3792a849f6e218efa5a4047e09b7649a.ppt
- Количество слайдов: 38
 Jia-Ming Chang 0508 Graph Algorithms and Their Applications to Bioinformatics GRAPH ALGORITHM IN NMR BACKBONE ASSIGNMENT 1/38
Jia-Ming Chang 0508 Graph Algorithms and Their Applications to Bioinformatics GRAPH ALGORITHM IN NMR BACKBONE ASSIGNMENT 1/38
 Determine Protein Structure X-ray 波長約 1 Å 長度接近原子間的距離 研究結晶的狀態的分子行為 定出其晶體結構,也包含蛋白質體結構 X-ray與結構生物學 利用 X-ray繞射法分析高度純化結晶的蛋白質的每 個基團和原子的空間定位。 Nuclear magnetic resonance (NMR) NMR是涉及原子核吸收的過程。因為對某些原子核而 言,具有自旋和磁矩的性質。因此,若暴露於強磁場 中原子核會吸收電磁輻射,這是由磁場誘導而發生能 階分裂的結果。科學家並發現,分子環境會影響在磁 場中原子核的無線電波的吸收,利用這種特性來分析 分子的結構 AVANCE 800 AV IBMS, Sinica 2/38
Determine Protein Structure X-ray 波長約 1 Å 長度接近原子間的距離 研究結晶的狀態的分子行為 定出其晶體結構,也包含蛋白質體結構 X-ray與結構生物學 利用 X-ray繞射法分析高度純化結晶的蛋白質的每 個基團和原子的空間定位。 Nuclear magnetic resonance (NMR) NMR是涉及原子核吸收的過程。因為對某些原子核而 言,具有自旋和磁矩的性質。因此,若暴露於強磁場 中原子核會吸收電磁輻射,這是由磁場誘導而發生能 階分裂的結果。科學家並發現,分子環境會影響在磁 場中原子核的無線電波的吸收,利用這種特性來分析 分子的結構 AVANCE 800 AV IBMS, Sinica 2/38
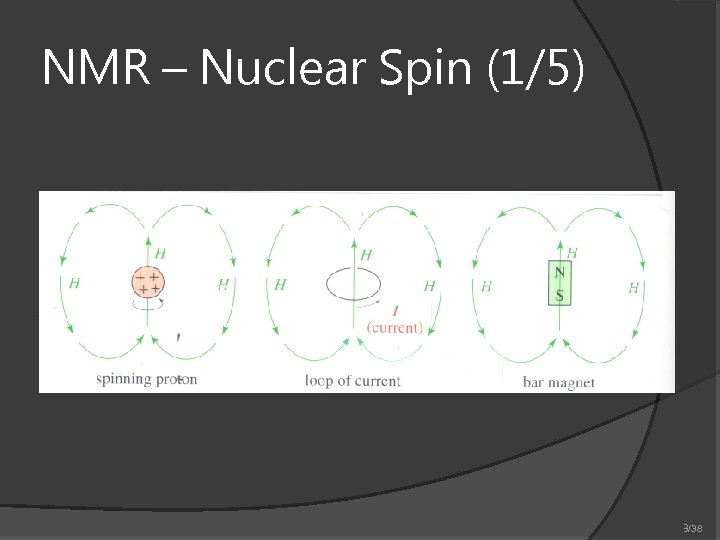 NMR – Nuclear Spin (1/5) 3/38
NMR – Nuclear Spin (1/5) 3/38
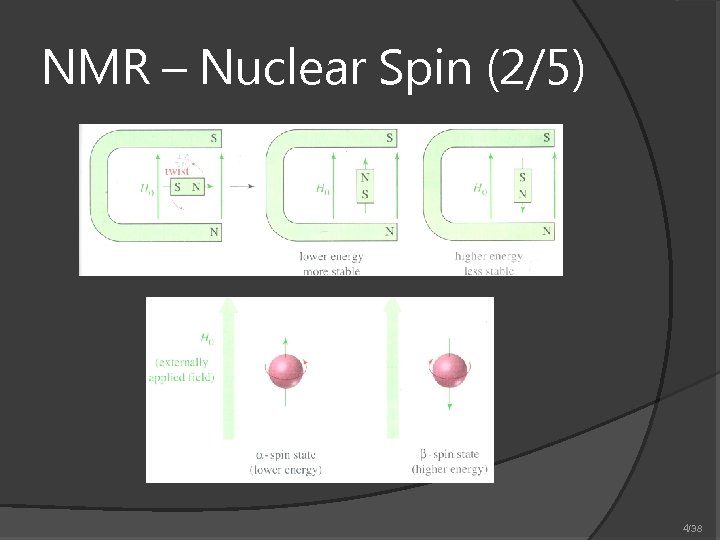 NMR – Nuclear Spin (2/5) 4/38
NMR – Nuclear Spin (2/5) 4/38
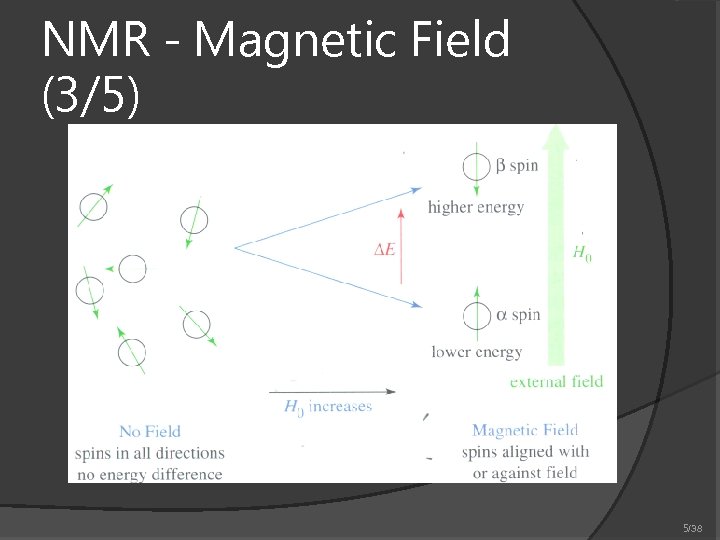 NMR - Magnetic Field (3/5) 5/38
NMR - Magnetic Field (3/5) 5/38
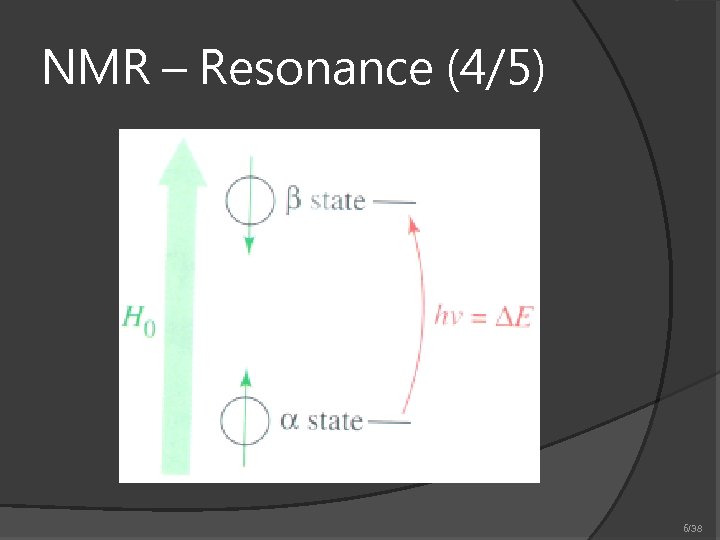 NMR – Resonance (4/5) 6/38
NMR – Resonance (4/5) 6/38
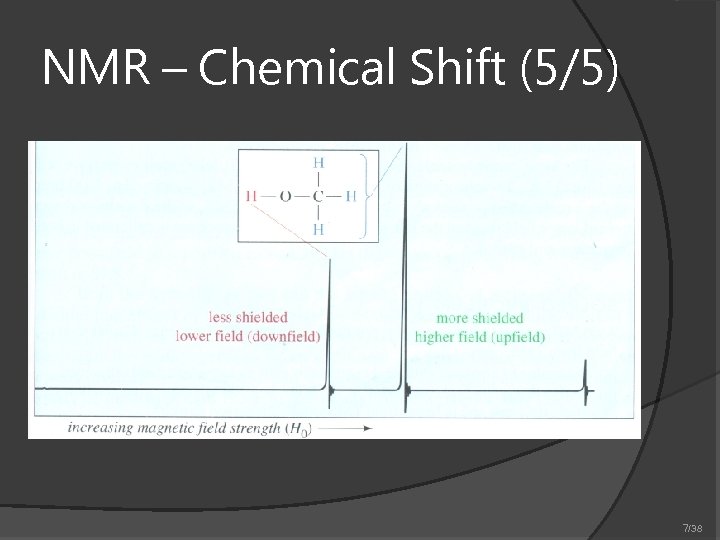 NMR – Chemical Shift (5/5) 7/38
NMR – Chemical Shift (5/5) 7/38
 Chemical Shift Assignment (1/2) Find out Chemical Shift for Each Atom • Backbone: Ca, Cb, C’, N, NH HSQC, CBCANH, CBCACONH Cd H 3 One amino acid Cg H 2 Cb H 2 N Ca H H CO 8/38
Chemical Shift Assignment (1/2) Find out Chemical Shift for Each Atom • Backbone: Ca, Cb, C’, N, NH HSQC, CBCANH, CBCACONH Cd H 3 One amino acid Cg H 2 Cb H 2 N Ca H H CO 8/38
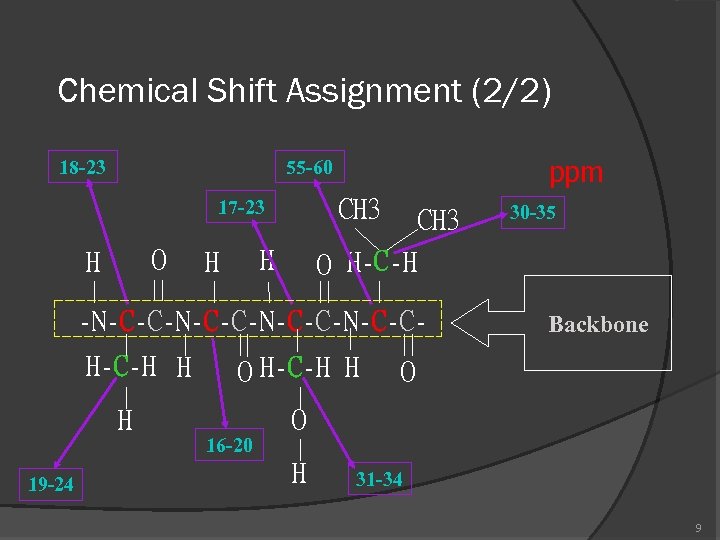 Chemical Shift Assignment (2/2) 18 -23 ppm 55 -60 CH 3 17 -23 O H H H CH 3 O H-C-H -N-C-C-N-C-CH-C-H H O H-C-H H H 30 -35 Backbone O O 16 -20 19 -24 H 31 -34 9
Chemical Shift Assignment (2/2) 18 -23 ppm 55 -60 CH 3 17 -23 O H H H CH 3 O H-C-H -N-C-C-N-C-CH-C-H H O H-C-H H H 30 -35 Backbone O O 16 -20 19 -24 H 31 -34 9
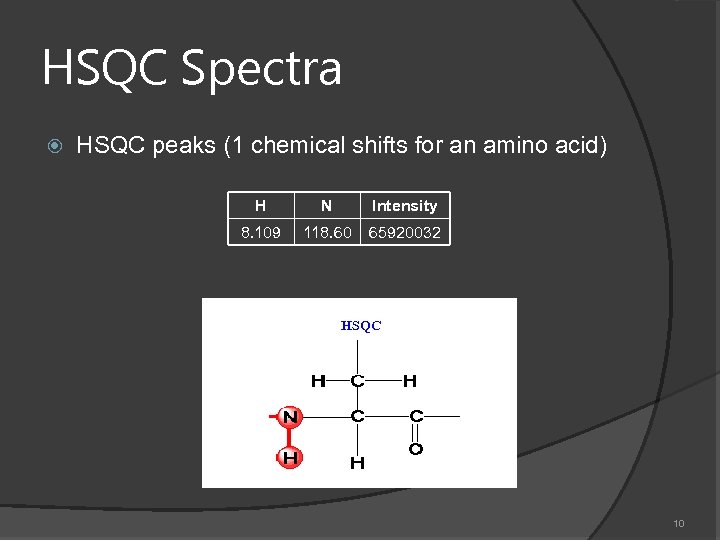 HSQC Spectra HSQC peaks (1 chemical shifts for an amino acid) H N Intensity 8. 109 118. 60 65920032 HSQC 10
HSQC Spectra HSQC peaks (1 chemical shifts for an amino acid) H N Intensity 8. 109 118. 60 65920032 HSQC 10
 CBCA(CO)NH Spectra CBCA(CO)NH peaks (2 chemical shifts for one amino acid) H N C Intensity 8. 116 118. 25 16. 37 79238811 8. 109 118. 60 36. 52 65920032 11
CBCA(CO)NH Spectra CBCA(CO)NH peaks (2 chemical shifts for one amino acid) H N C Intensity 8. 116 118. 25 16. 37 79238811 8. 109 118. 60 36. 52 65920032 11
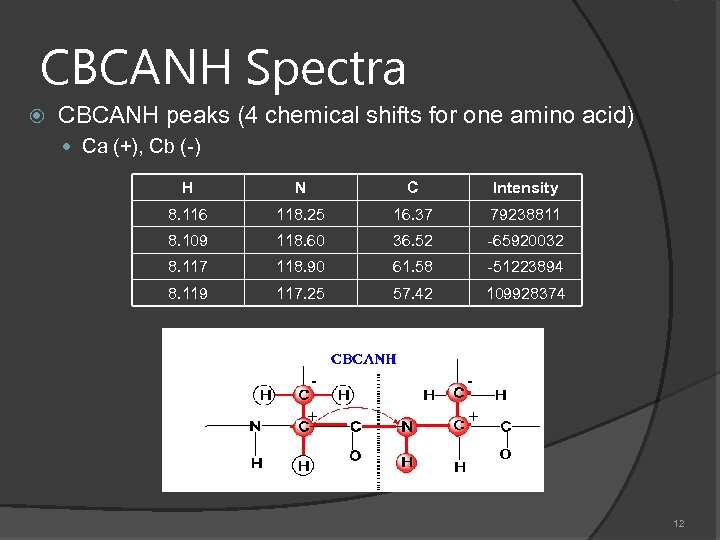 CBCANH Spectra CBCANH peaks (4 chemical shifts for one amino acid) Ca (+), Cb (-) H N C Intensity 8. 116 118. 25 16. 37 79238811 8. 109 118. 60 36. 52 -65920032 8. 117 118. 90 61. 58 -51223894 8. 119 117. 25 57. 42 109928374 - - + + 12
CBCANH Spectra CBCANH peaks (4 chemical shifts for one amino acid) Ca (+), Cb (-) H N C Intensity 8. 116 118. 25 16. 37 79238811 8. 109 118. 60 36. 52 -65920032 8. 117 118. 90 61. 58 -51223894 8. 119 117. 25 57. 42 109928374 - - + + 12
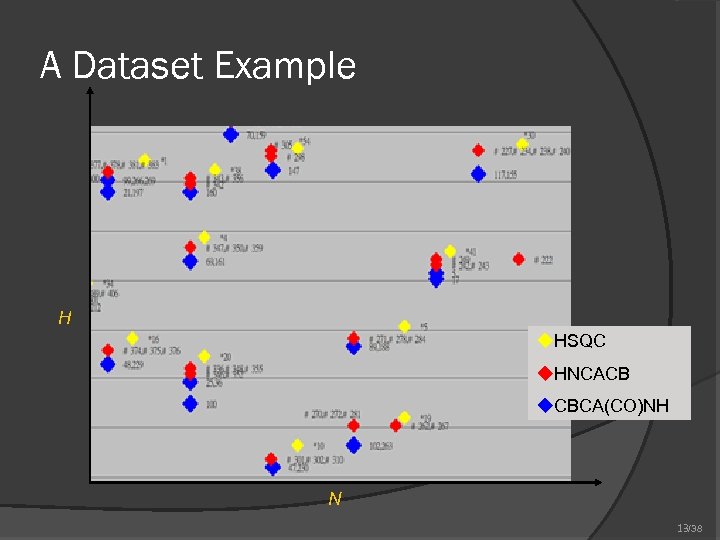 A Dataset Example H u. HSQC u. HNCACB u. CBCA(CO)NH N 13/38
A Dataset Example H u. HSQC u. HNCACB u. CBCA(CO)NH N 13/38
 A Perfect Spin System Group CBCA(CO)NH N H C Intensity 113. 293 7. 897 56. 294 1. 64325 e+008 i -1 113. 293 7. 897 27. 853 1. 08099 e+008 i -1 Cai-1 N H C Intensity 113. 293 7. 92 62. 544 8. 52851 e+007 7. 92 56. 294 4. 71331 e+007 7. 92 68. 483 -8. 54121 e+007 7. 92 28. 165 -3. 49346 e+007 62. 544 68. 483 Cb 113. 293 28. 165 Ca 113. 293 Cbi Ca 113. 293 Cai 56. 294 CBCANH Cbi-1 Cb 14
A Perfect Spin System Group CBCA(CO)NH N H C Intensity 113. 293 7. 897 56. 294 1. 64325 e+008 i -1 113. 293 7. 897 27. 853 1. 08099 e+008 i -1 Cai-1 N H C Intensity 113. 293 7. 92 62. 544 8. 52851 e+007 7. 92 56. 294 4. 71331 e+007 7. 92 68. 483 -8. 54121 e+007 7. 92 28. 165 -3. 49346 e+007 62. 544 68. 483 Cb 113. 293 28. 165 Ca 113. 293 Cbi Ca 113. 293 Cai 56. 294 CBCANH Cbi-1 Cb 14
 Coding Translate the target protein sequence and spin systems into coding sequences based on the following table. Atreya, H. S. , K. V. R. Chary, and G. Govil, Automated NMR assignments of proteins for high throughput structure determination: TATAPRO II. Current Science, 2002. 83(11): p. 1372 -1376. 15/38
Coding Translate the target protein sequence and spin systems into coding sequences based on the following table. Atreya, H. S. , K. V. R. Chary, and G. Govil, Automated NMR assignments of proteins for high throughput structure determination: TATAPRO II. Current Science, 2002. 83(11): p. 1372 -1376. 15/38
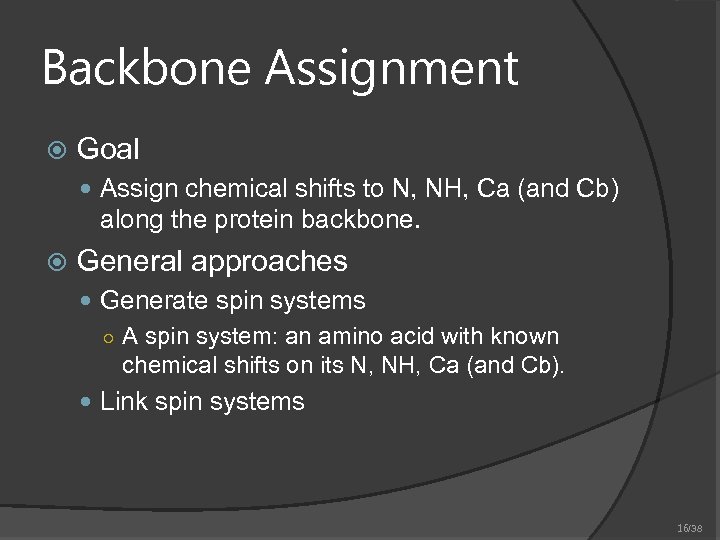 Backbone Assignment Goal Assign chemical shifts to N, NH, Ca (and Cb) along the protein backbone. General approaches Generate spin systems ○ A spin system: an amino acid with known chemical shifts on its N, NH, Ca (and Cb). Link spin systems 16/38
Backbone Assignment Goal Assign chemical shifts to N, NH, Ca (and Cb) along the protein backbone. General approaches Generate spin systems ○ A spin system: an amino acid with known chemical shifts on its N, NH, Ca (and Cb). Link spin systems 16/38
 Ambiguities All 4 point experiments are mixed together All 2 point experiments are mixed together Each spin system can be mapped to several amino acids in the protein sequence False positives, false negatives 17/38
Ambiguities All 4 point experiments are mixed together All 2 point experiments are mixed together Each spin system can be mapped to several amino acids in the protein sequence False positives, false negatives 17/38
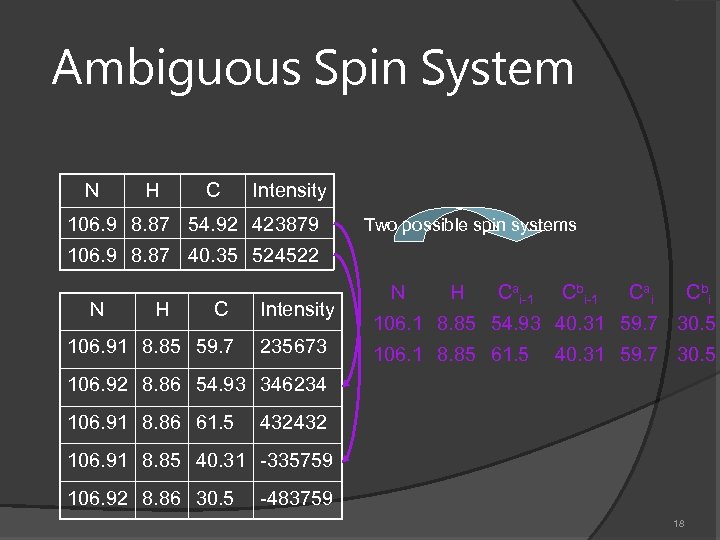 Ambiguous Spin System N H C Intensity 106. 9 8. 87 54. 92 423879 Two possible spin systems 106. 9 8. 87 40. 35 524522 N H C 106. 91 8. 85 59. 7 Intensity 235673 N H Cai-1 Cbi-1 Ca i Cb i 106. 1 8. 85 54. 93 40. 31 59. 7 30. 5 106. 1 8. 85 61. 5 40. 31 59. 7 30. 5 106. 92 8. 86 54. 93 346234 106. 91 8. 86 61. 5 432432 106. 91 8. 85 40. 31 -335759 106. 92 8. 86 30. 5 -483759 18
Ambiguous Spin System N H C Intensity 106. 9 8. 87 54. 92 423879 Two possible spin systems 106. 9 8. 87 40. 35 524522 N H C 106. 91 8. 85 59. 7 Intensity 235673 N H Cai-1 Cbi-1 Ca i Cb i 106. 1 8. 85 54. 93 40. 31 59. 7 30. 5 106. 1 8. 85 61. 5 40. 31 59. 7 30. 5 106. 92 8. 86 54. 93 346234 106. 91 8. 86 61. 5 432432 106. 91 8. 85 40. 31 -335759 106. 92 8. 86 30. 5 -483759 18
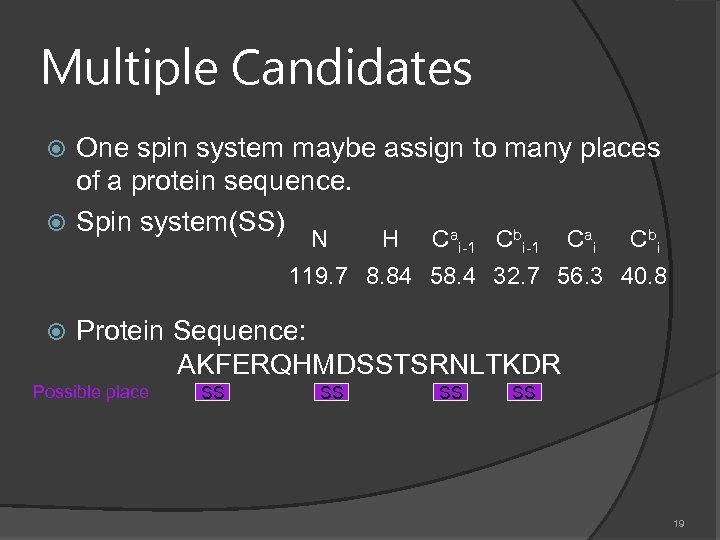 Multiple Candidates One spin system maybe assign to many places of a protein sequence. Spin system(SS) a b N H C i-1 C i 119. 7 8. 84 58. 4 32. 7 56. 3 40. 8 Protein Sequence: AKFERQHMDSSTSRNLTKDR Possible place SS SS 19
Multiple Candidates One spin system maybe assign to many places of a protein sequence. Spin system(SS) a b N H C i-1 C i 119. 7 8. 84 58. 4 32. 7 56. 3 40. 8 Protein Sequence: AKFERQHMDSSTSRNLTKDR Possible place SS SS 19
 False Positives and False Negatives False positives Noise with high intensity Produce fake spin systems False negatives Peaks with low intensity Missing peaks In real wet-lab data, nearly 50% are noises (false positive). 20/38
False Positives and False Negatives False positives Noise with high intensity Produce fake spin systems False negatives Peaks with low intensity Missing peaks In real wet-lab data, nearly 50% are noises (false positive). 20/38
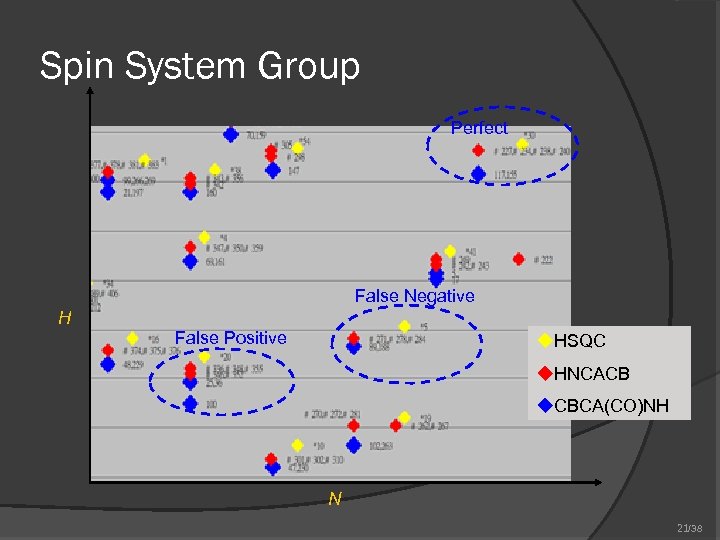 Spin System Group Perfect False Negative H False Positive u. HSQC u. HNCACB u. CBCA(CO)NH N 21/38
Spin System Group Perfect False Negative H False Positive u. HSQC u. HNCACB u. CBCA(CO)NH N 21/38
 Spin System Linking Goal Link spin system as long as possible. Constraints Each spin system is uniquely assigned to a position of the target protein sequence. Two spin systems are linked only if the chemical shift differences of their intra- and inter- residues are less than the predefined thresholds. 22/38
Spin System Linking Goal Link spin system as long as possible. Constraints Each spin system is uniquely assigned to a position of the target protein sequence. Two spin systems are linked only if the chemical shift differences of their intra- and inter- residues are less than the predefined thresholds. 22/38
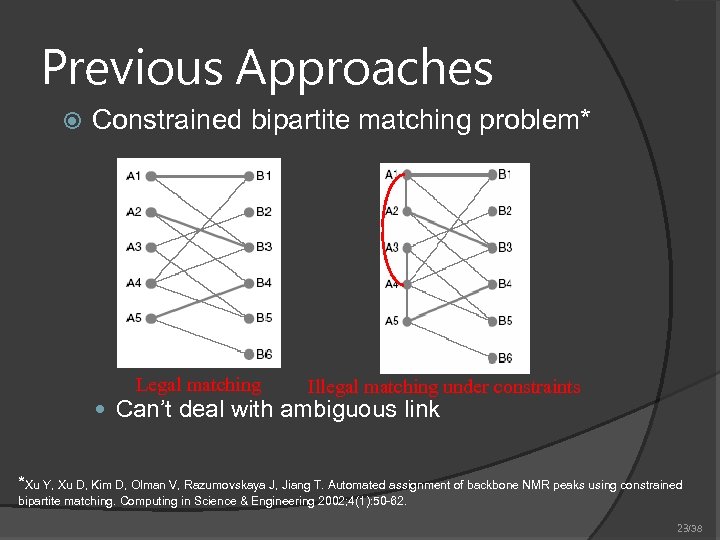 Previous Approaches Constrained bipartite matching problem* Legal matching Illegal matching under constraints Can’t deal with ambiguous link *Xu Y, Xu D, Kim D, Olman V, Razumovskaya J, Jiang T. Automated assignment of backbone NMR peaks using constrained bipartite matching. Computing in Science & Engineering 2002; 4(1): 50 -62. 23/38
Previous Approaches Constrained bipartite matching problem* Legal matching Illegal matching under constraints Can’t deal with ambiguous link *Xu Y, Xu D, Kim D, Olman V, Razumovskaya J, Jiang T. Automated assignment of backbone NMR peaks using constrained bipartite matching. Computing in Science & Engineering 2002; 4(1): 50 -62. 23/38
 Naatural Language Processing ─ Noises or Ambiguity ? Speech recognition:Homopone selection 台北市一位小孩走失了 台北市 小孩 台北 適宜 走失 事宜 一位 一味 移位 24/38
Naatural Language Processing ─ Noises or Ambiguity ? Speech recognition:Homopone selection 台北市一位小孩走失了 台北市 小孩 台北 適宜 走失 事宜 一位 一味 移位 24/38
 An Error-Tolerant Algorithm 25
An Error-Tolerant Algorithm 25
 Phrase, Sentence Combination 26
Phrase, Sentence Combination 26
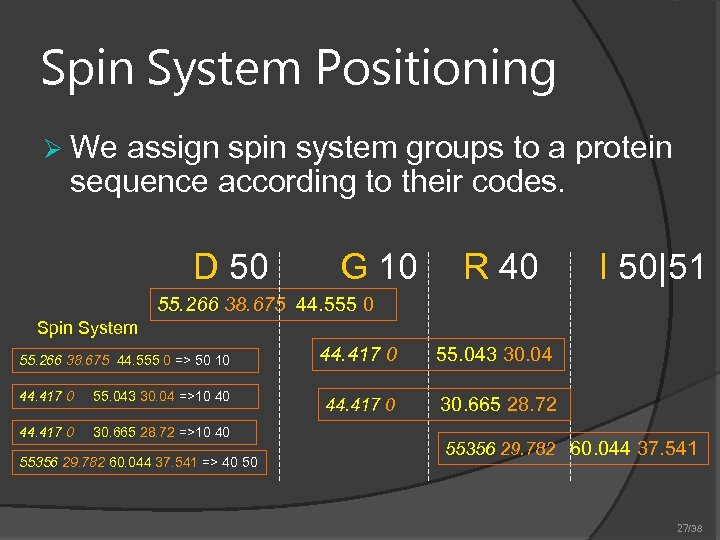 Spin System Positioning Ø We assign spin system groups to a protein sequence according to their codes. D 50 G 10 R 40 I 50|51 55. 266 38. 675 44. 555 0 Spin System 55. 266 38. 675 44. 555 0 => 50 10 44. 417 0 55. 043 30. 04 =>10 40 44. 417 0 30. 665 28. 72 =>10 40 55356 29. 782 60. 044 37. 541 => 40 50 44. 417 0 55. 043 30. 04 44. 417 0 30. 665 28. 72 55356 29. 782 60. 044 37. 541 27/38
Spin System Positioning Ø We assign spin system groups to a protein sequence according to their codes. D 50 G 10 R 40 I 50|51 55. 266 38. 675 44. 555 0 Spin System 55. 266 38. 675 44. 555 0 => 50 10 44. 417 0 55. 043 30. 04 =>10 40 44. 417 0 30. 665 28. 72 =>10 40 55356 29. 782 60. 044 37. 541 => 40 50 44. 417 0 55. 043 30. 04 44. 417 0 30. 665 28. 72 55356 29. 782 60. 044 37. 541 27/38
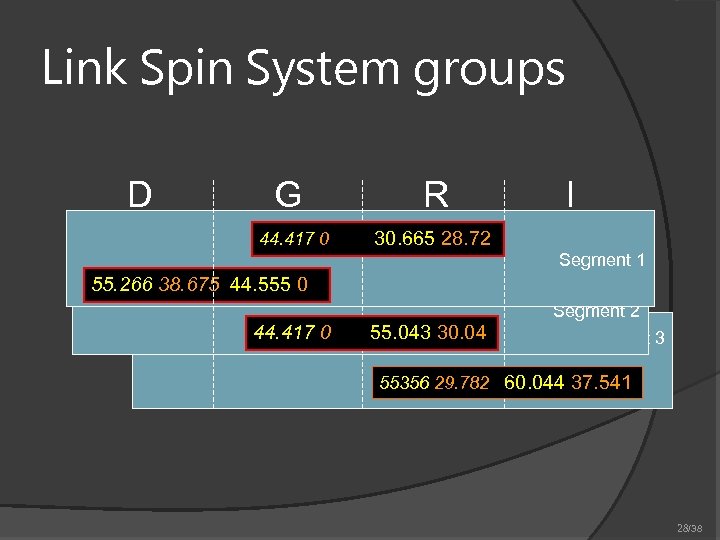 Link Spin System groups D G 44. 417 0 R I 30. 665 28. 72 Segment 1 55. 266 38. 675 44. 555 0 Segment 2 44. 417 0 55. 043 30. 04 Segment 3 55356 29. 782 60. 044 37. 541 28/38
Link Spin System groups D G 44. 417 0 R I 30. 665 28. 72 Segment 1 55. 266 38. 675 44. 555 0 Segment 2 44. 417 0 55. 043 30. 04 Segment 3 55356 29. 782 60. 044 37. 541 28/38
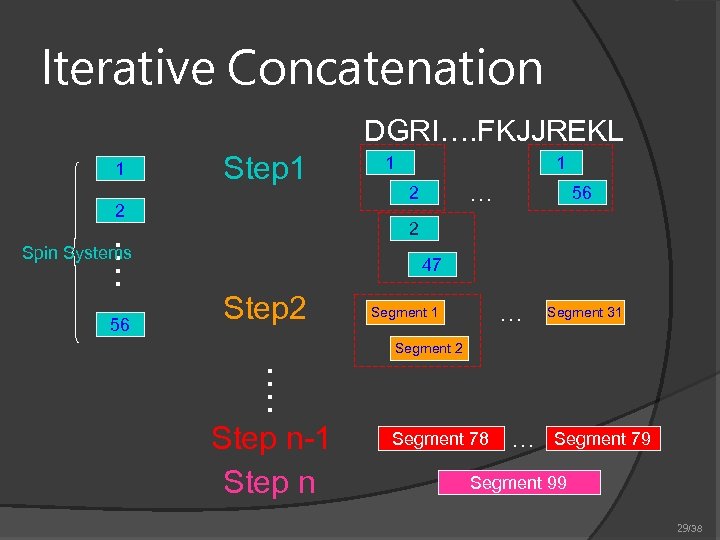 Iterative Concatenation DGRI…. FKJJREKL 1 Step 1 2 1 … 2 56 …. 2 Spin Systems 56 1 47 Step 2 … Segment 1 Segment 31 Segment 2 …. Step n-1 Step n Segment 78 … Segment 79 Segment 99 29/38
Iterative Concatenation DGRI…. FKJJREKL 1 Step 1 2 1 … 2 56 …. 2 Spin Systems 56 1 47 Step 2 … Segment 1 Segment 31 Segment 2 …. Step n-1 Step n Segment 78 … Segment 79 Segment 99 29/38
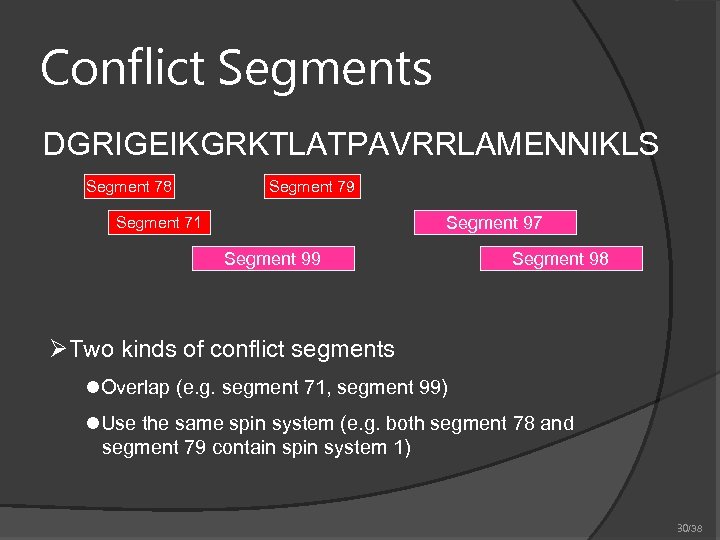 Conflict Segments DGRIGEIKGRKTLATPAVRRLAMENNIKLS Segment 78 Segment 79 Segment 97 Segment 71 Segment 99 Segment 98 ØTwo kinds of conflict segments l. Overlap (e. g. segment 71, segment 99) l. Use the same spin system (e. g. both segment 78 and segment 79 contain spin system 1) 30/38
Conflict Segments DGRIGEIKGRKTLATPAVRRLAMENNIKLS Segment 78 Segment 79 Segment 97 Segment 71 Segment 99 Segment 98 ØTwo kinds of conflict segments l. Overlap (e. g. segment 71, segment 99) l. Use the same spin system (e. g. both segment 78 and segment 79 contain spin system 1) 30/38
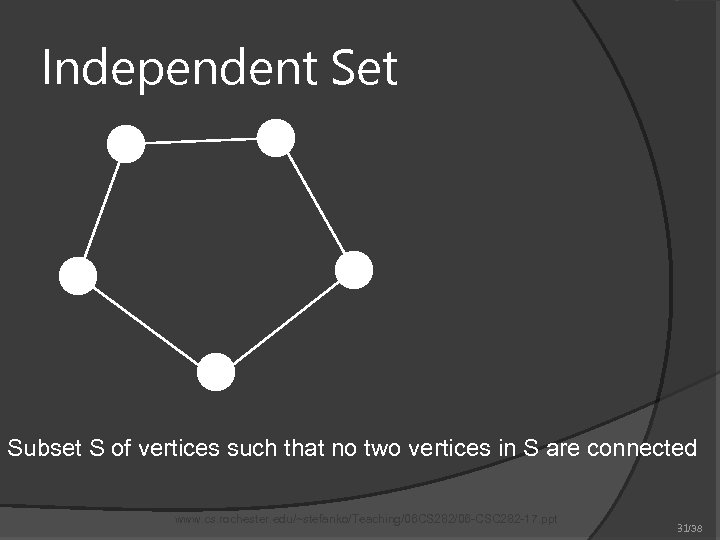 Independent Set Subset S of vertices such that no two vertices in S are connected www. cs. rochester. edu/~stefanko/Teaching/06 CS 282/06 -CSC 282 -17. ppt 31/38
Independent Set Subset S of vertices such that no two vertices in S are connected www. cs. rochester. edu/~stefanko/Teaching/06 CS 282/06 -CSC 282 -17. ppt 31/38
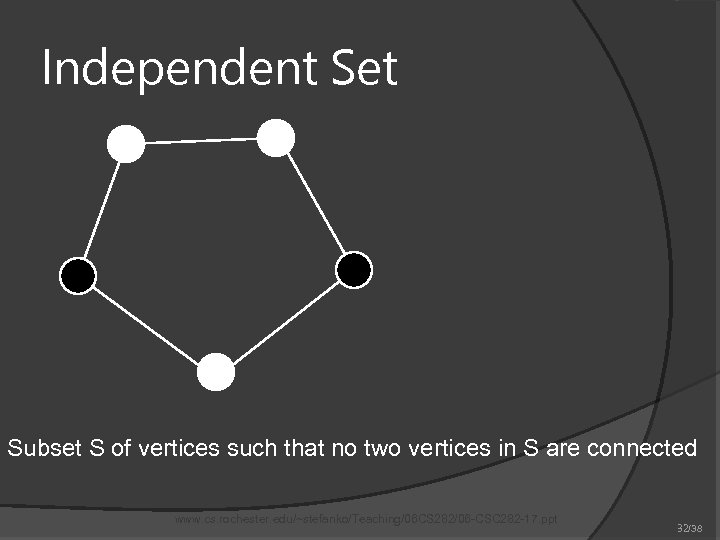 Independent Set Subset S of vertices such that no two vertices in S are connected www. cs. rochester. edu/~stefanko/Teaching/06 CS 282/06 -CSC 282 -17. ppt 32/38
Independent Set Subset S of vertices such that no two vertices in S are connected www. cs. rochester. edu/~stefanko/Teaching/06 CS 282/06 -CSC 282 -17. ppt 32/38
 A Graph Model for Spin System Linking G(V, E) V: a set of nodes (segments). E: (u, v), u, v V, u and v are conflict. Goal Assign as many non-conflict segments as possible => find the maximum independent set of G. 33
A Graph Model for Spin System Linking G(V, E) V: a set of nodes (segments). E: (u, v), u, v V, u and v are conflict. Goal Assign as many non-conflict segments as possible => find the maximum independent set of G. 33
 An Example of G Seg 1 SP 13 Seg 2 Segment 1: SP 12 ->SP 13 ->SP 14 Overlap Segment 3: SP 8 ->SP 15 ->SP 21 Overlap Segment 2: SP 9 ->SP 13 ->SP 20 ->SP 4 Segment 4: SP 7 ->SP 15 ->SP 3 Seg 4 SP 15 Seg 3 Ø Seq. : GEIKGRKTLATPAVRRLAMENNIKLSE Seg 1 Seg 3 Seg 4 Seg 2 34/38
An Example of G Seg 1 SP 13 Seg 2 Segment 1: SP 12 ->SP 13 ->SP 14 Overlap Segment 3: SP 8 ->SP 15 ->SP 21 Overlap Segment 2: SP 9 ->SP 13 ->SP 20 ->SP 4 Segment 4: SP 7 ->SP 15 ->SP 3 Seg 4 SP 15 Seg 3 Ø Seq. : GEIKGRKTLATPAVRRLAMENNIKLSE Seg 1 Seg 3 Seg 4 Seg 2 34/38
 Segment weight The larger length of segment is, the higher weight of segment is. The less frequency of segment is, the lower of segment is. 35/38
Segment weight The larger length of segment is, the higher weight of segment is. The less frequency of segment is, the lower of segment is. 35/38
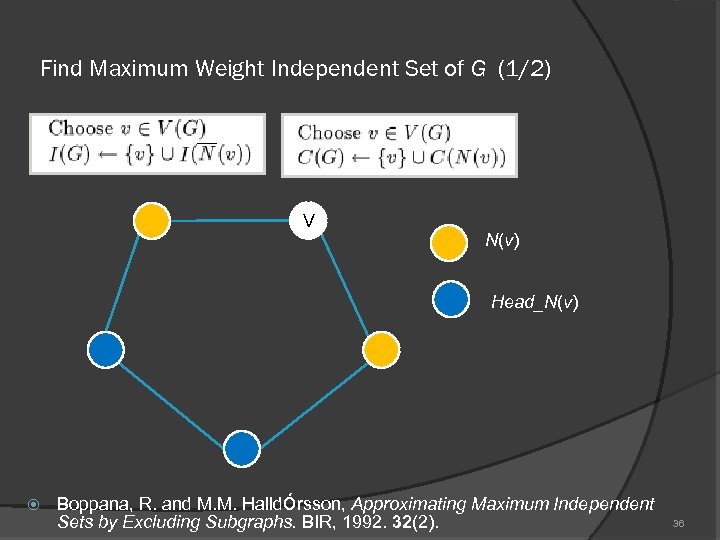 Find Maximum Weight Independent Set of G (1/2) V N(v) Head_N(v) Boppana, R. and M. M. Halldόrsson, Approximating Maximum Independent Sets by Excluding Subgraphs. BIR, 1992. 32(2). 36
Find Maximum Weight Independent Set of G (1/2) V N(v) Head_N(v) Boppana, R. and M. M. Halldόrsson, Approximating Maximum Independent Sets by Excluding Subgraphs. BIR, 1992. 32(2). 36
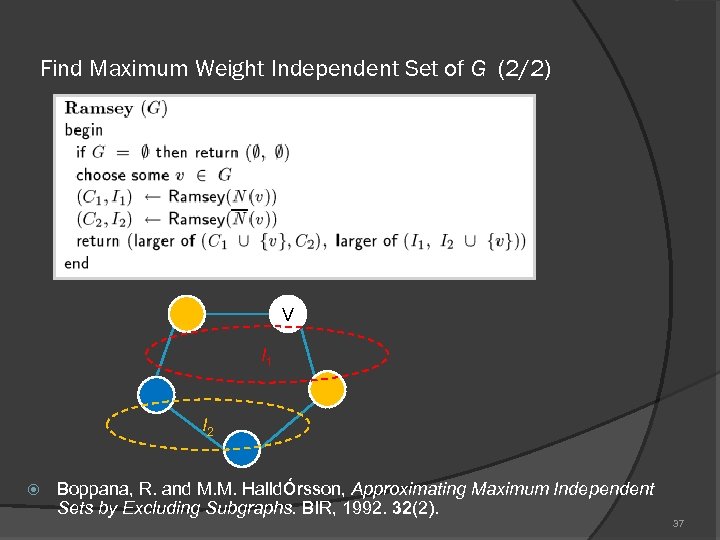 Find Maximum Weight Independent Set of G (2/2) V I 1 I 2 Boppana, R. and M. M. Halldόrsson, Approximating Maximum Independent Sets by Excluding Subgraphs. BIR, 1992. 32(2). 37
Find Maximum Weight Independent Set of G (2/2) V I 1 I 2 Boppana, R. and M. M. Halldόrsson, Approximating Maximum Independent Sets by Excluding Subgraphs. BIR, 1992. 32(2). 37
 An Iterative Approach We perform spin system generation and linking iteratively. Three stages. 38/38
An Iterative Approach We perform spin system generation and linking iteratively. Three stages. 38/38


