f8029be5dbd720f1413e02b90d5875d5.ppt
- Количество слайдов: 36
 Introduction to the Human Genome Alexei Fedorov Applications of Bioinformatics, Proteomics, and Genomics (BIPG 640)
Introduction to the Human Genome Alexei Fedorov Applications of Bioinformatics, Proteomics, and Genomics (BIPG 640)
 $3, 000, 000. 00 per single genome Human Genome Project Completed in 2003, the Human Genome Project (HGP) was a 13 -year project coordinated by the U. S. Department of Energy and the National Institutes of Health. During the early years of the HGP, the Wellcome Trust (U. K. ) became a major partner; additional contributions came from Japan, France, Germany, China, and others. http: //www. ornl. gov/sci/techresources/Human_ Genome/home. shtml
$3, 000, 000. 00 per single genome Human Genome Project Completed in 2003, the Human Genome Project (HGP) was a 13 -year project coordinated by the U. S. Department of Energy and the National Institutes of Health. During the early years of the HGP, the Wellcome Trust (U. K. ) became a major partner; additional contributions came from Japan, France, Germany, China, and others. http: //www. ornl. gov/sci/techresources/Human_ Genome/home. shtml
 International Team to Sequence 1000 Genomes J. Kaiser, Science. NOW Daily News. 01/22/2008 $30, 000. 00 per single genome Just a year after the first individual human genomes were sequenced, an international team announced today that it will probe the entire genomes of about 1000 people. The aim is to create the most detailed catalog yet of human genetic diversity to help biomedical researchers home in on disease genes. The 3 -year project, which will cost $30 million to $50 million, will take advantage of new technologies that have slashed the cost of sequencing. The work will be funded by the U. S. National Human Genome Research Institute (NHGRI) in Bethesda, Maryland, the Sanger Institute in Hinxton, U. K. , and the Beijing Genomics Institute in Shenzhen, China.
International Team to Sequence 1000 Genomes J. Kaiser, Science. NOW Daily News. 01/22/2008 $30, 000. 00 per single genome Just a year after the first individual human genomes were sequenced, an international team announced today that it will probe the entire genomes of about 1000 people. The aim is to create the most detailed catalog yet of human genetic diversity to help biomedical researchers home in on disease genes. The 3 -year project, which will cost $30 million to $50 million, will take advantage of new technologies that have slashed the cost of sequencing. The work will be funded by the U. S. National Human Genome Research Institute (NHGRI) in Bethesda, Maryland, the Sanger Institute in Hinxton, U. K. , and the Beijing Genomics Institute in Shenzhen, China.
 Realistic goal in three-five years Sequence the entire human genome in a few days for $1000 (Era of Personal Genomics) HOWEVER, speed of sequencing does not necessarily mean an understanding of the genetic information or DNA structure!
Realistic goal in three-five years Sequence the entire human genome in a few days for $1000 (Era of Personal Genomics) HOWEVER, speed of sequencing does not necessarily mean an understanding of the genetic information or DNA structure!
 Databases of nucleotide sequences • USA o NCBI (National Center for Biotechnology Information) o UCSC Genome Browser • Europe o EMBL Nucleotide Sequence Database • Japan o DDBJ DNA Data Bank of Japan
Databases of nucleotide sequences • USA o NCBI (National Center for Biotechnology Information) o UCSC Genome Browser • Europe o EMBL Nucleotide Sequence Database • Japan o DDBJ DNA Data Bank of Japan
 variation 116261 /allele="T" /allele="C" /db_xref="db. SNP: 699742" m. RNA complement(join(117904. . 118062, 133653. . 133779, 136940. . 137004)) /gene="LOC 126569" /product="hypothetical gene supported by XM_072587" /transcript_id="XM_072587. 1" /db_xref="Interim. ID: 126569" /note="Derived by automated computational analysis Genome. Scan. " gene complement(117904. . 137004) /gene="LOC 126569" /note="Located on Accession NT_019273" /db_xref="Interim. ID: 126569" CDS complement(join(117904. . 118062, 133653. . 133779, 136940. . 137004)) /gene="LOC 126569" /note="Located on Accession NT_019273" /codon_start=1 /protein_id="XP_072587. 1" /db_xref="GI: 17433766" variation 118483 /allele="G" /allele="A" /db_xref="db. SNP: 1342719"
variation 116261 /allele="T" /allele="C" /db_xref="db. SNP: 699742" m. RNA complement(join(117904. . 118062, 133653. . 133779, 136940. . 137004)) /gene="LOC 126569" /product="hypothetical gene supported by XM_072587" /transcript_id="XM_072587. 1" /db_xref="Interim. ID: 126569" /note="Derived by automated computational analysis Genome. Scan. " gene complement(117904. . 137004) /gene="LOC 126569" /note="Located on Accession NT_019273" /db_xref="Interim. ID: 126569" CDS complement(join(117904. . 118062, 133653. . 133779, 136940. . 137004)) /gene="LOC 126569" /note="Located on Accession NT_019273" /codon_start=1 /protein_id="XP_072587. 1" /db_xref="GI: 17433766" variation 118483 /allele="G" /allele="A" /db_xref="db. SNP: 1342719"
 variation 2845295 /note="WARNING: map location ambiguous" /allele="C" /allele="A" /db_xref="db. SNP: 1216906" 846302 a 578512 c 575805 g 843114 t 1703 others BASE COUNT ORIGIN 1 gaattcaaaa 61 ggacggcatt 121 aagattgttg 181 aacaagtagt 241 gactgtggga 301 gttcttttct 361 tttccaccta 421 aaaaa 481 acggggagat 541 aaaatttatt 601 gcacccattt 661 cctcttctaa 721 ttatttttaa 781 tttcaaaaat 841 ataaagagta 901 ctttatggca 961 ttgatgatta 1021 aaaaaattct 1081 gtgggcagat 1141 gtctctacta 1201 cttgggaggc 1261 agatcacacc 1321 aaaacttctg 1381 tatgacaccc aagaca tgagaaaatc aaaaagcaga gcagtgtgcc agccttacat ctctgctttc gcactaaaca gcagaacagc gaaagccaga tattcatcaa attactttca taaaattatc aatataggcc gtatgaaaaa aataaattaa tagatgaagg ccatcagaga gccaggcgcg cacctgaggt aaaatacaaa tgaggcagga attgcactcc gggaaatggt ttgggcagtc atgacttgta aggacagtgg cacagcactg tgaacatagg tccaggctta tatttttcac gtaactagtt tcagatcatc taagatggag tatttacata aaaattgaca tcactcaaat acttctactc tttcagttta agagctgtca caggaaatac tctgatatct gtggctcacg cagaagttcg atcagccggg gaatcacttg agcctgggca ggcctt atttatagag gctgaagcta tgtacttatc agtagcagca atgggaaatt gtggaataag gacctgaact cagactatat cagtgaagtg aagtaggaaa aatgtttatt atatacaagt tcatataact tattcatttt gtccccacca gaattaaaac tggctgaaaa cagggaagaa cctgtaatcc agaccagcct cgtggtggcg aacccaggag acaagggcga gtaacatcta tccttccctg tgatcaggaa aaataagaag tggagcagaa aggaaagata taaatattta cacctcccag atttaaaaaa gtgctactat tttacgaaac aattctaagt taataaaatc aaaaatacat tgcacttaac aatctcaatt actactacag ttttgtttat aagcctttca cagcactttg gaccaacatgcctgta gcagaggttg aactctgtct tgtgtcttag accagggaat aagatggggt atctgggcag aagcataagg aatggaggct aatctcatga tgaggagatg aaaaa tatactatta attttaaaag actatagtag atattagttt ttaataaatt attctcttgc tagaccccgg gtctccttca gtcaaagatt tataccactt ggaggctgag gagaaaccct atcccagcta cggtgagccg caaaaa agggccatgg catcctgcca
variation 2845295 /note="WARNING: map location ambiguous" /allele="C" /allele="A" /db_xref="db. SNP: 1216906" 846302 a 578512 c 575805 g 843114 t 1703 others BASE COUNT ORIGIN 1 gaattcaaaa 61 ggacggcatt 121 aagattgttg 181 aacaagtagt 241 gactgtggga 301 gttcttttct 361 tttccaccta 421 aaaaa 481 acggggagat 541 aaaatttatt 601 gcacccattt 661 cctcttctaa 721 ttatttttaa 781 tttcaaaaat 841 ataaagagta 901 ctttatggca 961 ttgatgatta 1021 aaaaaattct 1081 gtgggcagat 1141 gtctctacta 1201 cttgggaggc 1261 agatcacacc 1321 aaaacttctg 1381 tatgacaccc aagaca tgagaaaatc aaaaagcaga gcagtgtgcc agccttacat ctctgctttc gcactaaaca gcagaacagc gaaagccaga tattcatcaa attactttca taaaattatc aatataggcc gtatgaaaaa aataaattaa tagatgaagg ccatcagaga gccaggcgcg cacctgaggt aaaatacaaa tgaggcagga attgcactcc gggaaatggt ttgggcagtc atgacttgta aggacagtgg cacagcactg tgaacatagg tccaggctta tatttttcac gtaactagtt tcagatcatc taagatggag tatttacata aaaattgaca tcactcaaat acttctactc tttcagttta agagctgtca caggaaatac tctgatatct gtggctcacg cagaagttcg atcagccggg gaatcacttg agcctgggca ggcctt atttatagag gctgaagcta tgtacttatc agtagcagca atgggaaatt gtggaataag gacctgaact cagactatat cagtgaagtg aagtaggaaa aatgtttatt atatacaagt tcatataact tattcatttt gtccccacca gaattaaaac tggctgaaaa cagggaagaa cctgtaatcc agaccagcct cgtggtggcg aacccaggag acaagggcga gtaacatcta tccttccctg tgatcaggaa aaataagaag tggagcagaa aggaaagata taaatattta cacctcccag atttaaaaaa gtgctactat tttacgaaac aattctaagt taataaaatc aaaaatacat tgcacttaac aatctcaatt actactacag ttttgtttat aagcctttca cagcactttg gaccaacatgcctgta gcagaggttg aactctgtct tgtgtcttag accagggaat aagatggggt atctgggcag aagcataagg aatggaggct aatctcatga tgaggagatg aaaaa tatactatta attttaaaag actatagtag atattagttt ttaataaatt attctcttgc tagaccccgg gtctccttca gtcaaagatt tataccactt ggaggctgag gagaaaccct atcccagcta cggtgagccg caaaaa agggccatgg catcctgcca
 . . . after the first 50 pages. . 141601 141661 141721 141781 141841 141901 141961 142021 142081 142141 142201 142261 142321 142381 142441 142501 142561 142621 142681 142741 142801 142861 142921 142981 143041 143101 143161 143221 143281 143341 cagcaccaaa tgtccatgca cccacactat aaacttgaaa acagataacc cttaagtact gcatttatta aagaatgcta acctgaggaa acttaaaaac agagcagcat gcaattaggc ccacacgtgt ttagccaaaa gtatatcaaa agacctcaaa tacgtaatga cacttacatt cacccaggct caagcaattc tccagctaat tcgaactcct gcatcagccg ctgtctctacttggga ccgagatcac aaaaa tgttgatgct agttaaaatg agttgagaaa tcctctcatt atctgttgaa atatcaaaat atattgagat aacagaggaa tcaaaaaagt caaataattc agatcacatt aaaagctaac ctatcgaaat ttttccccat aatcttgtat gaatcctaaa ggaaaacgac atgatgaaat aatgcccaaa aacagaatac cagatttttt ggagggcagt tcctgcctca ttttgtattt ggcctcaagt ggtgcggtgg taaaatacaa ggctgaggga accactgtac aaaaaagaaa agtctattgt tatcaaaatgtaagca gcctttttaa aaatctggct aaacccaagt gaatattagt gtcagaaaac cattacaata agaaaaagga ttttaaaaag ctcacaagta aacgaagtgt tgtggaggga caaaaatctt acaattaaaa ctaaatgacg attttgcagc atatattaat agttgatcct tctttttgct ggcaccattc gcctcccaag ttagtagaga aatccacctg cttatgcctg aaaattagct tgagaattgc tccagcctgg aagaaaaaga gtaatttacc tatacacaaa aacatgaaga aaaatgttgt atttgcaaac gtataaaaga tagagctttg agtaatcatt cttaaaaacc tttatatccc tagctaaagg ttcaaccaaa ttggaaaatg gtgtgtaaat caaagtgttc gtatgaacat aatgatgtgc tttgaaaaggat tgaacaacgc ttttt tggctcacta tagctggaat cggagtttca cctcagcctc caatcccatc gagtgtggtg ttgaacctgg gcaacagagc aaaaggtatg accataaaat cacttagaga tgcagtatta ccaatttaac aaagaaaaaa gaaaatttta agtaggaaag tccttaatga ttacaacaat taataactaa ataatataaaataacc acaagattca tggtgtggtc ttactctttg atttttatgc aactgcatgg taattttgaa acaaaacttt tggtttgaac gagacgaagt caacctgcgt tacaggcgcc ccatgttggc ccaaagtgct ctggctaaca gcacatgcct gaggcagagg aagactccat ttatgaatgc atacacaggt tagtacatgg aatcataact atcaagacac tgtatagcct agtgaaacca gattttttga aaatacaaaa catgtggaaa agaagtgagg tgactaacag tcgagatacc aaatctggta tttctgaaaa atgaagaatt acaaagatgt ataaattgtt aaaactttaa attatttcac tgcactcgtc ctcactctgt ataccaggtt tgtcaccacg caggctggtc gggattacag cggtgaaacc atagttccag ttgcagtgag ctcaaaaaaa agaaagtata ctattataga tatcattccc gtataaaatt
. . . after the first 50 pages. . 141601 141661 141721 141781 141841 141901 141961 142021 142081 142141 142201 142261 142321 142381 142441 142501 142561 142621 142681 142741 142801 142861 142921 142981 143041 143101 143161 143221 143281 143341 cagcaccaaa tgtccatgca cccacactat aaacttgaaa acagataacc cttaagtact gcatttatta aagaatgcta acctgaggaa acttaaaaac agagcagcat gcaattaggc ccacacgtgt ttagccaaaa gtatatcaaa agacctcaaa tacgtaatga cacttacatt cacccaggct caagcaattc tccagctaat tcgaactcct gcatcagccg ctgtctctacttggga ccgagatcac aaaaa tgttgatgct agttaaaatg agttgagaaa tcctctcatt atctgttgaa atatcaaaat atattgagat aacagaggaa tcaaaaaagt caaataattc agatcacatt aaaagctaac ctatcgaaat ttttccccat aatcttgtat gaatcctaaa ggaaaacgac atgatgaaat aatgcccaaa aacagaatac cagatttttt ggagggcagt tcctgcctca ttttgtattt ggcctcaagt ggtgcggtgg taaaatacaa ggctgaggga accactgtac aaaaaagaaa agtctattgt tatcaaaatgtaagca gcctttttaa aaatctggct aaacccaagt gaatattagt gtcagaaaac cattacaata agaaaaagga ttttaaaaag ctcacaagta aacgaagtgt tgtggaggga caaaaatctt acaattaaaa ctaaatgacg attttgcagc atatattaat agttgatcct tctttttgct ggcaccattc gcctcccaag ttagtagaga aatccacctg cttatgcctg aaaattagct tgagaattgc tccagcctgg aagaaaaaga gtaatttacc tatacacaaa aacatgaaga aaaatgttgt atttgcaaac gtataaaaga tagagctttg agtaatcatt cttaaaaacc tttatatccc tagctaaagg ttcaaccaaa ttggaaaatg gtgtgtaaat caaagtgttc gtatgaacat aatgatgtgc tttgaaaaggat tgaacaacgc ttttt tggctcacta tagctggaat cggagtttca cctcagcctc caatcccatc gagtgtggtg ttgaacctgg gcaacagagc aaaaggtatg accataaaat cacttagaga tgcagtatta ccaatttaac aaagaaaaaa gaaaatttta agtaggaaag tccttaatga ttacaacaat taataactaa ataatataaaataacc acaagattca tggtgtggtc ttactctttg atttttatgc aactgcatgg taattttgaa acaaaacttt tggtttgaac gagacgaagt caacctgcgt tacaggcgcc ccatgttggc ccaaagtgct ctggctaaca gcacatgcct gaggcagagg aagactccat ttatgaatgc atacacaggt tagtacatgg aatcataact atcaagacac tgtatagcct agtgaaacca gattttttga aaatacaaaa catgtggaaa agaagtgagg tgactaacag tcgagatacc aaatctggta tttctgaaaa atgaagaatt acaaagatgt ataaattgtt aaaactttaa attatttcac tgcactcgtc ctcactctgt ataccaggtt tgtcaccacg caggctggtc gggattacag cggtgaaacc atagttccag ttgcagtgag ctcaaaaaaa agaaagtata ctattataga tatcattccc gtataaaatt
 . . . after next 200 pages 683041 683101 683161 683221 683281 683341 683401 683461 683521 683581 683641 683701 683761 683821 683881 683941 684001 684061 684121 684181 684241 684301 684361 684421 684481 684541 684601 684661 684721 684781 684841 684901 ggaggtgggg agccaccaac gaggagcacc agcgaccatc aatgtgggga ataggagact tctataacct aaatggatta aaaaaagaaa ctccaacact tttaaaggtt aagaatgttg cactgttagt ttttttcctt tcaaggagta ataggttggg tctcccagtc atatttcttg gctttatttc acatttcttg agtaccatta taatatgttg gaatgggtgg caattccact taaaacagtgtcatag tgtttgtccc gcactatttt taatcccagc ccaacgtggt atgcctataa cagaggttgt agcgcctctg ccatctggga tctgccgggc gagaatgggc aaagag ccattttgtt tacccccaaa agggcgatgc gagaaaaaaa tgtcacctaa ttcagcttaa aatattggcc ctgatggctt catttcaacc tctttgtggt gaagttctcc actttcaggt gaggctttgc attaagttag tcttttttgg cgctccgtga cctggtccag gtggttagat tactggtgag acaatgatat tgatcaggaa aatgtatatg atgaacttta actttgggag gaaaccacat tcccagctac ggtgagctga cccagccgcc agtgaggagc tgccccgtct catgatgacg agatcagatt ctgtactaag cccctgctct aagatgtgct aaatcattga tgaccaggga ctgttttgtc cccactctct ccctttgtgg atggtgaatc gttctctgta tggataatat acaccaatca tcattccttt tttatatttg gcctgataat ggacagggac agtagatact gaatggaatt aagccttgtc tgtttctgct taaagccagg gcagagggag aaatcctcat gccaaggcag ctctactaaa ttgggaggct gattgtgcca ccatctggga gcctctgcct gggaagtgtt atggtggttt gttactgtgt aaaaattctt ctgaaacatg ttgttaaaca aggattattt tcaataccca tcttaataaa tctggcttgt gtaacccagt tgacaattat tttcctgaat cctgaagagt aatgtaggtt tcattctttt actgtgcttt tactctgcaa tattttgttc catatataaa tgccttaatt taagtcttta accacaatgg gcttgaagca aaagaaaacc agcagggcca gcagatcact aatacaaaaa gaggcaggag ctgtactcca ggtggggagc ggccaccccg cccaacagct tgtcgaaaag ctgtgtagaa ctgccttggg tgctgtgtca gatgcttgaa atgccctatg caaatacagt tttttatata agagtttctg ctttct gtgtcttggt ttgaatattg gttttccaac tggtcttttc ttctctaatc atacttgaca gttaaaaagg attgttgcaa tacttgctga ttcaagatgg aaccttactt aaaaaaggac tctcctgatt gttgagtctt ggtgcagtgg tgaggtcagg ttagccaggc aaatgcttga gcctgggcaa gcctctgtcc tctgggaagt ctgaagagac aaaaggggga agaagtagac atgctgttaa actcagggtt gacagaaaaa gcatcccttt aagacctatt ggaaaa cagagagatc gcccttaaca gttgctcttc gcctgtgtgg ttggttccat acatagtccc ttgtcttcaa aagcactttc aaaaactcca cctaagcact ataaagggat attcaatttc tcctcatcta agaattactt cctagggcat aatctgtcag ctcacacctg accagcctgt gtggtggtgc acctgggagg cagaacaaga
. . . after next 200 pages 683041 683101 683161 683221 683281 683341 683401 683461 683521 683581 683641 683701 683761 683821 683881 683941 684001 684061 684121 684181 684241 684301 684361 684421 684481 684541 684601 684661 684721 684781 684841 684901 ggaggtgggg agccaccaac gaggagcacc agcgaccatc aatgtgggga ataggagact tctataacct aaatggatta aaaaaagaaa ctccaacact tttaaaggtt aagaatgttg cactgttagt ttttttcctt tcaaggagta ataggttggg tctcccagtc atatttcttg gctttatttc acatttcttg agtaccatta taatatgttg gaatgggtgg caattccact taaaacagtgtcatag tgtttgtccc gcactatttt taatcccagc ccaacgtggt atgcctataa cagaggttgt agcgcctctg ccatctggga tctgccgggc gagaatgggc aaagag ccattttgtt tacccccaaa agggcgatgc gagaaaaaaa tgtcacctaa ttcagcttaa aatattggcc ctgatggctt catttcaacc tctttgtggt gaagttctcc actttcaggt gaggctttgc attaagttag tcttttttgg cgctccgtga cctggtccag gtggttagat tactggtgag acaatgatat tgatcaggaa aatgtatatg atgaacttta actttgggag gaaaccacat tcccagctac ggtgagctga cccagccgcc agtgaggagc tgccccgtct catgatgacg agatcagatt ctgtactaag cccctgctct aagatgtgct aaatcattga tgaccaggga ctgttttgtc cccactctct ccctttgtgg atggtgaatc gttctctgta tggataatat acaccaatca tcattccttt tttatatttg gcctgataat ggacagggac agtagatact gaatggaatt aagccttgtc tgtttctgct taaagccagg gcagagggag aaatcctcat gccaaggcag ctctactaaa ttgggaggct gattgtgcca ccatctggga gcctctgcct gggaagtgtt atggtggttt gttactgtgt aaaaattctt ctgaaacatg ttgttaaaca aggattattt tcaataccca tcttaataaa tctggcttgt gtaacccagt tgacaattat tttcctgaat cctgaagagt aatgtaggtt tcattctttt actgtgcttt tactctgcaa tattttgttc catatataaa tgccttaatt taagtcttta accacaatgg gcttgaagca aaagaaaacc agcagggcca gcagatcact aatacaaaaa gaggcaggag ctgtactcca ggtggggagc ggccaccccg cccaacagct tgtcgaaaag ctgtgtagaa ctgccttggg tgctgtgtca gatgcttgaa atgccctatg caaatacagt tttttatata agagtttctg ctttct gtgtcttggt ttgaatattg gttttccaac tggtcttttc ttctctaatc atacttgaca gttaaaaagg attgttgcaa tacttgctga ttcaagatgg aaccttactt aaaaaaggac tctcctgatt gttgagtctt ggtgcagtgg tgaggtcagg ttagccaggc aaatgcttga gcctgggcaa gcctctgtcc tctgggaagt ctgaagagac aaaaggggga agaagtagac atgctgttaa actcagggtt gacagaaaaa gcatcccttt aagacctatt ggaaaa cagagagatc gcccttaaca gttgctcttc gcctgtgtgg ttggttccat acatagtccc ttgtcttcaa aagcactttc aaaaactcca cctaagcact ataaagggat attcaatttc tcctcatcta agaattactt cctagggcat aatctgtcag ctcacacctg accagcctgt gtggtggtgc acctgggagg cagaacaaga
 Human chromosome 1 4, 814, 628 lines = =100, 000 pages = 100 books (1000 pages each)
Human chromosome 1 4, 814, 628 lines = =100, 000 pages = 100 books (1000 pages each)
 GENOMICS Animal Genome Size Database http: //www. genomesize. com/ Go to Statistics How can I convert from picograms (pg) to base pairs (bp)? Number of base pairs = mass in pg x 0. 978 x 109 or simply: 1 pg = 978 Mb
GENOMICS Animal Genome Size Database http: //www. genomesize. com/ Go to Statistics How can I convert from picograms (pg) to base pairs (bp)? Number of base pairs = mass in pg x 0. 978 x 109 or simply: 1 pg = 978 Mb
 Advanced reading Gregory TR. Synergy between sequence and size in largescale genomics. Nat Rev Genet. 2005 Sep; 6(9): 699 -708. Vinogradov, A. E. Evolution of the genome size: multilevel selection, mutation bias or dynamical chaos? Curr. Opin. Genet. Dev. 2004, 14: 620 -626. Petrov, D. A. Evolution of genome size: new approaches to an old problem. Trends Genet. 2001, 17: 23 -28. Gregory T. R. Insertion-deletion biases and the evolution of genome size. Gene 2004, 324: 15 -34.
Advanced reading Gregory TR. Synergy between sequence and size in largescale genomics. Nat Rev Genet. 2005 Sep; 6(9): 699 -708. Vinogradov, A. E. Evolution of the genome size: multilevel selection, mutation bias or dynamical chaos? Curr. Opin. Genet. Dev. 2004, 14: 620 -626. Petrov, D. A. Evolution of genome size: new approaches to an old problem. Trends Genet. 2001, 17: 23 -28. Gregory T. R. Insertion-deletion biases and the evolution of genome size. Gene 2004, 324: 15 -34.
 We still do not even know the exact number of human genes! “The current genome sequence (Build 35) contains 2. 85 billion nucleotides interrupted by only 341 gaps. It covers ~99% of the euchromatic genome and is accurate to an error rate of ~1 event per 100, 000 bases. … Notably, the human genome seems to encode only 20, 00025, 000 protein-coding genes. ” Nature Oct 21, 2004, 431: 931 -945 13
We still do not even know the exact number of human genes! “The current genome sequence (Build 35) contains 2. 85 billion nucleotides interrupted by only 341 gaps. It covers ~99% of the euchromatic genome and is accurate to an error rate of ~1 event per 100, 000 bases. … Notably, the human genome seems to encode only 20, 00025, 000 protein-coding genes. ” Nature Oct 21, 2004, 431: 931 -945 13
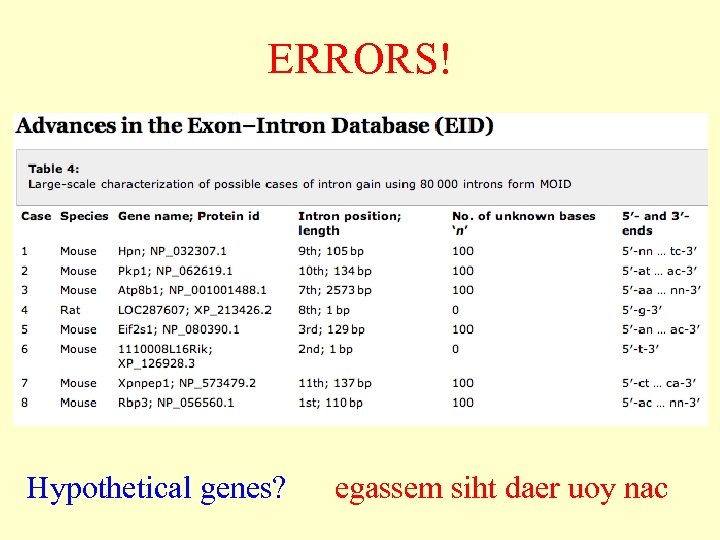 ERRORS! Hypothetical genes? egassem siht daer uoy nac
ERRORS! Hypothetical genes? egassem siht daer uoy nac
 Detailed investigation of the exon–intron structure in the rat homeobox 2 b gene. For the Figure legend see next slide Shepelev V , Fedorov A Brief Bioinform 2006; 7: 178 -185 © The Author 2006. Published by Oxford University Press. For Permissions, please email: journals. permissions@oxfordjournals. org
Detailed investigation of the exon–intron structure in the rat homeobox 2 b gene. For the Figure legend see next slide Shepelev V , Fedorov A Brief Bioinform 2006; 7: 178 -185 © The Author 2006. Published by Oxford University Press. For Permissions, please email: journals. permissions@oxfordjournals. org
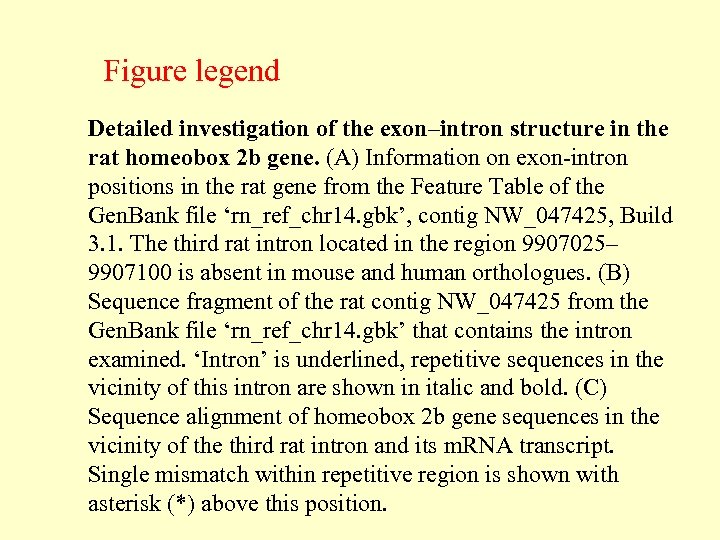 Figure legend Detailed investigation of the exon–intron structure in the rat homeobox 2 b gene. (A) Information on exon-intron positions in the rat gene from the Feature Table of the Gen. Bank file ‘rn_ref_chr 14. gbk’, contig NW_047425, Build 3. 1. The third rat intron located in the region 9907025– 9907100 is absent in mouse and human orthologues. (B) Sequence fragment of the rat contig NW_047425 from the Gen. Bank file ‘rn_ref_chr 14. gbk’ that contains the intron examined. ‘Intron’ is underlined, repetitive sequences in the vicinity of this intron are shown in italic and bold. (C) Sequence alignment of homeobox 2 b gene sequences in the vicinity of the third rat intron and its m. RNA transcript. Single mismatch within repetitive region is shown with asterisk (*) above this position.
Figure legend Detailed investigation of the exon–intron structure in the rat homeobox 2 b gene. (A) Information on exon-intron positions in the rat gene from the Feature Table of the Gen. Bank file ‘rn_ref_chr 14. gbk’, contig NW_047425, Build 3. 1. The third rat intron located in the region 9907025– 9907100 is absent in mouse and human orthologues. (B) Sequence fragment of the rat contig NW_047425 from the Gen. Bank file ‘rn_ref_chr 14. gbk’ that contains the intron examined. ‘Intron’ is underlined, repetitive sequences in the vicinity of this intron are shown in italic and bold. (C) Sequence alignment of homeobox 2 b gene sequences in the vicinity of the third rat intron and its m. RNA transcript. Single mismatch within repetitive region is shown with asterisk (*) above this position.
 It is impossible to keep in mind such enormous amount of information! Our brain is simply too small even to remember all gene names and their functions
It is impossible to keep in mind such enormous amount of information! Our brain is simply too small even to remember all gene names and their functions
 What is the most astonishing feature of the Human genome ?
What is the most astonishing feature of the Human genome ?
 super-perfection in reproduction
super-perfection in reproduction
 immense genetic variability
immense genetic variability
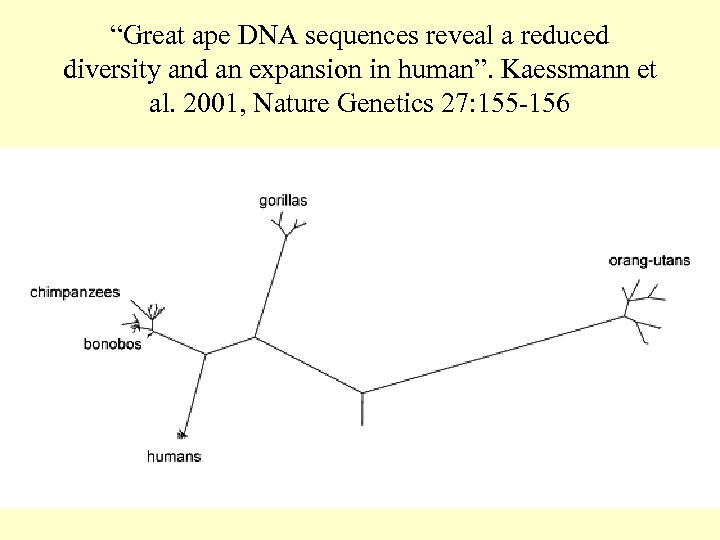 “Great ape DNA sequences reveal a reduced diversity and an expansion in human”. Kaessmann et al. 2001, Nature Genetics 27: 155 -156
“Great ape DNA sequences reveal a reduced diversity and an expansion in human”. Kaessmann et al. 2001, Nature Genetics 27: 155 -156
 Example for your Homework #1 Find a promoter region (10 Kb) of the human gene COPZ 2 m. RNA identifier: AB 037938
Example for your Homework #1 Find a promoter region (10 Kb) of the human gene COPZ 2 m. RNA identifier: AB 037938
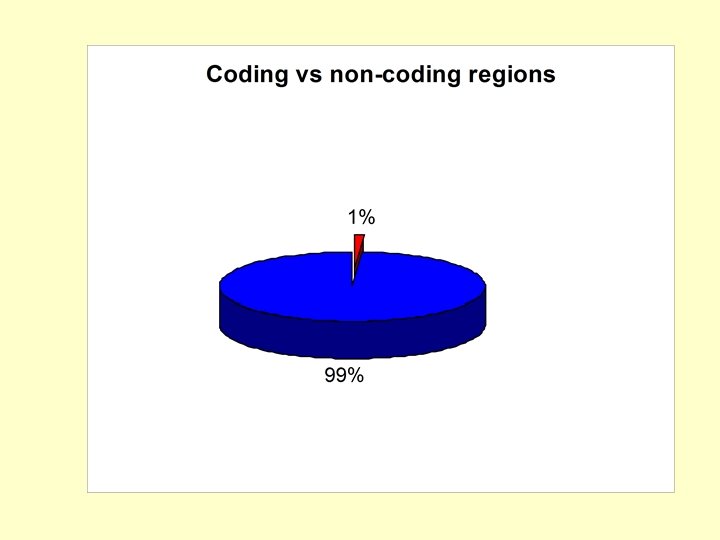
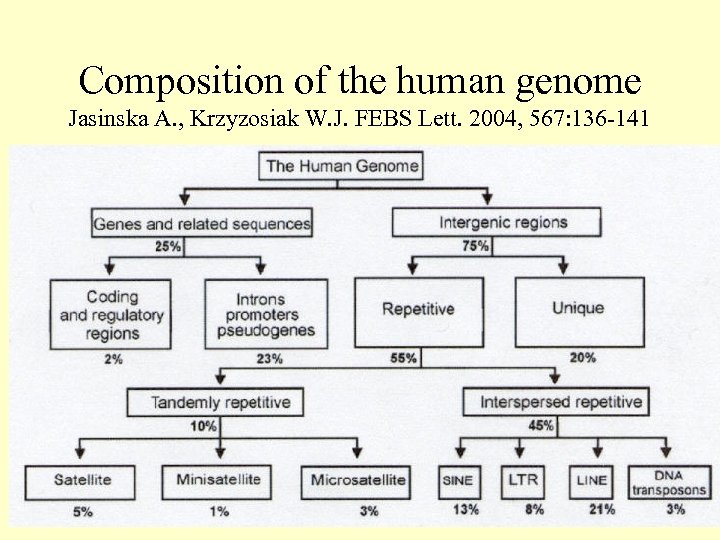 Composition of the human genome Jasinska A. , Krzyzosiak W. J. FEBS Lett. 2004, 567: 136 -141
Composition of the human genome Jasinska A. , Krzyzosiak W. J. FEBS Lett. 2004, 567: 136 -141
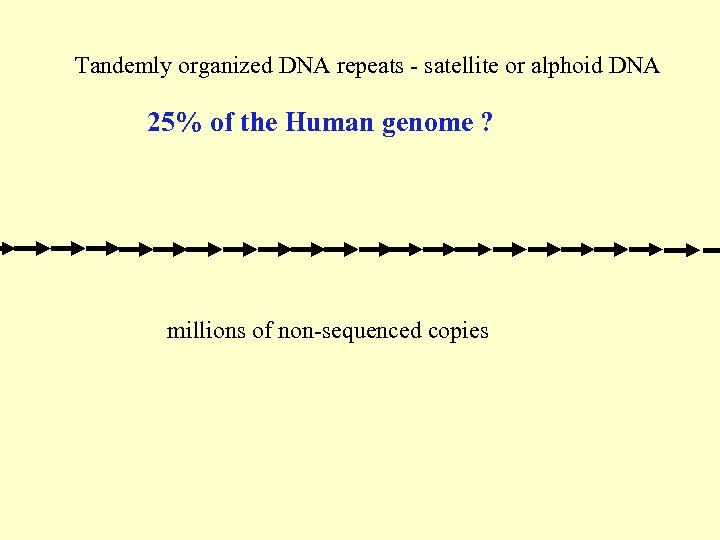 Tandemly organized DNA repeats - satellite or alphoid DNA 25% of the Human genome ? millions of non-sequenced copies
Tandemly organized DNA repeats - satellite or alphoid DNA 25% of the Human genome ? millions of non-sequenced copies
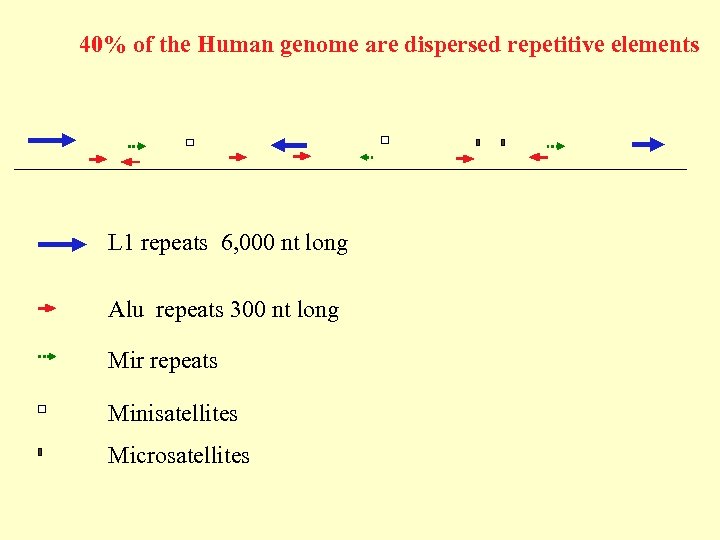 40% of the Human genome are dispersed repetitive elements L 1 repeats 6, 000 nt long Alu repeats 300 nt long Mir repeats Minisatellites Microsatellites
40% of the Human genome are dispersed repetitive elements L 1 repeats 6, 000 nt long Alu repeats 300 nt long Mir repeats Minisatellites Microsatellites
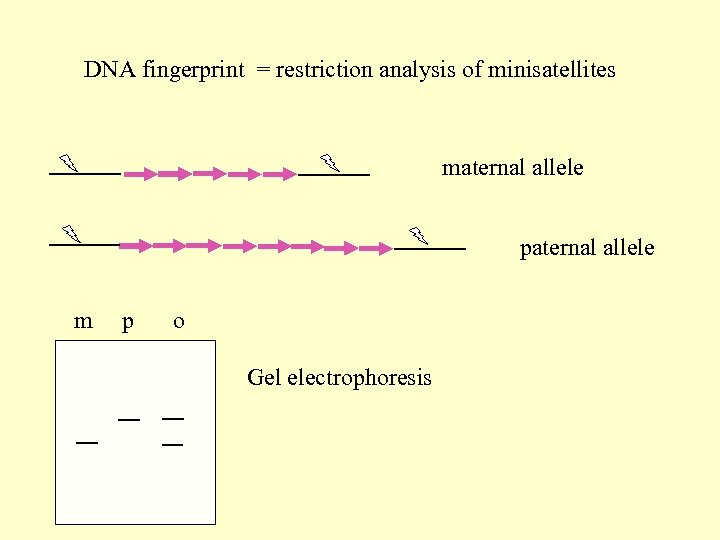 DNA fingerprint = restriction analysis of minisatellites maternal allele paternal allele m p o Gel electrophoresis
DNA fingerprint = restriction analysis of minisatellites maternal allele paternal allele m p o Gel electrophoresis
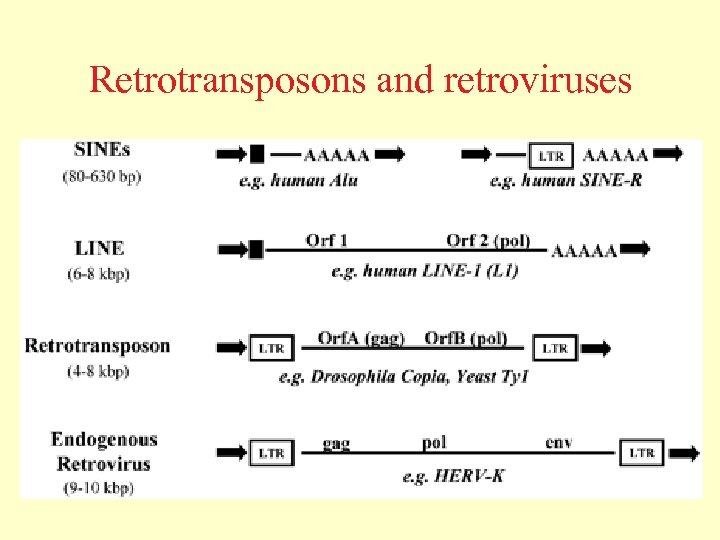 Retrotransposons and retroviruses
Retrotransposons and retroviruses
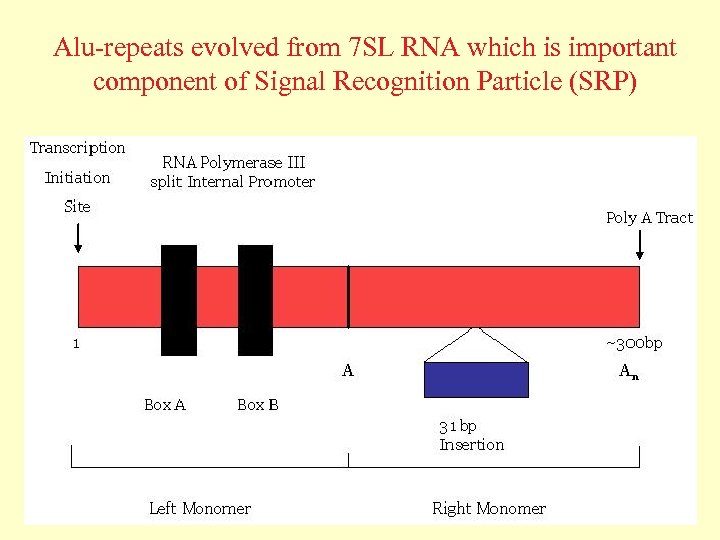 Alu-repeats evolved from 7 SL RNA which is important component of Signal Recognition Particle (SRP)
Alu-repeats evolved from 7 SL RNA which is important component of Signal Recognition Particle (SRP)
 “Mobile elements: Drivers of genome evolution” H. Kazazian Science, 2004, 303: 1626 -1632 Over millions of years of evolution, mobile elements have achieved a balance between detrimental effects on the individual and long-term beneficial effects on a species through genome modification. Indeed, we may soon learn that the shaping of the genome by mobile elements has played an important role in events leading to speciation. Whether these repeated sequences are now "junk DNA" is a complex issue. Some may have had an important function long ago, but have lost that role today. Others may never have had a function, yet the cluttering of our genomes with nonfunctional DNA was a small price to pay for the genome malleability they provided.
“Mobile elements: Drivers of genome evolution” H. Kazazian Science, 2004, 303: 1626 -1632 Over millions of years of evolution, mobile elements have achieved a balance between detrimental effects on the individual and long-term beneficial effects on a species through genome modification. Indeed, we may soon learn that the shaping of the genome by mobile elements has played an important role in events leading to speciation. Whether these repeated sequences are now "junk DNA" is a complex issue. Some may have had an important function long ago, but have lost that role today. Others may never have had a function, yet the cluttering of our genomes with nonfunctional DNA was a small price to pay for the genome malleability they provided.
 Homework #2 Read 2 reviews (Kazazian 2004; Richard et al. 2008) on the human DNA repeats. You will have several questions about them at your Exam In a short assay (~2 pages) • List all types and names of human DNA repetitive sequences • Are these DNA repeats harmful? • What positive functions could they have?
Homework #2 Read 2 reviews (Kazazian 2004; Richard et al. 2008) on the human DNA repeats. You will have several questions about them at your Exam In a short assay (~2 pages) • List all types and names of human DNA repetitive sequences • Are these DNA repeats harmful? • What positive functions could they have?
 Repeat Masker program http: //www. repeatmasker. org/ Institute for System Biology
Repeat Masker program http: //www. repeatmasker. org/ Institute for System Biology
 Libraries of DNA repeats • http: //www. molbiol. bbsrc. ac. uk/images/help /Repeat. Masker. html
Libraries of DNA repeats • http: //www. molbiol. bbsrc. ac. uk/images/help /Repeat. Masker. html
 Importance of repeat masking • Comparison of two or more human genomic DNA segments
Importance of repeat masking • Comparison of two or more human genomic DNA segments
 Homework #1 1) Get the nucleotide sequence of human beta-globin m. RNA by accession number NM_000518 2) Use on-line BLAST program to search entire human genome sequence with your query. 3) Get 50, 000 bp promoter region of the human beta hemoglobin gene. (You need to get it in a FASTA format and not in a Gen. Bank format!) 4) Get another 50 Kb promoter region of human AGT (angiotensinogen) gene. (Use search keyword ”human AGT”). 4) Use on-line Repeat. Masker program to mask all DNA-repeats in two promoter sequences (AGT and beta-hemoglobin genes). Create two files with masked sequences. Send to me these masked sequences by e-mail. 6) Make a thoughtful comparison of your results with the results obtained in this lecture (intron 3_heparanase 2, see in the folder Repeat. Masker_intron 3_HPSE 2) (write a short paragraph)
Homework #1 1) Get the nucleotide sequence of human beta-globin m. RNA by accession number NM_000518 2) Use on-line BLAST program to search entire human genome sequence with your query. 3) Get 50, 000 bp promoter region of the human beta hemoglobin gene. (You need to get it in a FASTA format and not in a Gen. Bank format!) 4) Get another 50 Kb promoter region of human AGT (angiotensinogen) gene. (Use search keyword ”human AGT”). 4) Use on-line Repeat. Masker program to mask all DNA-repeats in two promoter sequences (AGT and beta-hemoglobin genes). Create two files with masked sequences. Send to me these masked sequences by e-mail. 6) Make a thoughtful comparison of your results with the results obtained in this lecture (intron 3_heparanase 2, see in the folder Repeat. Masker_intron 3_HPSE 2) (write a short paragraph)
 Homework #2 Read 2 reviews (Kazazian 2004; Richard et al. 2008) on the human DNA repeats. You will have several questions about them at your Exam In a short assay (~2 pages) • List all types and names of human DNA repetitive sequences • Are these DNA repeats harmful? • What positive functions could they have?
Homework #2 Read 2 reviews (Kazazian 2004; Richard et al. 2008) on the human DNA repeats. You will have several questions about them at your Exam In a short assay (~2 pages) • List all types and names of human DNA repetitive sequences • Are these DNA repeats harmful? • What positive functions could they have?


