SSW3 TRanslation.pptx
- Количество слайдов: 29
 Initiation of translation in prokaryotes: initiation factors, initiator codons, 3'end of RNA small ribosomal subunit and the Shine-Dalgarno sequence in m. RNA» Done by: Maulenova R. Moldakozhayev A. Naizabayeva D.
Initiation of translation in prokaryotes: initiation factors, initiator codons, 3'end of RNA small ribosomal subunit and the Shine-Dalgarno sequence in m. RNA» Done by: Maulenova R. Moldakozhayev A. Naizabayeva D.
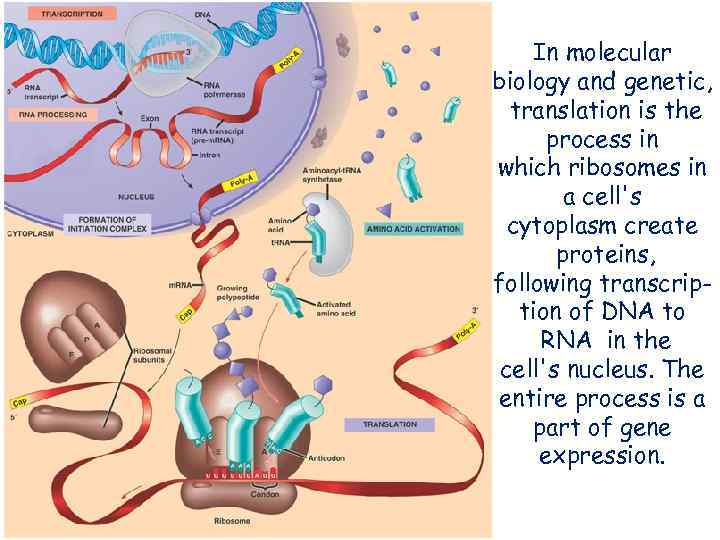 In molecular biology and genetic, translation is the process in which ribosomes in a cell's cytoplasm create proteins, following transcription of DNA to RNA in the cell's nucleus. The entire process is a part of gene expression.
In molecular biology and genetic, translation is the process in which ribosomes in a cell's cytoplasm create proteins, following transcription of DNA to RNA in the cell's nucleus. The entire process is a part of gene expression.
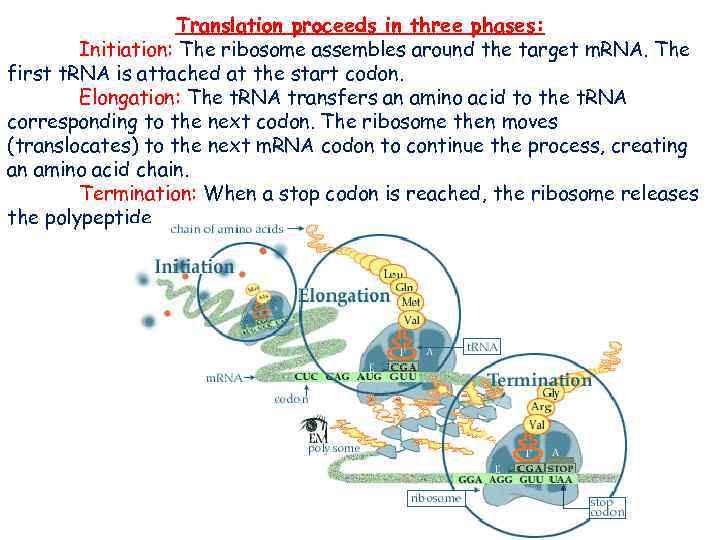 Translation proceeds in three phases: Initiation: The ribosome assembles around the target m. RNA. The first t. RNA is attached at the start codon. Elongation: The t. RNA transfers an amino acid to the t. RNA corresponding to the next codon. The ribosome then moves (translocates) to the next m. RNA codon to continue the process, creating an amino acid chain. Termination: When a stop codon is reached, the ribosome releases the polypeptide.
Translation proceeds in three phases: Initiation: The ribosome assembles around the target m. RNA. The first t. RNA is attached at the start codon. Elongation: The t. RNA transfers an amino acid to the t. RNA corresponding to the next codon. The ribosome then moves (translocates) to the next m. RNA codon to continue the process, creating an amino acid chain. Termination: When a stop codon is reached, the ribosome releases the polypeptide.
 In bacteria, translation occurs in the cytoplasm, where the large and small subunits of the ribosome bind to the m. RNA. In eukaryotes, translation occurs in the cytosol or across the membrane of the endoplasmic reticulum in a process called vectorial synthesis. In many instances, the entire ribosome/m. RNA complex binds to the outer membrane of the rough endoplasmic reticulum (ER); the newly created polypeptide is stored inside the ER for later vesicle transport and secretion outside of the cell. Many types of transcribed RNA, such as transfer RNA, ribosomal RNA, and small nuclear RNA, do not undergo translation into proteins.
In bacteria, translation occurs in the cytoplasm, where the large and small subunits of the ribosome bind to the m. RNA. In eukaryotes, translation occurs in the cytosol or across the membrane of the endoplasmic reticulum in a process called vectorial synthesis. In many instances, the entire ribosome/m. RNA complex binds to the outer membrane of the rough endoplasmic reticulum (ER); the newly created polypeptide is stored inside the ER for later vesicle transport and secretion outside of the cell. Many types of transcribed RNA, such as transfer RNA, ribosomal RNA, and small nuclear RNA, do not undergo translation into proteins.
 A number of antibiotics act by inhibiting translation. These include anisomycin, cycloheximide, chloramphenicol, tetracycline, streptomycin, erythromycin, and puromycin. Prokaryotic ribosomes have a different structure from that of eukaryotic ribosomes, and thus antibiotics can specifically target bacterial infections without any harm to a eukaryotic host's cells.
A number of antibiotics act by inhibiting translation. These include anisomycin, cycloheximide, chloramphenicol, tetracycline, streptomycin, erythromycin, and puromycin. Prokaryotic ribosomes have a different structure from that of eukaryotic ribosomes, and thus antibiotics can specifically target bacterial infections without any harm to a eukaryotic host's cells.
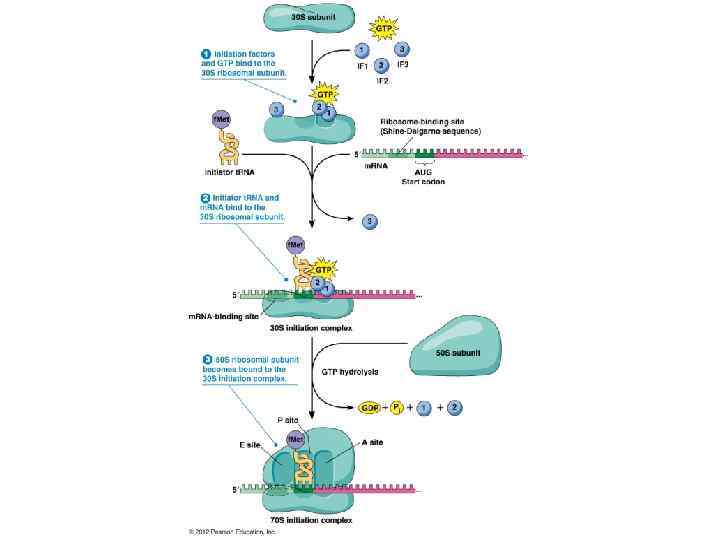
 Translation initiation: Initiation factors Prokaryotes require the use of three initiation factors: IF 1, IF 2, and IF 3, for translation. IF 1 associates with the 30 S ribosomal subunit in the A site and prevents an aminoacyl-t. RNA from entering. It modulates IF 2 binding to the ribosome by increasing its affinity. It may also prevent the 50 S subunit from binding, stopping the formation of the 70 S subunit. It also contains a β-domain fold common for nucleic acid binding proteins.
Translation initiation: Initiation factors Prokaryotes require the use of three initiation factors: IF 1, IF 2, and IF 3, for translation. IF 1 associates with the 30 S ribosomal subunit in the A site and prevents an aminoacyl-t. RNA from entering. It modulates IF 2 binding to the ribosome by increasing its affinity. It may also prevent the 50 S subunit from binding, stopping the formation of the 70 S subunit. It also contains a β-domain fold common for nucleic acid binding proteins.
 Translation initiation: Initiation factors IF 2 binds to an initiator t. RNA and controls the entry of t. RNA onto the ribosome. IF 2, bound to GTP, binds to the 30 S P site. After associating with the 30 S subunit, f. Mett. RNAf binds to the IF 2, then IF 2 transfers the t. RNA into the partial P site. When the 50 S subunit joins, it hydrolyzes GTP to GDP and Pi, causing a conformational change in the IF 2 that causes IF 2 to release and allow the 70 S subunit to form.
Translation initiation: Initiation factors IF 2 binds to an initiator t. RNA and controls the entry of t. RNA onto the ribosome. IF 2, bound to GTP, binds to the 30 S P site. After associating with the 30 S subunit, f. Mett. RNAf binds to the IF 2, then IF 2 transfers the t. RNA into the partial P site. When the 50 S subunit joins, it hydrolyzes GTP to GDP and Pi, causing a conformational change in the IF 2 that causes IF 2 to release and allow the 70 S subunit to form.
 Translation initiation: Initiation factors IF 3 is not universally found in all bacterial species but in E. coli it is required for the 30 S subunit to bind to the initiation site in m. RNA. In addition, it has several other jobs including the stabilization of free 30 S subunits, enables 30 S subunits to bind to m. RNA and checks for accuracy against the first aminoacyl-t. RNA. It also allows for rapid codon-anticodon pairing for the initiator t. RNA to bind quickly to. IF 3 is required by the small subunit to form initiation complexes, but has to be released to allow the 50 S subunit to bind.
Translation initiation: Initiation factors IF 3 is not universally found in all bacterial species but in E. coli it is required for the 30 S subunit to bind to the initiation site in m. RNA. In addition, it has several other jobs including the stabilization of free 30 S subunits, enables 30 S subunits to bind to m. RNA and checks for accuracy against the first aminoacyl-t. RNA. It also allows for rapid codon-anticodon pairing for the initiator t. RNA to bind quickly to. IF 3 is required by the small subunit to form initiation complexes, but has to be released to allow the 50 S subunit to bind.
 Ribosome The fact that cells typically contain many ribosomes reflects the central importance of protein synthesis in cell metabolism. E. coli, for example, contain about 20, 000 ribosomes, which account for approximately 25% of the dry weight of the cell, and rapidly growing mammalian cells contain about 10 million ribosomes.
Ribosome The fact that cells typically contain many ribosomes reflects the central importance of protein synthesis in cell metabolism. E. coli, for example, contain about 20, 000 ribosomes, which account for approximately 25% of the dry weight of the cell, and rapidly growing mammalian cells contain about 10 million ribosomes.
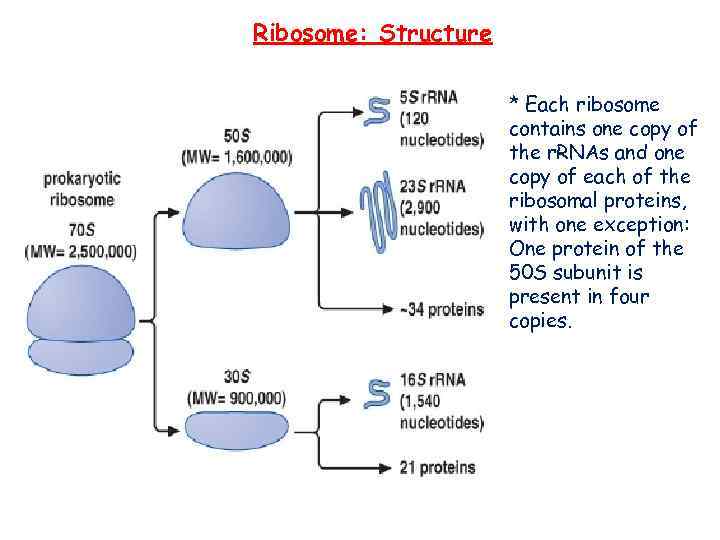 Ribosome: Structure * Each ribosome contains one copy of the r. RNAs and one copy of each of the ribosomal proteins, with one exception: One protein of the 50 S subunit is present in four copies.
Ribosome: Structure * Each ribosome contains one copy of the r. RNAs and one copy of each of the ribosomal proteins, with one exception: One protein of the 50 S subunit is present in four copies.
 Ribosome: r. RNA A noteworthy feature of ribosomes is that they can be formed in vitro by self-assembly of their RNA and protein constituents. As first described in 1968 by Masayasu Nomura, purified ribosomal proteins and r. RNAs can be mixed together and, under appropriate conditions, will reform a functional ribosome. Initially, r. RNAs were thought to play a structural role, providing a scaffold upon which ribosomal proteins assemble. However, with the discovery of the catalytic activity of other RNA molecules, the possible catalytic role of r. RNA became widely considered. Consistent with this hypothesis, r. RNAs were found to be absolutely required for the in vitro assembly of functional ribosomes.
Ribosome: r. RNA A noteworthy feature of ribosomes is that they can be formed in vitro by self-assembly of their RNA and protein constituents. As first described in 1968 by Masayasu Nomura, purified ribosomal proteins and r. RNAs can be mixed together and, under appropriate conditions, will reform a functional ribosome. Initially, r. RNAs were thought to play a structural role, providing a scaffold upon which ribosomal proteins assemble. However, with the discovery of the catalytic activity of other RNA molecules, the possible catalytic role of r. RNA became widely considered. Consistent with this hypothesis, r. RNAs were found to be absolutely required for the in vitro assembly of functional ribosomes.
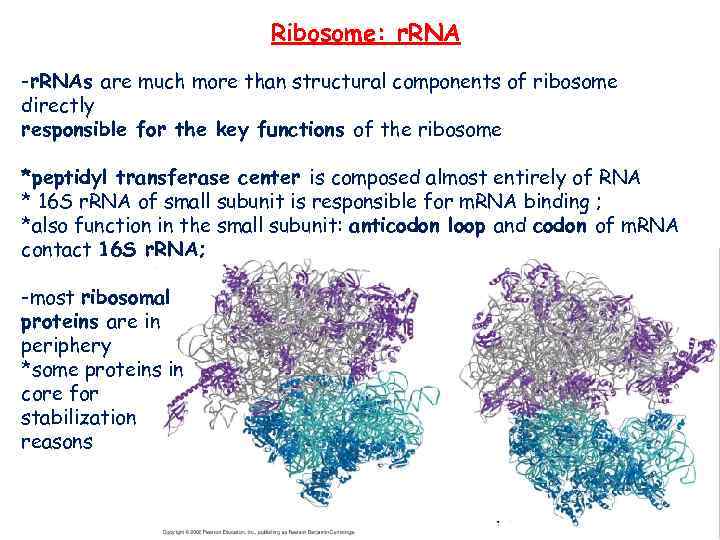 Ribosome: r. RNA -r. RNAs are much more than structural components of ribosome directly responsible for the key functions of the ribosome *peptidyl transferase center is composed almost entirely of RNA * 16 S r. RNA of small subunit is responsible for m. RNA binding ; *also function in the small subunit: anticodon loop and codon of m. RNA contact 16 S r. RNA; -most ribosomal proteins are in periphery *some proteins in core for stabilization reasons
Ribosome: r. RNA -r. RNAs are much more than structural components of ribosome directly responsible for the key functions of the ribosome *peptidyl transferase center is composed almost entirely of RNA * 16 S r. RNA of small subunit is responsible for m. RNA binding ; *also function in the small subunit: anticodon loop and codon of m. RNA contact 16 S r. RNA; -most ribosomal proteins are in periphery *some proteins in core for stabilization reasons
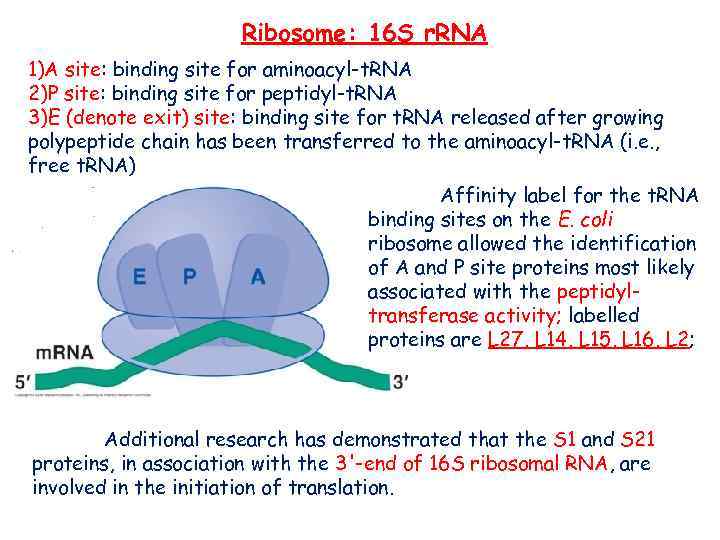 Ribosome: 16 S r. RNA 1)A site: binding site for aminoacyl-t. RNA 2)P site: binding site for peptidyl-t. RNA 3)E (denote exit) site: binding site for t. RNA released after growing polypeptide chain has been transferred to the aminoacyl-t. RNA (i. e. , free t. RNA) Affinity label for the t. RNA binding sites on the E. coli ribosome allowed the identification of A and P site proteins most likely associated with the peptidyltransferase activity; labelled proteins are L 27, L 14, L 15, L 16, L 2; Additional research has demonstrated that the S 1 and S 21 proteins, in association with the 3'-end of 16 S ribosomal RNA, are involved in the initiation of translation.
Ribosome: 16 S r. RNA 1)A site: binding site for aminoacyl-t. RNA 2)P site: binding site for peptidyl-t. RNA 3)E (denote exit) site: binding site for t. RNA released after growing polypeptide chain has been transferred to the aminoacyl-t. RNA (i. e. , free t. RNA) Affinity label for the t. RNA binding sites on the E. coli ribosome allowed the identification of A and P site proteins most likely associated with the peptidyltransferase activity; labelled proteins are L 27, L 14, L 15, L 16, L 2; Additional research has demonstrated that the S 1 and S 21 proteins, in association with the 3'-end of 16 S ribosomal RNA, are involved in the initiation of translation.
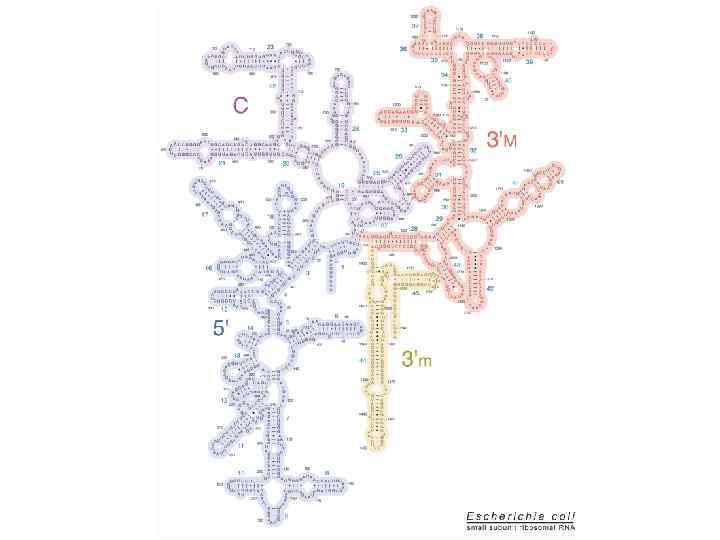
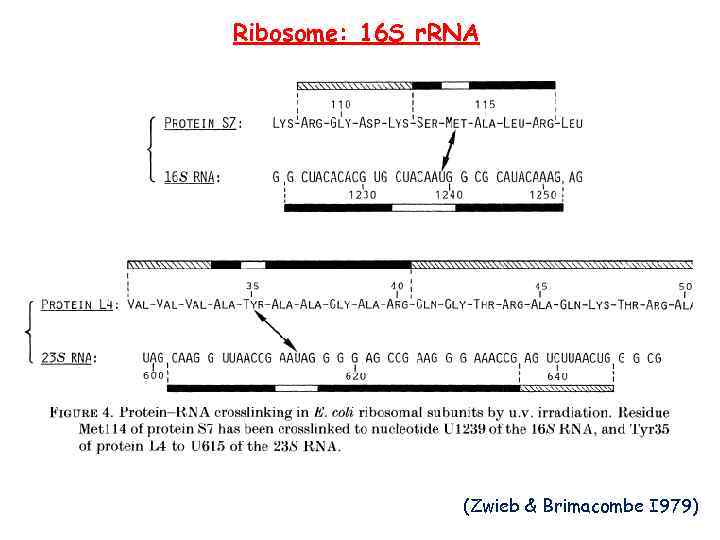 Ribosome: 16 S r. RNA (Zwieb & Brimacombe I 979)
Ribosome: 16 S r. RNA (Zwieb & Brimacombe I 979)
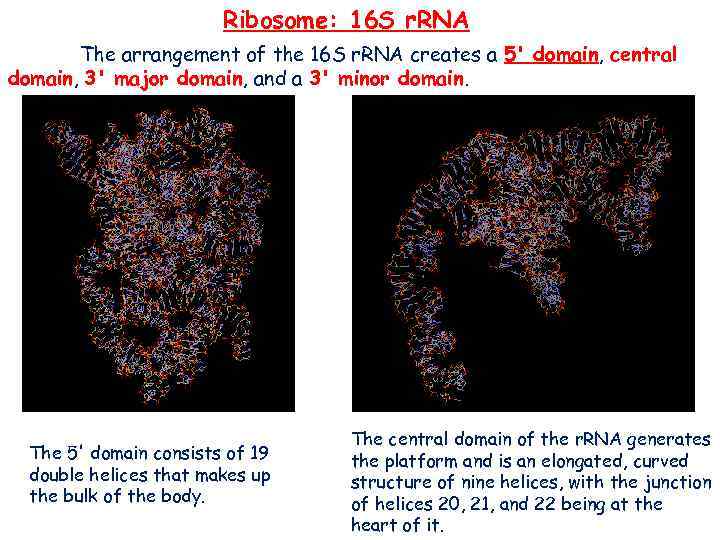 Ribosome: 16 S r. RNA The arrangement of the 16 S r. RNA creates a 5' domain, central domain, 3' major domain, and a 3' minor domain. The 5' domain consists of 19 double helices that makes up the bulk of the body. The central domain of the r. RNA generates the platform and is an elongated, curved structure of nine helices, with the junction of helices 20, 21, and 22 being at the heart of it.
Ribosome: 16 S r. RNA The arrangement of the 16 S r. RNA creates a 5' domain, central domain, 3' major domain, and a 3' minor domain. The 5' domain consists of 19 double helices that makes up the bulk of the body. The central domain of the r. RNA generates the platform and is an elongated, curved structure of nine helices, with the junction of helices 20, 21, and 22 being at the heart of it.
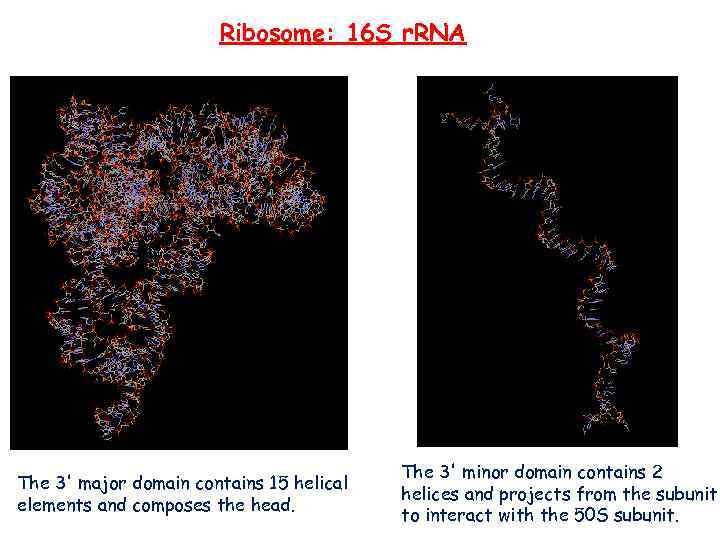 Ribosome: 16 S r. RNA The 3' major domain contains 15 helical elements and composes the head. The 3' minor domain contains 2 helices and projects from the subunit to interact with the 50 S subunit.
Ribosome: 16 S r. RNA The 3' major domain contains 15 helical elements and composes the head. The 3' minor domain contains 2 helices and projects from the subunit to interact with the 50 S subunit.
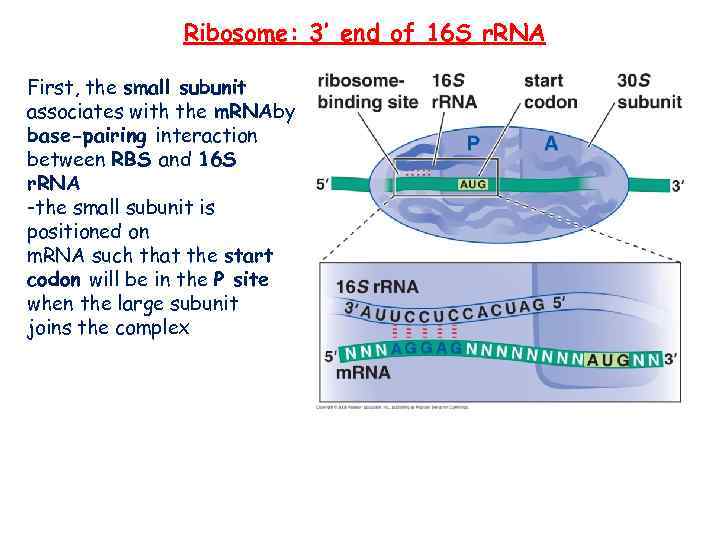 Ribosome: 3’ end of 16 S r. RNA First, the small subunit associates with the m. RNAby base-pairing interaction between RBS and 16 S r. RNA -the small subunit is positioned on m. RNA such that the start codon will be in the P site when the large subunit joins the complex
Ribosome: 3’ end of 16 S r. RNA First, the small subunit associates with the m. RNAby base-pairing interaction between RBS and 16 S r. RNA -the small subunit is positioned on m. RNA such that the start codon will be in the P site when the large subunit joins the complex
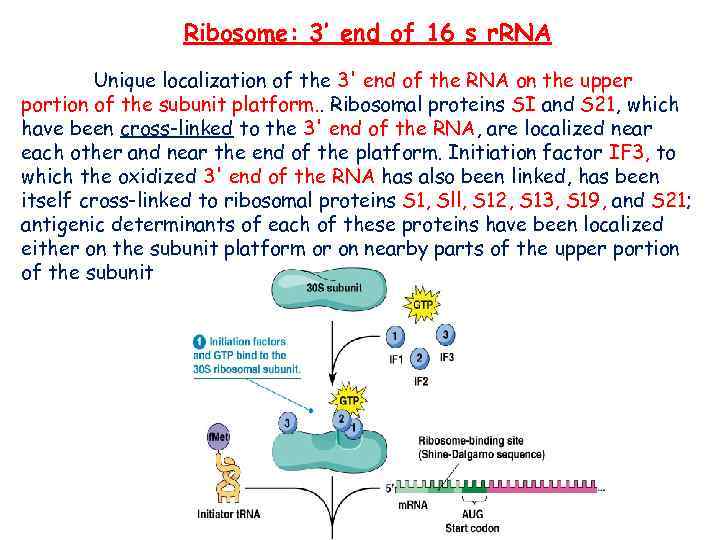 Ribosome: 3’ end of 16 s r. RNA Unique localization of the 3' end of the RNA on the upper portion of the subunit platform. . Ribosomal proteins SI and S 21, which have been cross-linked to the 3' end of the RNA, are localized near each other and near the end of the platform. Initiation factor IF 3, to which the oxidized 3' end of the RNA has also been linked, has been itself cross-linked to ribosomal proteins S 1, Sll, S 12, S 13, S 19, and S 21; antigenic determinants of each of these proteins have been localized either on the subunit platform or on nearby parts of the upper portion of the subunit
Ribosome: 3’ end of 16 s r. RNA Unique localization of the 3' end of the RNA on the upper portion of the subunit platform. . Ribosomal proteins SI and S 21, which have been cross-linked to the 3' end of the RNA, are localized near each other and near the end of the platform. Initiation factor IF 3, to which the oxidized 3' end of the RNA has also been linked, has been itself cross-linked to ribosomal proteins S 1, Sll, S 12, S 13, S 19, and S 21; antigenic determinants of each of these proteins have been localized either on the subunit platform or on nearby parts of the upper portion of the subunit
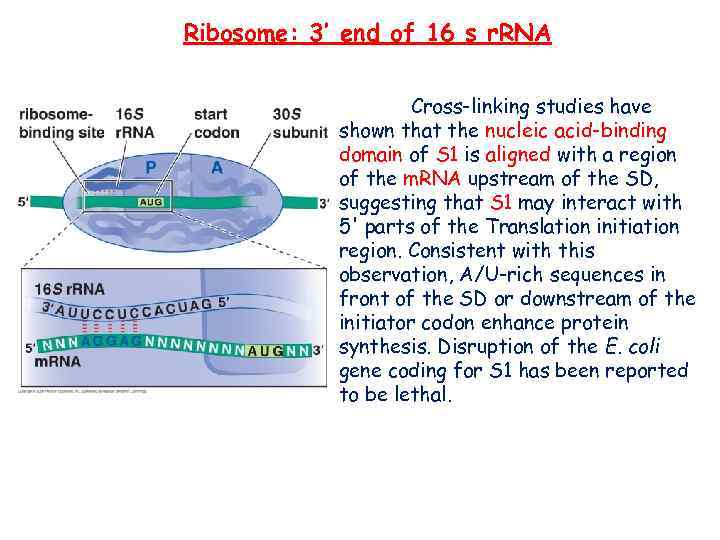 Ribosome: 3’ end of 16 s r. RNA Cross-linking studies have shown that the nucleic acid-binding domain of S 1 is aligned with a region of the m. RNA upstream of the SD, suggesting that S 1 may interact with 5' parts of the Translation initiation region. Consistent with this observation, A/U-rich sequences in front of the SD or downstream of the initiator codon enhance protein synthesis. Disruption of the E. coli gene coding for S 1 has been reported to be lethal.
Ribosome: 3’ end of 16 s r. RNA Cross-linking studies have shown that the nucleic acid-binding domain of S 1 is aligned with a region of the m. RNA upstream of the SD, suggesting that S 1 may interact with 5' parts of the Translation initiation region. Consistent with this observation, A/U-rich sequences in front of the SD or downstream of the initiator codon enhance protein synthesis. Disruption of the E. coli gene coding for S 1 has been reported to be lethal.
 Antibiotics affecting 16 S r. RNA Colicin E 3 (protein antibiotic from E. coli) makes a single cut in the 16 S r. RNA of 70 S ribosomes, these include the loss of activity. Pactamycin (Pct) was isolated from Streptomyces pactum as a potential new human antitumor drug, but in fact a potent inhibitor of translation in all three kingdoms, eukarya, bacteria, and archaea (Bhuyan et al. , 1961; Mankin, 1997). For this reason, the drug is expected to interact with highly conserved regions of 16 S RNA. Streptomycin and spectinomycin are typical examples which function by binding to specific sites on prokaryotic r. RNA and affecting the fidelity of protein synthesis. Binding of drug to the 16 S subunit near the A-site of the 30 S subunit leads to a decrease in translational accuracy and inhibition of the translocation of the ribosome.
Antibiotics affecting 16 S r. RNA Colicin E 3 (protein antibiotic from E. coli) makes a single cut in the 16 S r. RNA of 70 S ribosomes, these include the loss of activity. Pactamycin (Pct) was isolated from Streptomyces pactum as a potential new human antitumor drug, but in fact a potent inhibitor of translation in all three kingdoms, eukarya, bacteria, and archaea (Bhuyan et al. , 1961; Mankin, 1997). For this reason, the drug is expected to interact with highly conserved regions of 16 S RNA. Streptomycin and spectinomycin are typical examples which function by binding to specific sites on prokaryotic r. RNA and affecting the fidelity of protein synthesis. Binding of drug to the 16 S subunit near the A-site of the 30 S subunit leads to a decrease in translational accuracy and inhibition of the translocation of the ribosome.
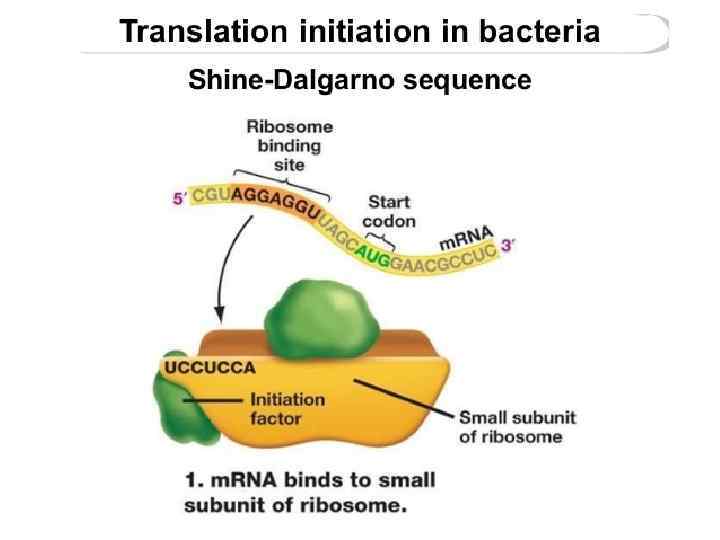
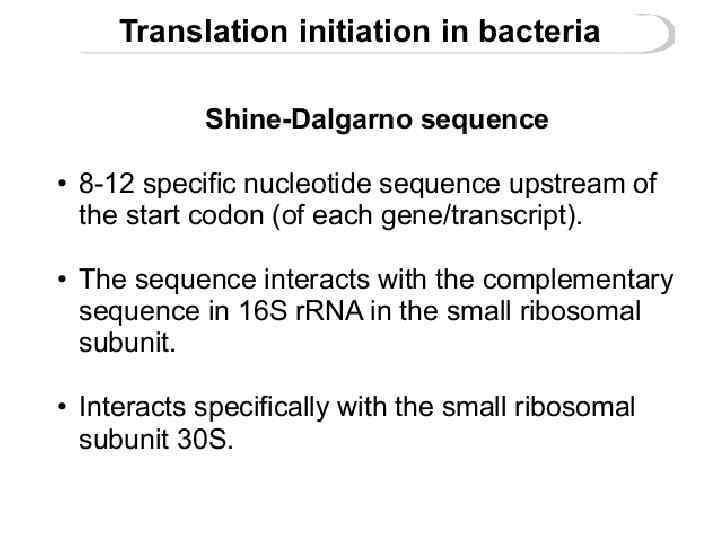
 Shine-Dalgarno sequence
Shine-Dalgarno sequence
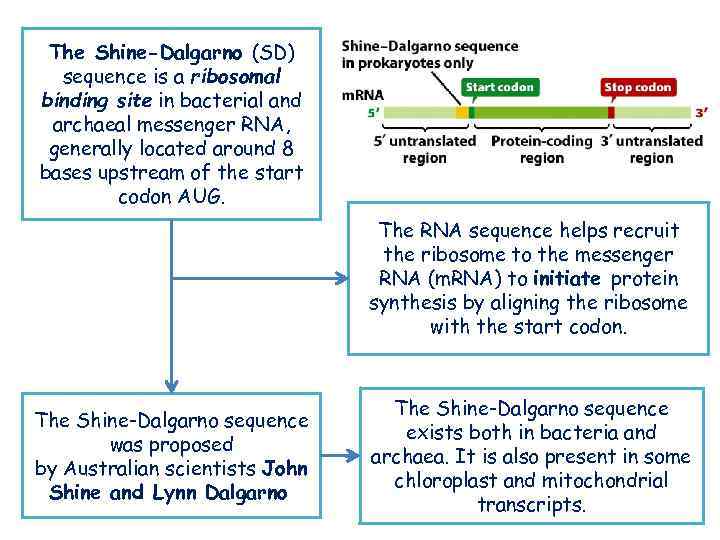 The Shine-Dalgarno (SD) sequence is a ribosomal binding site in bacterial and archaeal messenger RNA, generally located around 8 bases upstream of the start codon AUG. The RNA sequence helps recruit the ribosome to the messenger RNA (m. RNA) to initiate protein synthesis by aligning the ribosome with the start codon. The Shine-Dalgarno sequence was proposed by Australian scientists John Shine and Lynn Dalgarno The Shine-Dalgarno sequence exists both in bacteria and archaea. It is also present in some chloroplast and mitochondrial transcripts.
The Shine-Dalgarno (SD) sequence is a ribosomal binding site in bacterial and archaeal messenger RNA, generally located around 8 bases upstream of the start codon AUG. The RNA sequence helps recruit the ribosome to the messenger RNA (m. RNA) to initiate protein synthesis by aligning the ribosome with the start codon. The Shine-Dalgarno sequence was proposed by Australian scientists John Shine and Lynn Dalgarno The Shine-Dalgarno sequence exists both in bacteria and archaea. It is also present in some chloroplast and mitochondrial transcripts.
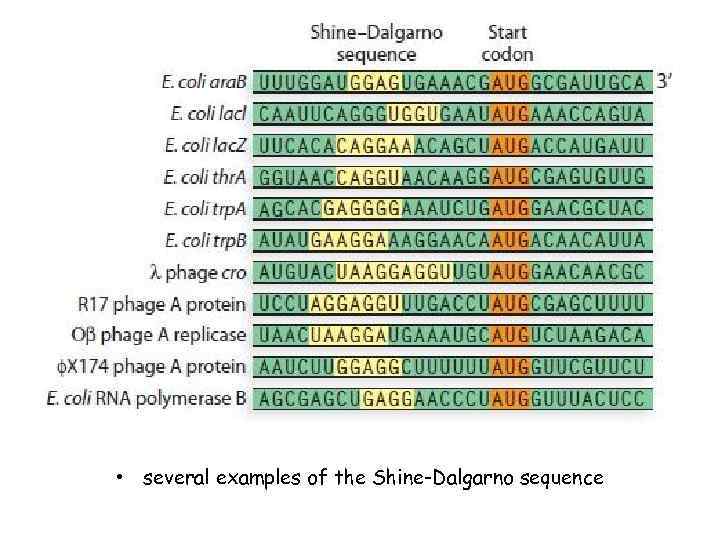 • several examples of the Shine-Dalgarno sequence
• several examples of the Shine-Dalgarno sequence
 REFERENCE 1. The Cell: A Molecular Approach. 2 nd edition. Cooper GM. Sunderland (MA): Sinauer Associates; 2000. 2. Watson J D, Baker T A , Bell S P, Gann A, Levine M, Losick R. Molecular Biology of the Gene. 5 th edition. Pearson education 2004. 3. J. E. KREBS, E. S. GOLDSTEIN, S. T. KILPATRICK. Lewin’s genes XI. Copyright© 2 0 1 4 by Jones & B artlett Learning, LLC, an Ascend Learning Company 4. Daniel H. Lackner et al. , Translational Control of Gene Expression: From Transcripts to Transcriptomes, International Review of Cell and Molecular Biology, 2008, 271, 200 -238. 5. The Cell: A Molecular Approach. 2 nd edition. Cooper GM. Sunderland (MA): Sinauer Associates; 2000. 6. Czernilofsky, A; Kurland, C. G. ; Stöffler, G. (1975). "30 S RIBOSOMALPROTEINS ASSOCIATED WITH 3'-TERMINUS OF 16 S RNA". FEBS Letters. ELSEVIER SCIENCE BV. 58 (1): 281– 284. 7. H. G. WITTMANN. Structure and evolution of ribosomes. Proc. R. Soc. Lond. B 216, 117 -135 (1982) 8. Zwieb, C. & Brimacombe, R. I 979 Nucl. Acids Res. 6, 1775 -1790. 9. HELEN MCKUSKIE OLSON* AND DOHN G. GLITZ. Ribosome structure: Localization of 3' end of RNA in small subunit by immunoelectronmicroscopy. Proc. Natl. Acad. Sci. USA. Vol. 76, No. 8, pp. 3769 -3773, August 1979 10. Heinz-Giinter WITTMANN. Structure, Function and Evolution of Ribosomes. Eur. J. Biochem. 61, 1 - 13 (1976)
REFERENCE 1. The Cell: A Molecular Approach. 2 nd edition. Cooper GM. Sunderland (MA): Sinauer Associates; 2000. 2. Watson J D, Baker T A , Bell S P, Gann A, Levine M, Losick R. Molecular Biology of the Gene. 5 th edition. Pearson education 2004. 3. J. E. KREBS, E. S. GOLDSTEIN, S. T. KILPATRICK. Lewin’s genes XI. Copyright© 2 0 1 4 by Jones & B artlett Learning, LLC, an Ascend Learning Company 4. Daniel H. Lackner et al. , Translational Control of Gene Expression: From Transcripts to Transcriptomes, International Review of Cell and Molecular Biology, 2008, 271, 200 -238. 5. The Cell: A Molecular Approach. 2 nd edition. Cooper GM. Sunderland (MA): Sinauer Associates; 2000. 6. Czernilofsky, A; Kurland, C. G. ; Stöffler, G. (1975). "30 S RIBOSOMALPROTEINS ASSOCIATED WITH 3'-TERMINUS OF 16 S RNA". FEBS Letters. ELSEVIER SCIENCE BV. 58 (1): 281– 284. 7. H. G. WITTMANN. Structure and evolution of ribosomes. Proc. R. Soc. Lond. B 216, 117 -135 (1982) 8. Zwieb, C. & Brimacombe, R. I 979 Nucl. Acids Res. 6, 1775 -1790. 9. HELEN MCKUSKIE OLSON* AND DOHN G. GLITZ. Ribosome structure: Localization of 3' end of RNA in small subunit by immunoelectronmicroscopy. Proc. Natl. Acad. Sci. USA. Vol. 76, No. 8, pp. 3769 -3773, August 1979 10. Heinz-Giinter WITTMANN. Structure, Function and Evolution of Ribosomes. Eur. J. Biochem. 61, 1 - 13 (1976)
 REFERENCE 11. A. P. CZERNILOFSKY, C. G. KURLAND and G. STOFFLER. 30 s RIBOSOMAL PROTEINS ASSOCIATED WITH THE 3’-TERMINUS OF 16 s RNA. FEBS LETTERS Volume 58, number 1, October 1975. 12. Vladimir Vimberg 1, Age Tats 2, Maido Remm 2 and Tanel Tenson. Translation initiation region sequence preferences in Escherichia coli. BMC Molecular Biology 2007, 8: 100 13. Weiling Hong, Jie Zeng, and Jianping Xie. Antibiotic drugs targeting bacterial RNAs. Acta Pharm Sin B. 2014 Aug; 4(4): 258– 265. 14. Ditlev E. Brodersen, * William M. Clemons, Jr. , *† Andrew P. Carter, * Robert J. Morgan-Warren, * Brian T. Wimberly, * and V. Ramakrishnan*. The Structural Basis for the Action of the Antibiotics Tetracycline, Pactamycin, and Hygromycin B on the 30 S Ribosomal Subunit. Cell, Vol. 103, 1143– 1154, December 22, 2000, Copyright 2000 by Cell Press. 15. Hale WG, Margham JP, Saunders VA, Collins Dictionary of Biology, (2 nd ed) Shine-Dalgarno (SD) sequence Links: 1. http: //www. biochem. umd. edu/biochem/kahn/bchm 46501/ribosome/16 Sr. RNA. html 2. http: //rna. ucsc. edu/rnacenter/ribosome_images. html 3. https: //en. wikipedia. org/wiki/Shine-Dalgarno_sequence 4. thermofisher. com/Ribosomal Binding Site Sequence Requirements
REFERENCE 11. A. P. CZERNILOFSKY, C. G. KURLAND and G. STOFFLER. 30 s RIBOSOMAL PROTEINS ASSOCIATED WITH THE 3’-TERMINUS OF 16 s RNA. FEBS LETTERS Volume 58, number 1, October 1975. 12. Vladimir Vimberg 1, Age Tats 2, Maido Remm 2 and Tanel Tenson. Translation initiation region sequence preferences in Escherichia coli. BMC Molecular Biology 2007, 8: 100 13. Weiling Hong, Jie Zeng, and Jianping Xie. Antibiotic drugs targeting bacterial RNAs. Acta Pharm Sin B. 2014 Aug; 4(4): 258– 265. 14. Ditlev E. Brodersen, * William M. Clemons, Jr. , *† Andrew P. Carter, * Robert J. Morgan-Warren, * Brian T. Wimberly, * and V. Ramakrishnan*. The Structural Basis for the Action of the Antibiotics Tetracycline, Pactamycin, and Hygromycin B on the 30 S Ribosomal Subunit. Cell, Vol. 103, 1143– 1154, December 22, 2000, Copyright 2000 by Cell Press. 15. Hale WG, Margham JP, Saunders VA, Collins Dictionary of Biology, (2 nd ed) Shine-Dalgarno (SD) sequence Links: 1. http: //www. biochem. umd. edu/biochem/kahn/bchm 46501/ribosome/16 Sr. RNA. html 2. http: //rna. ucsc. edu/rnacenter/ribosome_images. html 3. https: //en. wikipedia. org/wiki/Shine-Dalgarno_sequence 4. thermofisher. com/Ribosomal Binding Site Sequence Requirements


