91713a3836d6c4df68e1c8e4422dc6af.ppt
- Количество слайдов: 31

Integration of Fast Data Collection and Automated Probabilistic Assignment for Protein NMR Spectroscopy Arash Bahrami
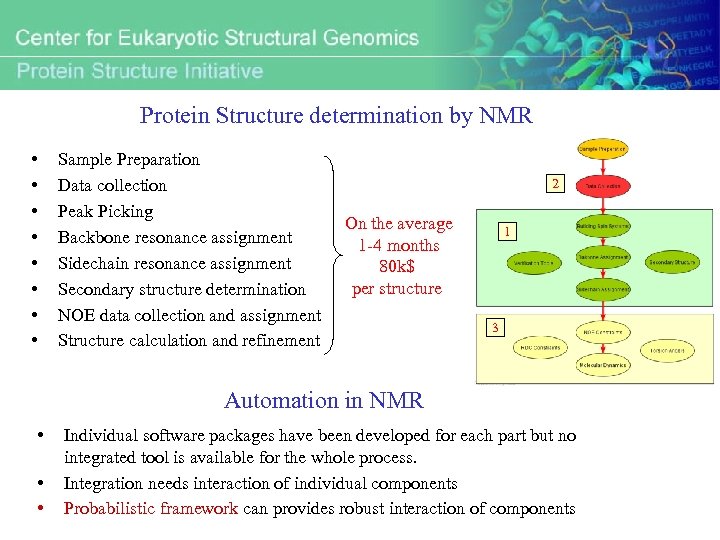
Protein Structure determination by NMR • • Sample Preparation Data collection Peak Picking Backbone resonance assignment Sidechain resonance assignment Secondary structure determination NOE data collection and assignment Structure calculation and refinement 2 On the average 1 -4 months 80 k$ per structure 1 3 Automation in NMR • • • Individual software packages have been developed for each part but no integrated tool is available for the whole process. Integration needs interaction of individual components Probabilistic framework can provides robust interaction of components
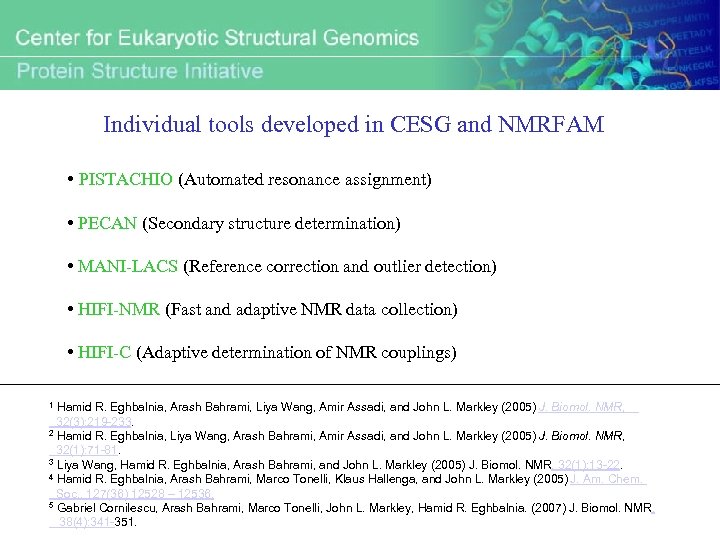
Individual tools developed in CESG and NMRFAM • PISTACHIO (Automated resonance assignment) • PECAN (Secondary structure determination) • MANI-LACS (Reference correction and outlier detection) • HIFI-NMR (Fast and adaptive NMR data collection) • HIFI-C (Adaptive determination of NMR couplings) Hamid R. Eghbalnia, Arash Bahrami, Liya Wang, Amir Assadi, and John L. Markley (2005) J. Biomol. NMR, 32(3): 219 -233. 2 Hamid R. Eghbalnia, Liya Wang, Arash Bahrami, Amir Assadi, and John L. Markley (2005) J. Biomol. NMR, 32(1): 71 -81. 3 Liya Wang, Hamid R. Eghbalnia, Arash Bahrami, and John L. Markley (2005) J. Biomol. NMR, 32(1): 13 -22. 4 Hamid R. Eghbalnia, Arash Bahrami, Marco Tonelli, Klaus Hallenga, and John L. Markley (2005) J. Am. Chem. Soc. , 127(36) 12528 – 12536. 5 Gabriel Cornilescu, Arash Bahrami, Marco Tonelli, John L. Markley, Hamid R. Eghbalnia. (2007) J. Biomol. NMR, 38(4): 341 -351. 1
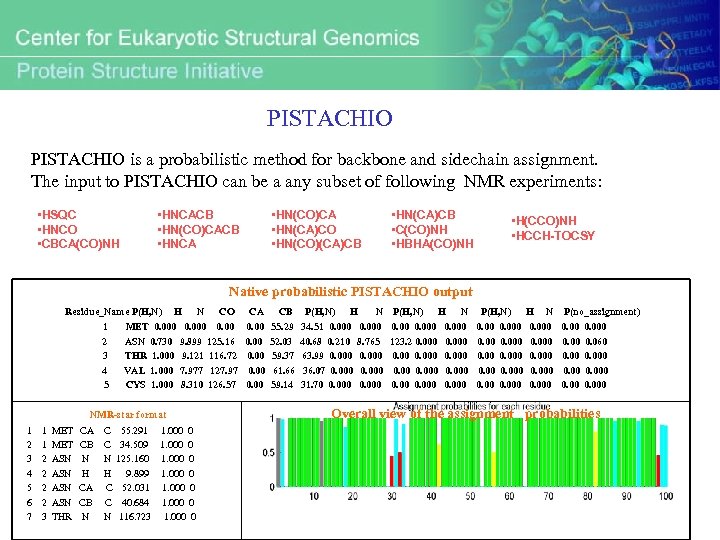
PISTACHIO is a probabilistic method for backbone and sidechain assignment. The input to PISTACHIO can be a any subset of following NMR experiments: • HSQC • HNCO • CBCA(CO)NH • HNCACB • HN(CO)CACB • HNCA • HN(CO)CA • HN(CA)CO • HN(CO)(CA)CB • HN(CA)CB • C(CO)NH • HBHA(CO)NH • H(CCO)NH • HCCH-TOCSY Native probabilistic PISTACHIO output Residue_Name P(H, N) 1 2 3 4 5 H MET 0. 000 ASN 0. 730 THR 1. 000 VAL 1. 000 CYS 1. 000 1 1 2 2 3 MET ASN ASN THR CA CB N H CA CB N C C N H C C N CO 0. 000 0. 00 9. 899 125. 16 9. 121 116. 72 7. 977 127. 97 8. 310 126. 57 NMR-star format 1 2 3 4 5 6 7 N 55. 291 1. 000 0 34. 509 1. 000 0 125. 160 1. 000 0 9. 899 1. 000 0 52. 031 1. 000 0 40. 684 1. 000 0 116. 723 1. 000 0 CA CB 0. 00 55. 29 52. 03 59. 37 61. 66 59. 14 P(H, N) H 34. 51 0. 000 40. 68 0. 210 63. 99 0. 000 36. 07 0. 000 31. 70 0. 000 N 0. 000 8. 765 0. 000 P(H, N) 0. 000 123. 2 0. 000 0. 000 H N 0. 000 P(H, N) 0. 000 0. 000 H N 0. 000 P(no_assignment) 0. 000 0. 060 0. 000 Overall view of the assignment probabilities
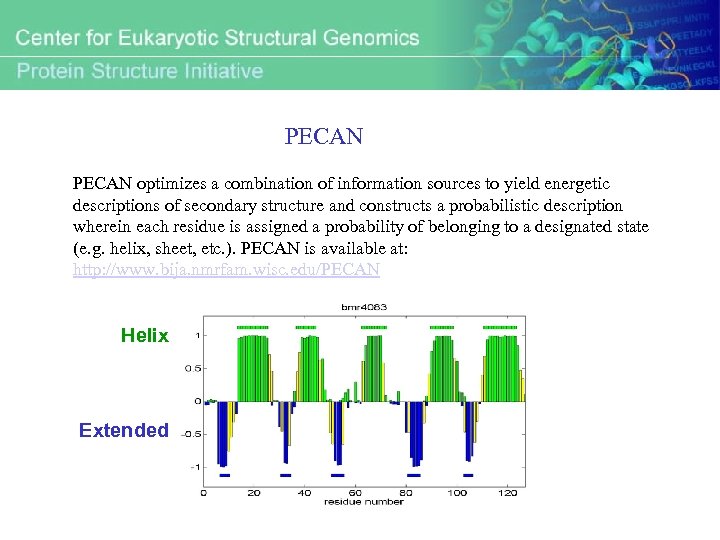
PECAN optimizes a combination of information sources to yield energetic descriptions of secondary structure and constructs a probabilistic description wherein each residue is assigned a probability of belonging to a designated state (e. g. helix, sheet, etc. ). PECAN is available at: http: //www. bija. nmrfam. wisc. edu/PECAN Helix Extended
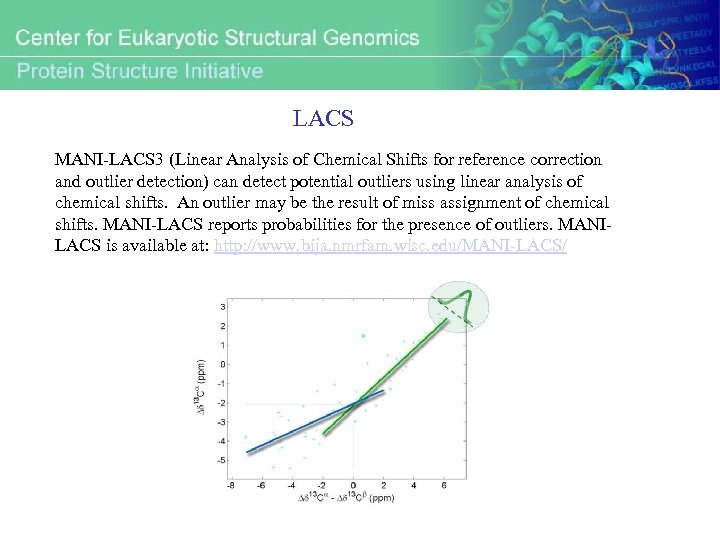
LACS MANI-LACS 3 (Linear Analysis of Chemical Shifts for reference correction and outlier detection) can detect potential outliers using linear analysis of chemical shifts. An outlier may be the result of miss assignment of chemical shifts. MANI-LACS reports probabilities for the presence of outliers. MANILACS is available at: http: //www. bija. nmrfam. wisc. edu/MANI-LACS/
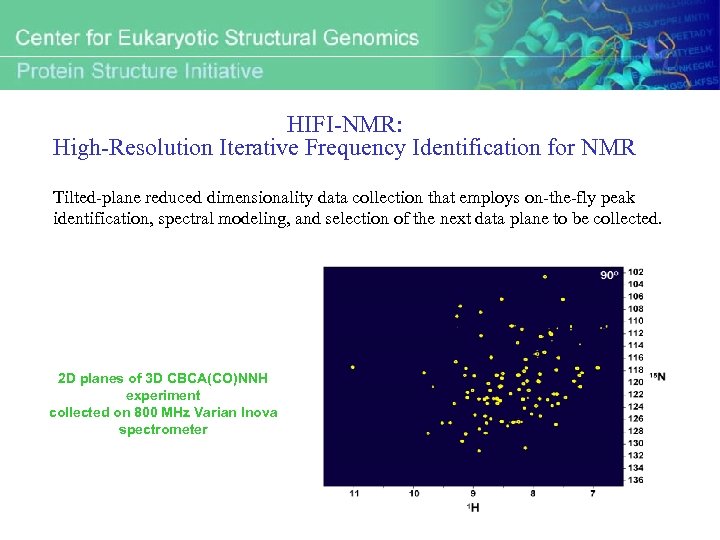
HIFI-NMR: High-Resolution Iterative Frequency Identification for NMR Tilted-plane reduced dimensionality data collection that employs on-the-fly peak identification, spectral modeling, and selection of the next data plane to be collected. 2 D planes of 3 D CBCA(CO)NNH experiment collected on 800 MHz Varian Inova spectrometer
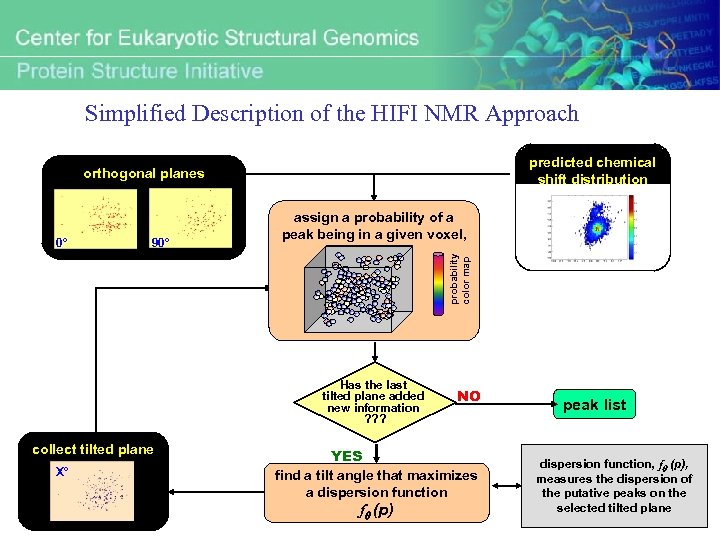
Simplified Description of the HIFI NMR Approach predicted chemical shift distribution orthogonal planes 90° p Has the last tilted plane added new information ? ? ? collect tilted plane X° probability color map 0° assign a probability of a peak being in a given voxel, NO YES find a tilt angle that maximizes a dispersion function fq (p) peak list dispersion function, fq (p), measures the dispersion of the putative peaks on the selected tilted plane
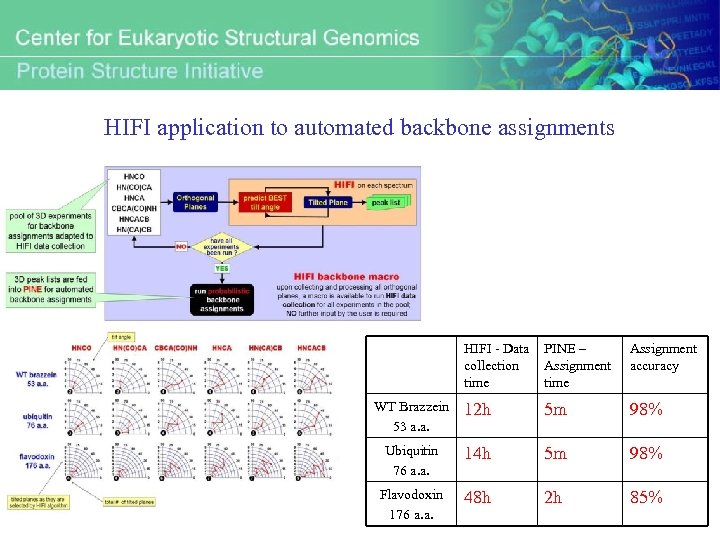
HIFI application to automated backbone assignments HIFI - Data collection time PINE – Assignment time Assignment accuracy WT Brazzein 53 a. a. 12 h 5 m 98% Ubiquitin 76 a. a. 14 h 5 m 98% Flavodoxin 176 a. a. 48 h 2 h 85%
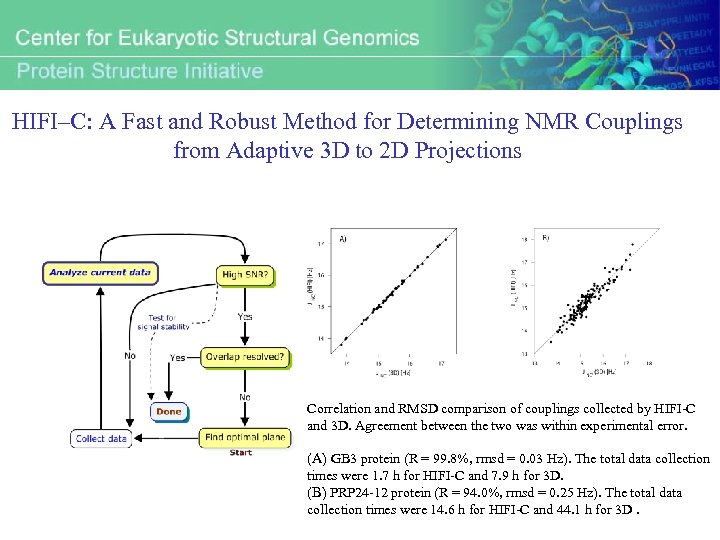
HIFI–C: A Fast and Robust Method for Determining NMR Couplings from Adaptive 3 D to 2 D Projections Correlation and RMSD comparison of couplings collected by HIFI-C and 3 D. Agreement between the two was within experimental error. (A) GB 3 protein (R = 99. 8%, rmsd = 0. 03 Hz). The total data collection times were 1. 7 h for HIFI-C and 7. 9 h for 3 D. (B) PRP 24 -12 protein (R = 94. 0%, rmsd = 0. 25 Hz). The total data collection times were 14. 6 h for HIFI-C and 44. 1 h for 3 D.
Back to Automation Steps in NMR Proteomics HIFI-NMR PISTACHIO MANI-LACS HIFI-C PECAN
Redesign the Individual Tools to Provide Robust Probabilistic Interaction: PINE PISTACHIO MANI-LACS PECAN
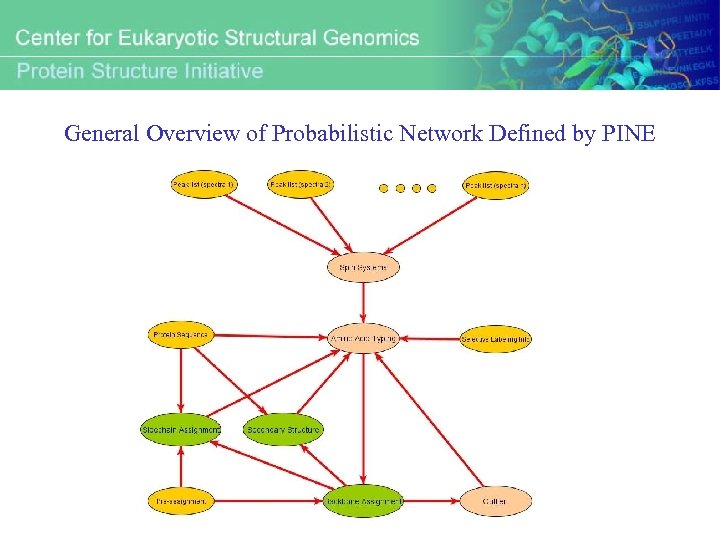
General Overview of Probabilistic Network Defined by PINE
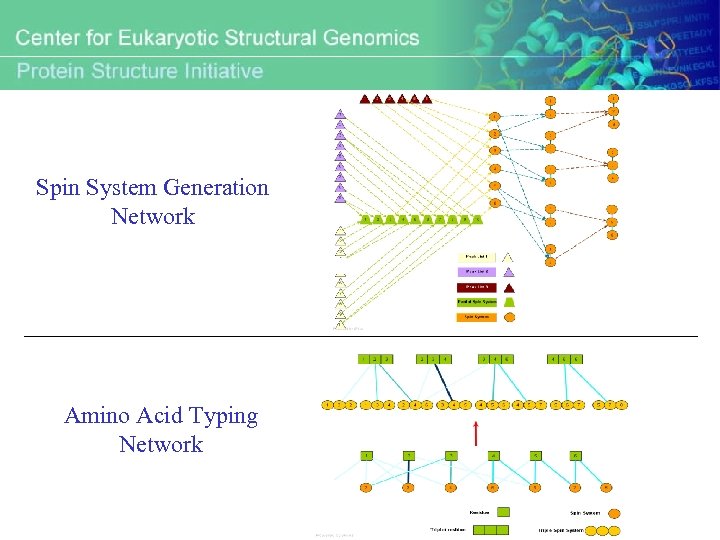
Spin System Generation Network Amino Acid Typing Network
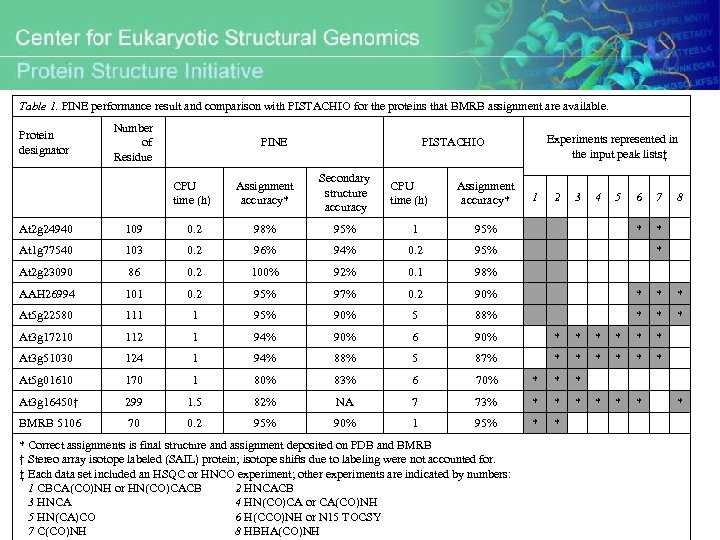
Table 1. PINE performance result and comparison with PISTACHIO for the proteins that BMRB assignment are available. Protein designator Number of Residue PINE Experiments represented in the input peak lists‡ PISTACHIO CPU time (h) Assignment accuracy* Secondary structure accuracy CPU time (h) Assignment accuracy* 1 2 3 4 5 6 7 * 8 * At 2 g 24940 109 0. 2 98% 95% 1 95% At 1 g 77540 103 0. 2 96% 94% 0. 2 95% At 2 g 23090 86 0. 2 100% 92% 0. 1 98% AAH 26994 101 0. 2 95% 97% 0. 2 90% * * * At 5 g 22580 111 1 95% 90% 5 88% * * * At 3 g 17210 112 1 94% 90% 6 90% * * * At 3 g 51030 124 1 94% 88% 5 87% * * * At 5 g 01610 170 1 80% 83% 6 70% * * * At 3 g 16450† 299 1. 5 82% NA 7 73% * * * BMRB 5106 70 0. 2 95% 90% 1 95% * * * Correct assignments is final structure and assignment deposited on PDB and BMRB † Stereo array isotope labeled (SAIL) protein; isotope shifts due to labeling were not accounted for. ‡ Each data set included an HSQC or HNCO experiment; other experiments are indicated by numbers: 1 CBCA(CO)NH or HN(CO)CACB 2 HNCACB 3 HNCA 4 HN(CO)CA or CA(CO)NH 5 HN(CA)CO 6 H(CCO)NH or N 15 TOCSY 7 C(CO)NH 8 HBHA(CO)NH * *
PINE Web Server
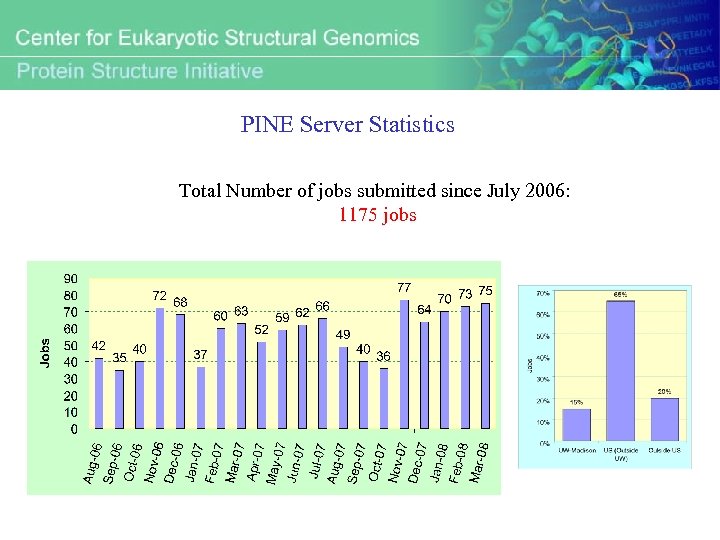
PINE Server Statistics Total Number of jobs submitted since July 2006: 1175 jobs
Iterative HCCH-TOCSY assignment HBHA(CO)NH HCCH-TOCSY H (CCO)NH C(CO)NH
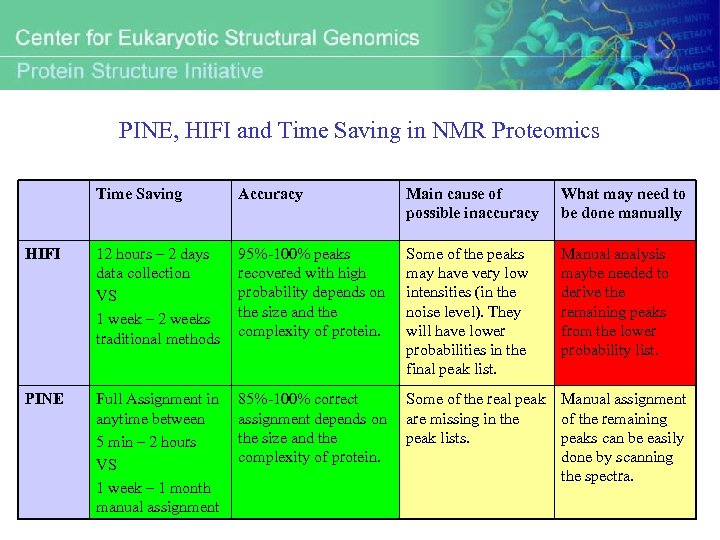
PINE, HIFI and Time Saving in NMR Proteomics Time Saving Accuracy Main cause of possible inaccuracy What may need to be done manually HIFI 12 hours – 2 days data collection VS 1 week – 2 weeks traditional methods 95%-100% peaks recovered with high probability depends on the size and the complexity of protein. Some of the peaks may have very low intensities (in the noise level). They will have lower probabilities in the final peak list. Manual analysis maybe needed to derive the remaining peaks from the lower probability list. PINE Full Assignment in anytime between 5 min – 2 hours VS 1 week – 1 month manual assignment 85%-100% correct assignment depends on the size and the complexity of protein. Some of the real peak Manual assignment are missing in the of the remaining peak lists. peaks can be easily done by scanning the spectra.
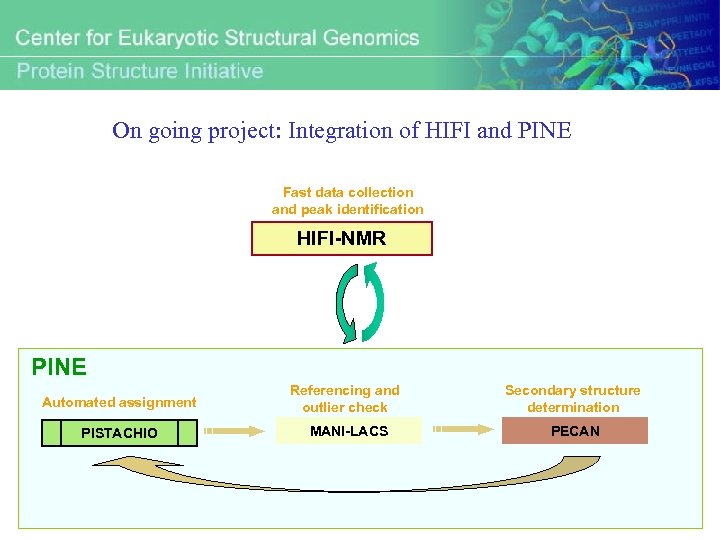
On going project: Integration of HIFI and PINE Fast data collection and peak identification HIFI-NMR PINE Automated assignment Referencing and outlier check Secondary structure determination PISTACHIO MANI-LACS PECAN
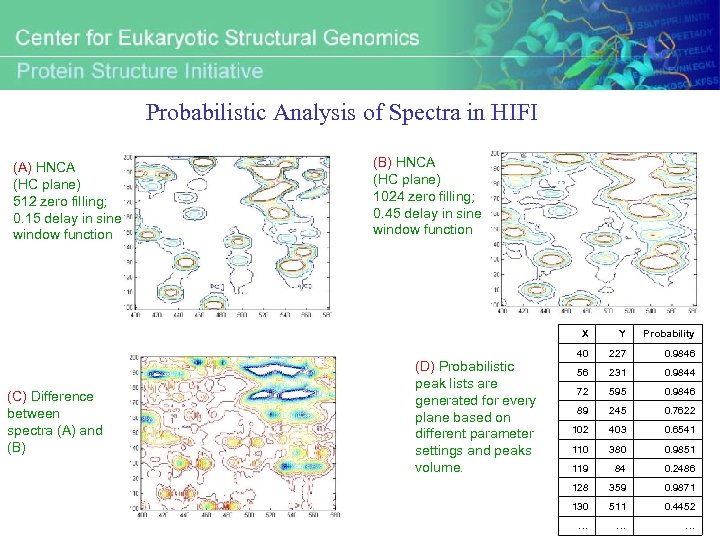
Probabilistic Analysis of Spectra in HIFI (A) HNCA (HC plane) 512 zero filling; 0. 15 delay in sine window function (B) HNCA (HC plane) 1024 zero filling; 0. 45 delay in sine window function X (C) Difference between spectra (A) and (B) (D) Probabilistic peak lists are generated for every plane based on different parameter settings and peaks volume. Y Probability 40 227 0. 9846 56 231 0. 9844 72 595 0. 9846 89 245 0. 7622 102 403 0. 6541 110 380 0. 9851 119 84 0. 2486 128 359 0. 9871 130 511 0. 4452 … … …
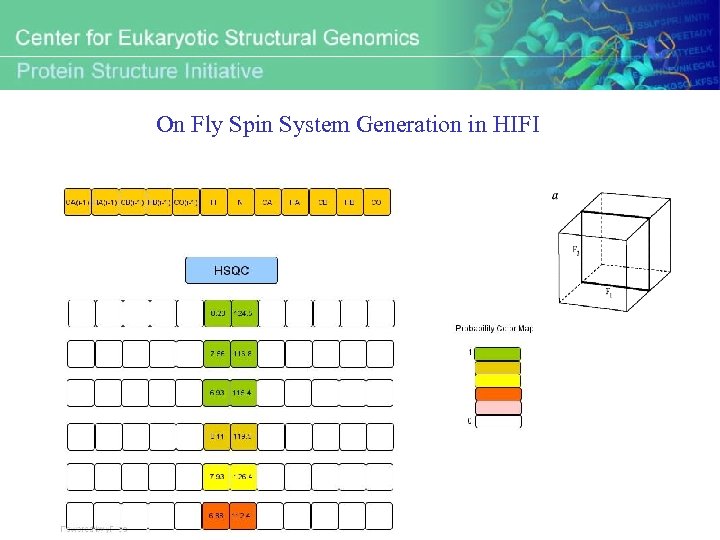
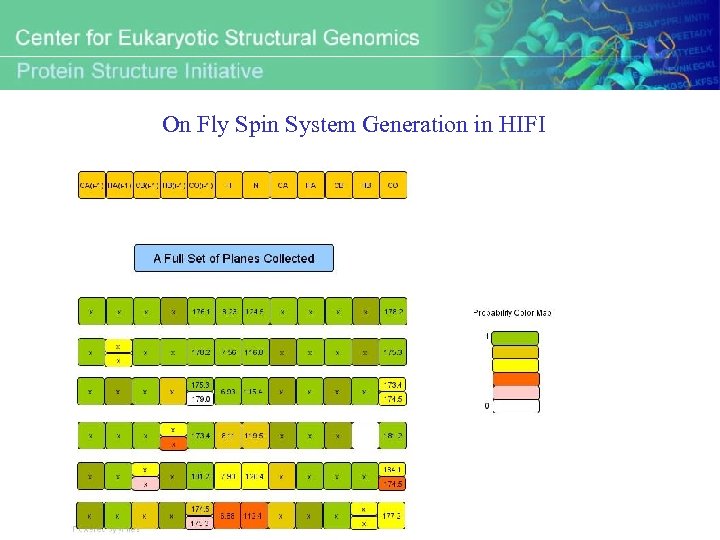
On Fly Spin System Generation in HIFI
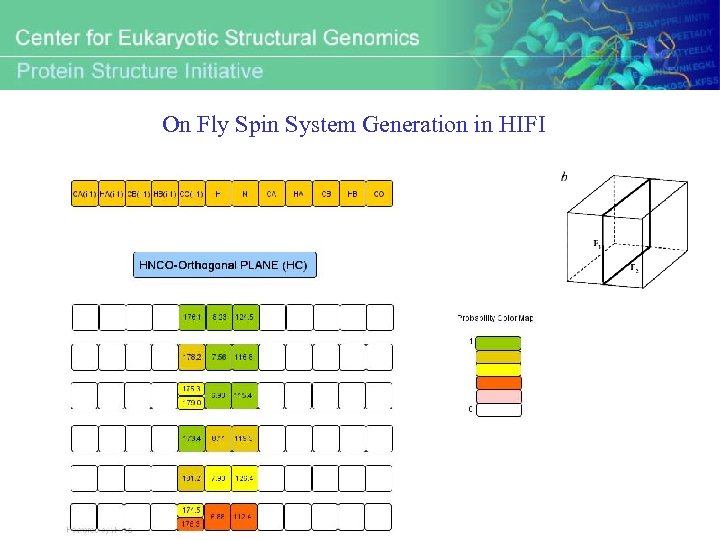
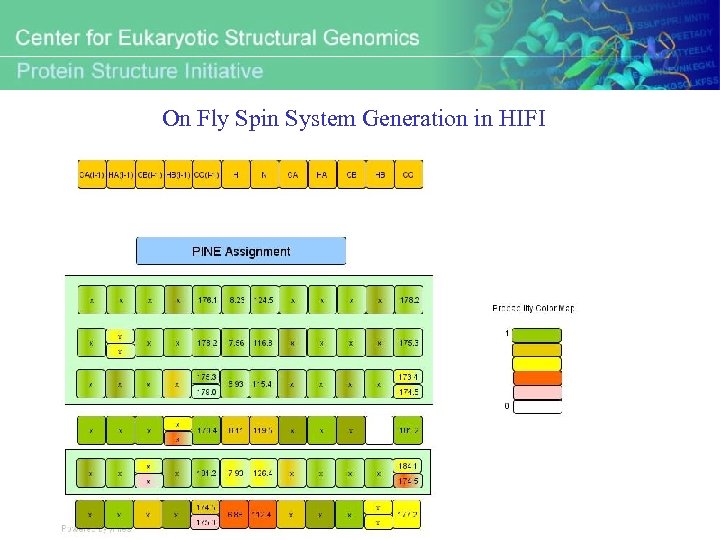
On Fly Spin System Generation in HIFI
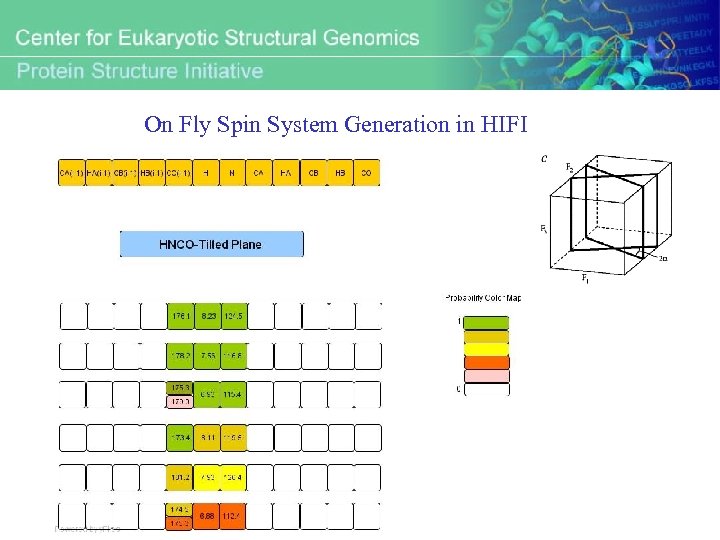
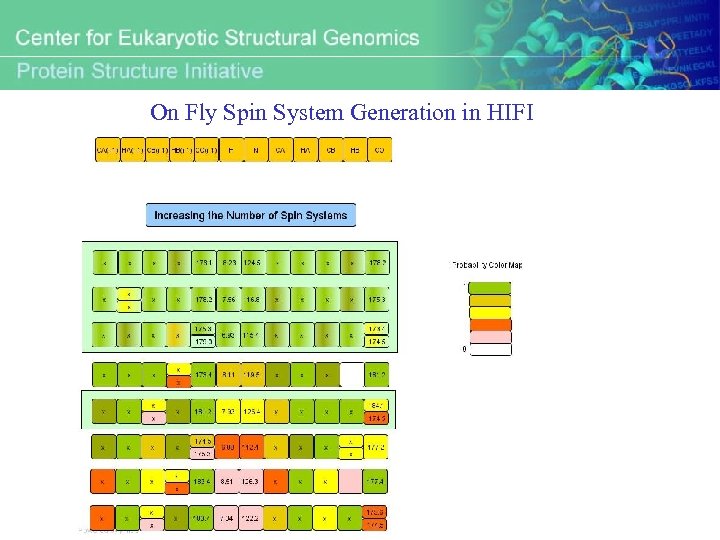
On Fly Spin System Generation in HIFI
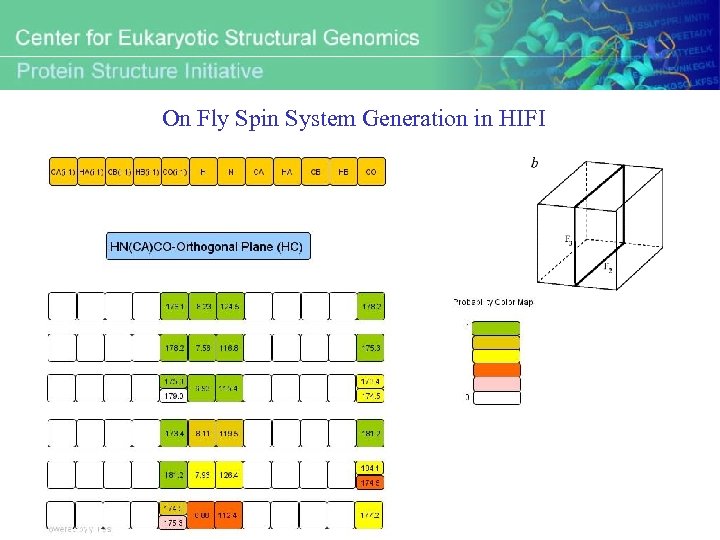
On Fly Spin System Generation in HIFI
On Fly Spin System Generation in HIFI
On Fly Spin System Generation in HIFI
On Fly Spin System Generation in HIFI
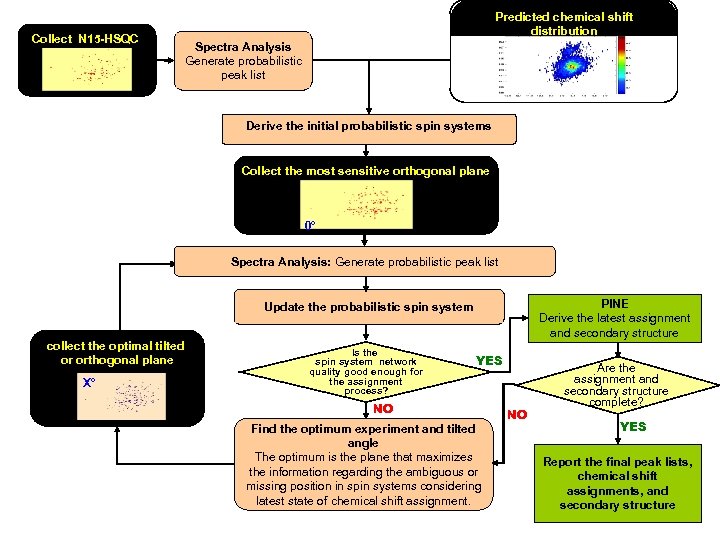
Collect N 15 -HSQC Predicted chemical shift distribution Spectra Analysis Generate probabilistic peak list Derive the initial probabilistic spin systems Collect the most sensitive orthogonal plane 0° Spectra Analysis: Generate probabilistic peak list PINE Derive the latest assignment and secondary structure Update the probabilistic spin system collect the optimal tilted or orthogonal plane X° Is the spin system network quality good enough for the assignment process? YES NO Find the optimum experiment and tilted angle The optimum is the plane that maximizes the information regarding the ambiguous or missing position in spin systems considering latest state of chemical shift assignment. NO Are the assignment and secondary structure complete? YES Report the final peak lists, chemical shift assignments, and secondary structure
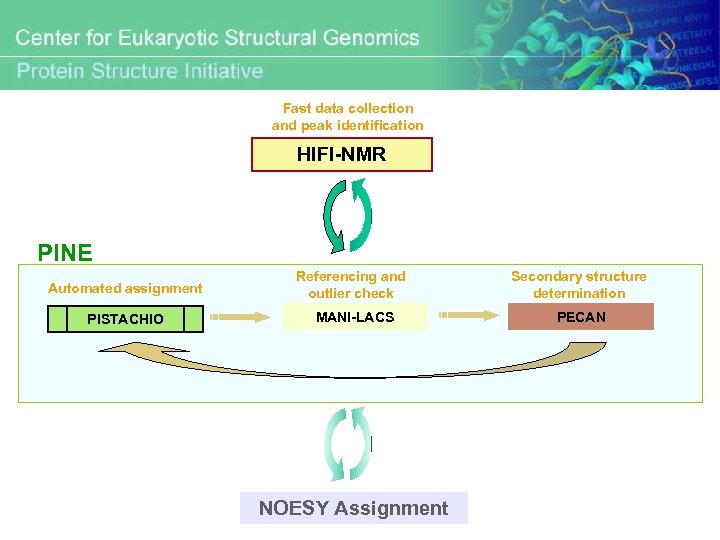
Fast data collection and peak identification HIFI-NMR PINE Automated assignment Referencing and outlier check Secondary structure determination PISTACHIO MANI-LACS PECAN NOESY Assignment
Acknowledgements • John Markley • Hamid Eghbalnia • Marco Tonelli • • • Ziqi Dai Gabriel Cornislescu Klaus Hallenga Milo Westler Liya Wang • Eldon Ulrich All CESG member providing data: • Claudia Cornilescu • Shanteri Singh • Jikui Song • Brian Volkman • Francis Peterson
91713a3836d6c4df68e1c8e4422dc6af.ppt