05060c02092ef3bbac0a6ac5f31470be.ppt
- Количество слайдов: 49
 Integative Genomic Approaches to Personalized Cancer Therapy Patrick Tan, MD Ph. D International Conference on Bioinformatics Singapore, Sept 09 2009
Integative Genomic Approaches to Personalized Cancer Therapy Patrick Tan, MD Ph. D International Conference on Bioinformatics Singapore, Sept 09 2009
 Genomic Oncology in Singapore : Translating Information into Knowledge Disease Genes Clinical Biomarkers Cancer Pathways
Genomic Oncology in Singapore : Translating Information into Knowledge Disease Genes Clinical Biomarkers Cancer Pathways
 Basic Science to Translation 1) Metastasis Genes - Network Structures 2) Cancer Classification - Pathway Signatures 3) Lung Cancer Outcome - Integrative Genomics
Basic Science to Translation 1) Metastasis Genes - Network Structures 2) Cancer Classification - Pathway Signatures 3) Lung Cancer Outcome - Integrative Genomics
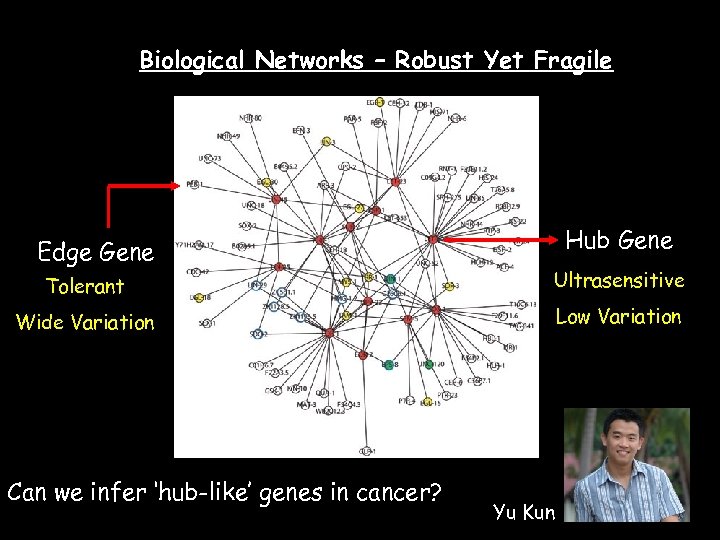 Biological Networks – Robust Yet Fragile Edge Gene Hub Gene Tolerant Ultrasensitive Wide Variation Low Variation Can we infer ‘hub-like’ genes in cancer? Yu Kun
Biological Networks – Robust Yet Fragile Edge Gene Hub Gene Tolerant Ultrasensitive Wide Variation Low Variation Can we infer ‘hub-like’ genes in cancer? Yu Kun
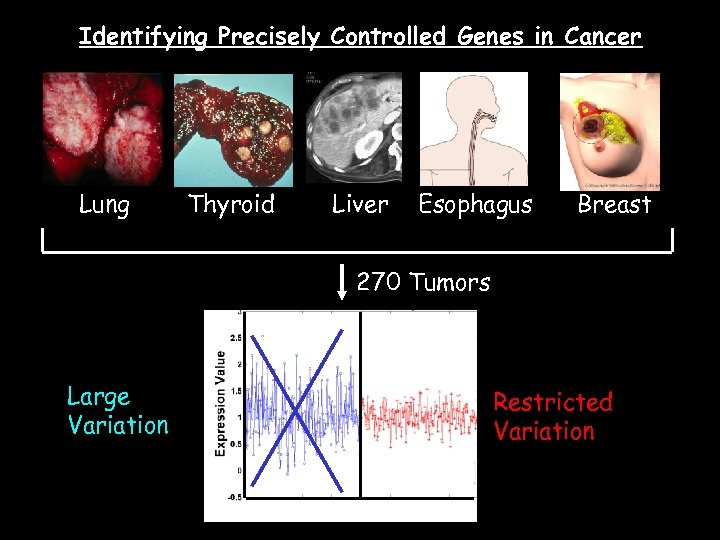 Identifying Precisely Controlled Genes in Cancer Lung Thyroid Liver Esophagus Breast 270 Tumors Large Variation Restricted Variation
Identifying Precisely Controlled Genes in Cancer Lung Thyroid Liver Esophagus Breast 270 Tumors Large Variation Restricted Variation
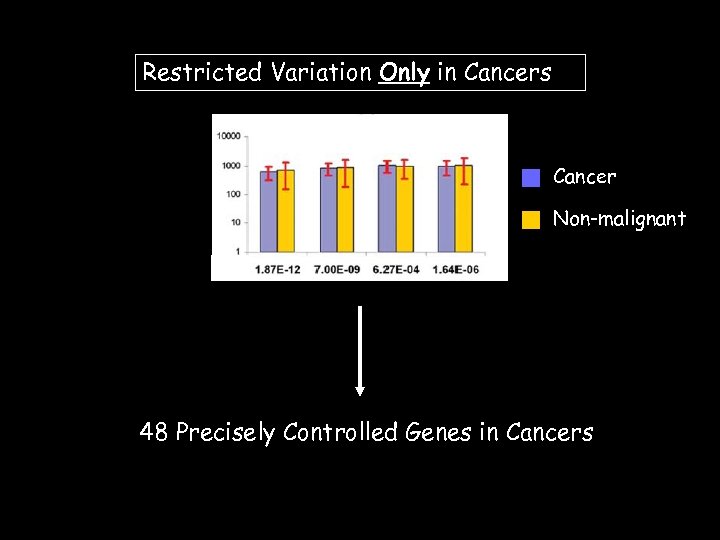 Restricted Variation Only in Cancers Cancer Non-malignant 48 Precisely Controlled Genes in Cancers
Restricted Variation Only in Cancers Cancer Non-malignant 48 Precisely Controlled Genes in Cancers
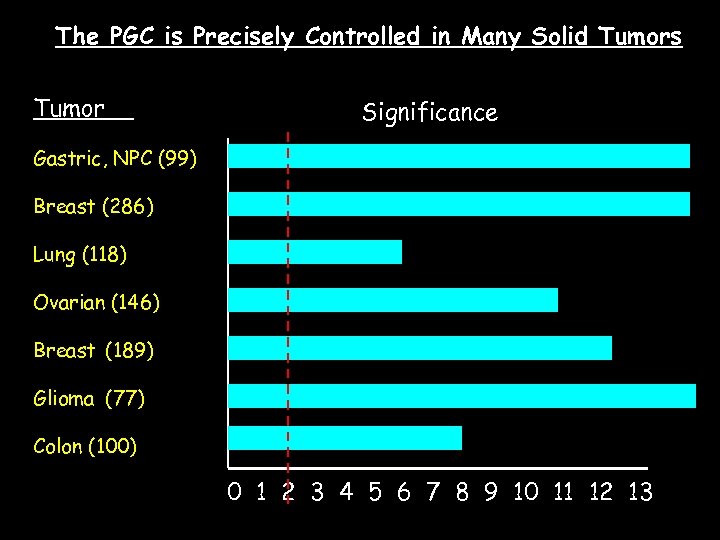 The PGC is Precisely Controlled in Many Solid Tumors Tumor Significance Gastric, NPC (99) Breast (286) Lung (118) Ovarian (146) Breast (189) Glioma (77) Colon (100) 0 1 2 3 4 5 6 7 8 9 10 11 12 13
The PGC is Precisely Controlled in Many Solid Tumors Tumor Significance Gastric, NPC (99) Breast (286) Lung (118) Ovarian (146) Breast (189) Glioma (77) Colon (100) 0 1 2 3 4 5 6 7 8 9 10 11 12 13
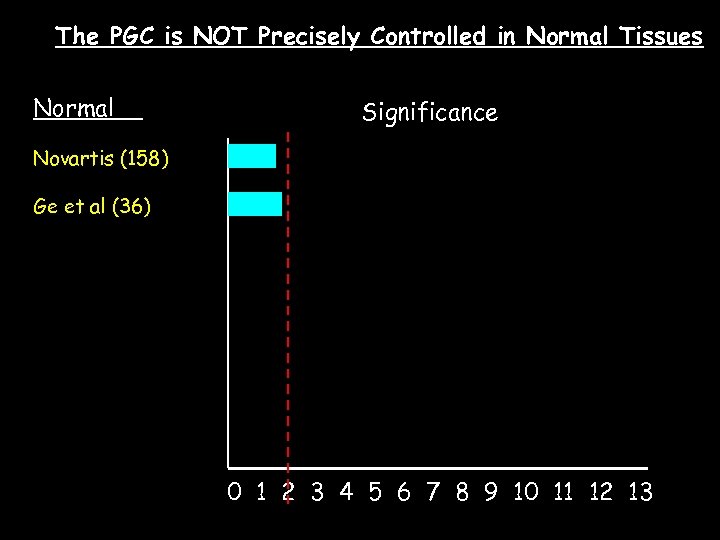 The PGC is NOT Precisely Controlled in Normal Tissues Normal Significance Novartis (158) Ge et al (36) 0 1 2 3 4 5 6 7 8 9 10 11 12 13
The PGC is NOT Precisely Controlled in Normal Tissues Normal Significance Novartis (158) Ge et al (36) 0 1 2 3 4 5 6 7 8 9 10 11 12 13
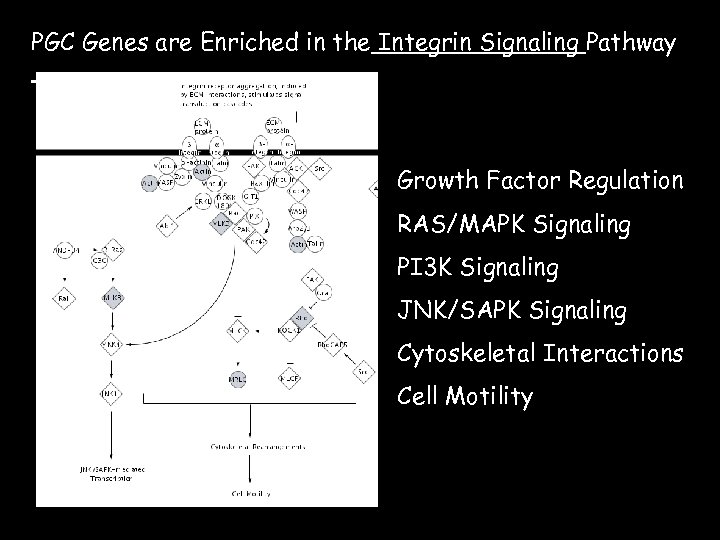 PGC Genes are Enriched in the Integrin Signaling Pathway Growth Factor Regulation RAS/MAPK Signaling PI 3 K Signaling JNK/SAPK Signaling Cytoskeletal Interactions Cell Motility
PGC Genes are Enriched in the Integrin Signaling Pathway Growth Factor Regulation RAS/MAPK Signaling PI 3 K Signaling JNK/SAPK Signaling Cytoskeletal Interactions Cell Motility
 Implications of Precise PGC Regulation Dedicated Cellular Mechanisms to Ensure Accurate Expression A Functional Requirement for Tight PGC Control in Tumors? Are Tumors Ultrasensitive to PGC Activity?
Implications of Precise PGC Regulation Dedicated Cellular Mechanisms to Ensure Accurate Expression A Functional Requirement for Tight PGC Control in Tumors? Are Tumors Ultrasensitive to PGC Activity?
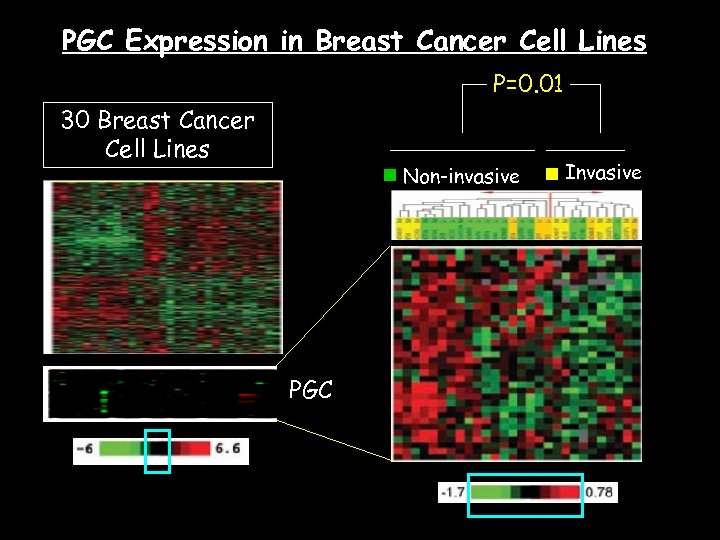 PGC Expression in Breast Cancer Cell Lines P=0. 01 30 Breast Cancer Cell Lines Non-invasive PGC Invasive
PGC Expression in Breast Cancer Cell Lines P=0. 01 30 Breast Cancer Cell Lines Non-invasive PGC Invasive
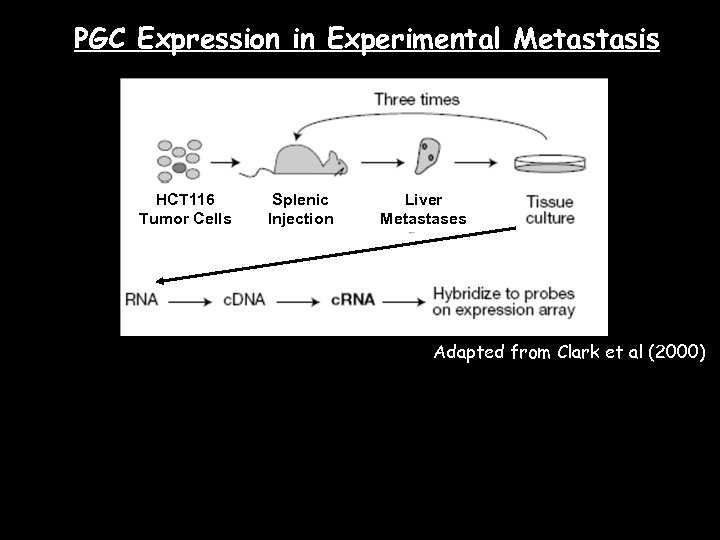 PGC Expression in Experimental Metastasis HCT 116 Tumor Cells Splenic Injection Liver Metastases Adapted from Clark et al (2000)
PGC Expression in Experimental Metastasis HCT 116 Tumor Cells Splenic Injection Liver Metastases Adapted from Clark et al (2000)
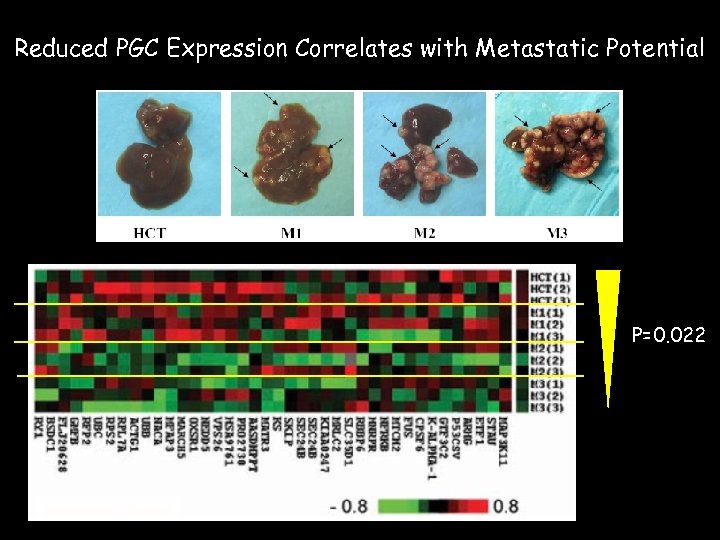 Reduced PGC Expression Correlates with Metastatic Potential P=0. 022
Reduced PGC Expression Correlates with Metastatic Potential P=0. 022
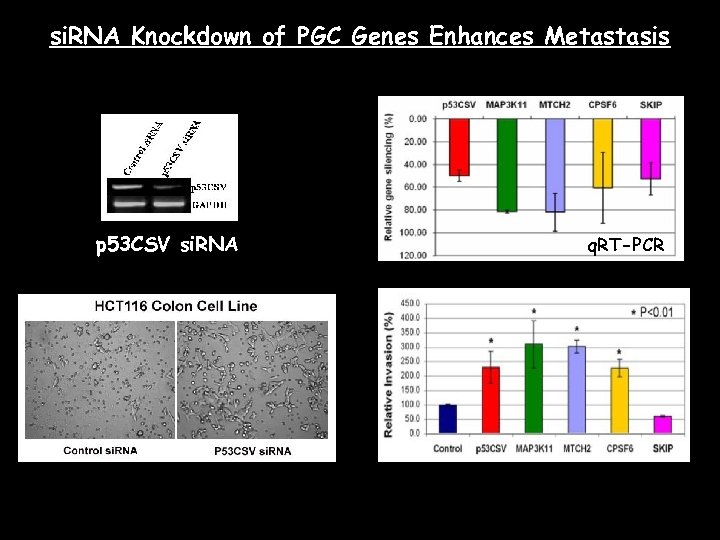 si. RNA Knockdown of PGC Genes Enhances Metastasis p 53 CSV si. RNA q. RT-PCR
si. RNA Knockdown of PGC Genes Enhances Metastasis p 53 CSV si. RNA q. RT-PCR
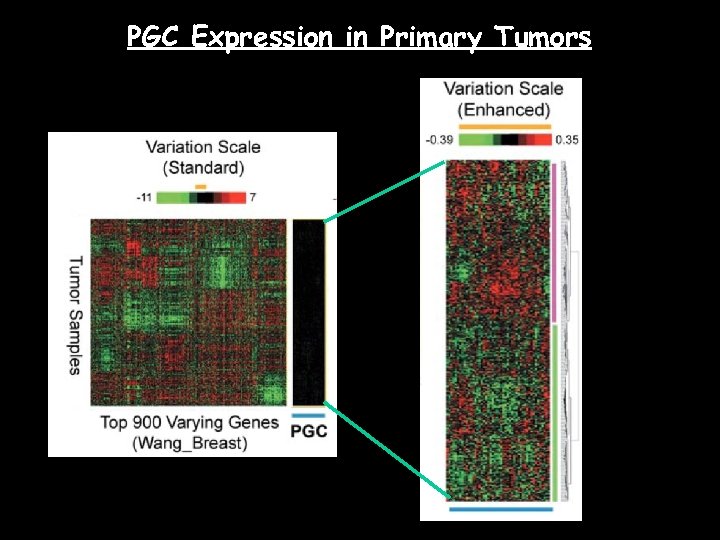 PGC Expression in Primary Tumors
PGC Expression in Primary Tumors
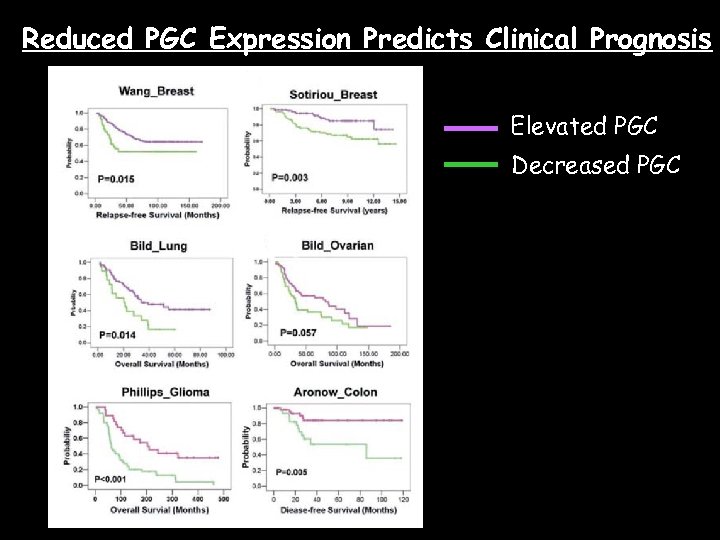 Reduced PGC Expression Predicts Clinical Prognosis Elevated PGC Decreased PGC
Reduced PGC Expression Predicts Clinical Prognosis Elevated PGC Decreased PGC
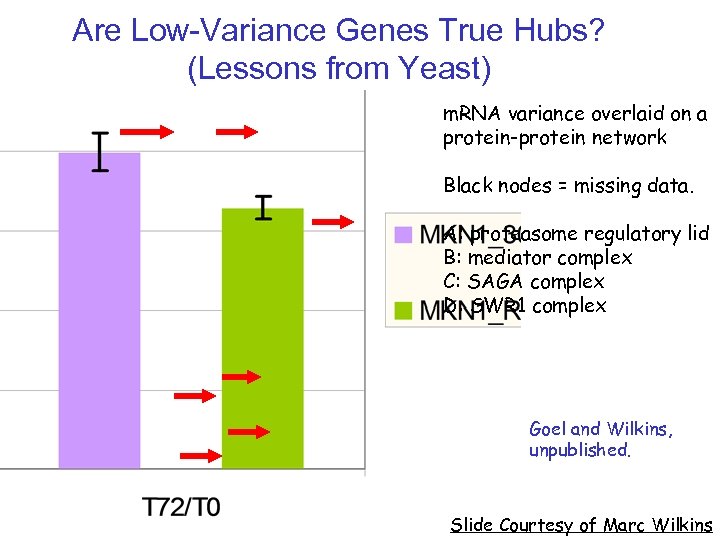 Are Low-Variance Genes True Hubs? (Lessons from Yeast) m. RNA variance overlaid on a protein-protein network Black nodes = missing data. A: proteasome regulatory lid B: mediator complex C: SAGA complex D: SWR 1 complex Goel and Wilkins, unpublished. Slide Courtesy of Marc Wilkins
Are Low-Variance Genes True Hubs? (Lessons from Yeast) m. RNA variance overlaid on a protein-protein network Black nodes = missing data. A: proteasome regulatory lid B: mediator complex C: SAGA complex D: SWR 1 complex Goel and Wilkins, unpublished. Slide Courtesy of Marc Wilkins
 Take Home Messages - A General Strategy for Identifying Tightly Regulated Genes - A Precisely Regulated Expression Cassette in Cancer - Fine-scale alterations potently modulate tumor behaviour and clinical outcome -Not discernible by conventional microarray analysis methods Yu et al (2008) PLOS Genetics
Take Home Messages - A General Strategy for Identifying Tightly Regulated Genes - A Precisely Regulated Expression Cassette in Cancer - Fine-scale alterations potently modulate tumor behaviour and clinical outcome -Not discernible by conventional microarray analysis methods Yu et al (2008) PLOS Genetics
 Basic Science to Translation 1) Metastasis Genes - Network Structures 2) Cancer Classification - Pathway Sigantures 3) Lung Cancer Outcome - Integrative Genomics
Basic Science to Translation 1) Metastasis Genes - Network Structures 2) Cancer Classification - Pathway Sigantures 3) Lung Cancer Outcome - Integrative Genomics
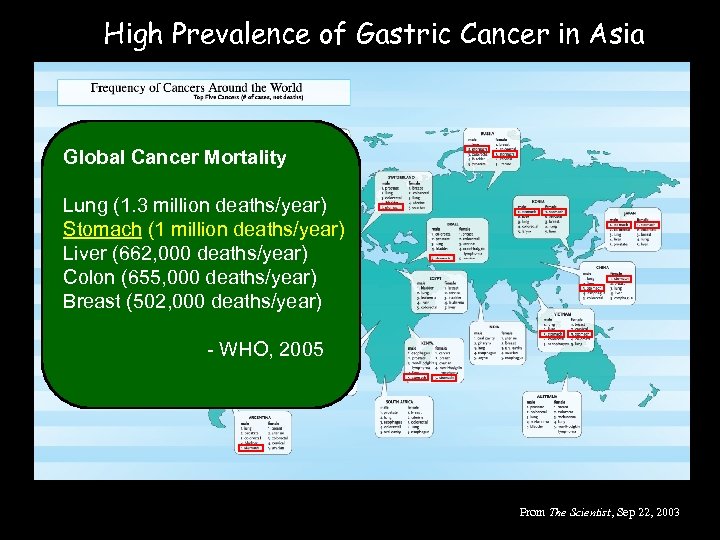 High Prevalence of Gastric Cancer in Asia Global Cancer Mortality Lung (1. 3 million deaths/year) Stomach (1 million deaths/year) Liver (662, 000 deaths/year) Colon (655, 000 deaths/year) Breast (502, 000 deaths/year) - WHO, 2005 From The Scientist, Sep 22, 2003
High Prevalence of Gastric Cancer in Asia Global Cancer Mortality Lung (1. 3 million deaths/year) Stomach (1 million deaths/year) Liver (662, 000 deaths/year) Colon (655, 000 deaths/year) Breast (502, 000 deaths/year) - WHO, 2005 From The Scientist, Sep 22, 2003
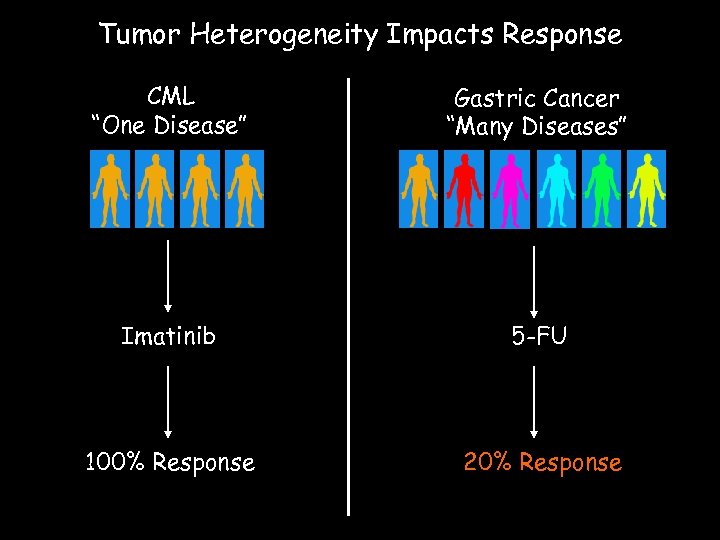 Tumor Heterogeneity Impacts Response CML “One Disease” Gastric Cancer “Many Diseases” Imatinib 5 -FU 100% Response 20% Response
Tumor Heterogeneity Impacts Response CML “One Disease” Gastric Cancer “Many Diseases” Imatinib 5 -FU 100% Response 20% Response
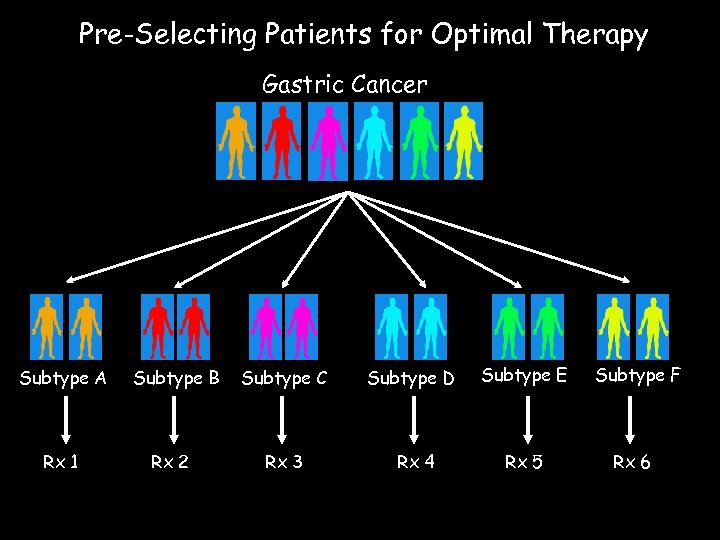 Pre-Selecting Patients for Optimal Therapy Gastric Cancer Subtype A Rx 1 Subtype B Rx 2 Subtype C Subtype D Subtype E Rx 3 Rx 4 Rx 5 Subtype F Rx 6
Pre-Selecting Patients for Optimal Therapy Gastric Cancer Subtype A Rx 1 Subtype B Rx 2 Subtype C Subtype D Subtype E Rx 3 Rx 4 Rx 5 Subtype F Rx 6
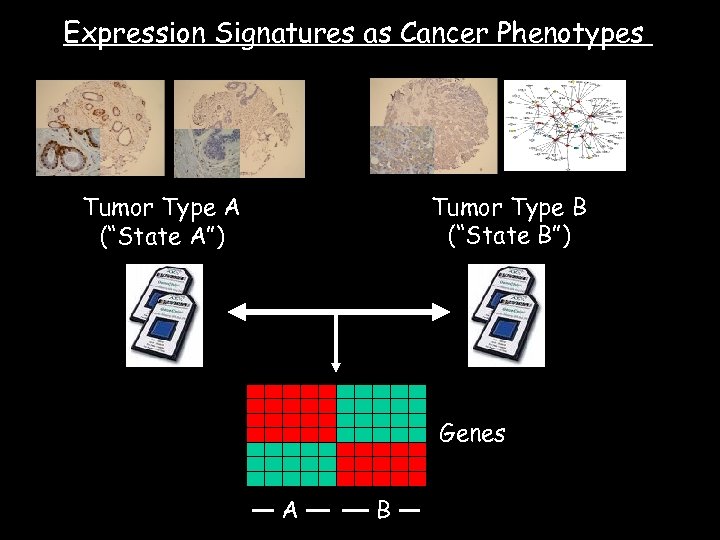 Expression Signatures as Cancer Phenotypes Tumor Type B (“State B”) Tumor Type A (“State A”) Genes A B
Expression Signatures as Cancer Phenotypes Tumor Type B (“State B”) Tumor Type A (“State A”) Genes A B
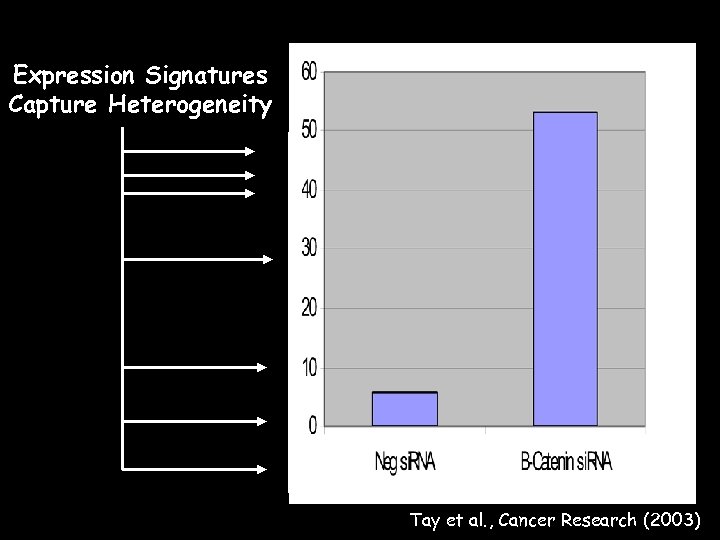 Expression Signatures Capture Heterogeneity Tay et al. , Cancer Research (2003)
Expression Signatures Capture Heterogeneity Tay et al. , Cancer Research (2003)
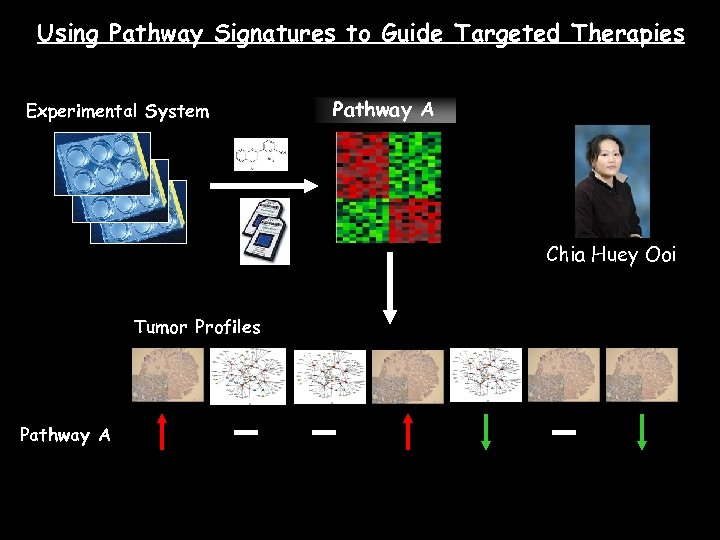 Using Pathway Signatures to Guide Targeted Therapies Experimental System Pathway A Chia Huey Ooi Tumor Profiles Pathway A
Using Pathway Signatures to Guide Targeted Therapies Experimental System Pathway A Chia Huey Ooi Tumor Profiles Pathway A
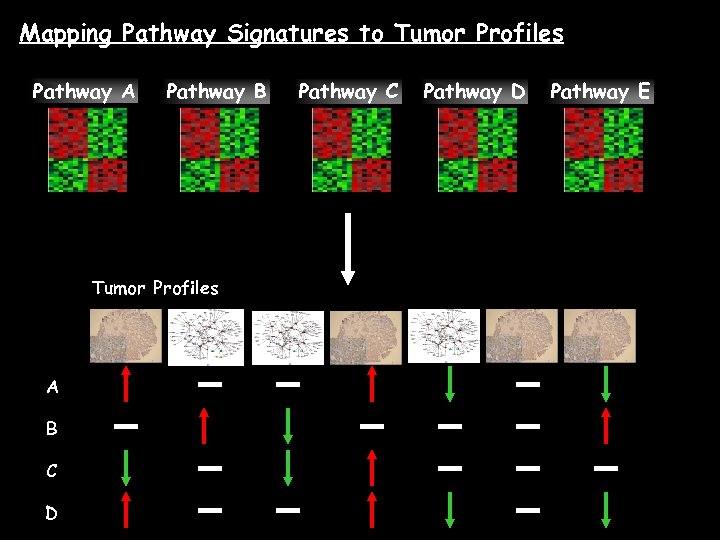 Mapping Pathway Signatures to Tumor Profiles Pathway A Pathway B Tumor Profiles A B C D Pathway C Pathway D Pathway E
Mapping Pathway Signatures to Tumor Profiles Pathway A Pathway B Tumor Profiles A B C D Pathway C Pathway D Pathway E
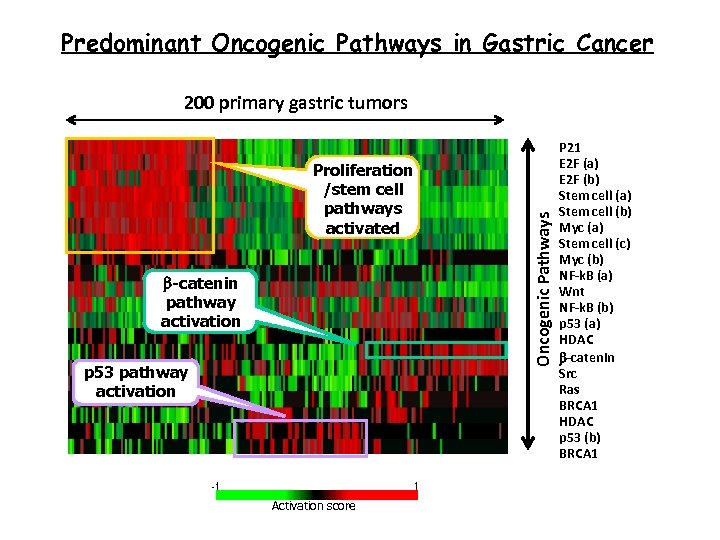 Predominant Oncogenic Pathways in Gastric Cancer Proliferation /stem cell pathways activated b-catenin pathway activation p 53 pathway activation Activation score Oncogenic Pathways 200 primary gastric tumors P 21 E 2 F (a) E 2 F (b) Stem cell (a) Stem cell (b) Myc (a) Stem cell (c) Myc (b) NF-k. B (a) Wnt NF-k. B (b) p 53 (a) HDAC b-catenin Src Ras BRCA 1 HDAC p 53 (b) BRCA 1
Predominant Oncogenic Pathways in Gastric Cancer Proliferation /stem cell pathways activated b-catenin pathway activation p 53 pathway activation Activation score Oncogenic Pathways 200 primary gastric tumors P 21 E 2 F (a) E 2 F (b) Stem cell (a) Stem cell (b) Myc (a) Stem cell (c) Myc (b) NF-k. B (a) Wnt NF-k. B (b) p 53 (a) HDAC b-catenin Src Ras BRCA 1 HDAC p 53 (b) BRCA 1
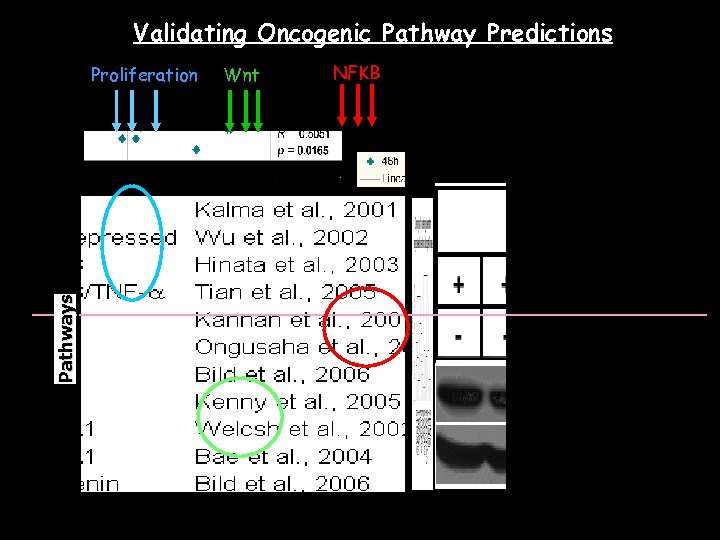 Validating Oncogenic Pathway Predictions Wnt Pathways Proliferation GC cell lines NFKB
Validating Oncogenic Pathway Predictions Wnt Pathways Proliferation GC cell lines NFKB
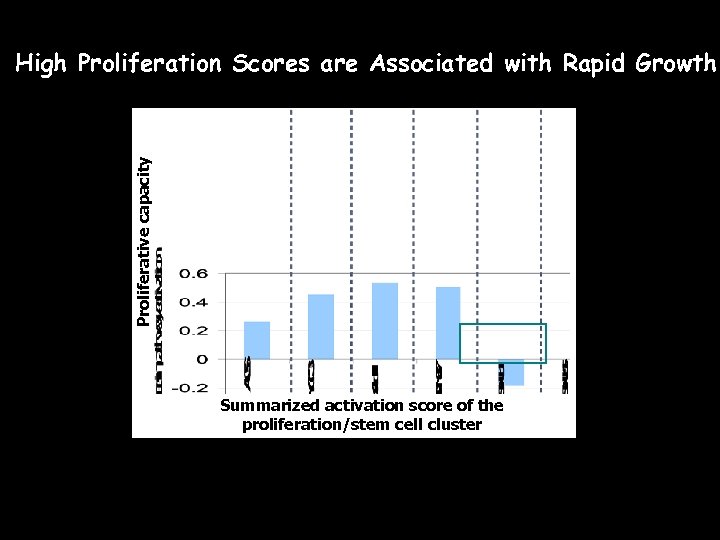 Proliferative capacity High Proliferation Scores are Associated with Rapid Growth Summarized activation score of the proliferation/stem cell cluster
Proliferative capacity High Proliferation Scores are Associated with Rapid Growth Summarized activation score of the proliferation/stem cell cluster
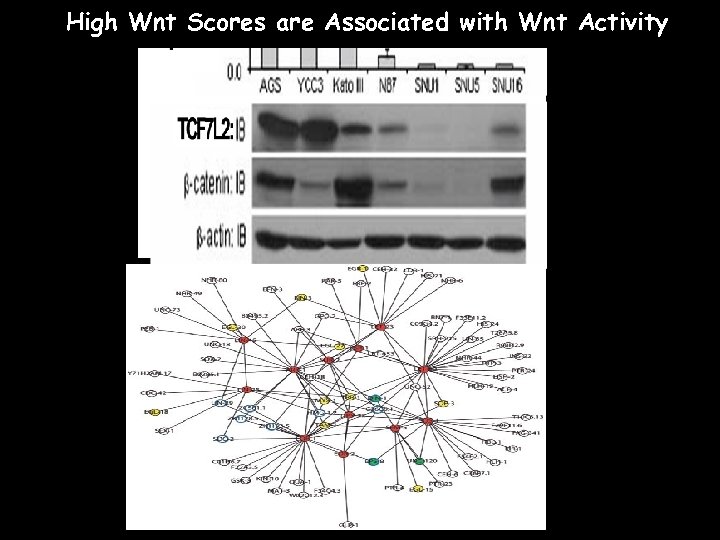 High Wnt Scores are Associated with Wnt Activity
High Wnt Scores are Associated with Wnt Activity
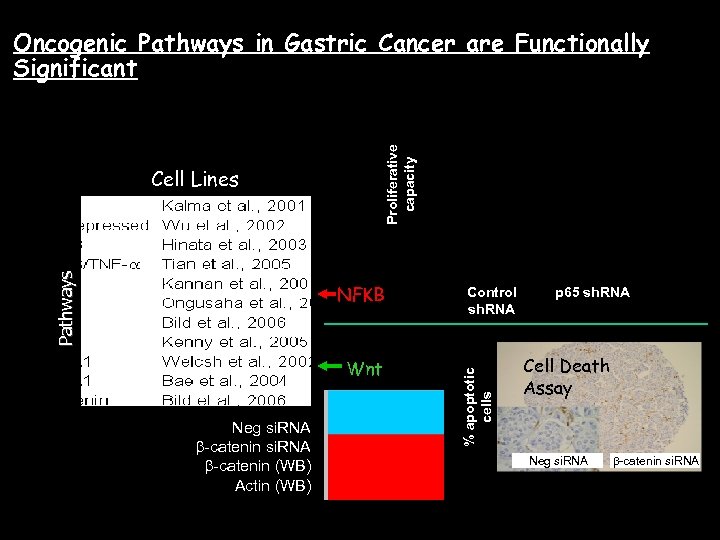 Oncogenic Pathways in Gastric Cancer are Functionally Significant Proliferative capacity p=4. 549 10 6 NFKB Wnt Neg si. RNA b-catenin (WB) GC cell Actin (WB)lines Control sh. RNA % apoptotic cells Pathways Cell Lines p 65 sh. RNA Cell Death Assay Neg si. RNA b-catenin si. RNA
Oncogenic Pathways in Gastric Cancer are Functionally Significant Proliferative capacity p=4. 549 10 6 NFKB Wnt Neg si. RNA b-catenin (WB) GC cell Actin (WB)lines Control sh. RNA % apoptotic cells Pathways Cell Lines p 65 sh. RNA Cell Death Assay Neg si. RNA b-catenin si. RNA
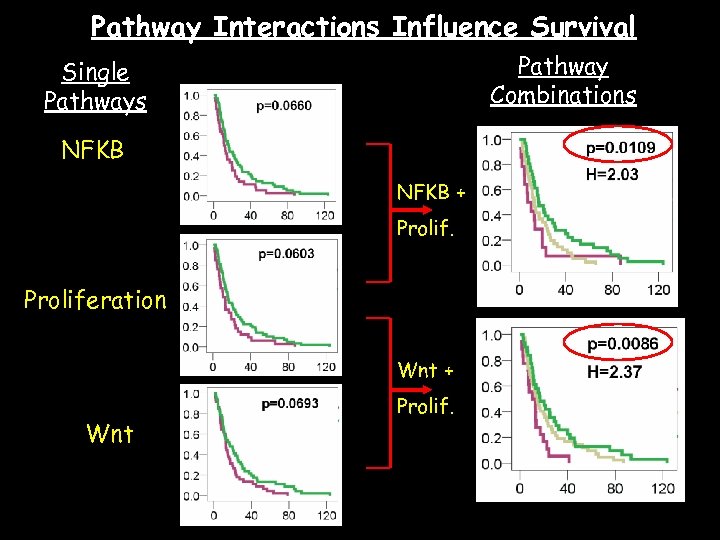 Pathway Interactions Influence Survival Pathway Combinations Single Pathways NFKB + Proliferation Wnt + Wnt Prolif.
Pathway Interactions Influence Survival Pathway Combinations Single Pathways NFKB + Proliferation Wnt + Wnt Prolif.
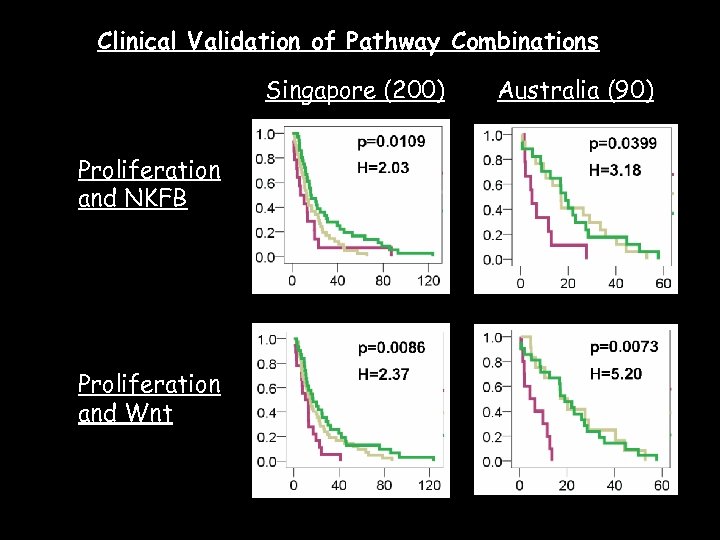 Clinical Validation of Pathway Combinations Singapore (200) Proliferation and NKFB Proliferation and Wnt Australia (90)
Clinical Validation of Pathway Combinations Singapore (200) Proliferation and NKFB Proliferation and Wnt Australia (90)
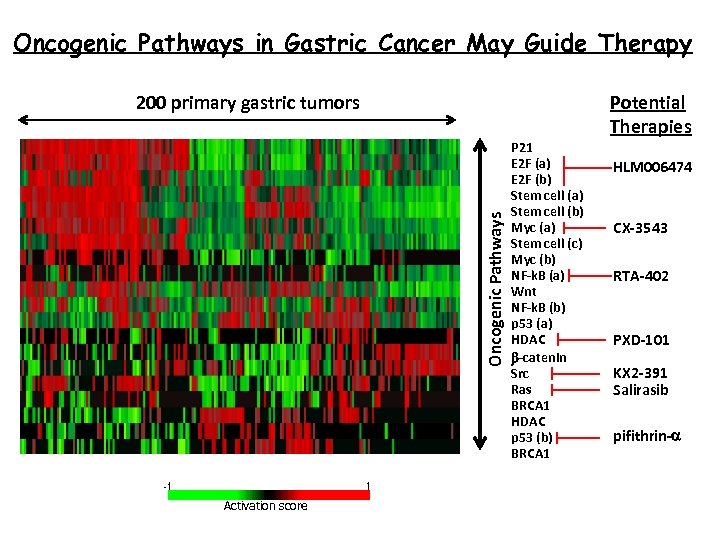 Oncogenic Pathways in Gastric Cancer May Guide Therapy Potential Therapies Oncogenic Pathways 200 primary gastric tumors Activation score P 21 E 2 F (a) E 2 F (b) Stem cell (a) Stem cell (b) Myc (a) Stem cell (c) Myc (b) NF-k. B (a) Wnt NF-k. B (b) p 53 (a) HDAC b-catenin Src Ras BRCA 1 HDAC p 53 (b) BRCA 1 HLM 006474 CX-3543 RTA-402 PXD-101 KX 2 -391 Salirasib pifithrin-a
Oncogenic Pathways in Gastric Cancer May Guide Therapy Potential Therapies Oncogenic Pathways 200 primary gastric tumors Activation score P 21 E 2 F (a) E 2 F (b) Stem cell (a) Stem cell (b) Myc (a) Stem cell (c) Myc (b) NF-k. B (a) Wnt NF-k. B (b) p 53 (a) HDAC b-catenin Src Ras BRCA 1 HDAC p 53 (b) BRCA 1 HLM 006474 CX-3543 RTA-402 PXD-101 KX 2 -391 Salirasib pifithrin-a
 Take Home Messages • A framework for mapping defined pathway signatures into complex tumor profiles • Signatures are transportable (in vitro to in vivo) • Gastric cancers can be subdivided by pathway activity into biologically and clinically relevant subgroups • “High-throughput pathway profiling” highlights the role of oncogenic pathway combinations in clinical behavior Ooi et al (2009) PLOS Genetics
Take Home Messages • A framework for mapping defined pathway signatures into complex tumor profiles • Signatures are transportable (in vitro to in vivo) • Gastric cancers can be subdivided by pathway activity into biologically and clinically relevant subgroups • “High-throughput pathway profiling” highlights the role of oncogenic pathway combinations in clinical behavior Ooi et al (2009) PLOS Genetics
 Basic Science to Translation 1) Metastasis Genes - Network Structures 2) Cancer Classification - Pathway Biology 3) Lung Cancer Outcome - Integrative Genomics
Basic Science to Translation 1) Metastasis Genes - Network Structures 2) Cancer Classification - Pathway Biology 3) Lung Cancer Outcome - Integrative Genomics
 Genomic Classification of Early Stage Lung Cancer Philippe and Sophine Broet INSERM U 472, Faculté de Médecine Paris-Sud Lance Miller Wake Forest University, USA Broet et al. , (2009) Cancer Research
Genomic Classification of Early Stage Lung Cancer Philippe and Sophine Broet INSERM U 472, Faculté de Médecine Paris-Sud Lance Miller Wake Forest University, USA Broet et al. , (2009) Cancer Research
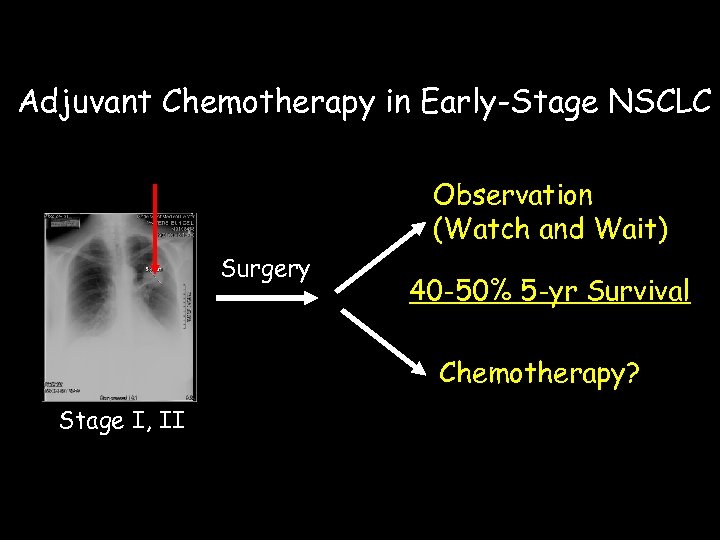 Adjuvant Chemotherapy in Early-Stage NSCLC Observation (Watch and Wait) Surgery 40 -50% 5 -yr Survival Chemotherapy? Stage I, II
Adjuvant Chemotherapy in Early-Stage NSCLC Observation (Watch and Wait) Surgery 40 -50% 5 -yr Survival Chemotherapy? Stage I, II
 Study Questions Clinical questions Can we use genomics to discriminate between low risk (pseudo-stage I) & high risk (pseudo-stage II) groups? Previous studies on NSCLC prognosis have been transcriptome centered, not incorporating genomic alterations
Study Questions Clinical questions Can we use genomics to discriminate between low risk (pseudo-stage I) & high risk (pseudo-stage II) groups? Previous studies on NSCLC prognosis have been transcriptome centered, not incorporating genomic alterations
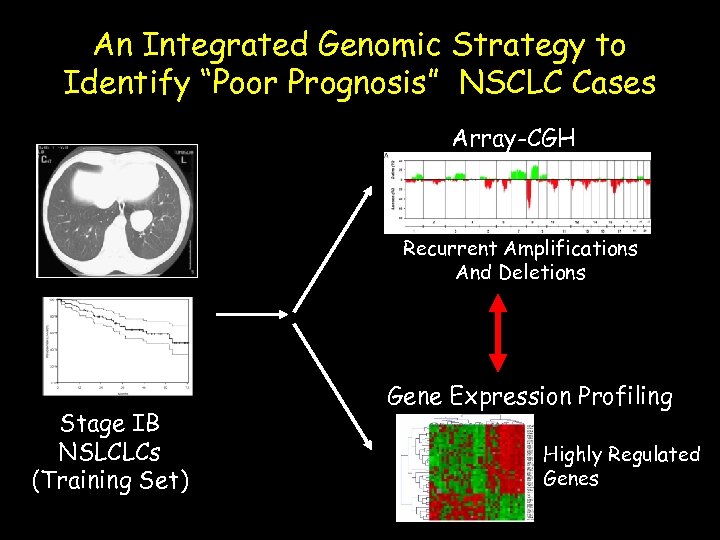 An Integrated Genomic Strategy to Identify “Poor Prognosis” NSCLC Cases Array-CGH Recurrent Amplifications And Deletions Stage IB NSLCLCs (Training Set) Gene Expression Profiling Highly Regulated Genes
An Integrated Genomic Strategy to Identify “Poor Prognosis” NSCLC Cases Array-CGH Recurrent Amplifications And Deletions Stage IB NSLCLCs (Training Set) Gene Expression Profiling Highly Regulated Genes
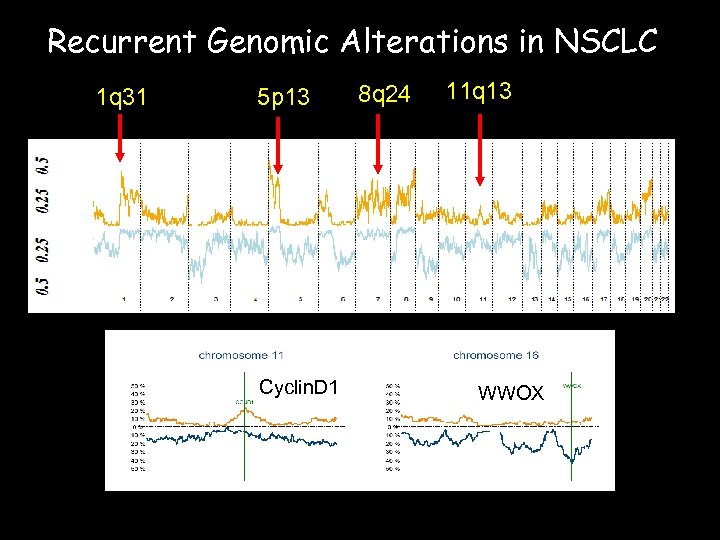 Recurrent Genomic Alterations in NSCLC 1 q 31 5 p 13 Cyclin. D 1 8 q 24 11 q 13 WWOX
Recurrent Genomic Alterations in NSCLC 1 q 31 5 p 13 Cyclin. D 1 8 q 24 11 q 13 WWOX
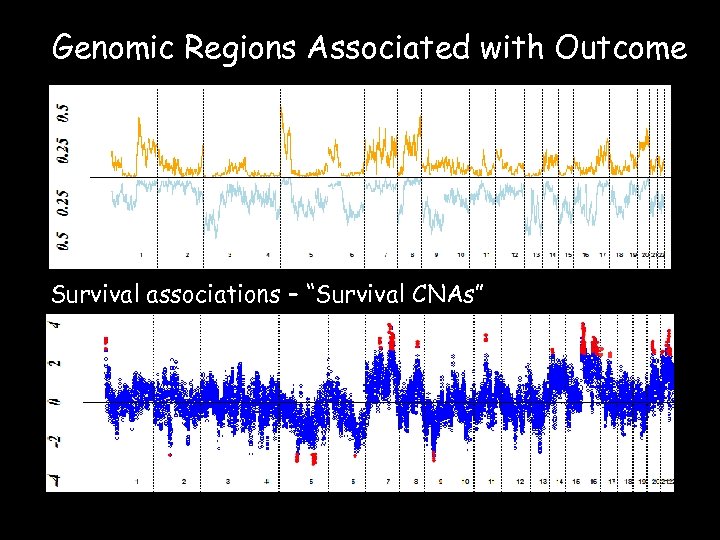 Genomic Regions Associated with Outcome Survival associations – “Survival CNAs”
Genomic Regions Associated with Outcome Survival associations – “Survival CNAs”
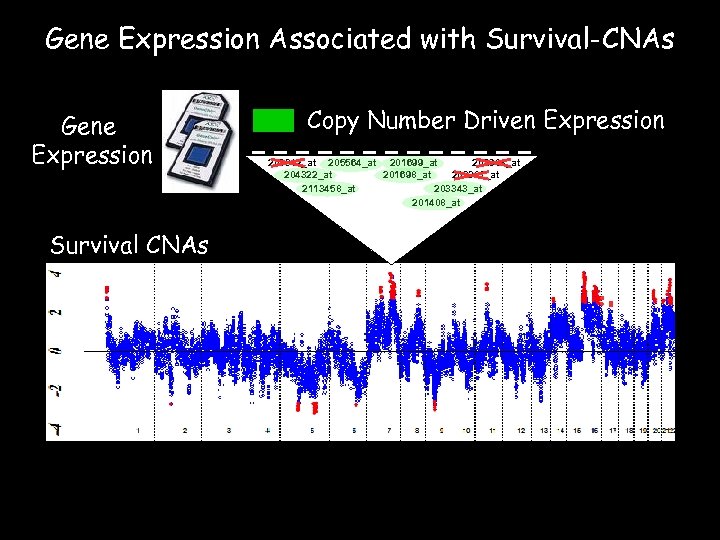 Gene Expression Associated with Survival-CNAs Gene Expression Survival CNAs Copy Number Driven Expression 203342_at 205564_at 201699_at 202988_at 204322_at 201698_at 203301_at 2113458_at 203343_at 201408_at
Gene Expression Associated with Survival-CNAs Gene Expression Survival CNAs Copy Number Driven Expression 203342_at 205564_at 201699_at 202988_at 204322_at 201698_at 203301_at 2113458_at 203343_at 201408_at
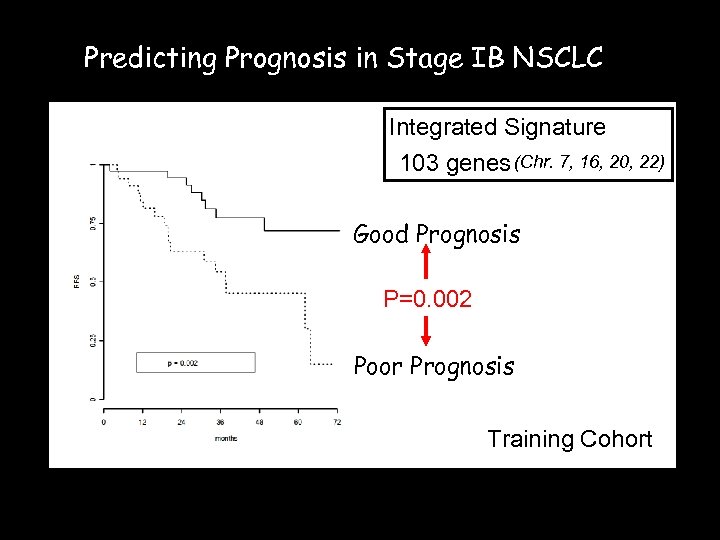 Predicting Prognosis in Stage IB NSCLC Integrated Signature 103 genes (Chr. 7, 16, 20, 22) Good Prognosis P=0. 002 Poor Prognosis Training Cohort
Predicting Prognosis in Stage IB NSCLC Integrated Signature 103 genes (Chr. 7, 16, 20, 22) Good Prognosis P=0. 002 Poor Prognosis Training Cohort
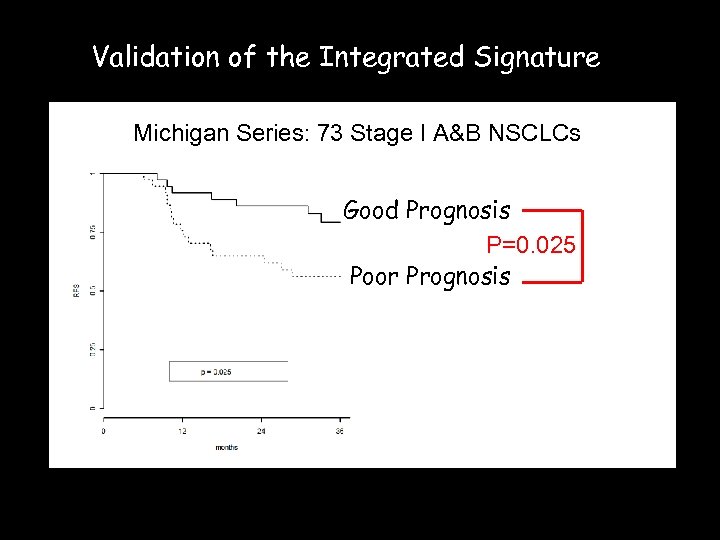 Validation of the Integrated Signature Michigan Series: 73 Stage I A&B NSCLCs Good Prognosis P=0. 025 Poor Prognosis
Validation of the Integrated Signature Michigan Series: 73 Stage I A&B NSCLCs Good Prognosis P=0. 025 Poor Prognosis
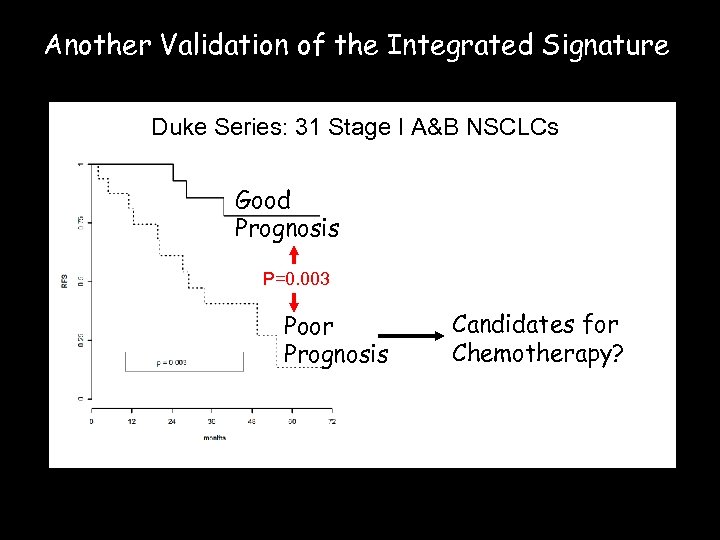 Another Validation of the Integrated Signature Duke Series: 31 Stage I A&B NSCLCs Good Prognosis P=0. 003 Poor Prognosis Candidates for Chemotherapy?
Another Validation of the Integrated Signature Duke Series: 31 Stage I A&B NSCLCs Good Prognosis P=0. 003 Poor Prognosis Candidates for Chemotherapy?
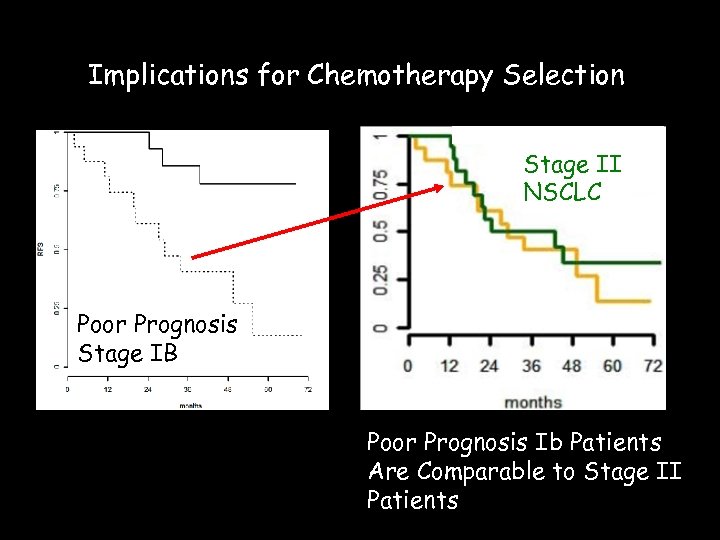 Implications for Chemotherapy Selection Stage II NSCLC Poor Prognosis Stage IB Poor Prognosis Ib Patients Are Comparable to Stage II Patients
Implications for Chemotherapy Selection Stage II NSCLC Poor Prognosis Stage IB Poor Prognosis Ib Patients Are Comparable to Stage II Patients
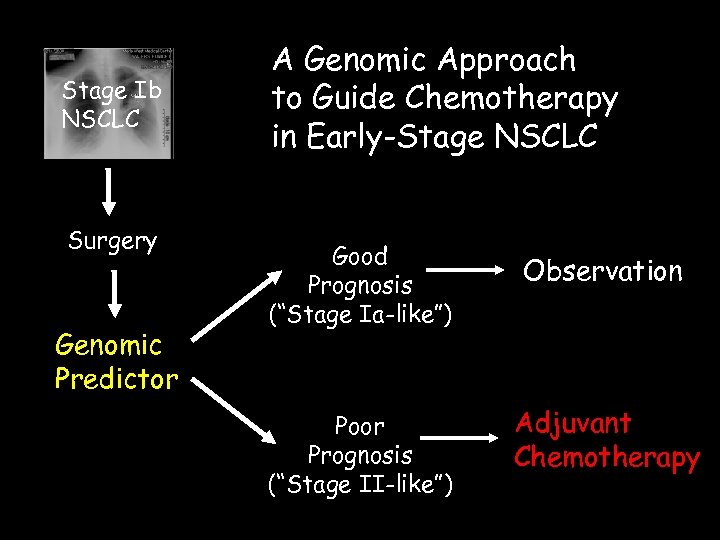 Stage Ib NSCLC Surgery Genomic Predictor A Genomic Approach to Guide Chemotherapy in Early-Stage NSCLC Good Prognosis (“Stage Ia-like”) Observation Poor Prognosis (“Stage II-like”) Adjuvant Chemotherapy
Stage Ib NSCLC Surgery Genomic Predictor A Genomic Approach to Guide Chemotherapy in Early-Stage NSCLC Good Prognosis (“Stage Ia-like”) Observation Poor Prognosis (“Stage II-like”) Adjuvant Chemotherapy
 Acknowledgements Kun Yu Kumaresan Ganesan Ooi Chia Huey Tatiana Ivanova Shenli Zhang Wu Yonghui Lai Ling Cheng Veena Gopalakrishnan Jun Hao Koo Julian Lee Ming Hui Lee Iain Tan Angie Tan Jiong Tao Jeanie Wu Yansong Zhu Philippe Broet (Paris) Sophine Broet (Paris) Lance Miller (GIS) Elaine Lim (NUH) Wei Chia Lin (GIS) Hooi Shing Chuan (NUS) Alex Boussioutas (Peter Mac, AU) David Bowtell (Peter Mac, AU) Sun Yong Rha (S. Korea) Heike Grabsch (Leeds) Support : French-Singapore MERLION program Singapore Cancer Syndicate Biomedical Research Council National Medical Research Council
Acknowledgements Kun Yu Kumaresan Ganesan Ooi Chia Huey Tatiana Ivanova Shenli Zhang Wu Yonghui Lai Ling Cheng Veena Gopalakrishnan Jun Hao Koo Julian Lee Ming Hui Lee Iain Tan Angie Tan Jiong Tao Jeanie Wu Yansong Zhu Philippe Broet (Paris) Sophine Broet (Paris) Lance Miller (GIS) Elaine Lim (NUH) Wei Chia Lin (GIS) Hooi Shing Chuan (NUS) Alex Boussioutas (Peter Mac, AU) David Bowtell (Peter Mac, AU) Sun Yong Rha (S. Korea) Heike Grabsch (Leeds) Support : French-Singapore MERLION program Singapore Cancer Syndicate Biomedical Research Council National Medical Research Council


