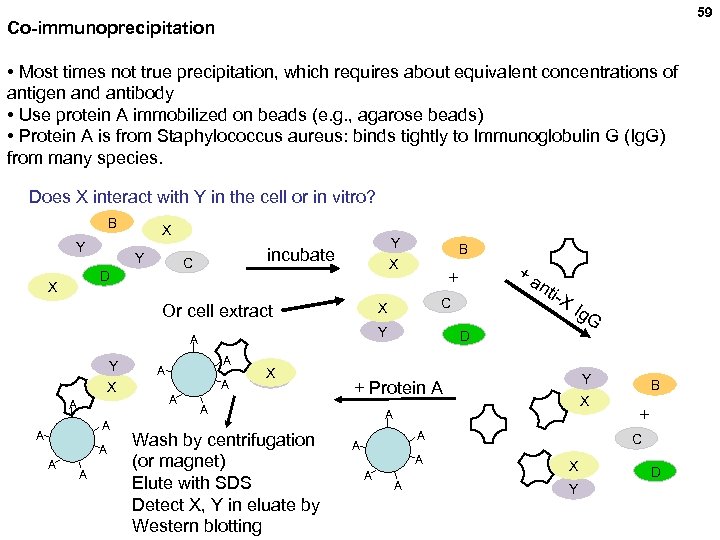
59 Co-immunoprecipitation • Most times not true precipitation, which requires about equivalent concentrations of antigen and antibody • Use protein A immobilized on beads (e. g. , agarose beads) • Protein A is from Staphylococcus aureus: binds tightly to Immunoglobulin G (Ig. G) from many species. Does X interact with Y in the cell or in vitro? B Y X incubate C D Y X X A A A A A X Y Ig. G D Y + Protein A A Wash by centrifugation (or magnet) Elute with SDS Detect X, Y in eluate by Western blotting nti -X C Y A +a + X Or cell extract Y B X A A A B + C X Y D
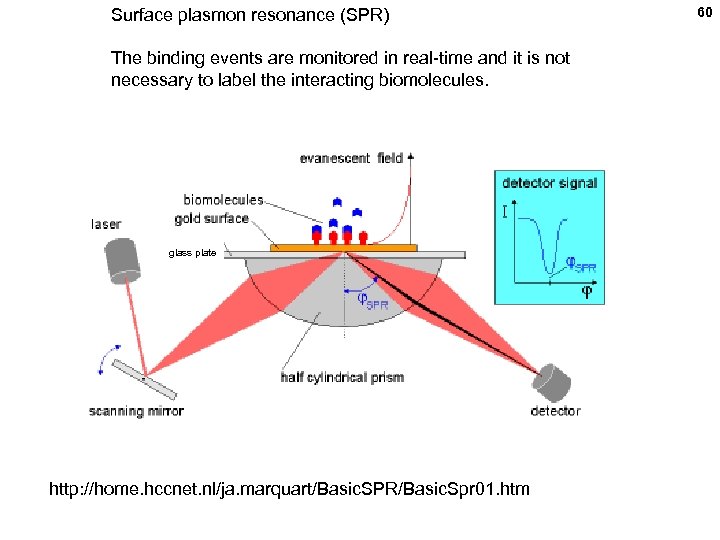
Surface plasmon resonance (SPR) The binding events are monitored in real-time and it is not necessary to label the interacting biomolecules. glass plate http: //home. hccnet. nl/ja. marquart/Basic. SPR/Basic. Spr 01. htm 60
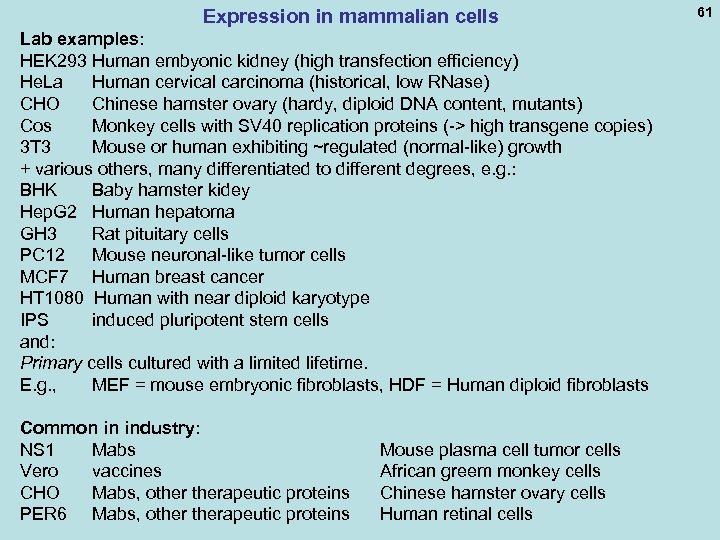
Expression in mammalian cells Lab examples: HEK 293 Human embyonic kidney (high transfection efficiency) He. La Human cervical carcinoma (historical, low RNase) CHO Chinese hamster ovary (hardy, diploid DNA content, mutants) Cos Monkey cells with SV 40 replication proteins (-> high transgene copies) 3 T 3 Mouse or human exhibiting ~regulated (normal-like) growth + various others, many differentiated to different degrees, e. g. : BHK Baby hamster kidey Hep. G 2 Human hepatoma GH 3 Rat pituitary cells PC 12 Mouse neuronal-like tumor cells MCF 7 Human breast cancer HT 1080 Human with near diploid karyotype IPS induced pluripotent stem cells and: Primary cells cultured with a limited lifetime. E. g. , MEF = mouse embryonic fibroblasts, HDF = Human diploid fibroblasts Common in industry: NS 1 Mabs Vero vaccines CHO Mabs, otherapeutic proteins PER 6 Mabs, otherapeutic proteins Mouse plasma cell tumor cells African greem monkey cells Chinese hamster ovary cells Human retinal cells 61

62
114208280b4a6dc3fa48b2f53a92873d.ppt