b62a039b505cf932cd337e28a480ea9c.ppt
- Количество слайдов: 28
Integrative Genomics Viewer (IGV) Satish C Bhalla, Ph. D Department of Computer Science and Engineering Johnson C Smith University Charlotte, NC
Integrative Genomics Viewer (IGV) • Visualization tool for exploring and analyzing large genome datasets • Work with prebuilt genomes or import a genome • View a variety of data such as expression data, NGS alignments, microarray, epigenomics, RNA-Seq, genomic annotations etc. • Run it locally on your desktop or launch it from the Broad Institute website. • Must register for launching or down loading. • IGV is a Java application 3/15/2018 2
Outline I. II. Launch or download IGV user interface A. B. Main Window Tool Bar III. Ex. 1 - Load a genome and define a region of interest IV. Ex. 2 – Search for Bladder Cancer Gene - DAPK 1 V. Ex. 3 - IGV Tools VI. Ex. 4 - SAMTOOLS 3/15/2018 3

Launch or Download IGV • • • Launch from Broad Institute website: Go to http: //www. broadinstitute. org/software/igv/download The first time you sign on, you will need to register to download IGV. • Click on the first launch icon (Launch with 750 MB); 750 MB is the smallest memory available and is sufficient for most applications. Download IGV Alternatively, you may download IGV zip file and install on your desktop. If you want to install and start IGV from command line (UNIX terminal) you will need the binary distribution. 3/15/2018 4
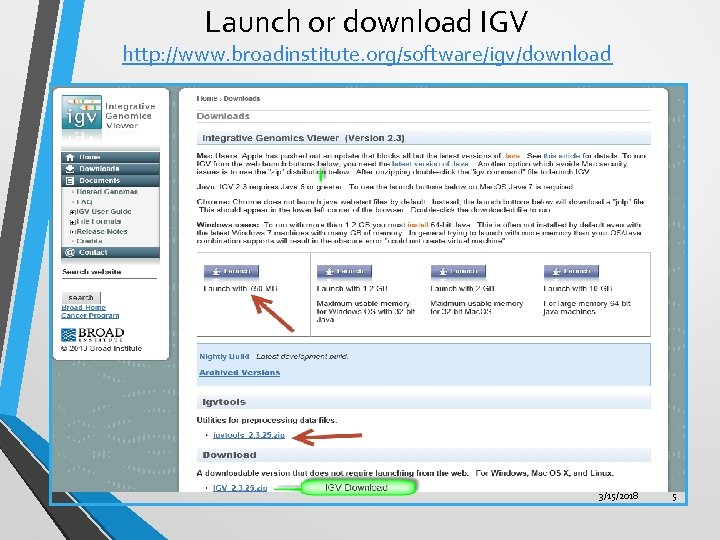
Launch or download IGV http: //www. broadinstitute. org/software/igv/download 3/15/2018 5
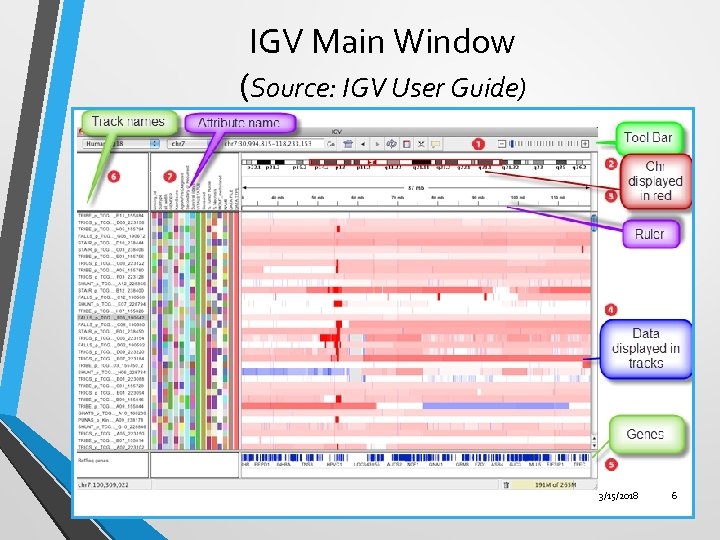
IGV Main Window (Source: IGV User Guide) 3/15/2018 6
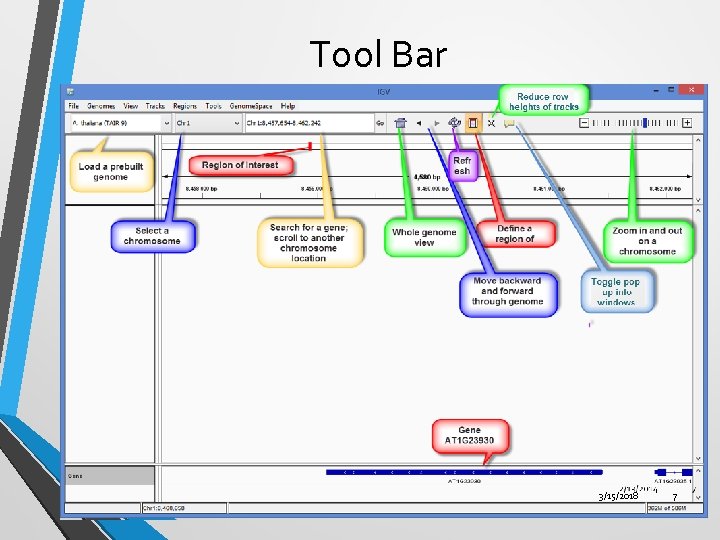
Tool Bar 3/15/2018 7
Ex. 1 - Load a genome and define a region of interest • Load a genome – Arabidopsis thaliana. • Select a chromosome – chr 1. • Define a region of interest. • Display the nucleotide sequence. 3/15/2018 8
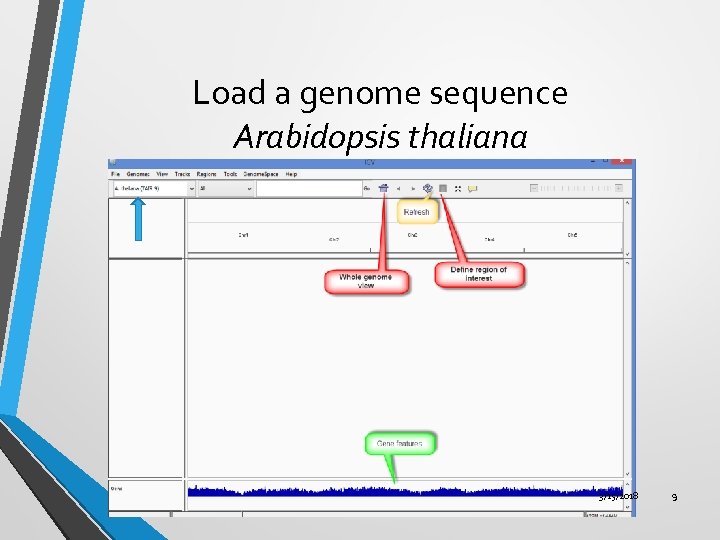
Load a genome sequence Arabidopsis thaliana 3/15/2018 9
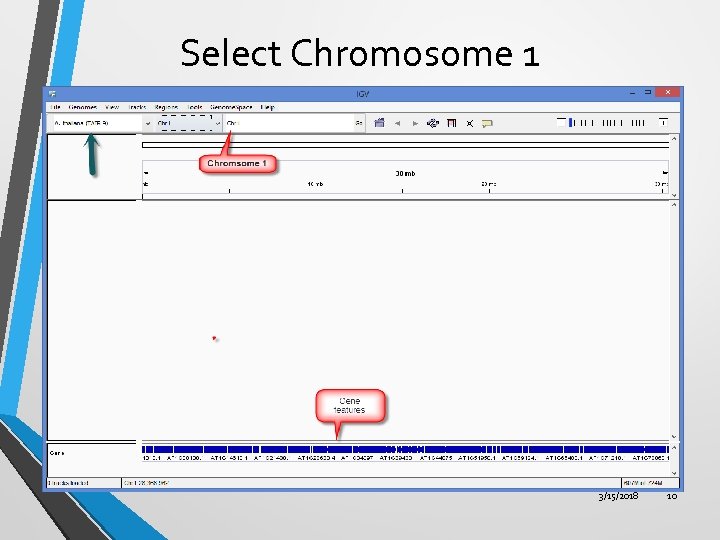
Select Chromosome 1 3/15/2018 10
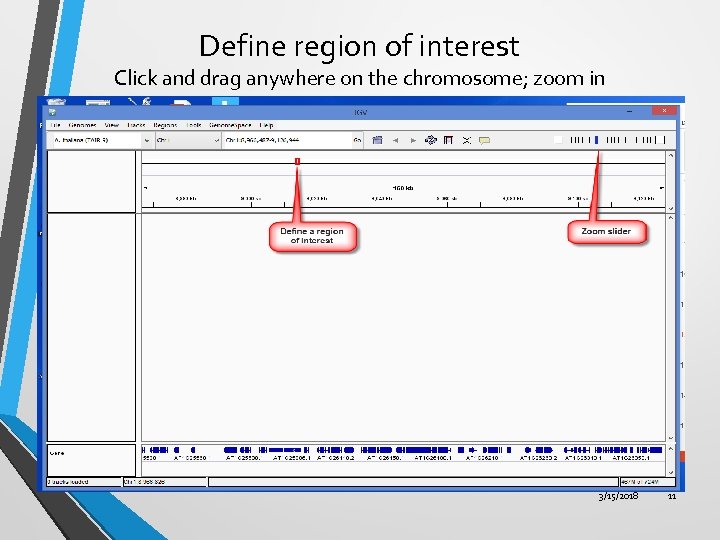
Define region of interest Click and drag anywhere on the chromosome; zoom in 3/15/2018 11
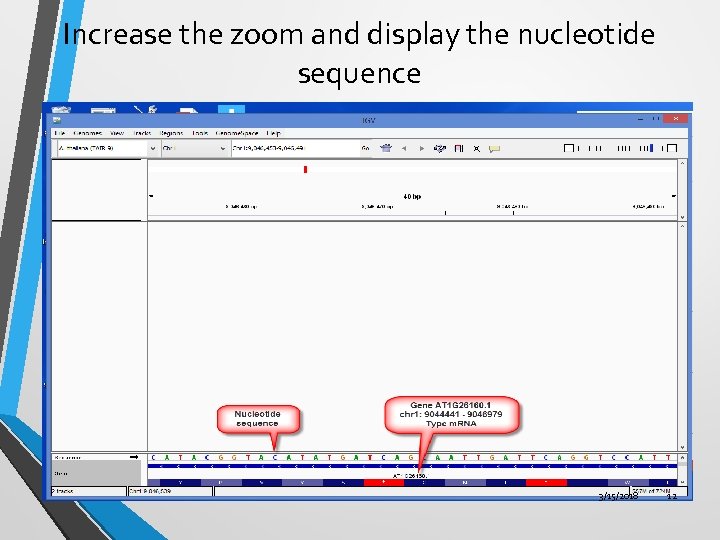
Increase the zoom and display the nucleotide sequence 3/15/2018 12
Ex. 2 – Search for Bladder Cancer Gene - DAPK 1 • Load a prebuilt genome – Human hg 19. • Search for a bladder cancer gene - DAPK 1. • View the gene sequence. • Translate the nucleotide sequence to an amino acid sequence. • Copy sequence and save it to your hard drive. 3/15/2018 13
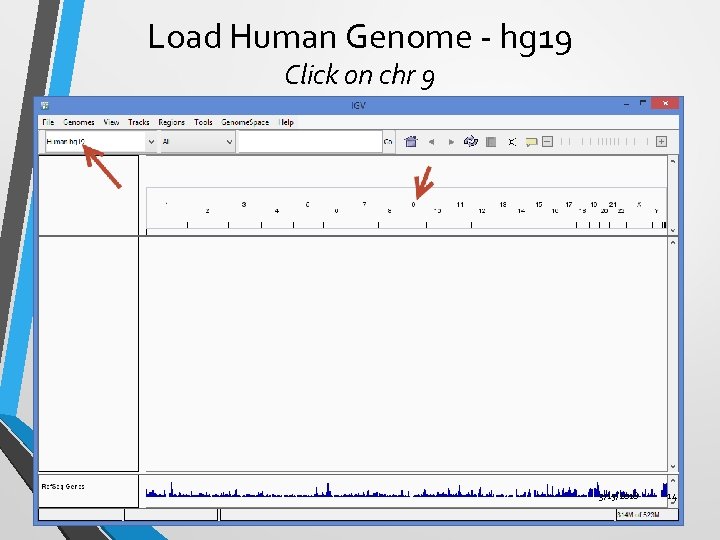
Load Human Genome - hg 19 Click on chr 9 3/15/2018 14
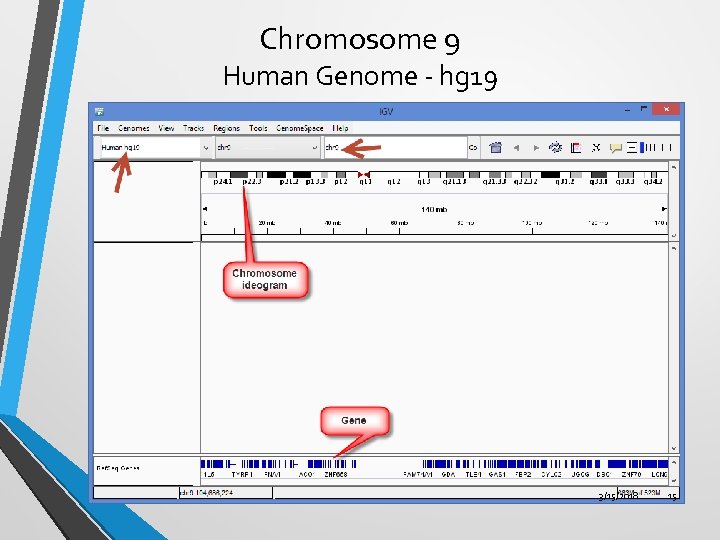
Chromosome 9 Human Genome - hg 19 3/15/2018 15
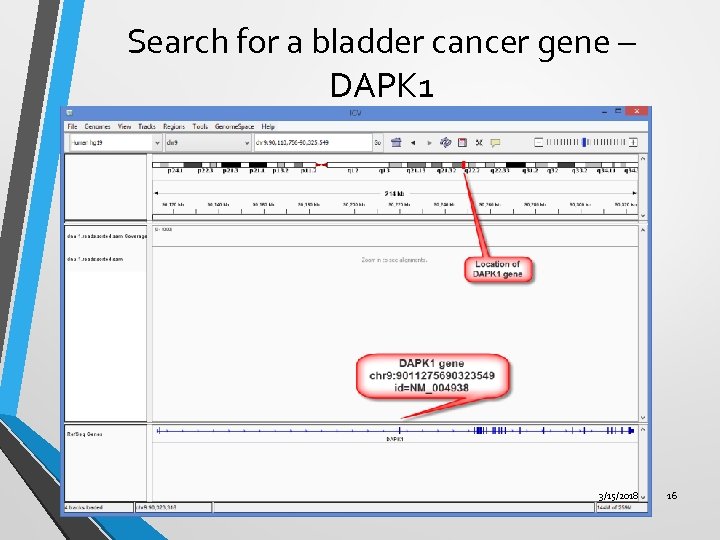
Search for a bladder cancer gene – DAPK 1 3/15/2018 16
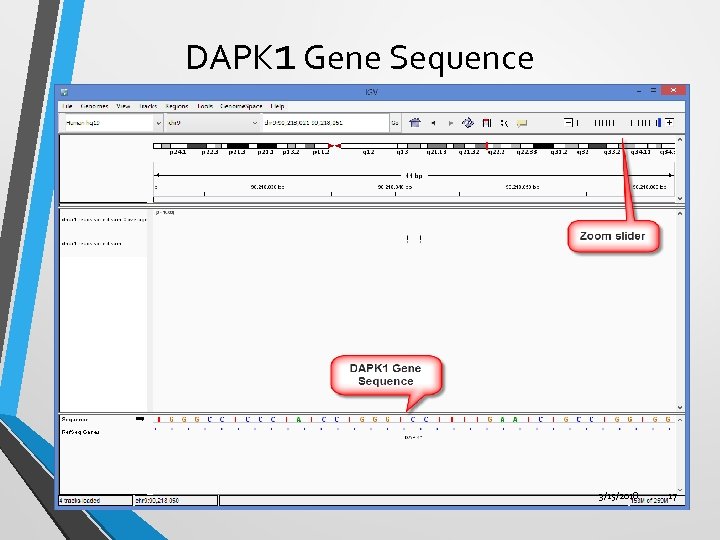
DAPK 1 Gene Sequence 3/15/2018 17
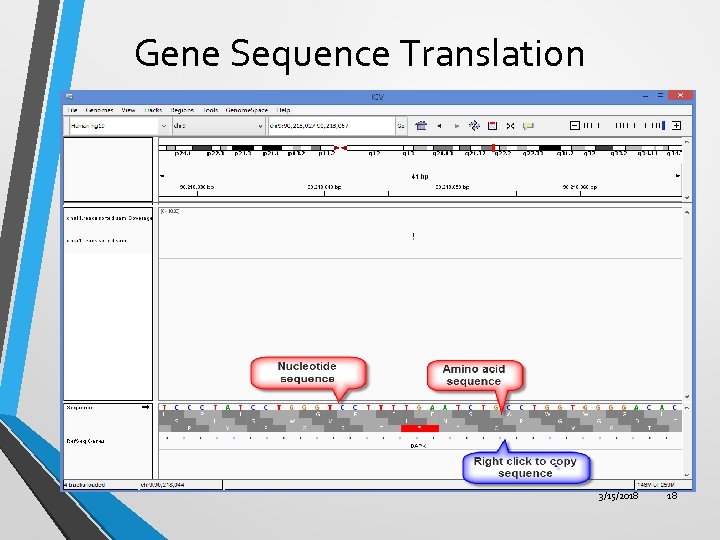
Gene Sequence Translation 3/15/2018 18

Ex. 3 IGV Tools • Use IGV Tools to • • • sort a file, (use SORT command) create an index file (use index command) create a. tdf file, (use ‘to TDF’ command) 3/15/2018 19
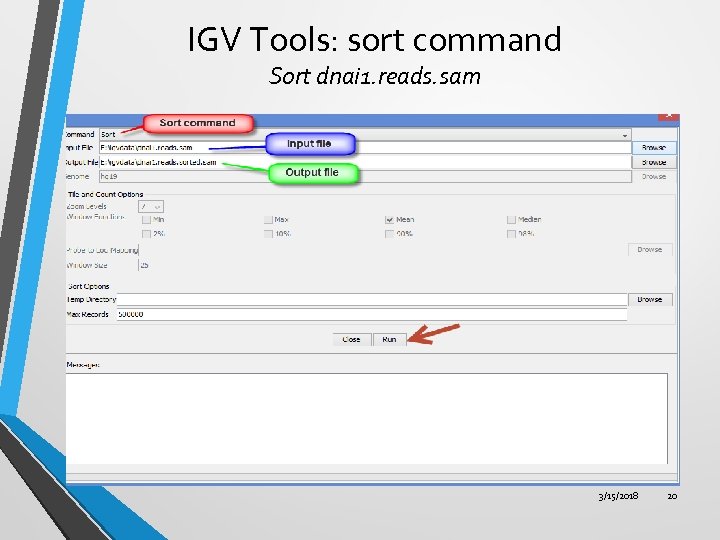
IGV Tools: sort command Sort dnai 1. reads. sam 3/15/2018 20
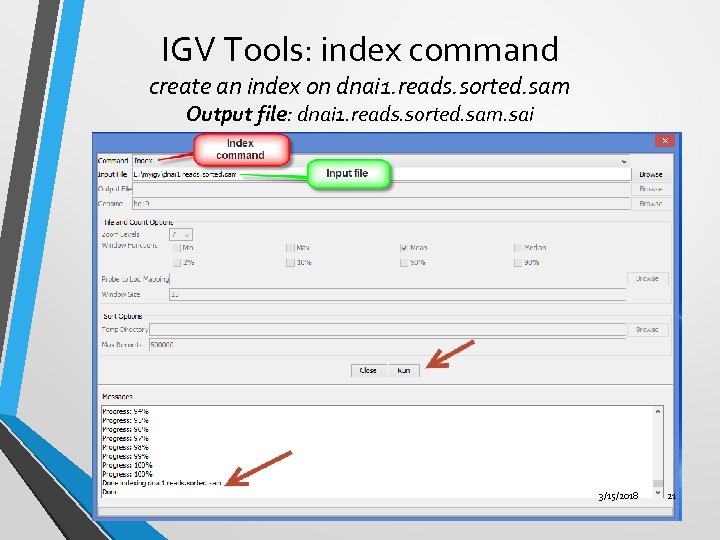
IGV Tools: index command create an index on dnai 1. reads. sorted. sam Output file: dnai 1. reads. sorted. sam. sai 3/15/2018 21
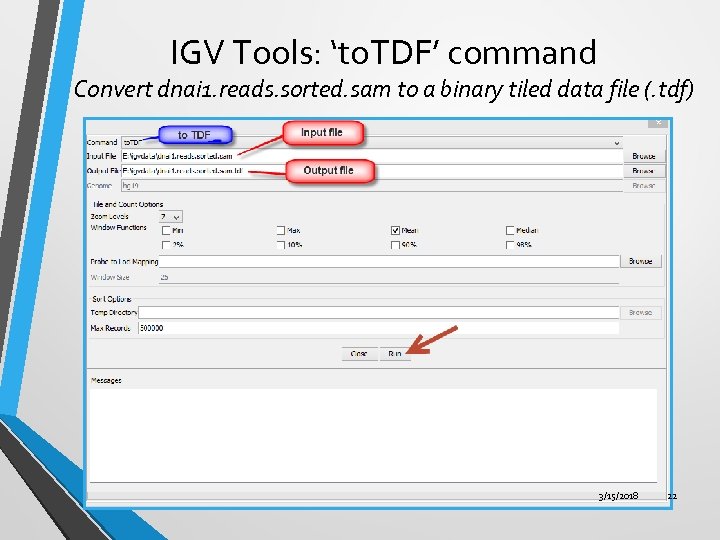
IGV Tools: ‘to. TDF’ command Convert dnai 1. reads. sorted. sam to a binary tiled data file (. tdf) 3/15/2018 22

Ex. 4 SAMTOOLS • • Install SAMTOOLS • Use SAMTOOLS to: Make SAMTOOLS executable (use make command) • • • Convert SAM file to a BAM file Sort the BAM file (use sort command) Create an index on BAM file (use index command) 3/15/2018 23
Install SAMTOOLS • SAMTOOLS is an open source program. • Download latest version of samtools-0. 1. 19. tar. bz 2 (514. 5 k. B) from sourceforge or samtools website • http: //sourceforge. net/projects/samtools/files/ • Unzip and install. • Commands are given on the command line in the UNIX environment. • Basic usage: samtools <command> [options] • If you happen to be using Windows computer, you may buy a KNOPPIX (LINUX) DVD or download Linux on a cd and make it executable. 3/15/2018 24

How to make SAMTOOLS executable? • Start SAMTOOLS and make it executable with make command • • • Point to SAMTOOLS-0. 1. 19 in the igvdata directory. Right click > open in terminal make 3/15/2018 25
Convert a SAM file to a BAM file with SAMTOOLS • Convert a SAM file to a BAM file • . /samtools view –b –S dnai 1. reads. sam > dnai 1_reads. bam • (Give the full path name of the input file such as: /media/sdb 1/igvdata/ dnai 1. reads. sam). • Note: You may convert SAM to BAM in Galaxy if you are familiar with it. 3/15/2018 26
Sort a BAM file • In order to create an index BAM file, it needs to be sorted first. • . /samtools sort –m 100000 dnai 1_reads. bam dnai 1_reads_sorted. bam • Note: Modifier –m indicates the maximum amount of memory that can be used 3/15/2018 27
Create a BAM index file • Create an index file on dnai 1_reads. sorted. bam • • . /samtools index dnai 1_reads_sorted. bam dnai 1_reads_sorted. sam. sai will be created and saved in the same directory as the input file. 3/15/2018 28
b62a039b505cf932cd337e28a480ea9c.ppt