d393712a94450dacb166e9ff0eafa309.ppt
- Количество слайдов: 21
 Integration of Mass Spectrometry with Highthroughput Protein Crystallography • Tarun Gheyi • SGX Pharmaceuticals, Inc. / NYSGXRC • April 14, 2008
Integration of Mass Spectrometry with Highthroughput Protein Crystallography • Tarun Gheyi • SGX Pharmaceuticals, Inc. / NYSGXRC • April 14, 2008
 Introduction In an effort to support various groups in the platform (protein production, crystallization and crystallography), we have made our goals simple: 1. Use as small amount of protein as we can 2. Provide as much information as possible on the “precious” protein sample 3. Reduce the analysis time to synchronize with the high-throughput activities in the platform
Introduction In an effort to support various groups in the platform (protein production, crystallization and crystallography), we have made our goals simple: 1. Use as small amount of protein as we can 2. Provide as much information as possible on the “precious” protein sample 3. Reduce the analysis time to synchronize with the high-throughput activities in the platform
 Introduction MS: established tool for analysis of bio-molecules in solution. MALDI-TOF MS (2000 ppm) + SDS-gel electrophoresis An accurate measure of sample purity ESI-MS (200 ppm) Mass accuracy At SGX, as part of the NYSGXRC activity, MALDI-MS and ESI-MS are routinely used to monitor the quality of proteins prior to initiation of crystallization trials
Introduction MS: established tool for analysis of bio-molecules in solution. MALDI-TOF MS (2000 ppm) + SDS-gel electrophoresis An accurate measure of sample purity ESI-MS (200 ppm) Mass accuracy At SGX, as part of the NYSGXRC activity, MALDI-MS and ESI-MS are routinely used to monitor the quality of proteins prior to initiation of crystallization trials
 MS Lab-Instrumentation • LC-ESI-MS (single quad; accurate mass) • MALDI-TOF (Voyager DE-RP; Linear Mode) • MALDI-TOF (Voyager DE-STR; Reflectron) • cap. LC-ESI-Q-Iontrap (MS/MS analysis)
MS Lab-Instrumentation • LC-ESI-MS (single quad; accurate mass) • MALDI-TOF (Voyager DE-RP; Linear Mode) • MALDI-TOF (Voyager DE-STR; Reflectron) • cap. LC-ESI-Q-Iontrap (MS/MS analysis)
 MS Criteria Intact MS analysis • Mass accuracy: ± 260 Da (~ 2 mutations) If d. M > 260 Da then protein characterization by tandem MS is performed to verify that it is intended protein. If yes, then DNA sequencing is done to identify all mutations • Purity: ≥ 80 % If contaminants are present with the intended protein then an extra purification step (Mono Qcolumn) is included if there is sufficient protein
MS Criteria Intact MS analysis • Mass accuracy: ± 260 Da (~ 2 mutations) If d. M > 260 Da then protein characterization by tandem MS is performed to verify that it is intended protein. If yes, then DNA sequencing is done to identify all mutations • Purity: ≥ 80 % If contaminants are present with the intended protein then an extra purification step (Mono Qcolumn) is included if there is sufficient protein
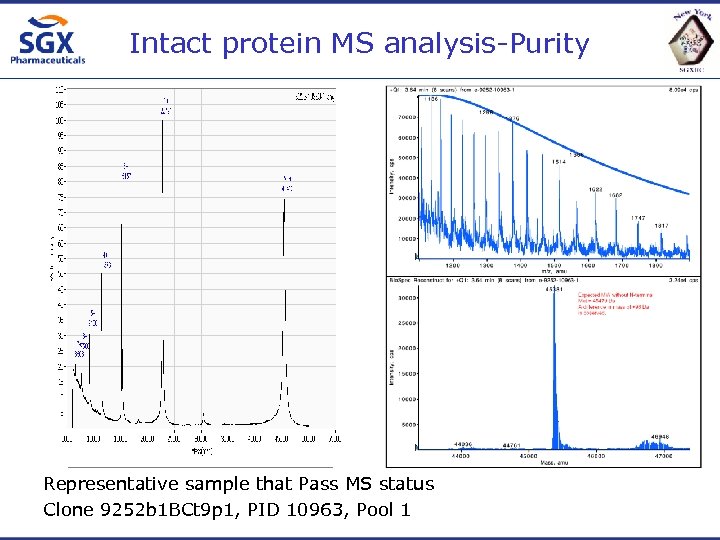 Intact protein MS analysis-Purity Representative sample that Pass MS status Clone 9252 b 1 BCt 9 p 1, PID 10963, Pool 1
Intact protein MS analysis-Purity Representative sample that Pass MS status Clone 9252 b 1 BCt 9 p 1, PID 10963, Pool 1
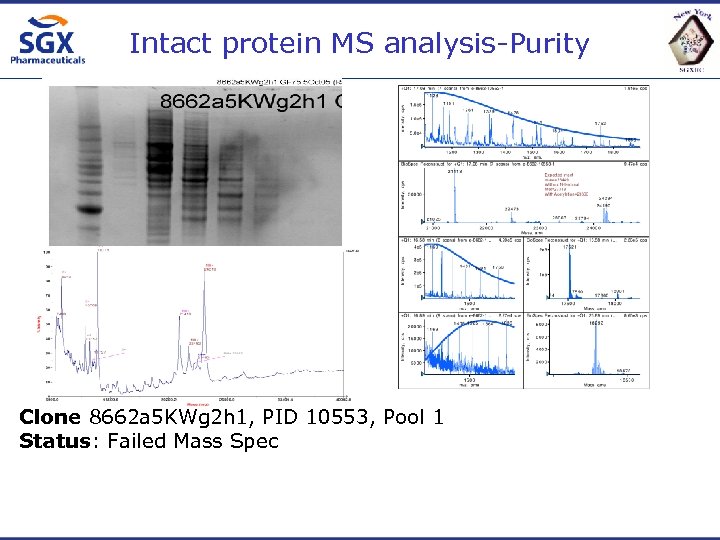 Intact protein MS analysis-Purity Clone 8662 a 5 KWg 2 h 1, PID 10553, Pool 1 Status: Failed Mass Spec
Intact protein MS analysis-Purity Clone 8662 a 5 KWg 2 h 1, PID 10553, Pool 1 Status: Failed Mass Spec
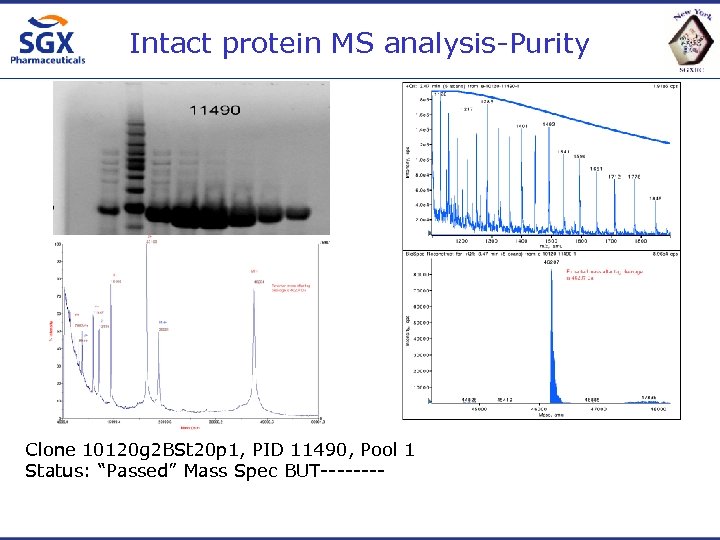 Intact protein MS analysis-Purity Clone 10120 g 2 BSt 20 p 1, PID 11490, Pool 1 Status: “Passed” Mass Spec BUT----
Intact protein MS analysis-Purity Clone 10120 g 2 BSt 20 p 1, PID 11490, Pool 1 Status: “Passed” Mass Spec BUT----
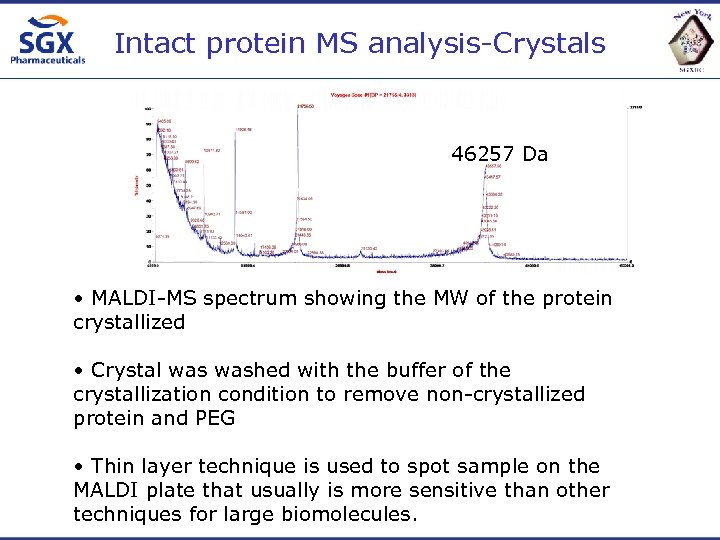 Intact protein MS analysis-Crystals 46257 Da • MALDI-MS spectrum showing the MW of the protein crystallized • Crystal washed with the buffer of the crystallization condition to remove non-crystallized protein and PEG • Thin layer technique is used to spot sample on the MALDI plate that usually is more sensitive than other techniques for large biomolecules.
Intact protein MS analysis-Crystals 46257 Da • MALDI-MS spectrum showing the MW of the protein crystallized • Crystal washed with the buffer of the crystallization condition to remove non-crystallized protein and PEG • Thin layer technique is used to spot sample on the MALDI plate that usually is more sensitive than other techniques for large biomolecules.
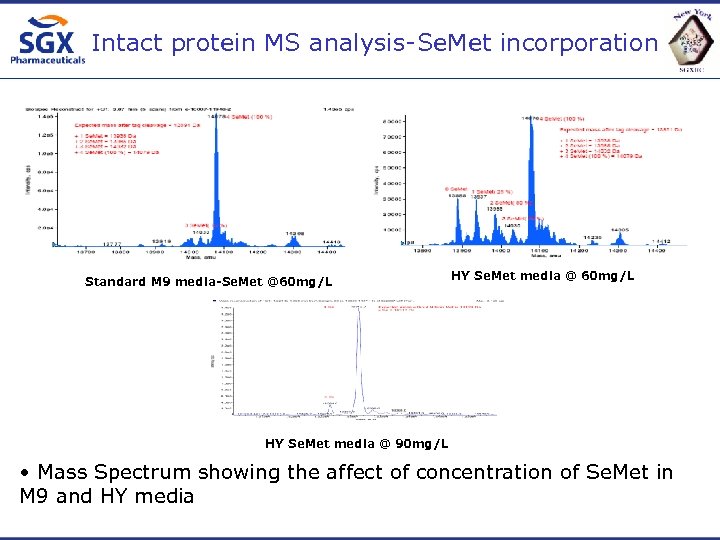 Intact protein MS analysis-Se. Met incorporation Standard M 9 media-Se. Met @60 mg/L HY Se. Met media @ 90 mg/L • Mass Spectrum showing the affect of concentration of Se. Met in M 9 and HY media
Intact protein MS analysis-Se. Met incorporation Standard M 9 media-Se. Met @60 mg/L HY Se. Met media @ 90 mg/L • Mass Spectrum showing the affect of concentration of Se. Met in M 9 and HY media
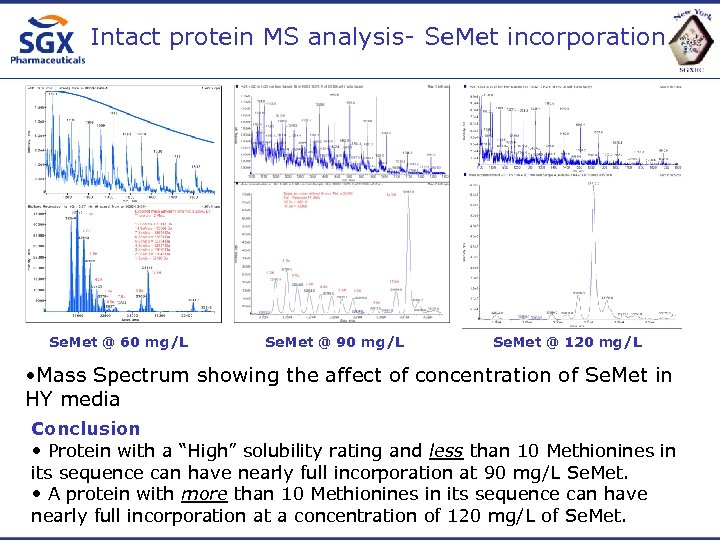 Intact protein MS analysis- Se. Met incorporation Se. Met @ 60 mg/L Se. Met @ 90 mg/L Se. Met @ 120 mg/L • Mass Spectrum showing the affect of concentration of Se. Met in HY media Conclusion • Protein with a “High” solubility rating and less than 10 Methionines in its sequence can have nearly full incorporation at 90 mg/L Se. Met. • A protein with more than 10 Methionines in its sequence can have nearly full incorporation at a concentration of 120 mg/L of Se. Met.
Intact protein MS analysis- Se. Met incorporation Se. Met @ 60 mg/L Se. Met @ 90 mg/L Se. Met @ 120 mg/L • Mass Spectrum showing the affect of concentration of Se. Met in HY media Conclusion • Protein with a “High” solubility rating and less than 10 Methionines in its sequence can have nearly full incorporation at 90 mg/L Se. Met. • A protein with more than 10 Methionines in its sequence can have nearly full incorporation at a concentration of 120 mg/L of Se. Met.
 Conclusions-Intact protein MS analysis • Sample purity and identity is routinely monitored on protein samples • If crystals are obtained on a sample where contaminants were also observed with the intended protein, MS analysis of the crystals can be performed (on request basis) to confirm it’s identity. • Percent Se. Met incorporation is routinely monitored. • Heterogeneity due to unknown PTMs are routinely monitored and pursued accordingly (Please check poster with title “Bottlenecks/Solutions for the Amidohydrolase Protein Superfamily” for more information)
Conclusions-Intact protein MS analysis • Sample purity and identity is routinely monitored on protein samples • If crystals are obtained on a sample where contaminants were also observed with the intended protein, MS analysis of the crystals can be performed (on request basis) to confirm it’s identity. • Percent Se. Met incorporation is routinely monitored. • Heterogeneity due to unknown PTMs are routinely monitored and pursued accordingly (Please check poster with title “Bottlenecks/Solutions for the Amidohydrolase Protein Superfamily” for more information)
 Tandem Mass Spectrometry (MS/MS) • MS/MS analysis is performed on all protein samples that have mass discrepancies, as observed by ESI-MS, to determine their true identity. • A batch of 15 samples (15 ug each sample) is simultaneously subjected to trypsin digestion (30: 1, protein: enzyme) for 14 hrs at 37°C. • MS/MS analysis is performed on the batch of 15 digested samples using ESI-quadrupole-ion trap mass spectrometer with online capillary-HPLC. • An example as a representative of different sources of mass discrepancies will be discussed further in this presentation.
Tandem Mass Spectrometry (MS/MS) • MS/MS analysis is performed on all protein samples that have mass discrepancies, as observed by ESI-MS, to determine their true identity. • A batch of 15 samples (15 ug each sample) is simultaneously subjected to trypsin digestion (30: 1, protein: enzyme) for 14 hrs at 37°C. • MS/MS analysis is performed on the batch of 15 digested samples using ESI-quadrupole-ion trap mass spectrometer with online capillary-HPLC. • An example as a representative of different sources of mass discrepancies will be discussed further in this presentation.
 High Performance Liquid Chromatography (HPLC)-Tandem Mass Spectrometry (MS/MS) HPLC instrument conditions • Instrument: (Agilent Technologies 1100 series). • Flow Rate: 5 u. L/min • Column: Zorbax 300 SB C-18; 3. 5 u. M particle size ; 150 X 0. 3 mm • Solvent A: 95% H 2 O, 5% Acetonitrile, 0. 1% Formic Acid • Solvent B: 5% H 2 O, 95% Acetonitrile, 0. 1% Formic Acid • Gradient: 0 -10 min: 100% A • 10 -60 min: 0 -100% B • 60 -70 min: 100% B • 70 -90 min: 100% A
High Performance Liquid Chromatography (HPLC)-Tandem Mass Spectrometry (MS/MS) HPLC instrument conditions • Instrument: (Agilent Technologies 1100 series). • Flow Rate: 5 u. L/min • Column: Zorbax 300 SB C-18; 3. 5 u. M particle size ; 150 X 0. 3 mm • Solvent A: 95% H 2 O, 5% Acetonitrile, 0. 1% Formic Acid • Solvent B: 5% H 2 O, 95% Acetonitrile, 0. 1% Formic Acid • Gradient: 0 -10 min: 100% A • 10 -60 min: 0 -100% B • 60 -70 min: 100% B • 70 -90 min: 100% A
 High Performance Liquid Chromatography (HPLC)-Tandem Mass Spectrometry (MS/MS) MS/MS instrument conditions • Instrument: Finnigan LCQDECA (Thermoquest, San Jose, CA, USA) ion-trap mass analyzer equipped with ESI source. • The experimental conditions were as follows: Duration of experiment Number of scan events MS mass range Default charge state Normalization collision energy Activation Q Activation time Activating gas ESI capillary tip voltage ESI source temperature 90 min 6 200 -2000 Da 2 35% (of the maximum) 25 e. V 30 msec Helium 4. 20 k. V 180ºC • Data dependent MS/MS mode is used. • Sequence-specific ions of the amino acids are used to search a non-redundant protein database with the Turbo. Sequest search engine (Bioworks 3. 3) to identify proteins. • The amino acid sequence of the identified protein is searched against in-house SGX_Gold database to identify the targets.
High Performance Liquid Chromatography (HPLC)-Tandem Mass Spectrometry (MS/MS) MS/MS instrument conditions • Instrument: Finnigan LCQDECA (Thermoquest, San Jose, CA, USA) ion-trap mass analyzer equipped with ESI source. • The experimental conditions were as follows: Duration of experiment Number of scan events MS mass range Default charge state Normalization collision energy Activation Q Activation time Activating gas ESI capillary tip voltage ESI source temperature 90 min 6 200 -2000 Da 2 35% (of the maximum) 25 e. V 30 msec Helium 4. 20 k. V 180ºC • Data dependent MS/MS mode is used. • Sequence-specific ions of the amino acids are used to search a non-redundant protein database with the Turbo. Sequest search engine (Bioworks 3. 3) to identify proteins. • The amino acid sequence of the identified protein is searched against in-house SGX_Gold database to identify the targets.
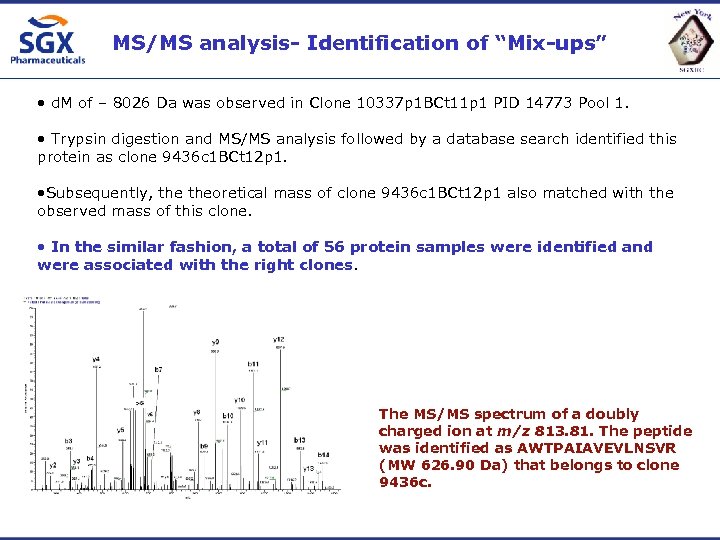 MS/MS analysis- Identification of “Mix-ups” • d. M of – 8026 Da was observed in Clone 10337 p 1 BCt 11 p 1 PID 14773 Pool 1. • Trypsin digestion and MS/MS analysis followed by a database search identified this protein as clone 9436 c 1 BCt 12 p 1. • Subsequently, theoretical mass of clone 9436 c 1 BCt 12 p 1 also matched with the observed mass of this clone. • In the similar fashion, a total of 56 protein samples were identified and were associated with the right clones. The MS/MS spectrum of a doubly charged ion at m/z 813. 81. The peptide was identified as AWTPAIAVEVLNSVR (MW 626. 90 Da) that belongs to clone 9436 c.
MS/MS analysis- Identification of “Mix-ups” • d. M of – 8026 Da was observed in Clone 10337 p 1 BCt 11 p 1 PID 14773 Pool 1. • Trypsin digestion and MS/MS analysis followed by a database search identified this protein as clone 9436 c 1 BCt 12 p 1. • Subsequently, theoretical mass of clone 9436 c 1 BCt 12 p 1 also matched with the observed mass of this clone. • In the similar fashion, a total of 56 protein samples were identified and were associated with the right clones. The MS/MS spectrum of a doubly charged ion at m/z 813. 81. The peptide was identified as AWTPAIAVEVLNSVR (MW 626. 90 Da) that belongs to clone 9436 c.
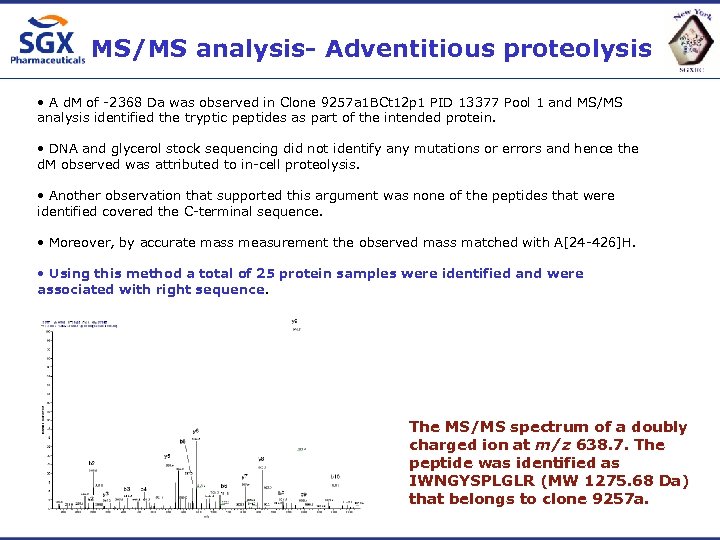 MS/MS analysis- Adventitious proteolysis • A d. M of -2368 Da was observed in Clone 9257 a 1 BCt 12 p 1 PID 13377 Pool 1 and MS/MS analysis identified the tryptic peptides as part of the intended protein. • DNA and glycerol stock sequencing did not identify any mutations or errors and hence the d. M observed was attributed to in-cell proteolysis. • Another observation that supported this argument was none of the peptides that were identified covered the C-terminal sequence. • Moreover, by accurate mass measurement the observed mass matched with A[24 -426]H. • Using this method a total of 25 protein samples were identified and were associated with right sequence. The MS/MS spectrum of a doubly charged ion at m/z 638. 7. The peptide was identified as IWNGYSPLGLR (MW 1275. 68 Da) that belongs to clone 9257 a.
MS/MS analysis- Adventitious proteolysis • A d. M of -2368 Da was observed in Clone 9257 a 1 BCt 12 p 1 PID 13377 Pool 1 and MS/MS analysis identified the tryptic peptides as part of the intended protein. • DNA and glycerol stock sequencing did not identify any mutations or errors and hence the d. M observed was attributed to in-cell proteolysis. • Another observation that supported this argument was none of the peptides that were identified covered the C-terminal sequence. • Moreover, by accurate mass measurement the observed mass matched with A[24 -426]H. • Using this method a total of 25 protein samples were identified and were associated with right sequence. The MS/MS spectrum of a doubly charged ion at m/z 638. 7. The peptide was identified as IWNGYSPLGLR (MW 1275. 68 Da) that belongs to clone 9257 a.
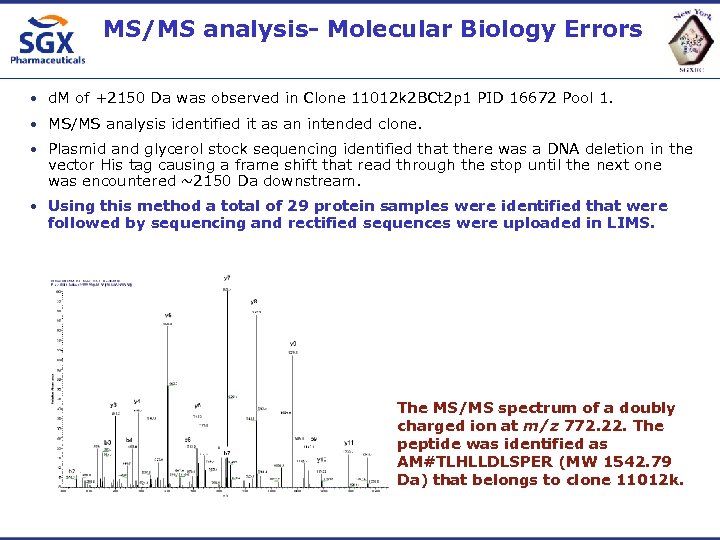 MS/MS analysis- Molecular Biology Errors • d. M of +2150 Da was observed in Clone 11012 k 2 BCt 2 p 1 PID 16672 Pool 1. • MS/MS analysis identified it as an intended clone. • Plasmid and glycerol stock sequencing identified that there was a DNA deletion in the vector His tag causing a frame shift that read through the stop until the next one was encountered ~2150 Da downstream. • Using this method a total of 29 protein samples were identified that were followed by sequencing and rectified sequences were uploaded in LIMS. The MS/MS spectrum of a doubly charged ion at m/z 772. 22. The peptide was identified as AM#TLHLLDLSPER (MW 1542. 79 Da) that belongs to clone 11012 k.
MS/MS analysis- Molecular Biology Errors • d. M of +2150 Da was observed in Clone 11012 k 2 BCt 2 p 1 PID 16672 Pool 1. • MS/MS analysis identified it as an intended clone. • Plasmid and glycerol stock sequencing identified that there was a DNA deletion in the vector His tag causing a frame shift that read through the stop until the next one was encountered ~2150 Da downstream. • Using this method a total of 29 protein samples were identified that were followed by sequencing and rectified sequences were uploaded in LIMS. The MS/MS spectrum of a doubly charged ion at m/z 772. 22. The peptide was identified as AM#TLHLLDLSPER (MW 1542. 79 Da) that belongs to clone 11012 k.
 Conclusions-Tandem Mass Spectrometry • In 2007, ~1400 protein samples were analyzed by ESI and MALDI-MS. • Out of these 120 samples of questionable identity were further analyzed by MS/MS. • Using mass spectrometry we have identified the following common problems that can lead to mass discrepancies and number of structures thus benefited from it: • Results indicated that these fit into the above groups as follows: • 1. Clone “mix-up”: 56 of which 8 resulted in structures; • 2. Cloning, etc. artifacts: 29 of which 3 resulted in structures; • 3. Truncation by proteolysis: 25 of which 3 resulted in structures; • 4. Unwanted E. coli contaminant: 10, none of which were pursued for structure determination. • In conclusion, our standard MS quality control analysis procedures contributed substantially to the success of 14 of a total 158 NYSGXRC PDB depositions during calendar 2007 and allowed us to avoid wasting effort on 10 inadvertently purified protein samples.
Conclusions-Tandem Mass Spectrometry • In 2007, ~1400 protein samples were analyzed by ESI and MALDI-MS. • Out of these 120 samples of questionable identity were further analyzed by MS/MS. • Using mass spectrometry we have identified the following common problems that can lead to mass discrepancies and number of structures thus benefited from it: • Results indicated that these fit into the above groups as follows: • 1. Clone “mix-up”: 56 of which 8 resulted in structures; • 2. Cloning, etc. artifacts: 29 of which 3 resulted in structures; • 3. Truncation by proteolysis: 25 of which 3 resulted in structures; • 4. Unwanted E. coli contaminant: 10, none of which were pursued for structure determination. • In conclusion, our standard MS quality control analysis procedures contributed substantially to the success of 14 of a total 158 NYSGXRC PDB depositions during calendar 2007 and allowed us to avoid wasting effort on 10 inadvertently purified protein samples.
 In the end------ • In an effort to support various groups in the platform (protein production, crystallization and crystallography), we have made our goals simple: • Use as less sample as we can-Never used more than 100 ug of sample under study • Provide as much information possible on the “precious” protein sample-Protein purity, Identity, PTM study, crystal analysis, Se. Met incorporation, In-gel trypsinization MS/MS analysis, Identification of mix-ups, In-cell proteolysis, Molecular Biology errors etc • Reduce the analysis time to synchronize with the high- throughput activities in the platform-use of high throughput MS approach helps us to reduce analysis time
In the end------ • In an effort to support various groups in the platform (protein production, crystallization and crystallography), we have made our goals simple: • Use as less sample as we can-Never used more than 100 ug of sample under study • Provide as much information possible on the “precious” protein sample-Protein purity, Identity, PTM study, crystal analysis, Se. Met incorporation, In-gel trypsinization MS/MS analysis, Identification of mix-ups, In-cell proteolysis, Molecular Biology errors etc • Reduce the analysis time to synchronize with the high- throughput activities in the platform-use of high throughput MS approach helps us to reduce analysis time
 Acknowledgement • This work was supported by SGX Pharmaceuticals, Inc. and NIH Grant U 54 GM 074945 (Principal Investigator: Stephen K. Burley)
Acknowledgement • This work was supported by SGX Pharmaceuticals, Inc. and NIH Grant U 54 GM 074945 (Principal Investigator: Stephen K. Burley)


