9e974e83569bfbb5d7aba9f77c0aee6f.ppt
- Количество слайдов: 68
 Introduction to micro. RNA mi. RNA Workshop Ravi K. Gutti Division of Newborn Medicine Children´s Hospital Boston and Harvard Medical School Ravi. Gutti@childrens. harvard. edu May 18, 2010 http: //catalyst. harvard. edu
Introduction to micro. RNA mi. RNA Workshop Ravi K. Gutti Division of Newborn Medicine Children´s Hospital Boston and Harvard Medical School Ravi. Gutti@childrens. harvard. edu May 18, 2010 http: //catalyst. harvard. edu
 Agenda – Introduction and History – Workflow in mi. RNA experiments – Finding mi. RNA targets by target search databases – mi. RNA validation and experimental considerations – Applications 1
Agenda – Introduction and History – Workflow in mi. RNA experiments – Finding mi. RNA targets by target search databases – mi. RNA validation and experimental considerations – Applications 1
 Why study mi. RNA? 2
Why study mi. RNA? 2
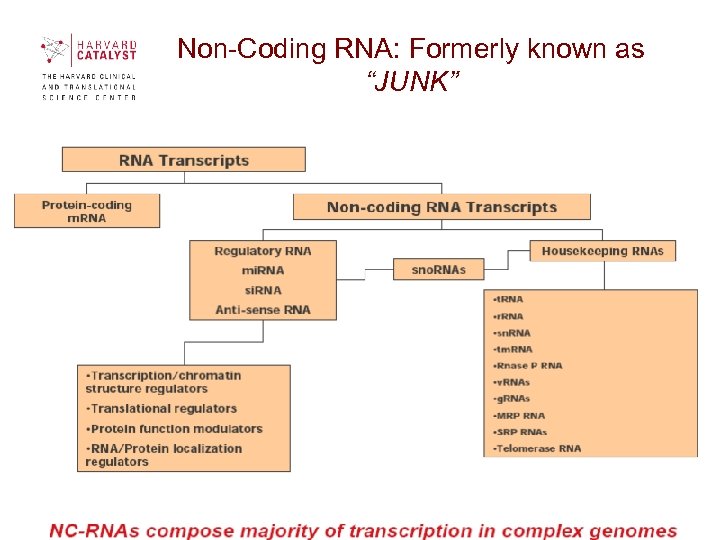 Non-Coding RNA: Formerly known as “JUNK”
Non-Coding RNA: Formerly known as “JUNK”
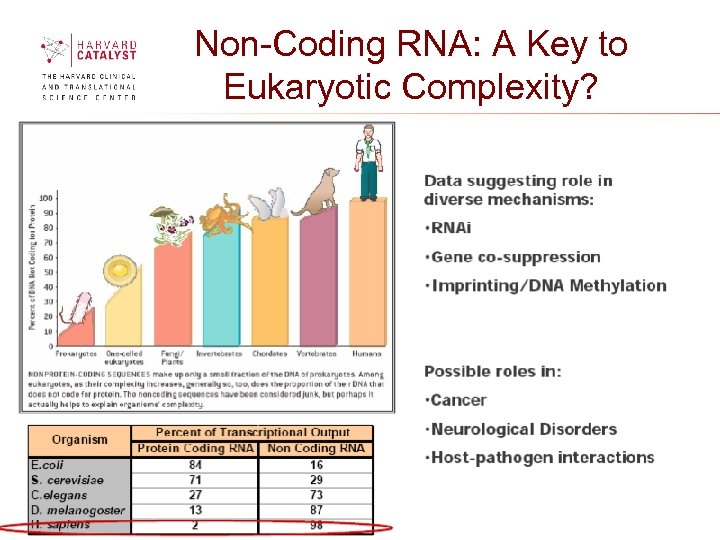 Non-Coding RNA: A Key to Eukaryotic Complexity?
Non-Coding RNA: A Key to Eukaryotic Complexity?
 What is mi. RNA? 5
What is mi. RNA? 5
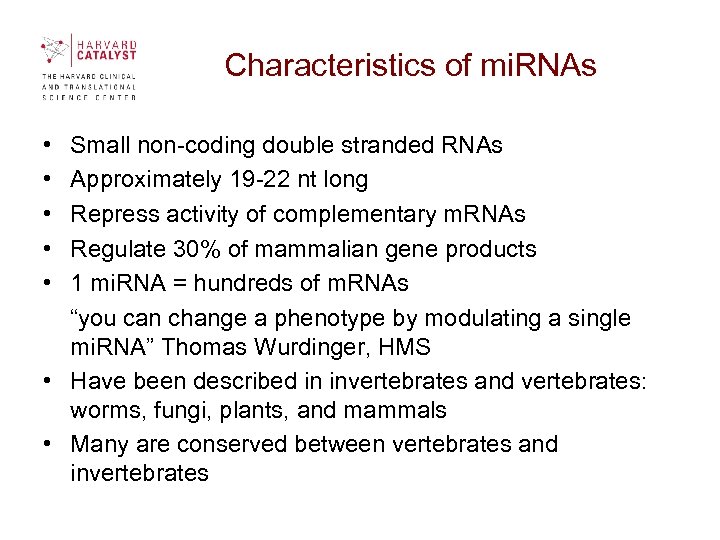 Characteristics of mi. RNAs • Small non-coding double stranded RNAs • Approximately 19 -22 nt long • Repress activity of complementary m. RNAs • Regulate 30% of mammalian gene products • 1 mi. RNA = hundreds of m. RNAs “you can change a phenotype by modulating a single mi. RNA” Thomas Wurdinger, HMS • Have been described in invertebrates and vertebrates: worms, fungi, plants, and mammals • Many are conserved between vertebrates and invertebrates
Characteristics of mi. RNAs • Small non-coding double stranded RNAs • Approximately 19 -22 nt long • Repress activity of complementary m. RNAs • Regulate 30% of mammalian gene products • 1 mi. RNA = hundreds of m. RNAs “you can change a phenotype by modulating a single mi. RNA” Thomas Wurdinger, HMS • Have been described in invertebrates and vertebrates: worms, fungi, plants, and mammals • Many are conserved between vertebrates and invertebrates
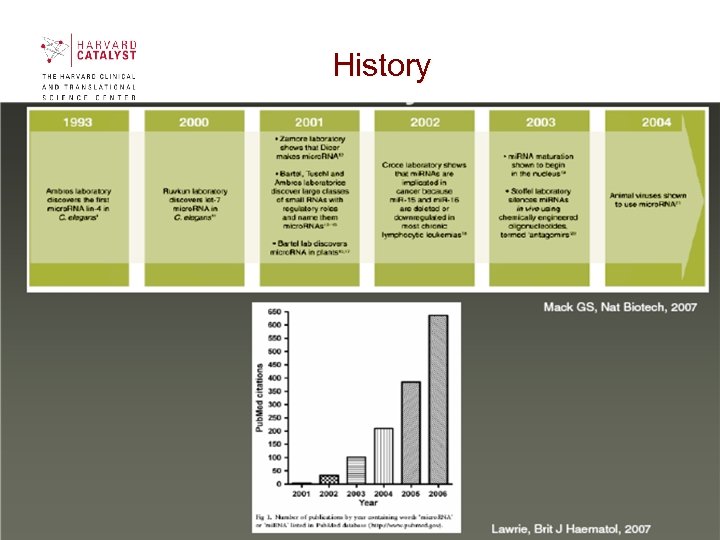 History
History
 History • lin-4, first mi. RNA to be described in C. elegans; important in development of the worm from larva to adult. • let-7, was also described in C. elegans (Reinhard BJ et al, 2000) as critical to stop the stem-cell-like divisions of seam cells and induce their fully differentiated state. Reduced let-7 expression is associated with human cancers and cancer stem cells, thus suggesting that let-7 in humans also promotes terminal differentiation and is a tumor suppressor. • 1998 -Fire and Mello, experiments in C. elegans, first to show that ds. RNA is much more potent at inhibiting gene expression than antisense RNA. Set the stage for understanding the role of mi. RNAs in development and gene regulation. (Nobel Prize in Physiology and Medicine, 2007).
History • lin-4, first mi. RNA to be described in C. elegans; important in development of the worm from larva to adult. • let-7, was also described in C. elegans (Reinhard BJ et al, 2000) as critical to stop the stem-cell-like divisions of seam cells and induce their fully differentiated state. Reduced let-7 expression is associated with human cancers and cancer stem cells, thus suggesting that let-7 in humans also promotes terminal differentiation and is a tumor suppressor. • 1998 -Fire and Mello, experiments in C. elegans, first to show that ds. RNA is much more potent at inhibiting gene expression than antisense RNA. Set the stage for understanding the role of mi. RNAs in development and gene regulation. (Nobel Prize in Physiology and Medicine, 2007).
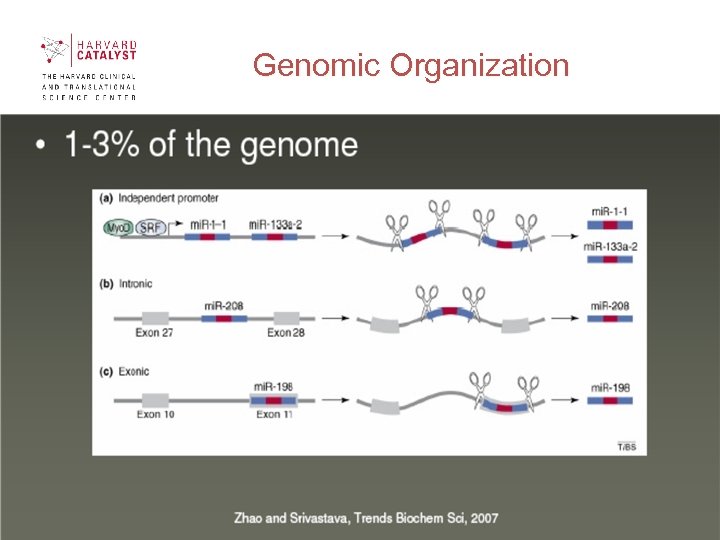 Genomic Organization
Genomic Organization
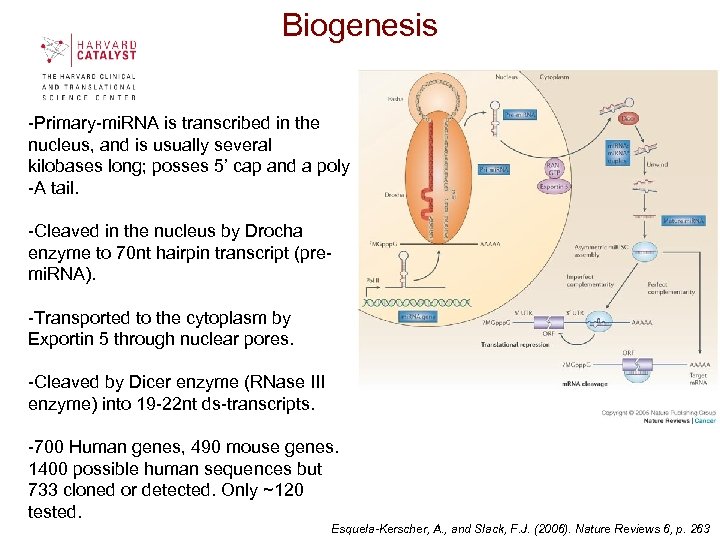 Biogenesis -Primary-mi. RNA is transcribed in the nucleus, and is usually several kilobases long; posses 5’ cap and a poly -A tail. -Cleaved in the nucleus by Drocha enzyme to 70 nt hairpin transcript (premi. RNA). -Transported to the cytoplasm by Exportin 5 through nuclear pores. -Cleaved by Dicer enzyme (RNase III enzyme) into 19 -22 nt ds-transcripts. -700 Human genes, 490 mouse genes. 1400 possible human sequences but 733 cloned or detected. Only ~120 tested. Esquela-Kerscher, A. , and Slack, F. J. (2006). Nature Reviews 6, p. 263
Biogenesis -Primary-mi. RNA is transcribed in the nucleus, and is usually several kilobases long; posses 5’ cap and a poly -A tail. -Cleaved in the nucleus by Drocha enzyme to 70 nt hairpin transcript (premi. RNA). -Transported to the cytoplasm by Exportin 5 through nuclear pores. -Cleaved by Dicer enzyme (RNase III enzyme) into 19 -22 nt ds-transcripts. -700 Human genes, 490 mouse genes. 1400 possible human sequences but 733 cloned or detected. Only ~120 tested. Esquela-Kerscher, A. , and Slack, F. J. (2006). Nature Reviews 6, p. 263
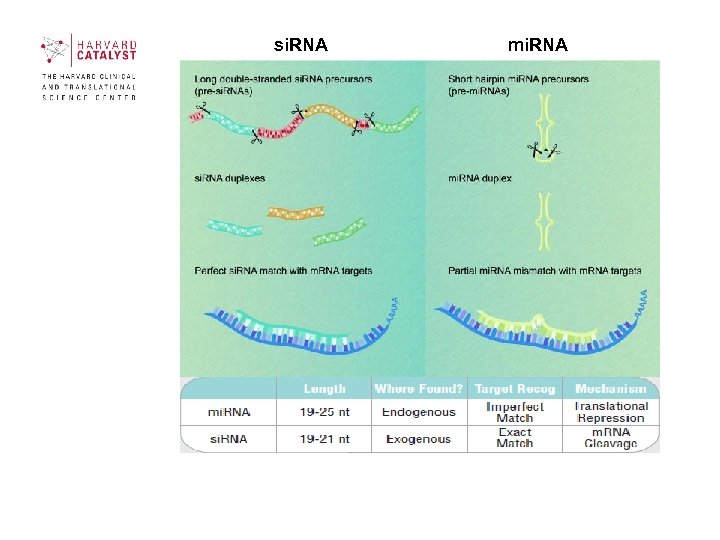 si. RNA mi. RNA
si. RNA mi. RNA
 Why analyze mi. RNA patterns? 12
Why analyze mi. RNA patterns? 12
 Why Analyze mi. RNA Expression Patterns? • Complete role played by each sequence not yet determined • Functional analysis tools not yet widely available Knockdown, Over-Expression, Reporter Assays • Correlate mi. RNA expression profiles with: Biological Phenotypes, Protein & Gene Expression Levels • Discover mi. RNA sequences regulating genes involved in the biological process of interest
Why Analyze mi. RNA Expression Patterns? • Complete role played by each sequence not yet determined • Functional analysis tools not yet widely available Knockdown, Over-Expression, Reporter Assays • Correlate mi. RNA expression profiles with: Biological Phenotypes, Protein & Gene Expression Levels • Discover mi. RNA sequences regulating genes involved in the biological process of interest
 Relevance of mi. RNA to Human Biology • Cancer. mi. RNAs have been found to be downregulated in a number of tumors , and in some cases the reintroduction of these mi. RNAs has been shown to impair the viability of cancer cells. The value of mi. RNA profiles in tumor diagnostics is well established. For instance, strong up and down regulations of 16 mi. RNAs have been shown in primary breast tumors, and these markers may aid in the development of drug-resistance and treatment-selection tests. Underlining the important role mi. RNA plays in oncology is the formation of several new companies which seek to expand development of mi. RNA-based therapeutics. • Age-Related Diseases. Evidence is accumulating that many age-related diseases are associated with a decreased control of cell signaling that occurs in mid-life. The mi. RNA control of such systems as the cell cycle, DNA repair, oxidative stress responses and apoptosis, has been shown to become abnormally expressed in mid-life. It is highly likely that continued research will reveal important associations with the aging process, and may lead to therapeutics that can improve the quality of life. • Heart Disease. Two heart-specific mi. RNAs were deleted in mouse models resulting in abnormal heart development in a large proportion of the offspring. While these lethal effects were expected, other studies show a more subtle role for mi. RNA in the heart. When mi. R-208 was eliminated, the mice appeared normal. Defects were revealed only when their hearts were stressed. These results show that comprehensive mi. RNA studies may be valuable in the diagnosis of heart disease. • Neurological Diseases. Numerous reports have demonstrated the role of mi. RNAs in neural development. Evidence for a role in Parkinsons disease comes from animal model studies published last year, showing that loss of mi. RNAs may be involved in the development and progession of the disease. In cell culture experiments, transfer of small RNA fragments partially preserved mi. RNA deficient nerve cells. While these results and others point to an important role for mi. RNA in neurodegenerative disorders, much more work is needed to delineate the exact role of mi. RNAs in this important area. • Immune Function Disorders. Recent mi. RNA deletion studies have revealed a central role in the regulation of the immune response. The deletion of mi. RNA-155 impaired T and B cell differentiation in germinal centers, and greatly decreased antibody and cytokine production. Two additional studies deleting mi. RNA-181 and 223 were found to control T cell response and granulocyte production, respectively. As more roles for mi. RNAs in the immune response are found, the list of immune function disorders with a mi. RNA component is certain to expand also. 14
Relevance of mi. RNA to Human Biology • Cancer. mi. RNAs have been found to be downregulated in a number of tumors , and in some cases the reintroduction of these mi. RNAs has been shown to impair the viability of cancer cells. The value of mi. RNA profiles in tumor diagnostics is well established. For instance, strong up and down regulations of 16 mi. RNAs have been shown in primary breast tumors, and these markers may aid in the development of drug-resistance and treatment-selection tests. Underlining the important role mi. RNA plays in oncology is the formation of several new companies which seek to expand development of mi. RNA-based therapeutics. • Age-Related Diseases. Evidence is accumulating that many age-related diseases are associated with a decreased control of cell signaling that occurs in mid-life. The mi. RNA control of such systems as the cell cycle, DNA repair, oxidative stress responses and apoptosis, has been shown to become abnormally expressed in mid-life. It is highly likely that continued research will reveal important associations with the aging process, and may lead to therapeutics that can improve the quality of life. • Heart Disease. Two heart-specific mi. RNAs were deleted in mouse models resulting in abnormal heart development in a large proportion of the offspring. While these lethal effects were expected, other studies show a more subtle role for mi. RNA in the heart. When mi. R-208 was eliminated, the mice appeared normal. Defects were revealed only when their hearts were stressed. These results show that comprehensive mi. RNA studies may be valuable in the diagnosis of heart disease. • Neurological Diseases. Numerous reports have demonstrated the role of mi. RNAs in neural development. Evidence for a role in Parkinsons disease comes from animal model studies published last year, showing that loss of mi. RNAs may be involved in the development and progession of the disease. In cell culture experiments, transfer of small RNA fragments partially preserved mi. RNA deficient nerve cells. While these results and others point to an important role for mi. RNA in neurodegenerative disorders, much more work is needed to delineate the exact role of mi. RNAs in this important area. • Immune Function Disorders. Recent mi. RNA deletion studies have revealed a central role in the regulation of the immune response. The deletion of mi. RNA-155 impaired T and B cell differentiation in germinal centers, and greatly decreased antibody and cytokine production. Two additional studies deleting mi. RNA-181 and 223 were found to control T cell response and granulocyte production, respectively. As more roles for mi. RNAs in the immune response are found, the list of immune function disorders with a mi. RNA component is certain to expand also. 14
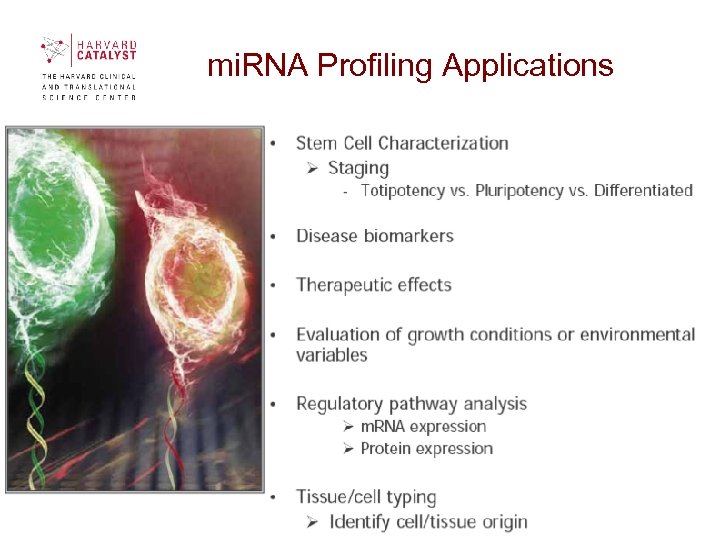 mi. RNA Profiling Applications
mi. RNA Profiling Applications
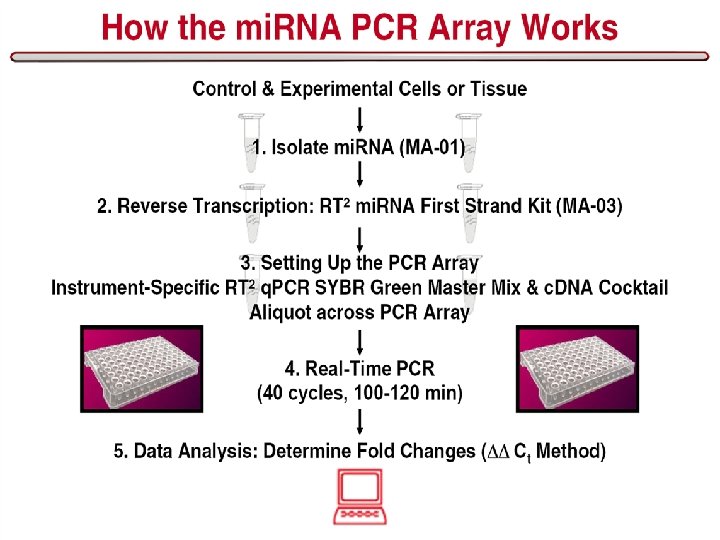
 Work flow in mi. RNA Experiments 16
Work flow in mi. RNA Experiments 16
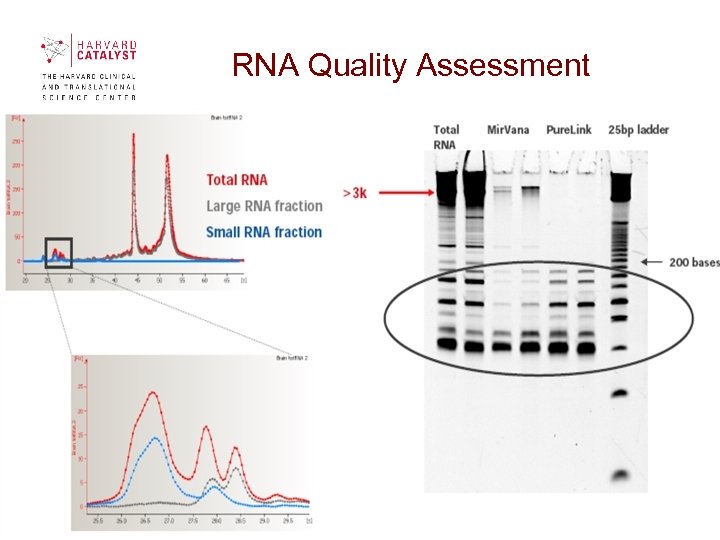 RNA Quality Assessment
RNA Quality Assessment
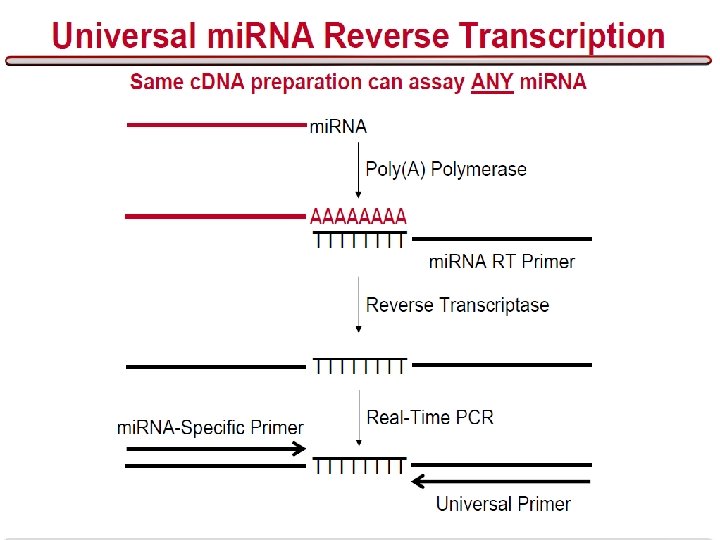
 Problems • How do we find micro. RNA genes? • Given a micro. RNA gene, how do we find its targets?
Problems • How do we find micro. RNA genes? • Given a micro. RNA gene, how do we find its targets?
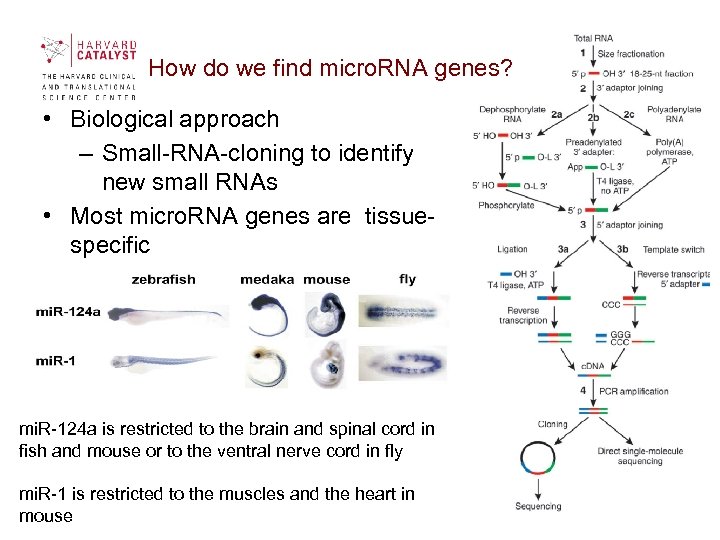 How do we find micro. RNA genes? • Biological approach – Small-RNA-cloning to identify new small RNAs • Most micro. RNA genes are tissuespecific mi. R-124 a is restricted to the brain and spinal cord in fish and mouse or to the ventral nerve cord in fly mi. R-1 is restricted to the muscles and the heart in mouse
How do we find micro. RNA genes? • Biological approach – Small-RNA-cloning to identify new small RNAs • Most micro. RNA genes are tissuespecific mi. R-124 a is restricted to the brain and spinal cord in fish and mouse or to the ventral nerve cord in fly mi. R-1 is restricted to the muscles and the heart in mouse
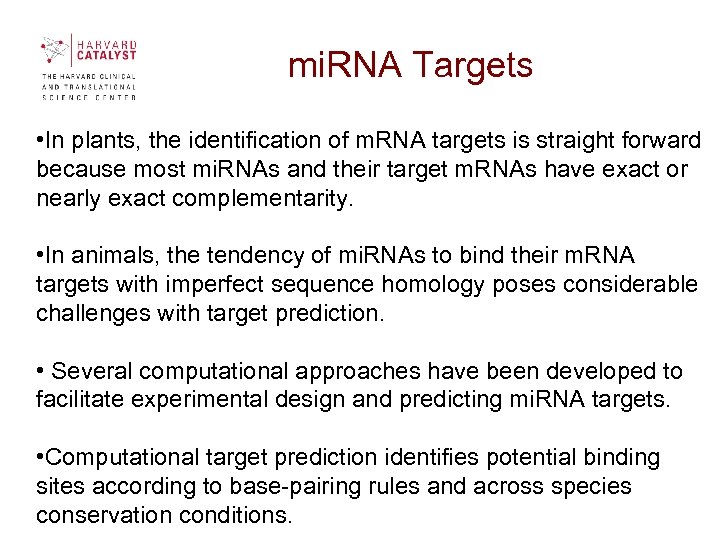 mi. RNA Targets • In plants, the identification of m. RNA targets is straight forward because most mi. RNAs and their target m. RNAs have exact or nearly exact complementarity. • In animals, the tendency of mi. RNAs to bind their m. RNA targets with imperfect sequence homology poses considerable challenges with target prediction. • Several computational approaches have been developed to facilitate experimental design and predicting mi. RNA targets. • Computational target prediction identifies potential binding sites according to base-pairing rules and across species conservation conditions.
mi. RNA Targets • In plants, the identification of m. RNA targets is straight forward because most mi. RNAs and their target m. RNAs have exact or nearly exact complementarity. • In animals, the tendency of mi. RNAs to bind their m. RNA targets with imperfect sequence homology poses considerable challenges with target prediction. • Several computational approaches have been developed to facilitate experimental design and predicting mi. RNA targets. • Computational target prediction identifies potential binding sites according to base-pairing rules and across species conservation conditions.
 Micro. RNA Targets Prediction and Analysis
Micro. RNA Targets Prediction and Analysis
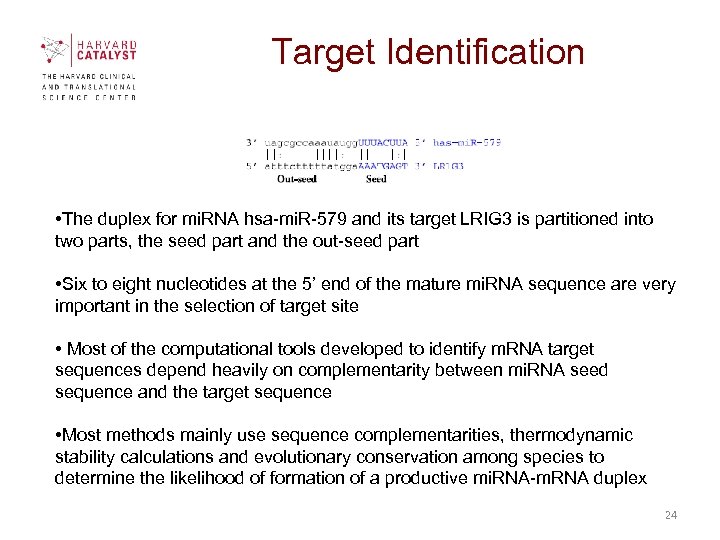 Target Identification • The duplex for mi. RNA hsa-mi. R-579 and its target LRIG 3 is partitioned into two parts, the seed part and the out-seed part • Six to eight nucleotides at the 5’ end of the mature mi. RNA sequence are very important in the selection of target site • Most of the computational tools developed to identify m. RNA target sequences depend heavily on complementarity between mi. RNA seed sequence and the target sequence • Most methods mainly use sequence complementarities, thermodynamic stability calculations and evolutionary conservation among species to determine the likelihood of formation of a productive mi. RNA-m. RNA duplex 24
Target Identification • The duplex for mi. RNA hsa-mi. R-579 and its target LRIG 3 is partitioned into two parts, the seed part and the out-seed part • Six to eight nucleotides at the 5’ end of the mature mi. RNA sequence are very important in the selection of target site • Most of the computational tools developed to identify m. RNA target sequences depend heavily on complementarity between mi. RNA seed sequence and the target sequence • Most methods mainly use sequence complementarities, thermodynamic stability calculations and evolutionary conservation among species to determine the likelihood of formation of a productive mi. RNA-m. RNA duplex 24
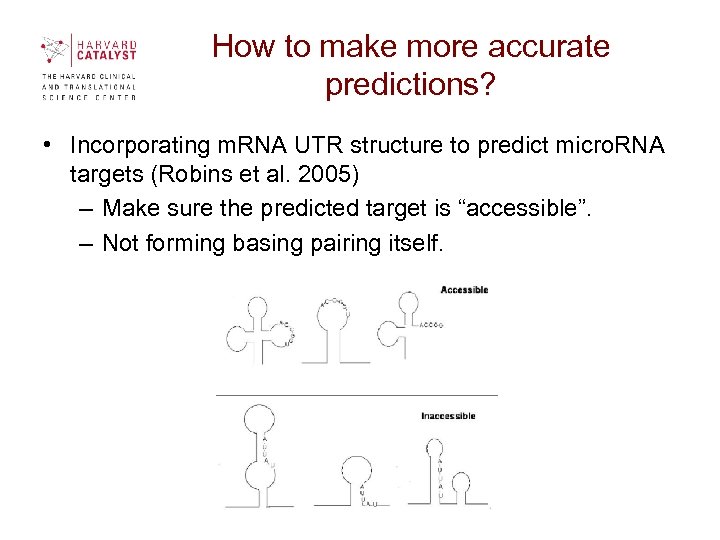 How to make more accurate predictions? • Incorporating m. RNA UTR structure to predict micro. RNA targets (Robins et al. 2005) – Make sure the predicted target is “accessible”. – Not forming basing pairing itself.
How to make more accurate predictions? • Incorporating m. RNA UTR structure to predict micro. RNA targets (Robins et al. 2005) – Make sure the predicted target is “accessible”. – Not forming basing pairing itself.
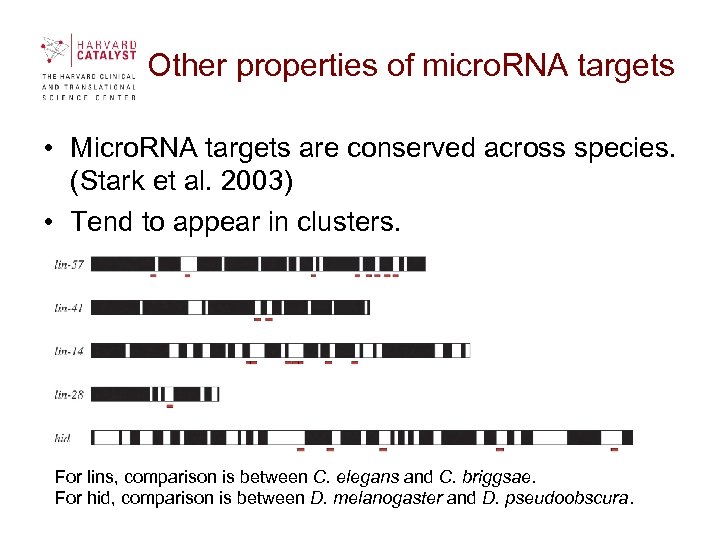 Other properties of micro. RNA targets • Micro. RNA targets are conserved across species. (Stark et al. 2003) • Tend to appear in clusters. For lins, comparison is between C. elegans and C. briggsae. For hid, comparison is between D. melanogaster and D. pseudoobscura.
Other properties of micro. RNA targets • Micro. RNA targets are conserved across species. (Stark et al. 2003) • Tend to appear in clusters. For lins, comparison is between C. elegans and C. briggsae. For hid, comparison is between D. melanogaster and D. pseudoobscura.
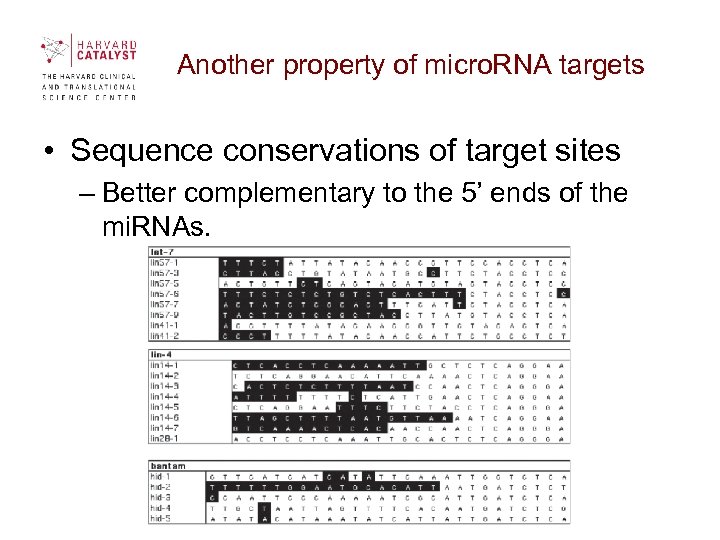 Another property of micro. RNA targets • Sequence conservations of target sites – Better complementary to the 5’ ends of the mi. RNAs.
Another property of micro. RNA targets • Sequence conservations of target sites – Better complementary to the 5’ ends of the mi. RNAs.
 Interesting properties of micro. RNA targets • Clusters of micro. RNA targets – Extensive co-occurrence of the sites for different micro. RNAs in target 3’ UTRs.
Interesting properties of micro. RNA targets • Clusters of micro. RNA targets – Extensive co-occurrence of the sites for different micro. RNAs in target 3’ UTRs.
 Micro. RNA Targets Prediction Databases
Micro. RNA Targets Prediction Databases
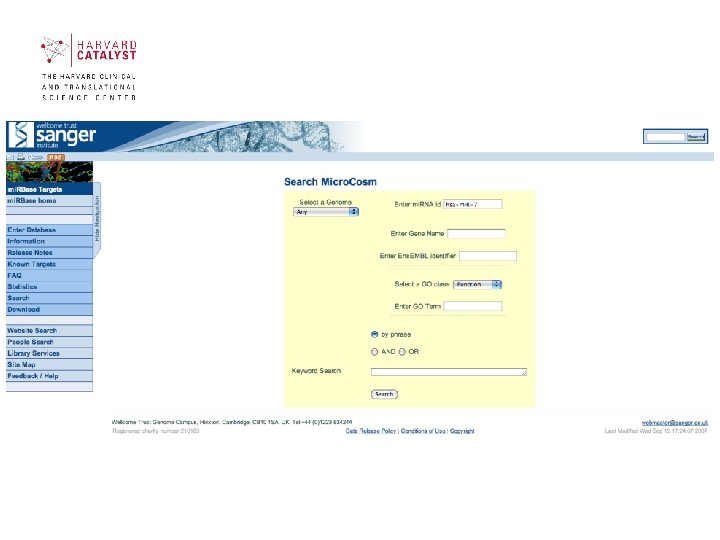
 Scientific Use Case Scenarios • What are the target genes of a given mi. RNA? • What are the mi. RNAs targeting a given gene and what are their binding sites? • What is the tissue expression profile of a given mi. RNA (or set of mi. RNAs)? • What is the mi. RNA expression profile in a given tissue (or set of tissues)? 30
Scientific Use Case Scenarios • What are the target genes of a given mi. RNA? • What are the mi. RNAs targeting a given gene and what are their binding sites? • What is the tissue expression profile of a given mi. RNA (or set of mi. RNAs)? • What is the mi. RNA expression profile in a given tissue (or set of tissues)? 30
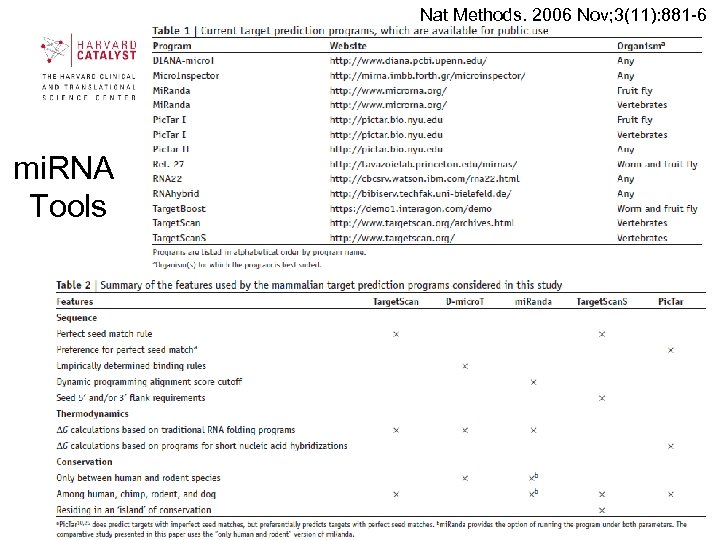 Nat Methods. 2006 Nov; 3(11): 881 -6 mi. RNA Tools
Nat Methods. 2006 Nov; 3(11): 881 -6 mi. RNA Tools
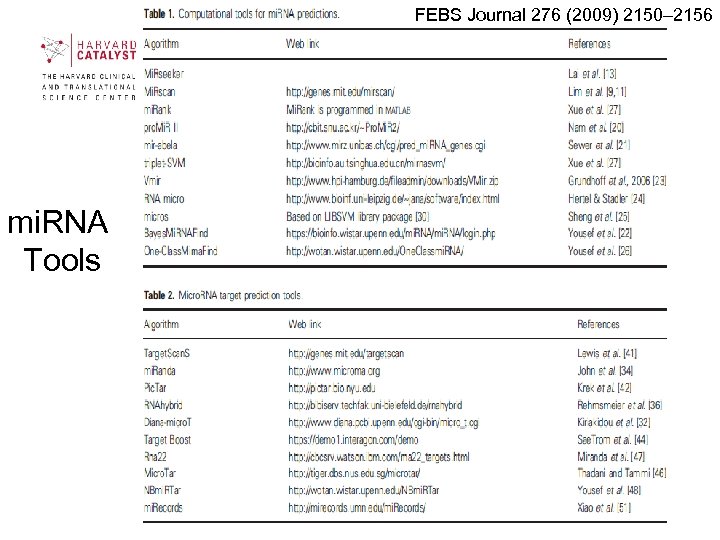 FEBS Journal 276 (2009) 2150– 2156 mi. RNA Tools
FEBS Journal 276 (2009) 2150– 2156 mi. RNA Tools
 Target prediction tools covered in today’s workshop • Targetscan • mi. RBase • Argonaute
Target prediction tools covered in today’s workshop • Targetscan • mi. RBase • Argonaute
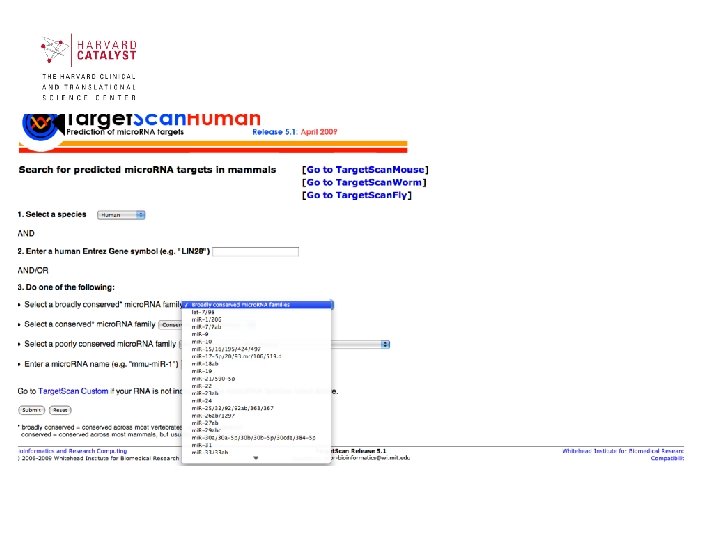
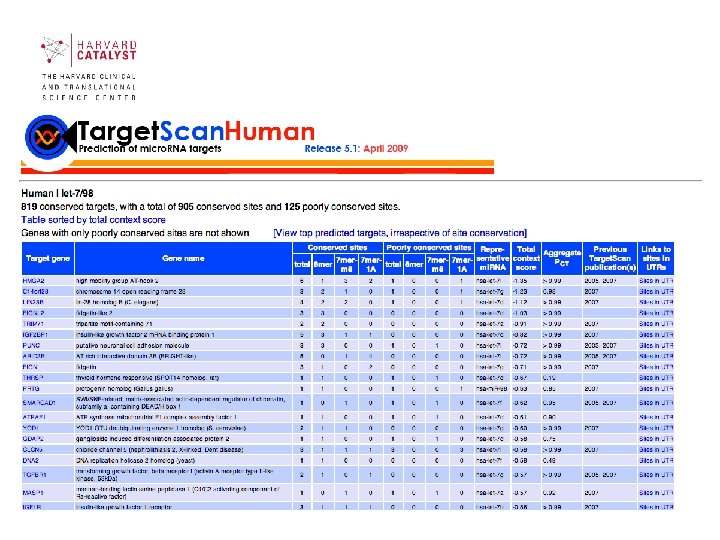
 Target. Scan (Lewis et al. , Cell 2003) • Target. Scan predicts biological targets of mi. RNAs by searching for the presence of conserved 8 mer and 7 mer sites that match the seed region of each mi. RNA. As an option, nonconserved sites are also predicted. • Also identified are sites with mismatches in the seed region that are compensated by conserved 3' pairing. • In mammals, predictions are ranked based on the predicted efficacy of targeting as calculated using the context scores of the sites. Target. Scan. Human considers matches to annotated human UTRs and their orthologs, as defined by UCSC wholegenome alignments. http: //www. targetscan. org/
Target. Scan (Lewis et al. , Cell 2003) • Target. Scan predicts biological targets of mi. RNAs by searching for the presence of conserved 8 mer and 7 mer sites that match the seed region of each mi. RNA. As an option, nonconserved sites are also predicted. • Also identified are sites with mismatches in the seed region that are compensated by conserved 3' pairing. • In mammals, predictions are ranked based on the predicted efficacy of targeting as calculated using the context scores of the sites. Target. Scan. Human considers matches to annotated human UTRs and their orthologs, as defined by UCSC wholegenome alignments. http: //www. targetscan. org/
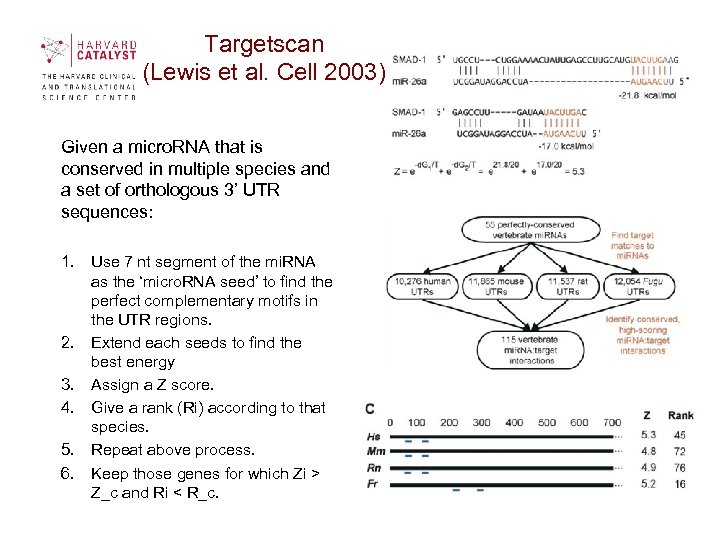 Targetscan (Lewis et al. Cell 2003) Given a micro. RNA that is conserved in multiple species and a set of orthologous 3’ UTR sequences: 1. Use 7 nt segment of the mi. RNA as the ‘micro. RNA seed’ to find the perfect complementary motifs in the UTR regions. 2. Extend each seeds to find the best energy 3. Assign a Z score. 4. Give a rank (Ri) according to that species. 5. Repeat above process. 6. Keep those genes for which Zi > Z_c and Ri < R_c.
Targetscan (Lewis et al. Cell 2003) Given a micro. RNA that is conserved in multiple species and a set of orthologous 3’ UTR sequences: 1. Use 7 nt segment of the mi. RNA as the ‘micro. RNA seed’ to find the perfect complementary motifs in the UTR regions. 2. Extend each seeds to find the best energy 3. Assign a Z score. 4. Give a rank (Ri) according to that species. 5. Repeat above process. 6. Keep those genes for which Zi > Z_c and Ri < R_c.
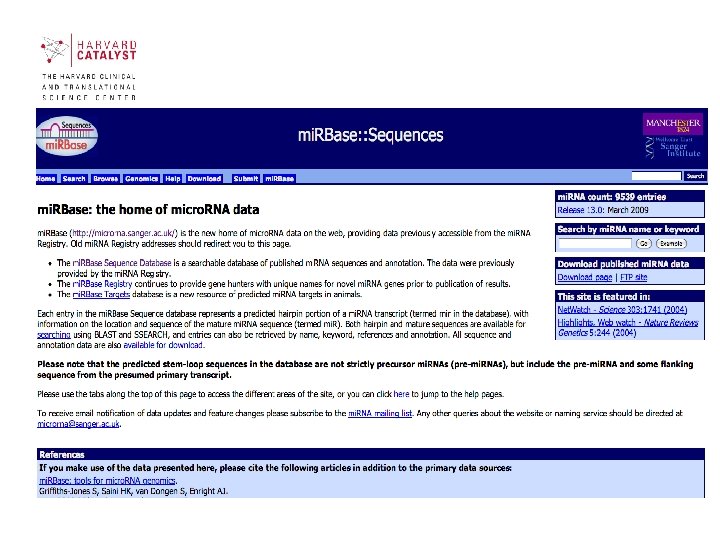
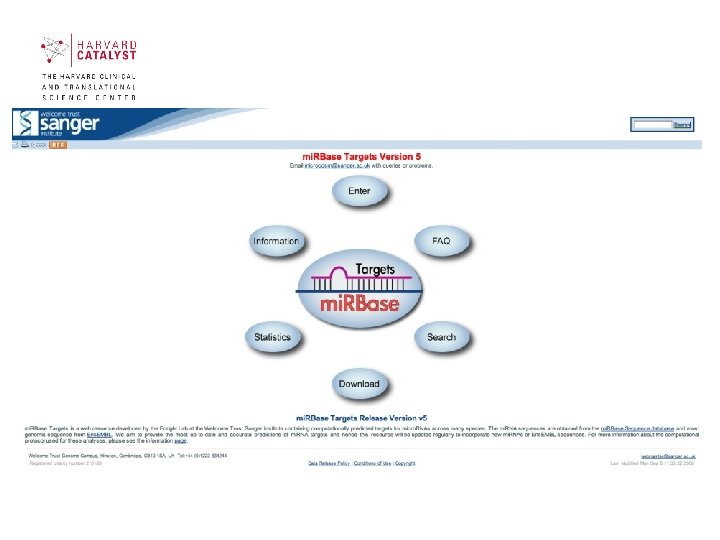
 mi. RBase • Aims to provide integrated interfaces to comprehensive mi. RNA sequence data, annotation and predicted gene targets. • mi. RBase takes over functionality from the mi. RNA Registry and fulfils three main roles: mi. RBase Sequences, mi. RBase Targets and mi. RBase Registry. http: //microrna. sanger. ac. uk/ 39
mi. RBase • Aims to provide integrated interfaces to comprehensive mi. RNA sequence data, annotation and predicted gene targets. • mi. RBase takes over functionality from the mi. RNA Registry and fulfils three main roles: mi. RBase Sequences, mi. RBase Targets and mi. RBase Registry. http: //microrna. sanger. ac. uk/ 39
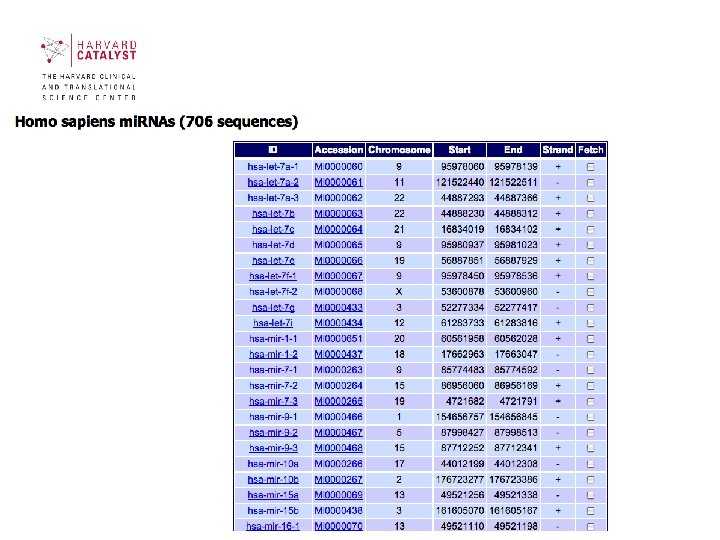
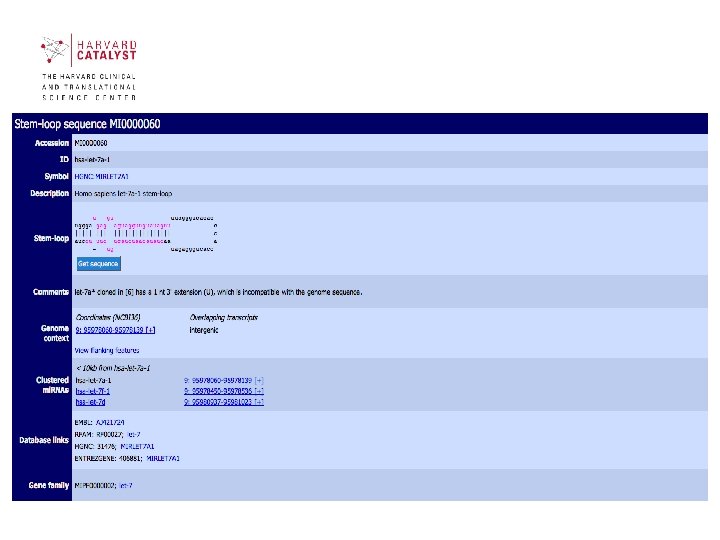
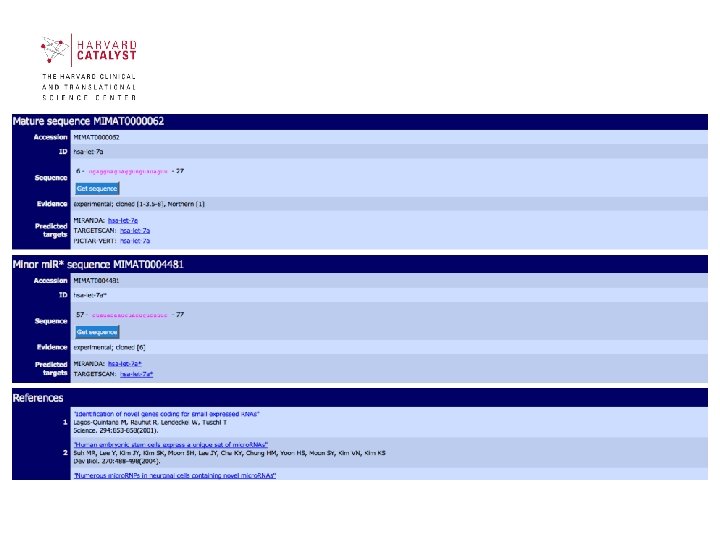
 Sections of mi. RBase contains 3 main sections: • mi. RBase Sequences contains all published mi. RNA sequences, genomic locations and associated annotation. • mi. RBase Targets is a newly developed database of predicted mi. RNA target genes. • mi. RBase Registry provides a confidential service assigning official names for novel mi. RNA genes prior to publication of their discovery 40
Sections of mi. RBase contains 3 main sections: • mi. RBase Sequences contains all published mi. RNA sequences, genomic locations and associated annotation. • mi. RBase Targets is a newly developed database of predicted mi. RNA target genes. • mi. RBase Registry provides a confidential service assigning official names for novel mi. RNA genes prior to publication of their discovery 40


 Argonaute (mi. RWalk) Nucleic Acids Res. 2006 Jan 1; 34(Database issue): D 115 -8 • Curated database • Argonaute collects latest information from both literature and other databases. In contrast to databases on mi. RNAs like mi. RBase: : Sequences, NONCODE or RNAdb, Argonaute hosts additional information on the origin of an mi. RNA, i. e. in which host gene it is encoded, its expression in different tissues and its known or proposed function, its potential target genes including Gene Ontology annotation, as well as mi. RNA families and proteins known to be involved in mi. RNA processing. • Additionally, target genes are linked to an information retrieval system that provides comprehensive information from sequence databases and a simultaneous search of MEDLINE with all synonyms of a given gene. The web interface allows the user to get information for a single or multiple mi. RNAs, either selected or uploaded through a text file. Argonaute currently has information on mi. RNAs from human, mouse and rat. http: //www. ma. uni-heidelberg. de/apps/zmf/argonaute/interface 49
Argonaute (mi. RWalk) Nucleic Acids Res. 2006 Jan 1; 34(Database issue): D 115 -8 • Curated database • Argonaute collects latest information from both literature and other databases. In contrast to databases on mi. RNAs like mi. RBase: : Sequences, NONCODE or RNAdb, Argonaute hosts additional information on the origin of an mi. RNA, i. e. in which host gene it is encoded, its expression in different tissues and its known or proposed function, its potential target genes including Gene Ontology annotation, as well as mi. RNA families and proteins known to be involved in mi. RNA processing. • Additionally, target genes are linked to an information retrieval system that provides comprehensive information from sequence databases and a simultaneous search of MEDLINE with all synonyms of a given gene. The web interface allows the user to get information for a single or multiple mi. RNAs, either selected or uploaded through a text file. Argonaute currently has information on mi. RNAs from human, mouse and rat. http: //www. ma. uni-heidelberg. de/apps/zmf/argonaute/interface 49
 http: //www. ma. uni-heidelberg. de/apps/zmf/argonaute/interface
http: //www. ma. uni-heidelberg. de/apps/zmf/argonaute/interface
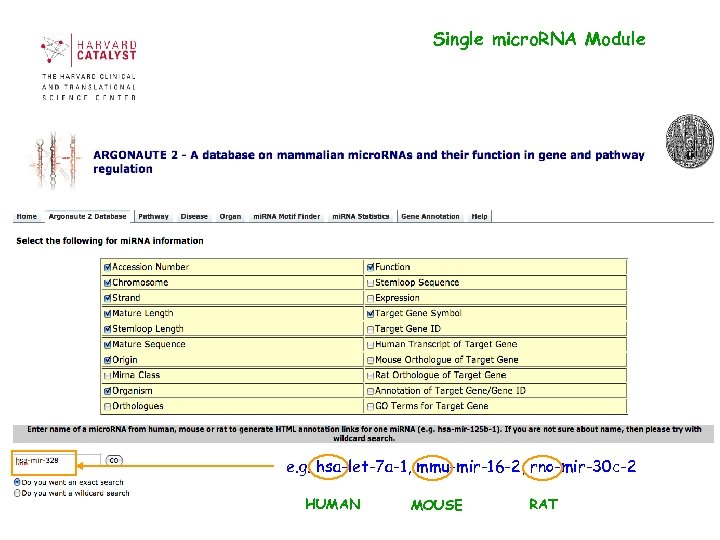 Single micro. RNA Module e. g. hsa-let-7 a-1, mmu-mir-16 -2, rno-mir-30 c-2 HUMAN MOUSE RAT
Single micro. RNA Module e. g. hsa-let-7 a-1, mmu-mir-16 -2, rno-mir-30 c-2 HUMAN MOUSE RAT
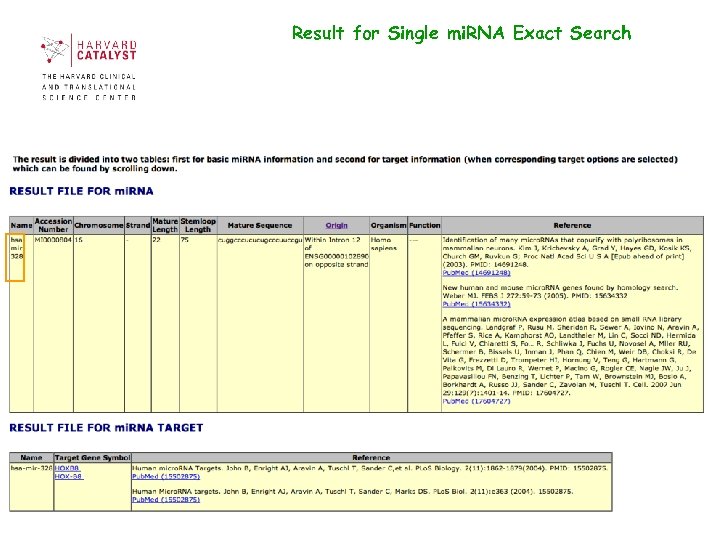 Result for Single mi. RNA Exact Search
Result for Single mi. RNA Exact Search
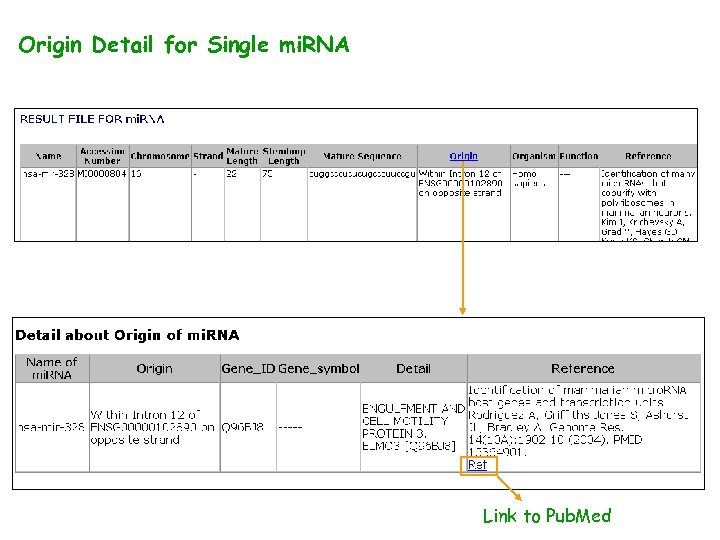 Origin Detail for Single mi. RNA Link to Pub. Med
Origin Detail for Single mi. RNA Link to Pub. Med
 Link to Pub. Med for each Reference
Link to Pub. Med for each Reference
 mi. RNA Target Validation and Considerations in RNAi Experiments 55
mi. RNA Target Validation and Considerations in RNAi Experiments 55
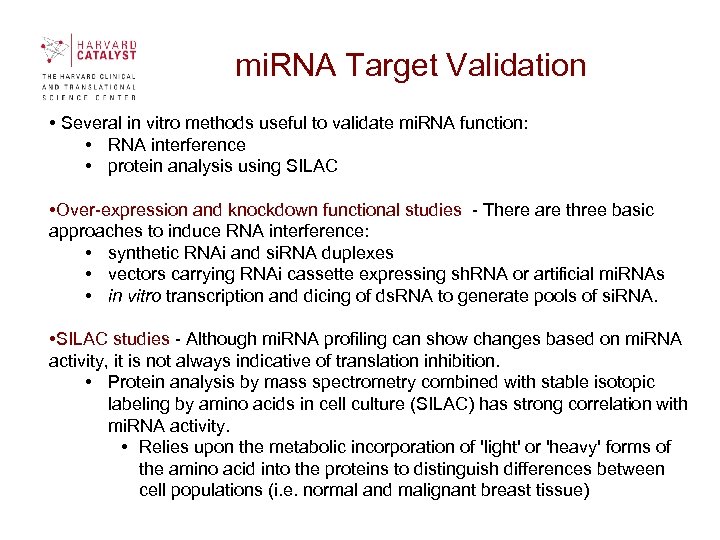 mi. RNA Target Validation • Several in vitro methods useful to validate mi. RNA function: • RNA interference • protein analysis using SILAC • Over-expression and knockdown functional studies - There are three basic approaches to induce RNA interference: • synthetic RNAi and si. RNA duplexes • vectors carrying RNAi cassette expressing sh. RNA or artificial mi. RNAs • in vitro transcription and dicing of ds. RNA to generate pools of si. RNA. • SILAC studies - Although mi. RNA profiling can show changes based on mi. RNA activity, it is not always indicative of translation inhibition. • Protein analysis by mass spectrometry combined with stable isotopic labeling by amino acids in cell culture (SILAC) has strong correlation with mi. RNA activity. • Relies upon the metabolic incorporation of 'light' or 'heavy' forms of the amino acid into the proteins to distinguish differences between cell populations (i. e. normal and malignant breast tissue)
mi. RNA Target Validation • Several in vitro methods useful to validate mi. RNA function: • RNA interference • protein analysis using SILAC • Over-expression and knockdown functional studies - There are three basic approaches to induce RNA interference: • synthetic RNAi and si. RNA duplexes • vectors carrying RNAi cassette expressing sh. RNA or artificial mi. RNAs • in vitro transcription and dicing of ds. RNA to generate pools of si. RNA. • SILAC studies - Although mi. RNA profiling can show changes based on mi. RNA activity, it is not always indicative of translation inhibition. • Protein analysis by mass spectrometry combined with stable isotopic labeling by amino acids in cell culture (SILAC) has strong correlation with mi. RNA activity. • Relies upon the metabolic incorporation of 'light' or 'heavy' forms of the amino acid into the proteins to distinguish differences between cell populations (i. e. normal and malignant breast tissue)
 Considerations for RNAi Experiments • Method of si. RNA production. If a good target sequence is unknown, or if you have had difficulty knocking down your gene using an oligonucleotide approach, use effective method such as transfecting a diced pool of si. RNA and validate data using Stealth™ RNAi , si. RNA or RNAi vectors. • Delivery method(s)/cell type to use. Options for delivering si. RNA include transfection and viral delivery. It is very important that the transfection efficiency be optimized. Even if an si. RNA were 100% effective (and most are only 70 -95% effective), if only 50% of the cells are transfected you will never see better than a 50% knockdown. • Select appropriate controls.
Considerations for RNAi Experiments • Method of si. RNA production. If a good target sequence is unknown, or if you have had difficulty knocking down your gene using an oligonucleotide approach, use effective method such as transfecting a diced pool of si. RNA and validate data using Stealth™ RNAi , si. RNA or RNAi vectors. • Delivery method(s)/cell type to use. Options for delivering si. RNA include transfection and viral delivery. It is very important that the transfection efficiency be optimized. Even if an si. RNA were 100% effective (and most are only 70 -95% effective), if only 50% of the cells are transfected you will never see better than a 50% knockdown. • Select appropriate controls.
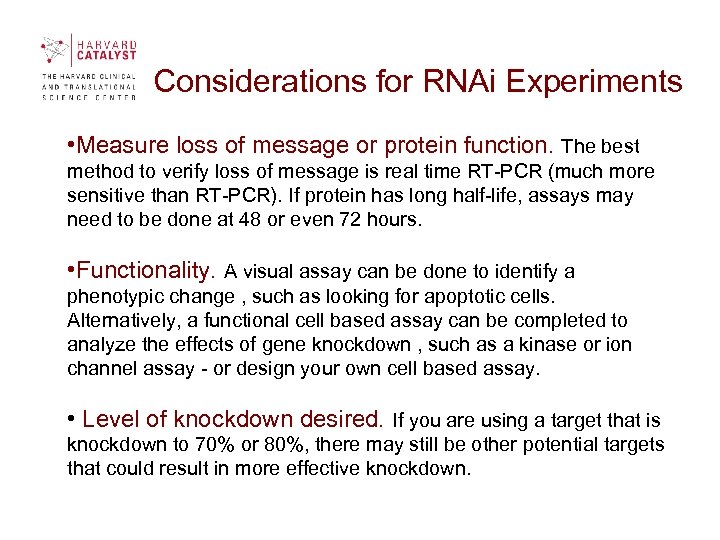 Considerations for RNAi Experiments • Measure loss of message or protein function. The best method to verify loss of message is real time RT-PCR (much more sensitive than RT-PCR). If protein has long half-life, assays may need to be done at 48 or even 72 hours. • Functionality. A visual assay can be done to identify a phenotypic change , such as looking for apoptotic cells. Alternatively, a functional cell based assay can be completed to analyze the effects of gene knockdown , such as a kinase or ion channel assay - or design your own cell based assay. • Level of knockdown desired. If you are using a target that is knockdown to 70% or 80%, there may still be other potential targets that could result in more effective knockdown.
Considerations for RNAi Experiments • Measure loss of message or protein function. The best method to verify loss of message is real time RT-PCR (much more sensitive than RT-PCR). If protein has long half-life, assays may need to be done at 48 or even 72 hours. • Functionality. A visual assay can be done to identify a phenotypic change , such as looking for apoptotic cells. Alternatively, a functional cell based assay can be completed to analyze the effects of gene knockdown , such as a kinase or ion channel assay - or design your own cell based assay. • Level of knockdown desired. If you are using a target that is knockdown to 70% or 80%, there may still be other potential targets that could result in more effective knockdown.
 mi. RNA Applications 59
mi. RNA Applications 59
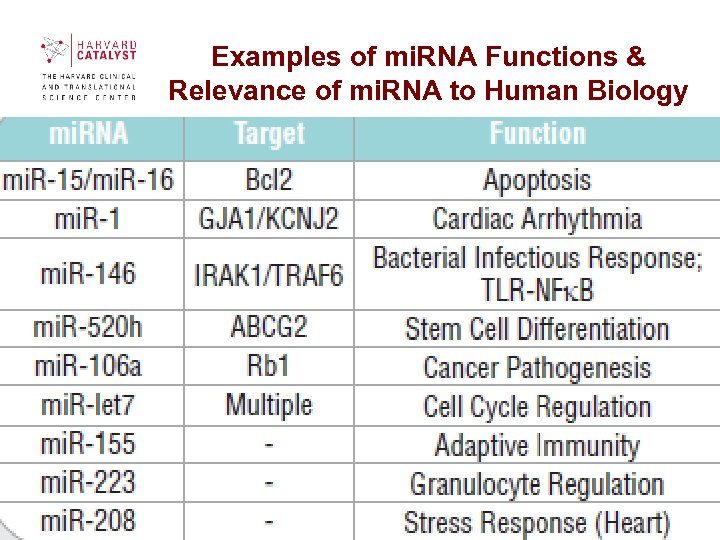 Examples of mi. RNA Functions & Relevance of mi. RNA to Human Biology 60
Examples of mi. RNA Functions & Relevance of mi. RNA to Human Biology 60
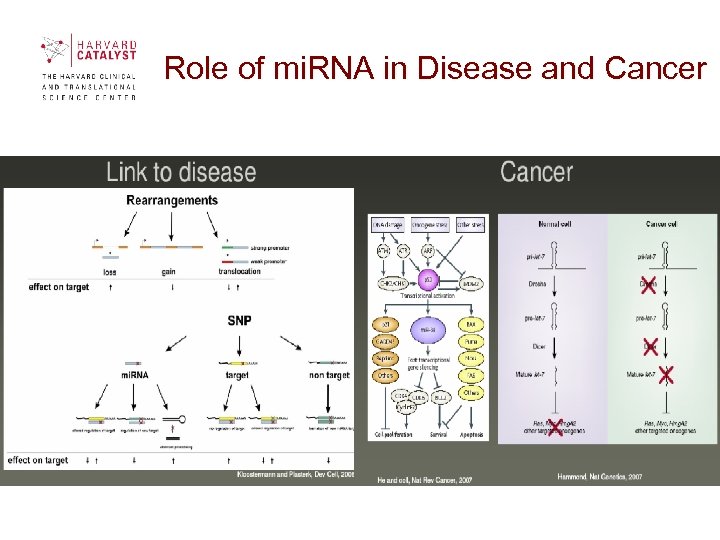 Role of mi. RNA in Disease and Cancer
Role of mi. RNA in Disease and Cancer
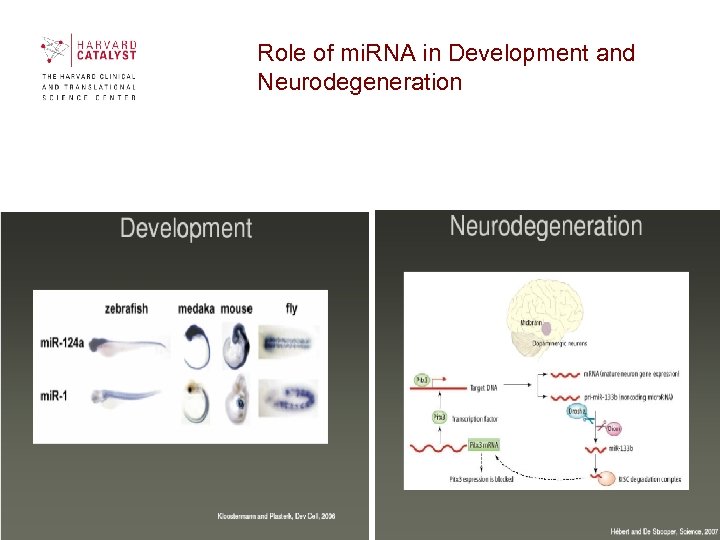 Role of mi. RNA in Development and Neurodegeneration
Role of mi. RNA in Development and Neurodegeneration
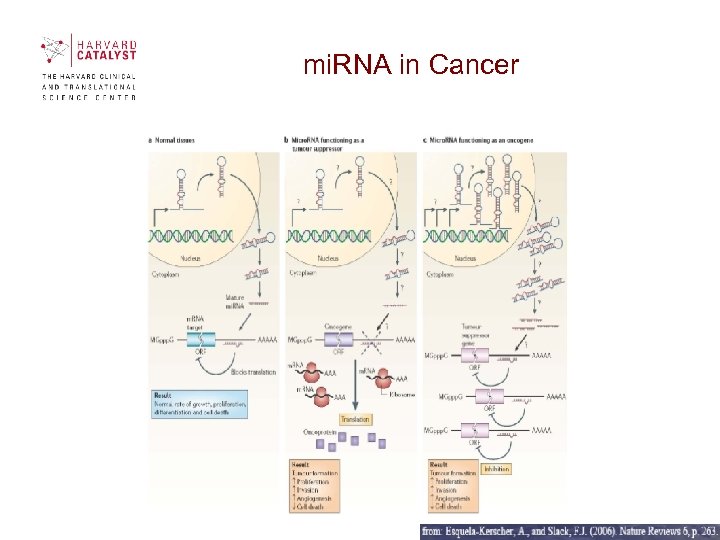 mi. RNA in Cancer
mi. RNA in Cancer
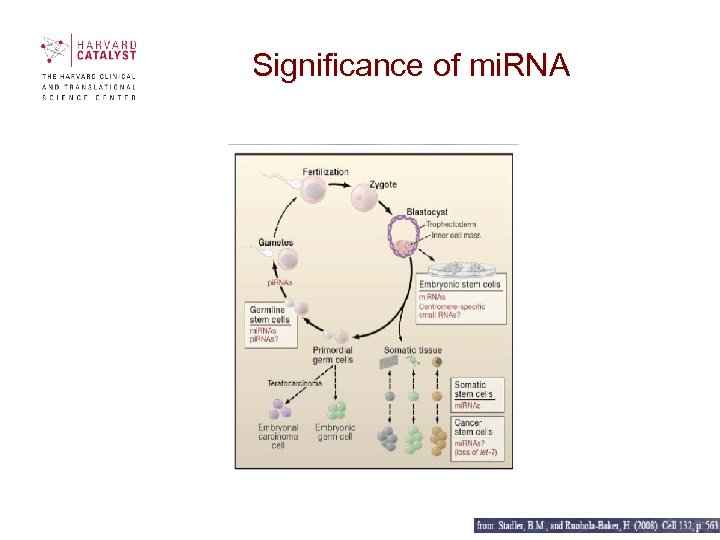 Significance of mi. RNA
Significance of mi. RNA
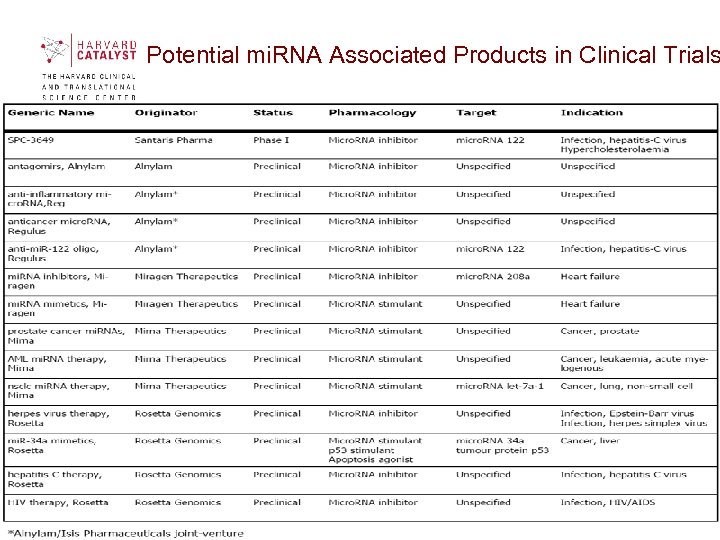 Potential mi. RNA Associated Products in Clinical Trials
Potential mi. RNA Associated Products in Clinical Trials
 Thank You
Thank You
 http: //catalyst. harvard. edu 67
http: //catalyst. harvard. edu 67


