084c758a9c7527093045c4c31cbc826c.ppt
- Количество слайдов: 37

Kolchanov N. A. , IC&G, BGRS 2000 NUCLEOSOME FORMATION SITES: CODING, ORGANIZATION AND FUNCTION 1 Institute of Cytology and Genetics of SB RAS, Novosibirsk, Russia N. A. Kolchanov and V. G. Levitsky kol@bionet. nsc. ru levitsky@bionet. nsc. ru
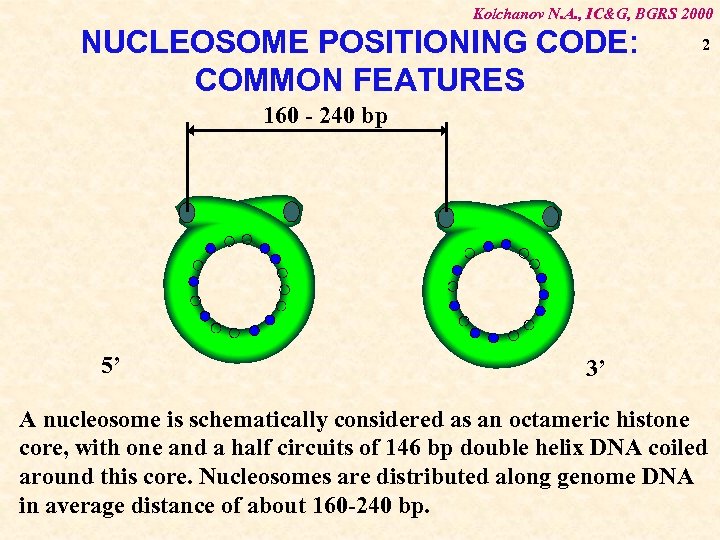
Kolchanov N. A. , IC&G, BGRS 2000 NUCLEOSOME POSITIONING CODE: COMMON FEATURES 2 160 - 240 bp 5’ 3’ A nucleosome is schematically considered as an octameric histone core, with one and a half circuits of 146 bp double helix DNA coiled around this core. Nucleosomes are distributed along genome DNA in average distance of about 160 -240 bp.
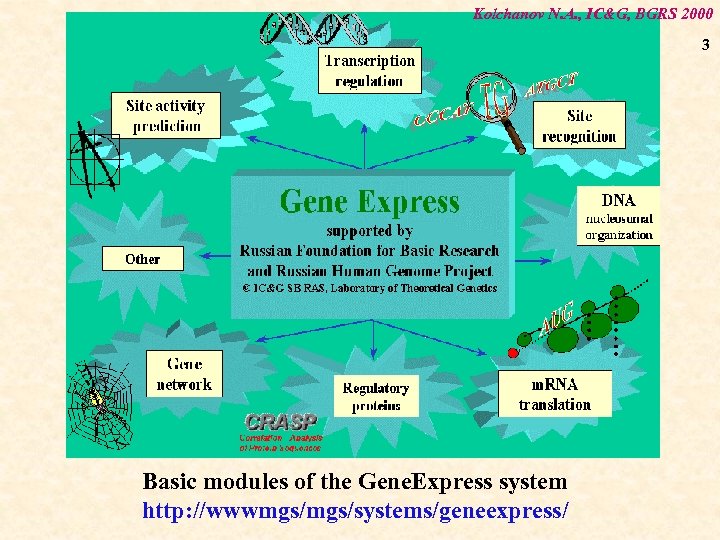
Kolchanov N. A. , IC&G, BGRS 2000 3 Basic modules of the Gene. Express system http: //wwwmgs/systems/geneexpress/
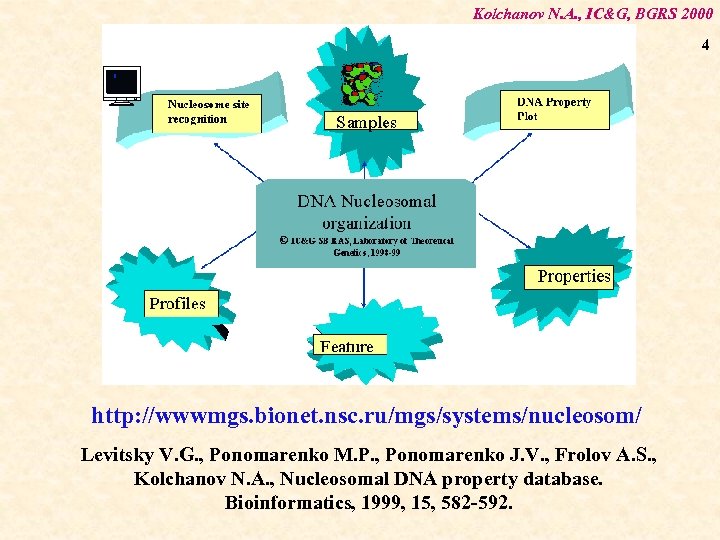
Kolchanov N. A. , IC&G, BGRS 2000 4 http: //wwwmgs. bionet. nsc. ru/mgs/systems/nucleosom/ Levitsky V. G. , Ponomarenko M. P. , Ponomarenko J. V. , Frolov A. S. , Kolchanov N. A. , Nucleosomal DNA property database. Bioinformatics, 1999, 15, 582 -592.
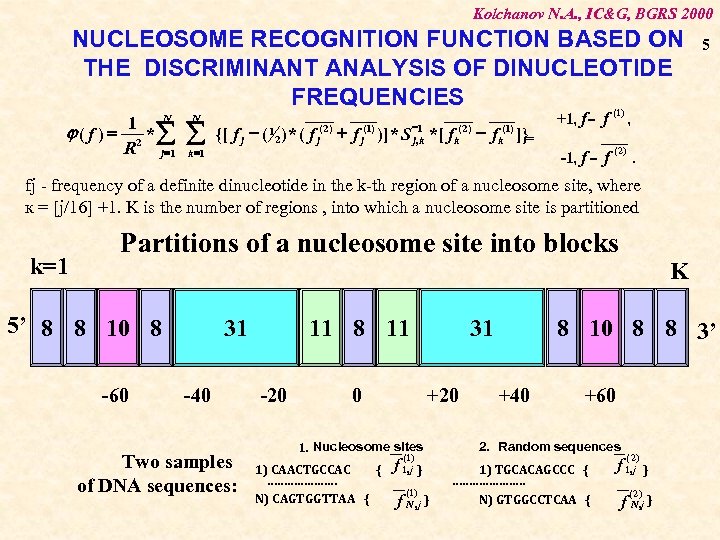
Kolchanov N. A. , IC&G, BGRS 2000 NUCLEOSOME RECOGNITION FUNCTION BASED ON THE DISCRIMINANT ANALYSIS OF DINUCLEOTIDE FREQUENCIES (1) 1 N j( f ) = 2 *å R j =1 N å k =1 {[ f j - ( ) * ( f 1 2 ( 2) j -1 j , k + f )] * S *[ f (1) j ( 2) k - f ]}= (1) k +1, f = f -1, f = f 5 , ( 2) . fj - frequency of a definite dinucleotide in the k-th region of a nucleosome site, where к = [j/16] +1. K is the number of regions , into which a nucleosome site is partitioned k=1 Partitions of a nucleosome site into blocks 5’ 8 8 10 8 -60 31 -40 Two samples of DNA sequences: -20 0 +20 1. Nucleosome sites 1) CAACTGCCAC { ………………… N) CAGTGGTTAA { f (1) 1, j } (1) 8 10 8 8 3’ 31 11 8 11 f N , j } K +40 +60 2. Random sequences 1) TGCACAGCCC { …………………. N) GTGGCCTCAA { ( 2) f 1, j (2) } f N , j }
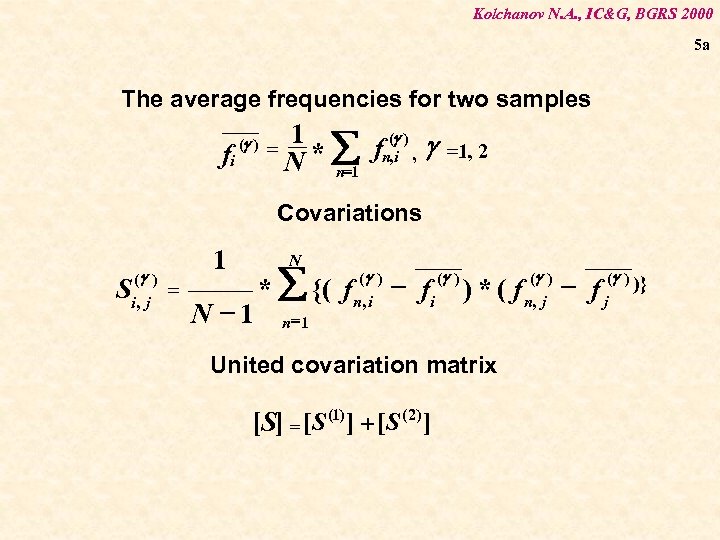
Kolchanov N. A. , IC&G, BGRS 2000 5 a The average frequencies for two samples fi (g ) = 1 (g ) * å fn, i , g N n=1 =1, 2 Covariations (g ) i, j S 1 = N N -1 * å {( f n =1 (g ) n, i - fi (g ) ) * ( f n(, gj ) - f j(g ) )} United covariation matrix [S] = [S (1) ] + [S (2) ]
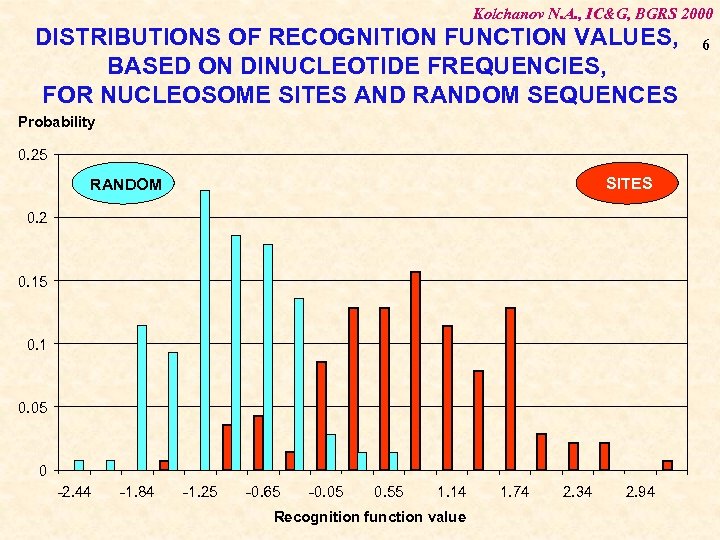
Kolchanov N. A. , IC&G, BGRS 2000 DISTRIBUTIONS OF RECOGNITION FUNCTION VALUES, BASED ON DINUCLEOTIDE FREQUENCIES, FOR NUCLEOSOME SITES AND RANDOM SEQUENCES Probability 0. 25 SITES RANDOM 0. 2 0. 15 0. 1 0. 05 0 -2. 44 -1. 84 -1. 25 -0. 65 -0. 05 0. 55 1. 14 Recognition function value 1. 74 2. 34 2. 94 6
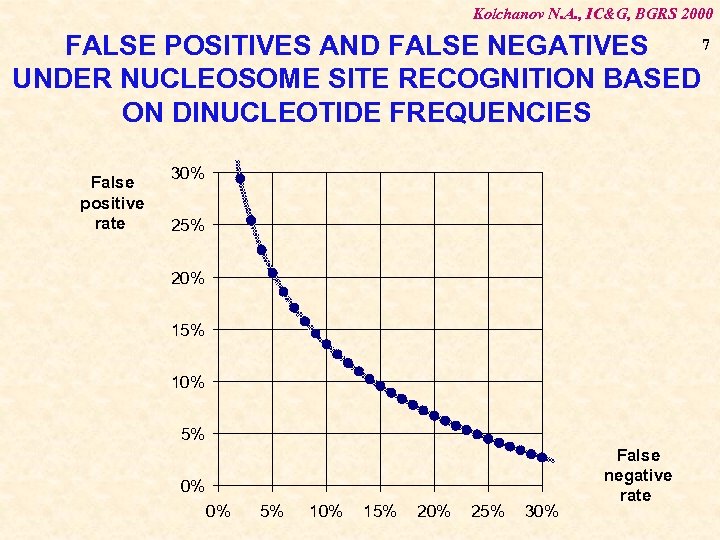
Kolchanov N. A. , IC&G, BGRS 2000 7 FALSE POSITIVES AND FALSE NEGATIVES UNDER NUCLEOSOME SITE RECOGNITION BASED ON DINUCLEOTIDE FREQUENCIES False positive rate 30% 25% 20% 15% 10% 5% 0% 0% 5% 10% 15% 20% 25% 30% False negative rate
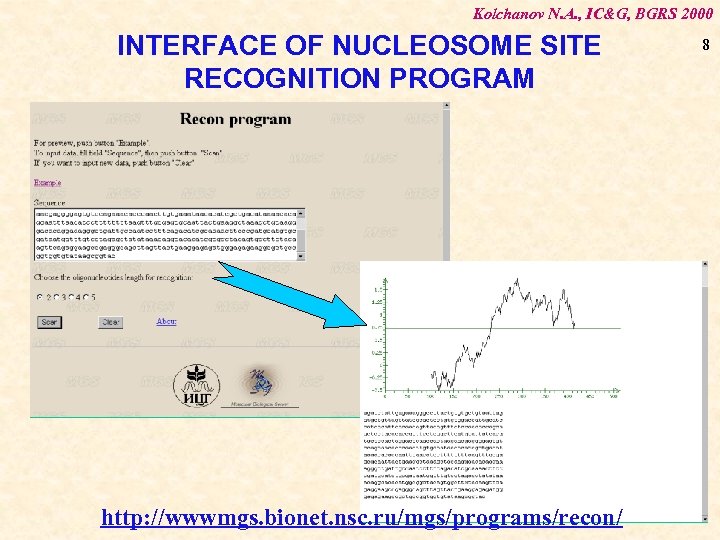
Kolchanov N. A. , IC&G, BGRS 2000 INTERFACE OF NUCLEOSOME SITE RECOGNITION PROGRAM http: //wwwmgs. bionet. nsc. ru/mgs/programs/recon/ 8
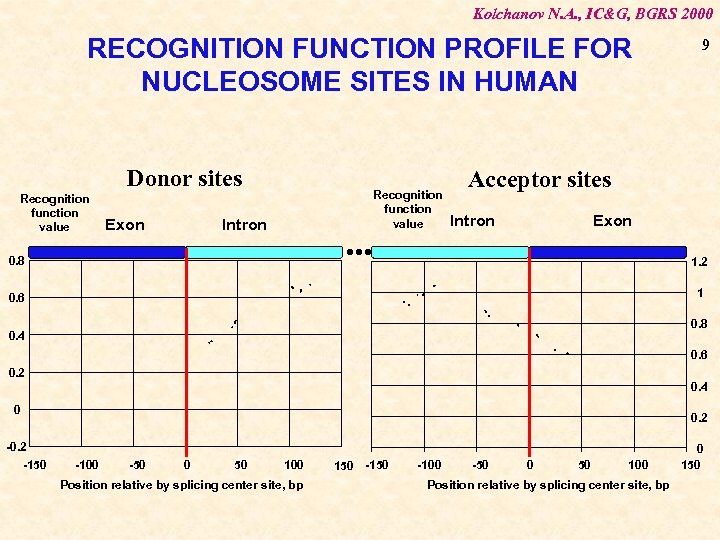
Kolchanov N. A. , IC&G, BGRS 2000 RECOGNITION FUNCTION PROFILE FOR NUCLEOSOME SITES IN HUMAN Donor sites Recognition function value Exon Intron . . . 0. 8 Recognition function value 9 Acceptor sites Intron Exon 1. 2 1 0. 6 0. 8 0. 4 0. 6 0. 2 0. 4 0 -0. 2 -150 0. 2 -100 -50 0 50 100 Position relative by splicing center site, bp 150 -100 -50 0 50 100 Position relative by splicing center site, bp 0 150
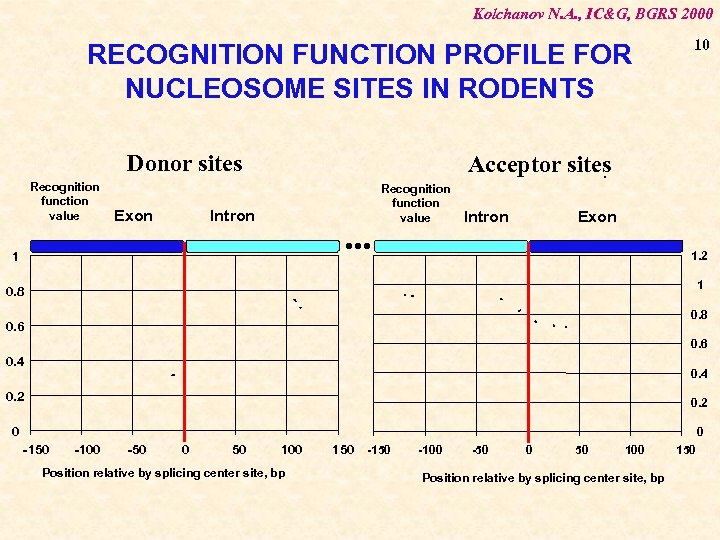
Kolchanov N. A. , IC&G, BGRS 2000 RECOGNITION FUNCTION PROFILE FOR NUCLEOSOME SITES IN RODENTS Donor sites Recognition function value Exon 10 Acceptor sites. Intron . . . 1 Recognition function value Intron Exon 1. 2 1 0. 8 0. 6 0. 4 0. 2 0 0 -150 -100 -50 0 50 100 Position relative by splicing center site, bp 150
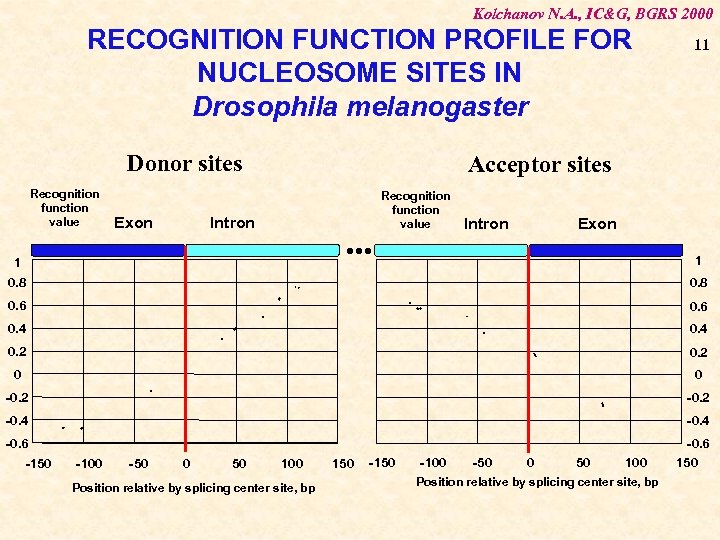
Kolchanov N. A. , IC&G, BGRS 2000 RECOGNITION FUNCTION PROFILE FOR NUCLEOSOME SITES IN Drosophila melanogaster Donor sites Recognition function value Exon 11 Acceptor sites Intron . . . 1 Recognition function value Intron Exon 1 0. 8 0. 6 0. 4 0. 2 0 0 -0. 2 -0. 4 -0. 6 150 -150 -100 -50 0 50 100 Position relative by splicing center site, bp
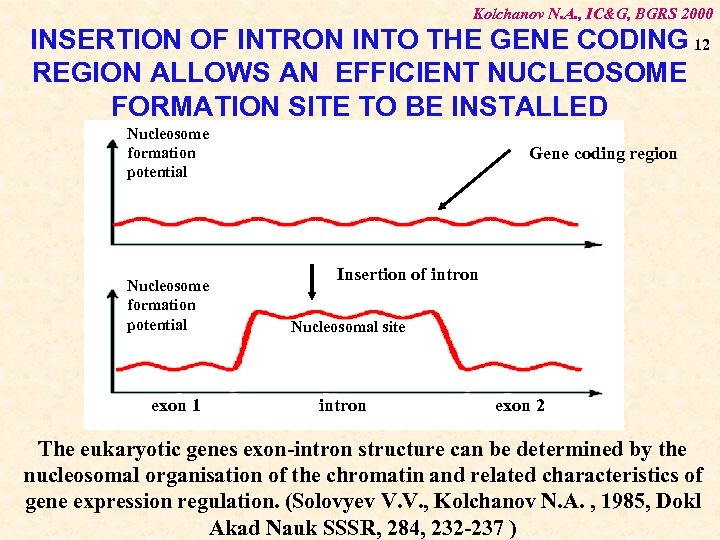
Kolchanov N. A. , IC&G, BGRS 2000 INSERTION OF INTRON INTO THE GENE CODING 12 REGION ALLOWS AN EFFICIENT NUCLEOSOME FORMATION SITE TO BE INSTALLED Nucleosome formation potential exon 1 Gene coding region Insertion of intron Nucleosomal site intron exon 2 The eukaryotic genes exon-intron structure can be determined by the nucleosomal organisation of the chromatin and related characteristics of gene expression regulation. (Solovyev V. V. , Kolchanov N. A. , 1985, Dokl Akad Nauk SSSR, 284, 232 -237 )
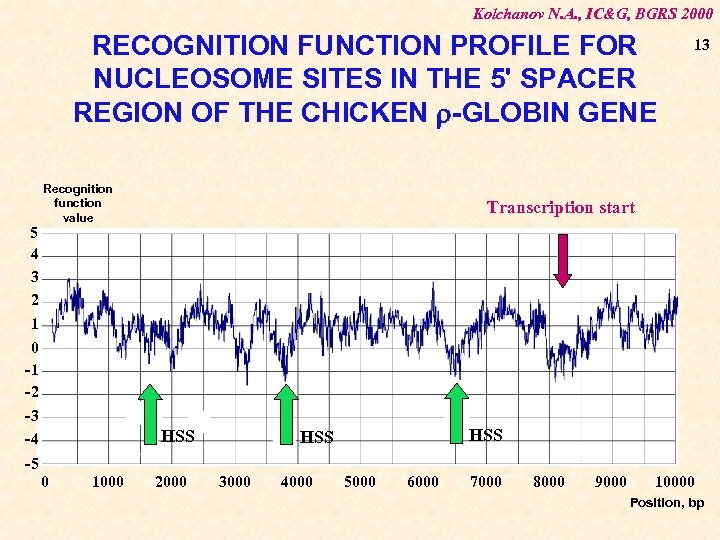
Kolchanov N. A. , IC&G, BGRS 2000 RECOGNITION FUNCTION PROFILE FOR NUCLEOSOME SITES IN THE 5' SPACER REGION OF THE CHICKEN -GLOBIN GENE 5 4 3 2 1 0 -1 -2 Recognition function value -3 -4 13 Transcription start HSS HSS -5 0 1000 2000 3000 4000 5000 6000 7000 8000 9000 10000 Position, bp
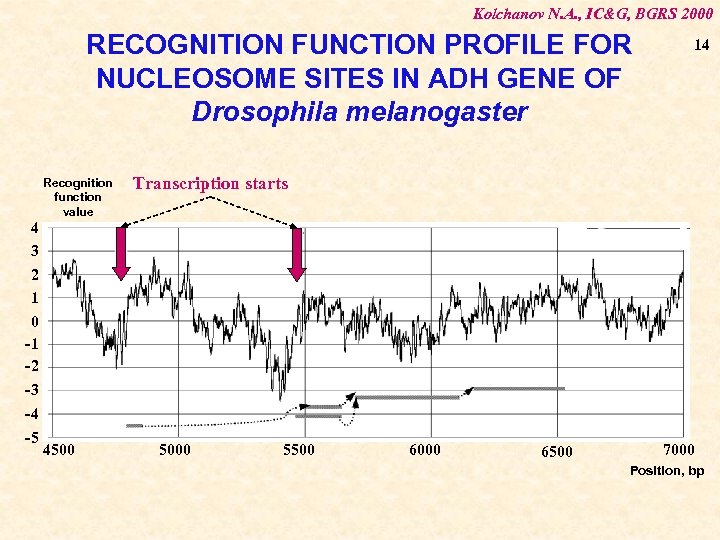
Kolchanov N. A. , IC&G, BGRS 2000 RECOGNITION FUNCTION PROFILE FOR NUCLEOSOME SITES IN ADH GENE OF Drosophila melanogaster Recognition function value 14 Transcription starts 4 3 2 1 0 -1 -2 -3 -4 -5 4500 5000 5500 6000 6500 7000 Position, bp
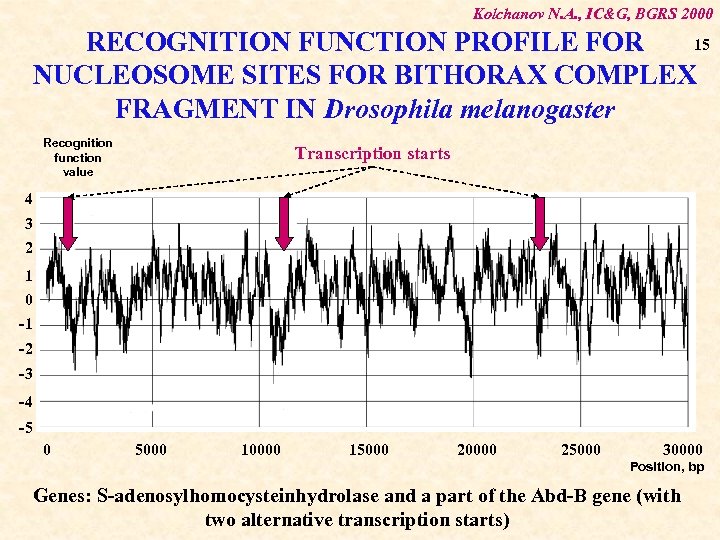
Kolchanov N. A. , IC&G, BGRS 2000 15 RECOGNITION FUNCTION PROFILE FOR NUCLEOSOME SITES FOR BITHORAX COMPLEX FRAGMENT IN Drosophila melanogaster Recognition function value Transcription starts 4 3 2 1 0 -1 -2 -3 -4 -5 0 5000 10000 15000 20000 25000 30000 Position, bp Genes: S-adenosylhomocysteinhydrolase and a part of the Abd-B gene (with two alternative transcription starts)
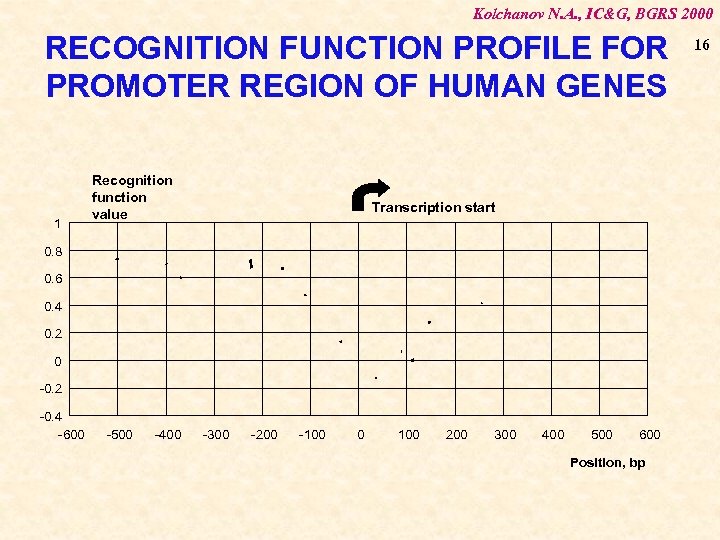
Kolchanov N. A. , IC&G, BGRS 2000 RECOGNITION FUNCTION PROFILE FOR PROMOTER REGION OF HUMAN GENES 1 Recognition function value Transcription start 0. 8 0. 6 0. 4 0. 2 0 -0. 2 -0. 4 -600 -500 -400 -300 -200 -100 0 100 200 300 400 500 600 Position, bp 16
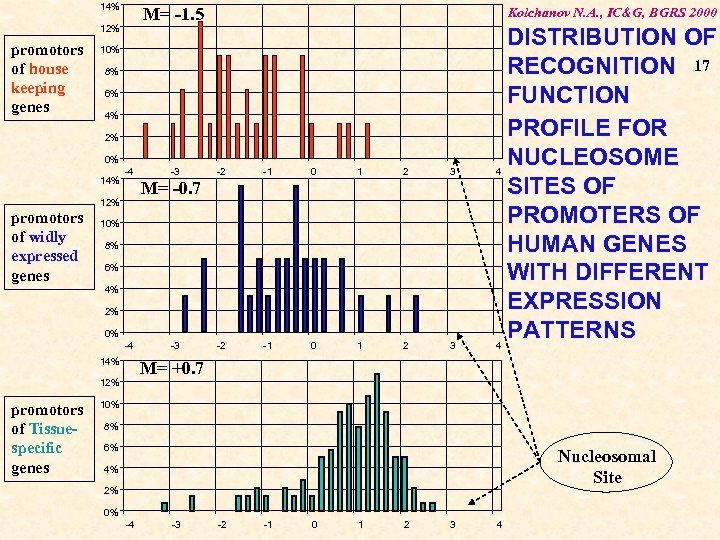
14% M= -1. 5 12% promotors of house keeping genes Kolchanov N. A. , IC&G, BGRS 2000 10% 8% 6% 4% 2% 0% 14% -4 -2 -1 0 1 2 3 M= -0. 7 12% promotors of widly expressed genes -3 10% 8% 6% 4% 2% 0% -4 14% M= +0. 7 12% promotors of Tissuespecific genes -3 DISTRIBUTION OF RECOGNITION 17 FUNCTION PROFILE FOR NUCLEOSOME 4 SITES OF PROMOTERS OF HUMAN GENES WITH DIFFERENT EXPRESSION PATTERNS 4 10% 8% 6% Nucleosomal Site 4% 2% 0% -4 -3 -2 -1 0 1 2 3 4
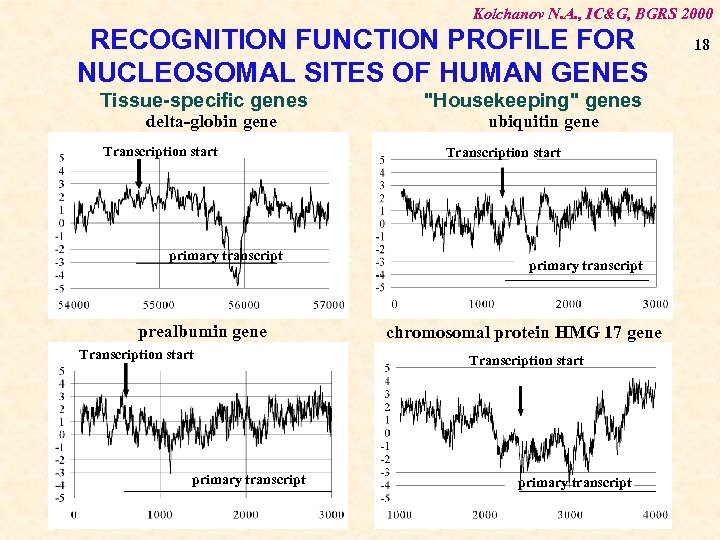
Kolchanov N. A. , IC&G, BGRS 2000 RECOGNITION FUNCTION PROFILE FOR NUCLEOSOMAL SITES OF HUMAN GENES Tissue-specific genes delta-globin gene Transcription start primary transcript prealbumin gene Transcription start primary transcript "Housekeeping" genes ubiquitin gene Transcription start primary transcript chromosomal protein HMG 17 gene Transcription start primary transcript 18
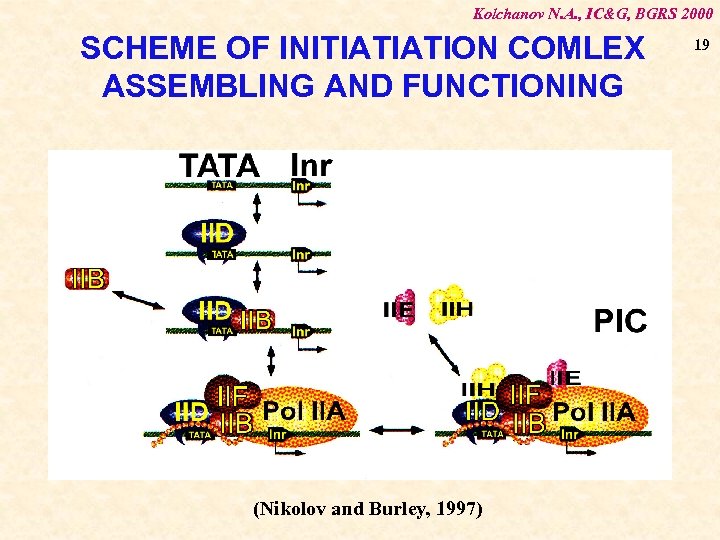
Kolchanov N. A. , IC&G, BGRS 2000 SCHEME OF INITIATIATION COMLEX ASSEMBLING AND FUNCTIONING (Nikolov and Burley, 1997) 19
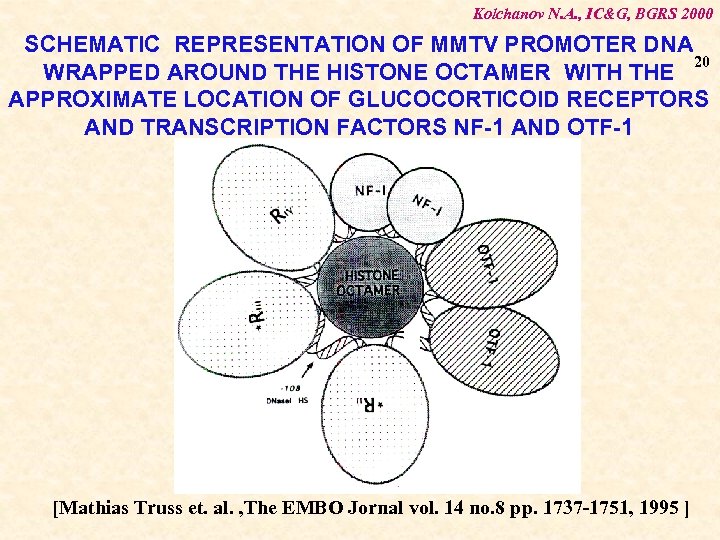
Kolchanov N. A. , IC&G, BGRS 2000 SCHEMATIC REPRESENTATION OF MMTV PROMOTER DNA 20 WRAPPED AROUND THE HISTONE OCTAMER WITH THE APPROXIMATE LOCATION OF GLUCOCORTICOID RECEPTORS AND TRANSCRIPTION FACTORS NF-1 AND OTF-1 [Mathias Truss et. al. , The EMBO Jornal vol. 14 no. 8 pp. 1737 -1751, 1995 ]
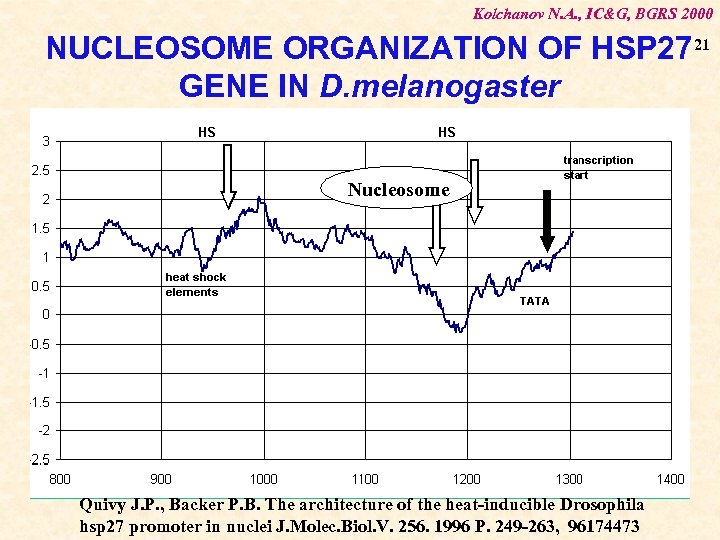
Kolchanov N. A. , IC&G, BGRS 2000 NUCLEOSOME ORGANIZATION OF HSP 27 21 GENE IN D. melanogaster Nucleosome Quivy J. P. , Backer P. B. The architecture of the heat-inducible Drosophila hsp 27 promoter in nuclei J. Molec. Biol. V. 256. 1996 P. 249 -263, 96174473
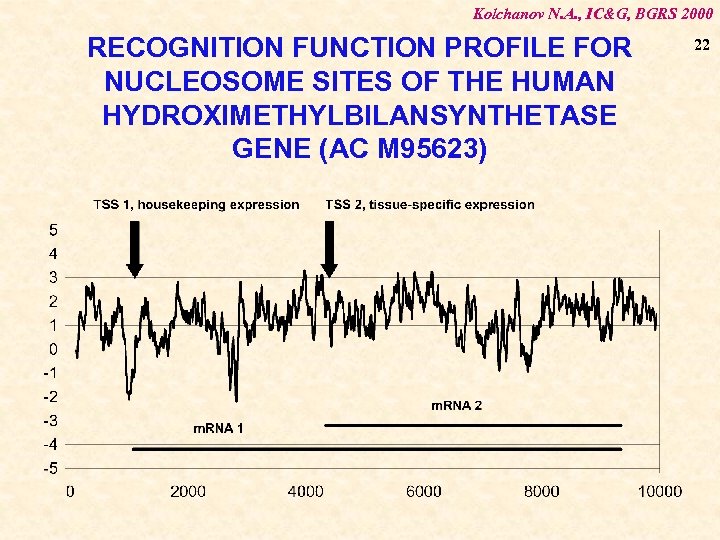
Kolchanov N. A. , IC&G, BGRS 2000 RECOGNITION FUNCTION PROFILE FOR NUCLEOSOME SITES OF THE HUMAN HYDROXIMETHYLBILANSYNTHETASE GENE (AC M 95623) 22
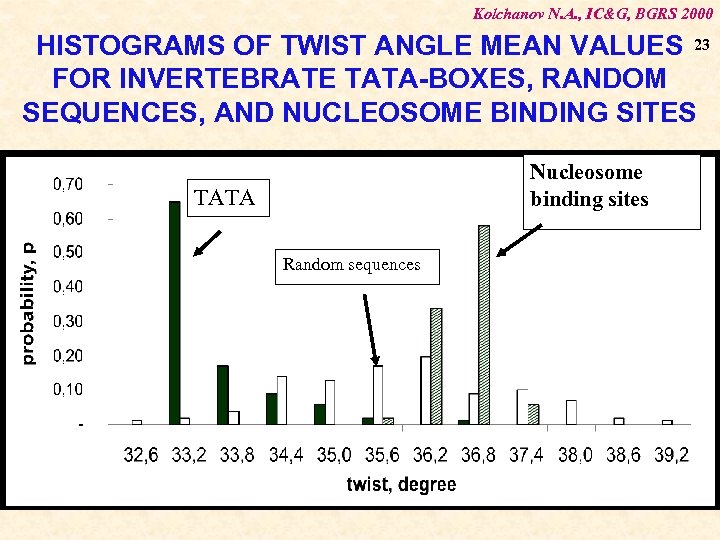
Kolchanov N. A. , IC&G, BGRS 2000 HISTOGRAMS OF TWIST ANGLE MEAN VALUES 23 FOR INVERTEBRATE TATA-BOXES, RANDOM SEQUENCES, AND NUCLEOSOME BINDING SITES Nucleosome binding sites TATA Random sequences
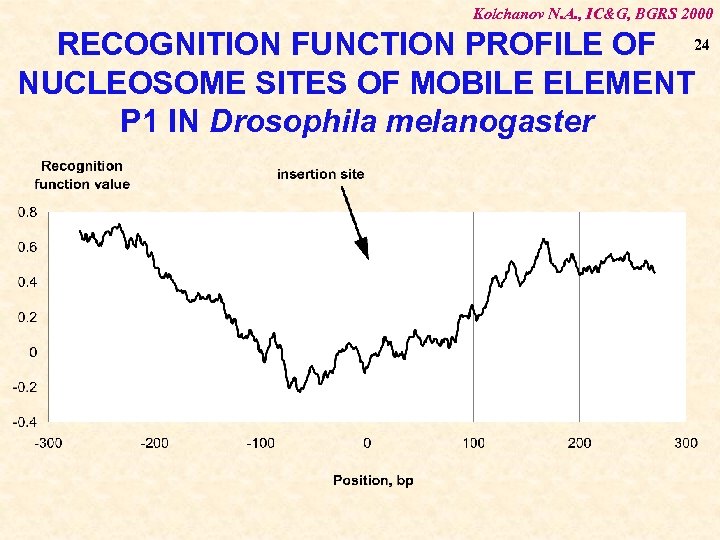
Kolchanov N. A. , IC&G, BGRS 2000 RECOGNITION FUNCTION PROFILE OF 24 NUCLEOSOME SITES OF MOBILE ELEMENT P 1 IN Drosophila melanogaster
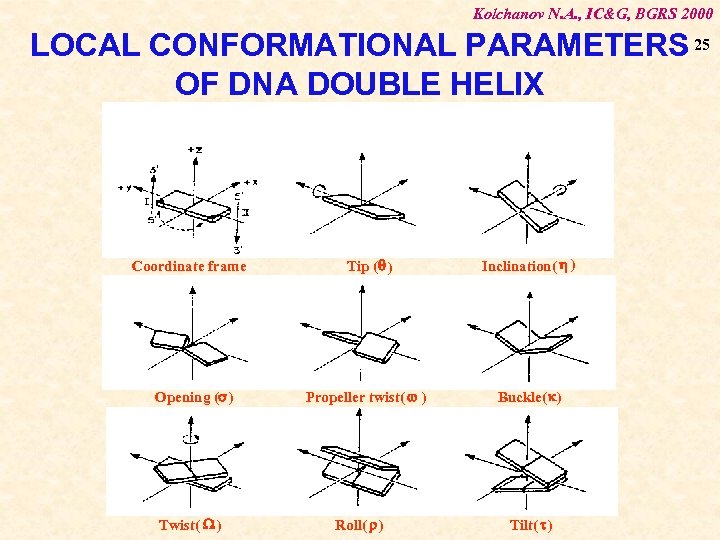
Kolchanov N. A. , IC&G, BGRS 2000 LOCAL CONFORMATIONAL PARAMETERS 25 OF DNA DOUBLE HELIX Coordinate frame Opening (s) Twist( W ) Tip (q ) Inclination( h ) Propeller twist( w ) Buckle(k) Roll( ) Tilt( t )
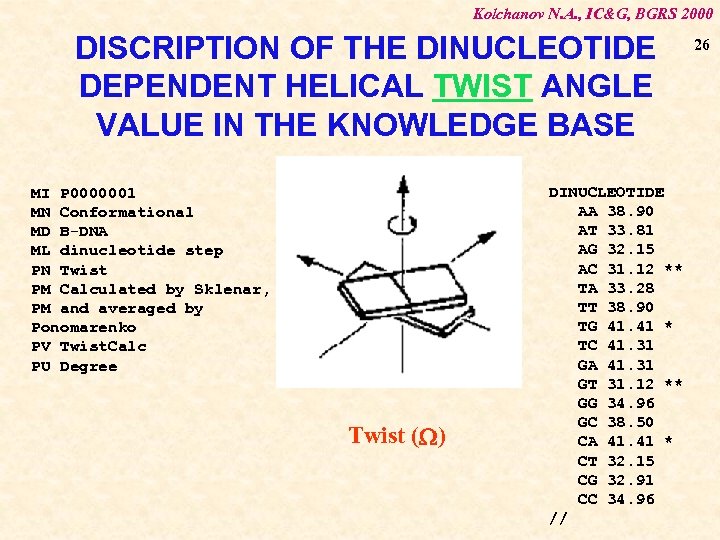
Kolchanov N. A. , IC&G, BGRS 2000 DISCRIPTION OF THE DINUCLEOTIDE DEPENDENT HELICAL TWIST ANGLE VALUE IN THE KNOWLEDGE BASE MI P 0000001 MN Conformational MD B-DNA ML dinucleotide step PN Twist PM Calculated by Sklenar, PM and averaged by Ponomarenko PV Twist. Calc PU Degree Twist (W) DINUCLEOTIDE AA 38. 90 AT 33. 81 AG 32. 15 AC 31. 12 ** TA 33. 28 TT 38. 90 TG 41. 41 * TC 41. 31 GA 41. 31 GT 31. 12 ** GG 34. 96 GC 38. 50 CA 41. 41 * CT 32. 15 CG 32. 91 CC 34. 96 // 26
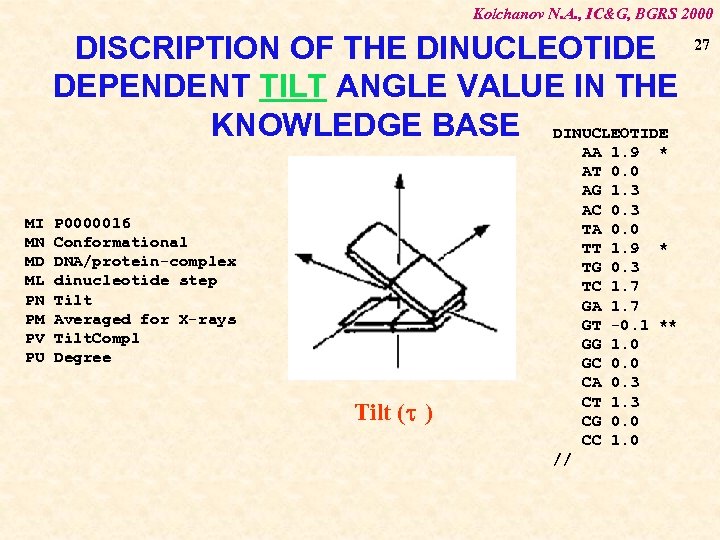
Kolchanov N. A. , IC&G, BGRS 2000 DISCRIPTION OF THE DINUCLEOTIDE DEPENDENT TILT ANGLE VALUE IN THE KNOWLEDGE BASE DINUCLEOTIDE MI MN MD ML PN PM PV PU AA AT AG AC TA TT TG TC GA GT GG GC CA CT CG CC P 0000016 Conformational DNA/protein-complex dinucleotide step Tilt Averaged for X-rays Tilt. Compl Degree Tilt (t ) // 1. 9 * 0. 0 1. 3 0. 0 1. 9 * 0. 3 1. 7 -0. 1 ** 1. 0 0. 3 1. 3 0. 0 1. 0 27
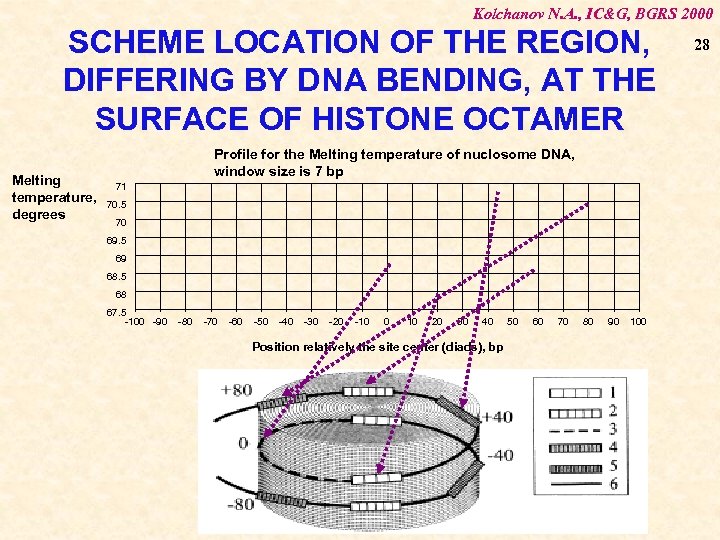
Kolchanov N. A. , IC&G, BGRS 2000 SCHEME LOCATION OF THE REGION, DIFFERING BY DNA BENDING, AT THE SURFACE OF HISTONE OCTAMER Melting temperature, degrees Profile for the Melting temperature of nuclosome DNA, window size is 7 bp 71 70. 5 70 69. 5 69 68. 5 68 67. 5 -100 -90 -80 -70 -60 -50 -40 -30 -20 -10 0 10 20 30 40 Position relatively the site center (diads), bp 50 60 70 80 90 100 28
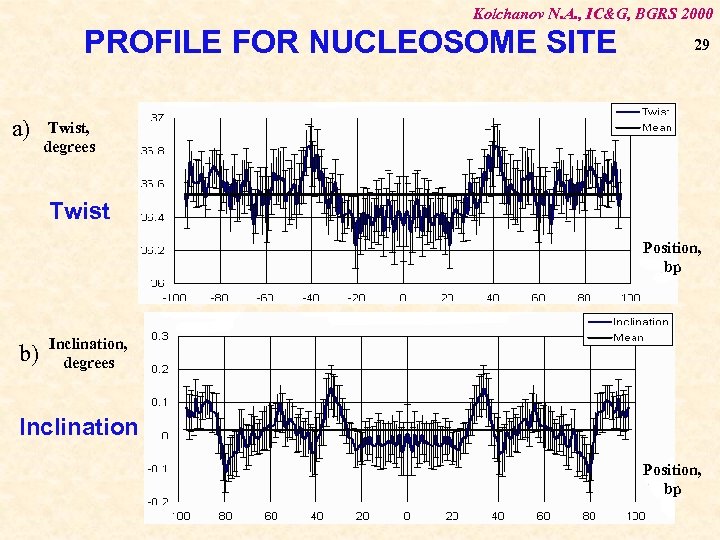
Kolchanov N. A. , IC&G, BGRS 2000 PROFILE FOR NUCLEOSOME SITE a) 29 Twist, degrees Twist Position, bp b) Inclination, degrees Inclination Position, bp
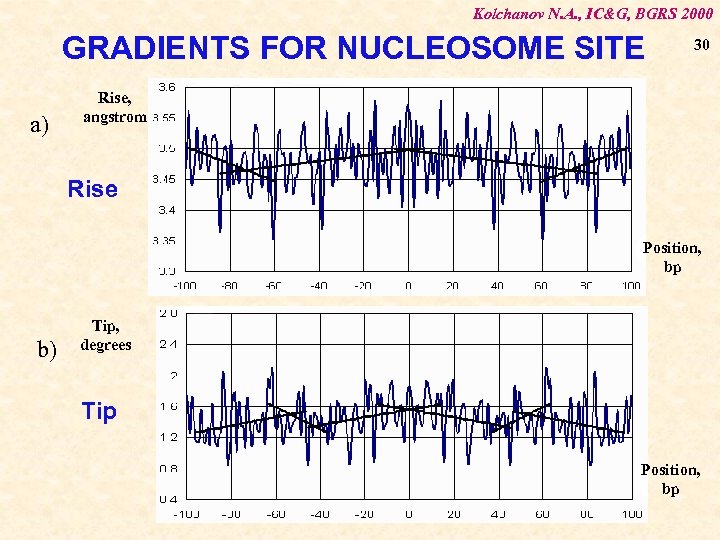
Kolchanov N. A. , IC&G, BGRS 2000 GRADIENTS FOR NUCLEOSOME SITE a) 30 Rise, angstrom Rise Position, bp b) Tip, degrees Tip Position, bp
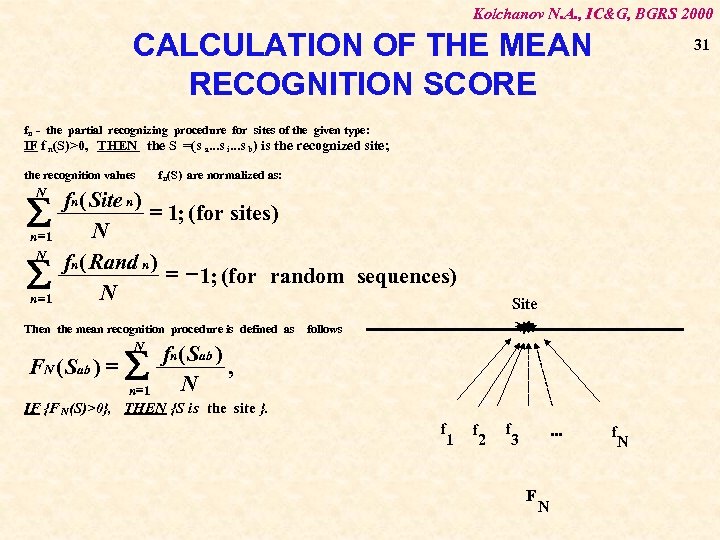
Kolchanov N. A. , IC&G, BGRS 2000 CALCULATION OF THE MEAN RECOGNITION SCORE 31 f n - the partial recognizing procedure for sites of the given type: IF f n(S)>0, THEN the S =(s a. . . s i. . . s b) is the recognized site; the recognition values N å n =1 f n(S) are normalized as: fn ( Site n ) = 1; (for sites) N fn ( Rand n ) = - 1; (for random sequences) N Then the mean recognition procedure is defined as FN ( Sab ) = N å n =1 Site follows fn ( Sab ) , N IF {F N(S)>0}, THEN {S is the site }. f 1 f 2 f . . . 3 F N f N
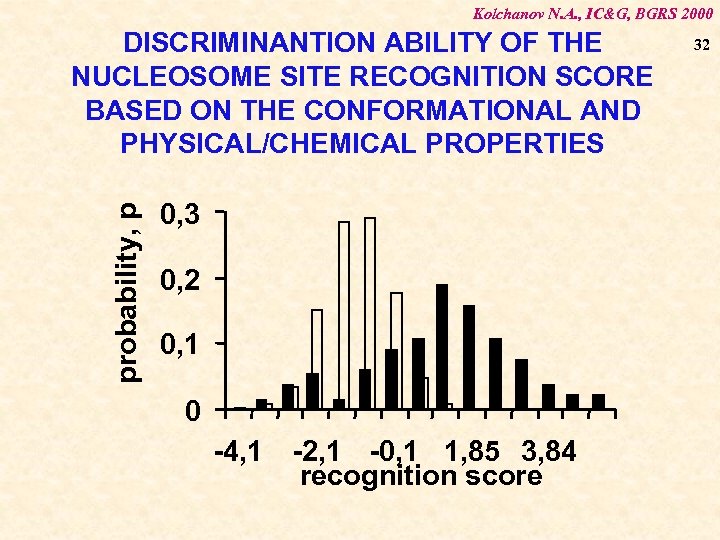
Kolchanov N. A. , IC&G, BGRS 2000 probability, p DISCRIMINANTION ABILITY OF THE NUCLEOSOME SITE RECOGNITION SCORE BASED ON THE CONFORMATIONAL AND PHYSICAL/CHEMICAL PROPERTIES 0, 3 0, 2 0, 1 0 -4, 1 -2, 1 -0, 1 1, 85 3, 84 recognition score 32
Kolchanov N. A. , IC&G, BGRS 2000 NUCLEOSOME POSITIONING CODE: COMMON FEATURES 33 The elements (or the signals) of nucleosomal code are dinucleotides (oligonucleotides) determining local conformational (physicochemical) nucleosome site features, necessary for interaction with core histones Nucleosomal code is point-wise: only particular groups of positions are used for coding genetic messages on local conformational DNA features, significant for interaction of nucleosome sites with core histone. Besides, these groups located in a definite (unfixed) distance.

Kolchanov N. A. , IC&G, BGRS 2000 NUCLEOSOME POSITIONING CODE: COMMON FEATURES Nucleosomal code is extremely degenerated: very differing DNA sequences that interact with histone octamer are recognized Nucleosomal code is point-wise: it provides the possibility of overlapping with the other types of codes 34
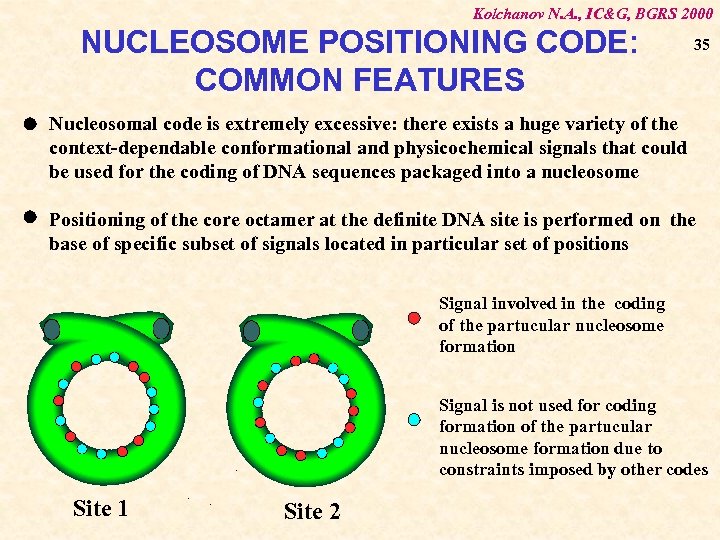
Kolchanov N. A. , IC&G, BGRS 2000 NUCLEOSOME POSITIONING CODE: COMMON FEATURES 35 Nucleosomal code is extremely excessive: there exists a huge variety of the context-dependable conformational and physicochemical signals that could be used for the coding of DNA sequences packaged into a nucleosome Positioning of the core octamer at the definite DNA site is performed on the base of specific subset of signals located in particular set of positions Signal involved in the coding of the partucular nucleosome formation Signal is not used for coding formation of the partucular nucleosome formation due to constraints imposed by other codes Site 1 Site 2

Kolchanov N. A. , IC&G, BGRS 2000 ACNOWLEDGEMENT The authors are grateful to Dr. M. P. Ponomarenko, Dr. O. A. Podkolodnaya, J. V. Ponomarenko for participance and help in work 36
084c758a9c7527093045c4c31cbc826c.ppt