3bac144a66ee497a3178ce5aeb5ae7a6.ppt
- Количество слайдов: 48
 IIIM-CSIR Challenges in new drug discovery in South Asia Ram Vishwakarma Indian Institute of Integrative Medicine (Council of Scientific & Industrial Research) Jammu and Srinagar, India
IIIM-CSIR Challenges in new drug discovery in South Asia Ram Vishwakarma Indian Institute of Integrative Medicine (Council of Scientific & Industrial Research) Jammu and Srinagar, India
 Drug Discovery and Development IIIM-CSIR (Current Business Model) • New drugs: Small organic molecules and therapeutic proteins (biologics) Big Pharma and Biotech companies (US, Europe, Japan) • Generic drugs: API formulation of off-patent drugs for domestic/world market Indian and Chinese companies • Contract research, API manufacturing and Clinical trials services Indian and Chinese companies
Drug Discovery and Development IIIM-CSIR (Current Business Model) • New drugs: Small organic molecules and therapeutic proteins (biologics) Big Pharma and Biotech companies (US, Europe, Japan) • Generic drugs: API formulation of off-patent drugs for domestic/world market Indian and Chinese companies • Contract research, API manufacturing and Clinical trials services Indian and Chinese companies
 IIIM-CSIR Time tested model for new drug discovery • Natural products (plants and microbial species) and traditional system of medicine and ethno-biology (Opiates, statins, antibiotics, rapamycin, ephedrine, quinine, artemisinin, amphotericin, vincritine, taxol, podophyllotoxin (almost all anticancer and antiinfective drugs are natural products derived) • Medicinal chemistry of know bioactive compounds (drugs) to decipher structure-activity relationship (SAR) and discovery of new pharmacological activity A single chemical motif gave rise to antibiotics, hypoglycemic agents, diuretics, and antihypertensive drugs
IIIM-CSIR Time tested model for new drug discovery • Natural products (plants and microbial species) and traditional system of medicine and ethno-biology (Opiates, statins, antibiotics, rapamycin, ephedrine, quinine, artemisinin, amphotericin, vincritine, taxol, podophyllotoxin (almost all anticancer and antiinfective drugs are natural products derived) • Medicinal chemistry of know bioactive compounds (drugs) to decipher structure-activity relationship (SAR) and discovery of new pharmacological activity A single chemical motif gave rise to antibiotics, hypoglycemic agents, diuretics, and antihypertensive drugs
 IIIM-CSIR • Random screening of compound libraries by HTS against clinically validated targets (enzymes, receptors etc) relevant to the disease Hits – lead – drug candidate (medicinal chemistry) • Mechanism based drug design and discovery (antimetabolites, methotrexate, trimethoprim, antidepressants, steroids like contraceptives and anti-inflammatory drugs) • Structure based drug design (X-ray crystallography, NMR spectroscopy, molecular modeling) (Imatinib) • Observational pharmacology (off-target optimization) Sidnafil citrate (Viagra), Cialis and other PDE 5 blocker)
IIIM-CSIR • Random screening of compound libraries by HTS against clinically validated targets (enzymes, receptors etc) relevant to the disease Hits – lead – drug candidate (medicinal chemistry) • Mechanism based drug design and discovery (antimetabolites, methotrexate, trimethoprim, antidepressants, steroids like contraceptives and anti-inflammatory drugs) • Structure based drug design (X-ray crystallography, NMR spectroscopy, molecular modeling) (Imatinib) • Observational pharmacology (off-target optimization) Sidnafil citrate (Viagra), Cialis and other PDE 5 blocker)
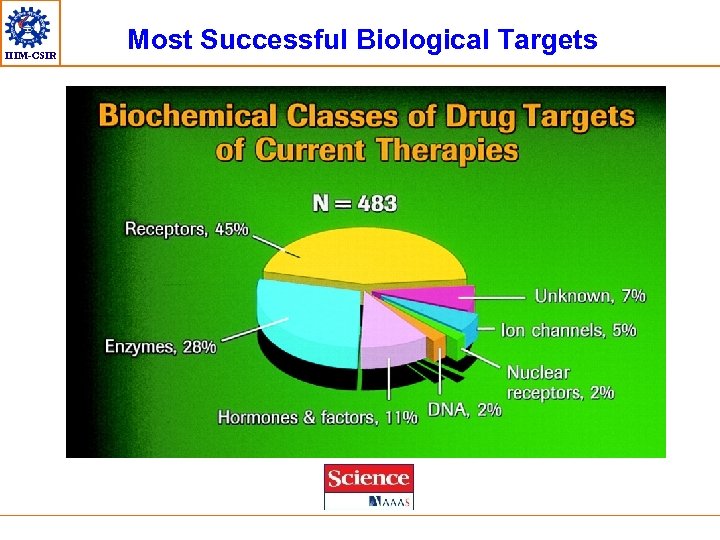 IIIM-CSIR Most Successful Biological Targets
IIIM-CSIR Most Successful Biological Targets
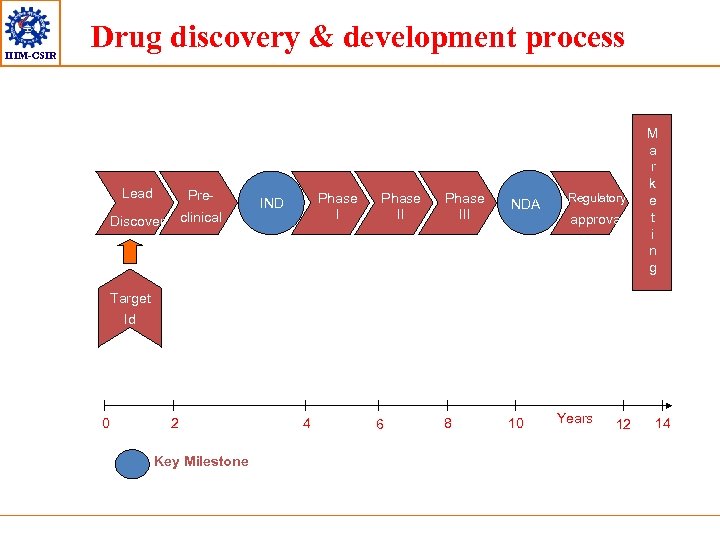 IIIM-CSIR Drug discovery & development process Lead Pre- Discover clinical Phase I IND Phase III NDA 8 10 Regulatory approval y M a r k e t i n g Target Id 0 2 Key Milestone 4 6 Years 12 14
IIIM-CSIR Drug discovery & development process Lead Pre- Discover clinical Phase I IND Phase III NDA 8 10 Regulatory approval y M a r k e t i n g Target Id 0 2 Key Milestone 4 6 Years 12 14
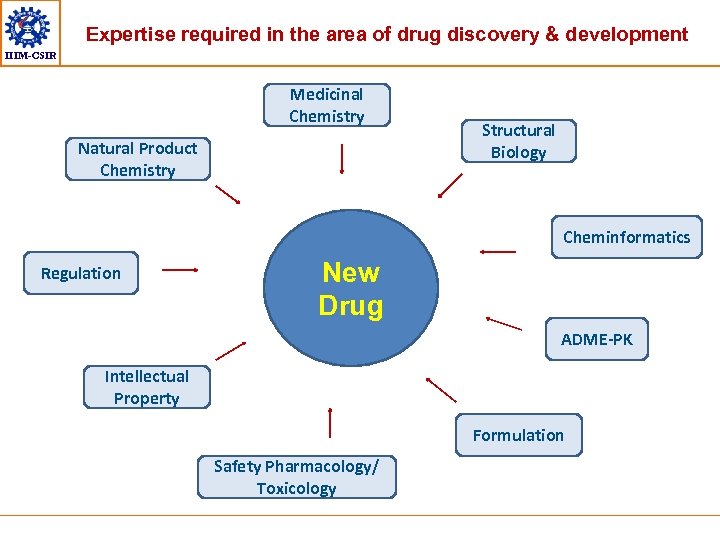 Expertise required in the area of drug discovery & development IIIM-CSIR Medicinal Chemistry Natural Product Chemistry Structural Biology Cheminformatics Regulation New Drug ADME-PK Intellectual Property Formulation Safety Pharmacology/ Toxicology
Expertise required in the area of drug discovery & development IIIM-CSIR Medicinal Chemistry Natural Product Chemistry Structural Biology Cheminformatics Regulation New Drug ADME-PK Intellectual Property Formulation Safety Pharmacology/ Toxicology
 IIIM-CSIR Uniqueness of drug discovery • Most regulated industry FDA and country-specific multiple agencies Risk of post-approval failure (Vioxx and Glitazones) Balance between profits and public-health Patent expiry and generic competition (India) • Goal-posts keep changing Current state of knowledge in Science & Technology Biological targets and approaches change significantly and R&D has to rapidly change (stem cells, RNAi, antibodies) Entirely new opportunities are created by new Science
IIIM-CSIR Uniqueness of drug discovery • Most regulated industry FDA and country-specific multiple agencies Risk of post-approval failure (Vioxx and Glitazones) Balance between profits and public-health Patent expiry and generic competition (India) • Goal-posts keep changing Current state of knowledge in Science & Technology Biological targets and approaches change significantly and R&D has to rapidly change (stem cells, RNAi, antibodies) Entirely new opportunities are created by new Science
 IIIM-CSIR Key discovery “ecosystem” issues ! • Drug discovery is a team work (chemistry, pharmacology, clinical sciences) with moments of brilliant thinking and months of painstakingly detailed work • Spotting, inspiring, nurturing and retaining leaders who are also accomplished scientists • Segregation of “first-in-class” and “best-in-class’ drug discovery teams (cultural issues) with appropriate goals and reward system • Outsourcing non-essential functions to remain focused on key core objectives and expertise • Partnership with innovator pharma companies in clinical development phases • Identify areas of strategic and future interests and initiate collaboration with leading academic groups (new ideas) • Answers to public health problems are “drugs” not “publications”
IIIM-CSIR Key discovery “ecosystem” issues ! • Drug discovery is a team work (chemistry, pharmacology, clinical sciences) with moments of brilliant thinking and months of painstakingly detailed work • Spotting, inspiring, nurturing and retaining leaders who are also accomplished scientists • Segregation of “first-in-class” and “best-in-class’ drug discovery teams (cultural issues) with appropriate goals and reward system • Outsourcing non-essential functions to remain focused on key core objectives and expertise • Partnership with innovator pharma companies in clinical development phases • Identify areas of strategic and future interests and initiate collaboration with leading academic groups (new ideas) • Answers to public health problems are “drugs” not “publications”
 IIIM-CSIR Our efforts in new drug discovery Targets: clinically validated kinases
IIIM-CSIR Our efforts in new drug discovery Targets: clinically validated kinases
 IIIM-CSIR Phosphatidylinositol 3 -kinase (PI 3 K) pathway • • • Inhibition of PI-3 K kinase (isoform specific inhibitors) PI 3 Ka for cancer and PI 3 Kg for inflammation Up-regulation of PTEN phosphatase Inhibition of AKT isoforms Inhibition of m. TOR
IIIM-CSIR Phosphatidylinositol 3 -kinase (PI 3 K) pathway • • • Inhibition of PI-3 K kinase (isoform specific inhibitors) PI 3 Ka for cancer and PI 3 Kg for inflammation Up-regulation of PTEN phosphatase Inhibition of AKT isoforms Inhibition of m. TOR
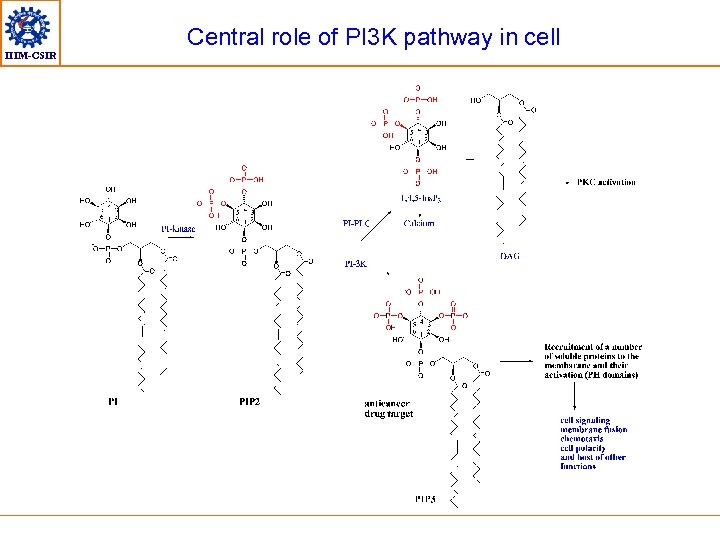 IIIM-CSIR Central role of PI 3 K pathway in cell
IIIM-CSIR Central role of PI 3 K pathway in cell
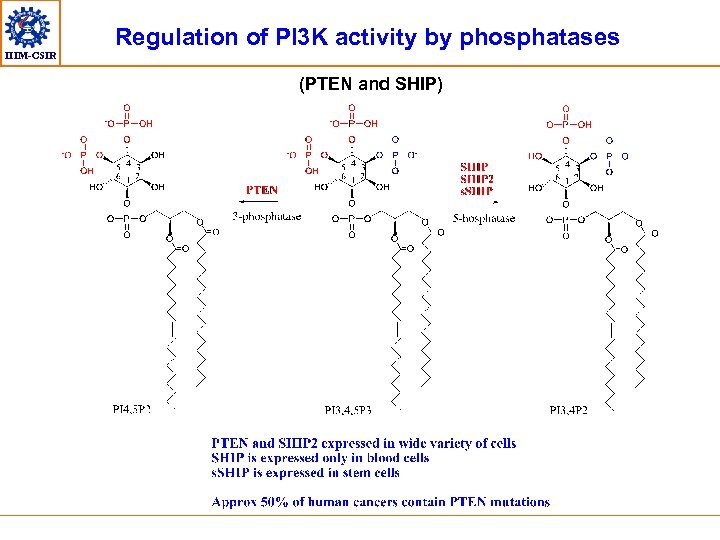 IIIM-CSIR Regulation of PI 3 K activity by phosphatases (PTEN and SHIP)
IIIM-CSIR Regulation of PI 3 K activity by phosphatases (PTEN and SHIP)
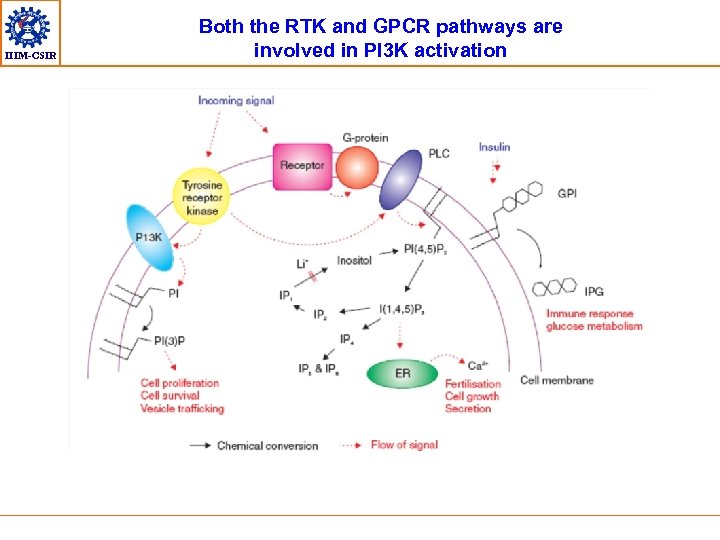 IIIM-CSIR Both the RTK and GPCR pathways are involved in PI 3 K activation
IIIM-CSIR Both the RTK and GPCR pathways are involved in PI 3 K activation
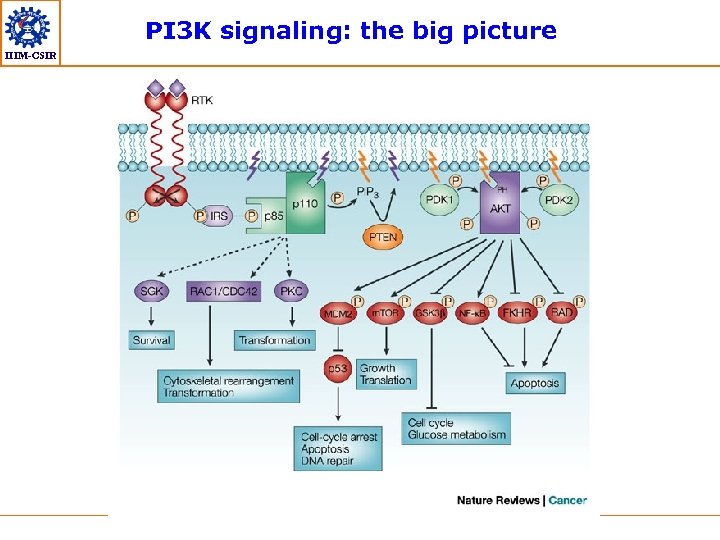 PI 3 K signaling: the big picture IIIM-CSIR
PI 3 K signaling: the big picture IIIM-CSIR
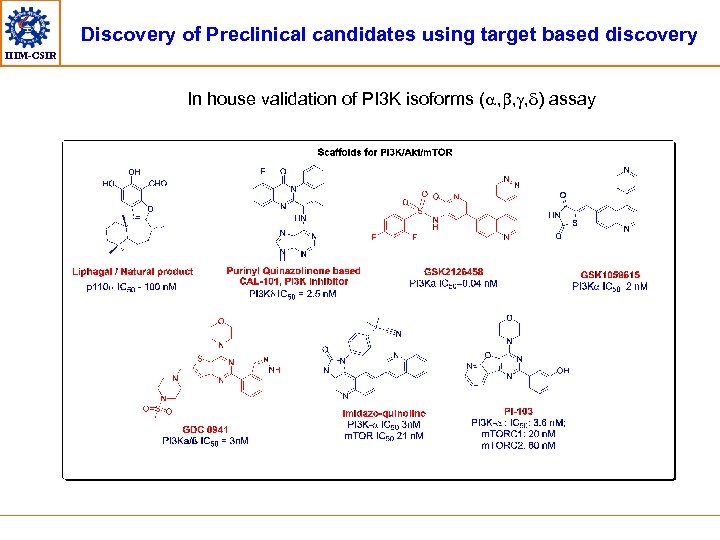 Discovery of Preclinical candidates using target based discovery IIIM-CSIR In house validation of PI 3 K isoforms (a, b, g, ) assay
Discovery of Preclinical candidates using target based discovery IIIM-CSIR In house validation of PI 3 K isoforms (a, b, g, ) assay
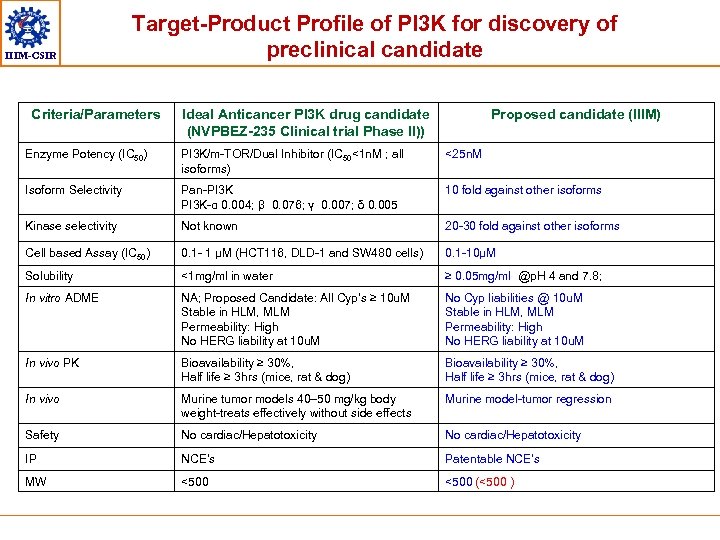 IIIM-CSIR Target-Product Profile of PI 3 K for discovery of preclinical candidate Criteria/Parameters Ideal Anticancer PI 3 K drug candidate (NVPBEZ-235 Clinical trial Phase II)) Proposed candidate (IIIM) Enzyme Potency (IC 50) PI 3 K/m-TOR/Dual Inhibitor (IC 50<1 n. M ; all isoforms) <25 n. M Isoform Selectivity Pan-PI 3 K-α 0. 004; β 0. 076; ү 0. 007; δ 0. 005 10 fold against other isoforms Kinase selectivity Not known 20 -30 fold against other isoforms Cell based Assay (IC 50) 0. 1 - 1 μM (HCT 116, DLD-1 and SW 480 cells) 0. 1 -10μM Solubility <1 mg/ml in water ≥ 0. 05 mg/ml @p. H 4 and 7. 8; In vitro ADME NA; Proposed Candidate: All Cyp’s ≥ 10 u. M Stable in HLM, MLM Permeability: High No HERG liability at 10 u. M No Cyp liabilities @ 10 u. M Stable in HLM, MLM Permeability: High No HERG liability at 10 u. M In vivo PK Bioavailability ≥ 30%, Half life ≥ 3 hrs (mice, rat & dog) In vivo Murine tumor models 40– 50 mg/kg body weight-treats effectively without side effects Murine model-tumor regression Safety No cardiac/Hepatotoxicity IP NCE’s Patentable NCE’s MW <500 (<500 )
IIIM-CSIR Target-Product Profile of PI 3 K for discovery of preclinical candidate Criteria/Parameters Ideal Anticancer PI 3 K drug candidate (NVPBEZ-235 Clinical trial Phase II)) Proposed candidate (IIIM) Enzyme Potency (IC 50) PI 3 K/m-TOR/Dual Inhibitor (IC 50<1 n. M ; all isoforms) <25 n. M Isoform Selectivity Pan-PI 3 K-α 0. 004; β 0. 076; ү 0. 007; δ 0. 005 10 fold against other isoforms Kinase selectivity Not known 20 -30 fold against other isoforms Cell based Assay (IC 50) 0. 1 - 1 μM (HCT 116, DLD-1 and SW 480 cells) 0. 1 -10μM Solubility <1 mg/ml in water ≥ 0. 05 mg/ml @p. H 4 and 7. 8; In vitro ADME NA; Proposed Candidate: All Cyp’s ≥ 10 u. M Stable in HLM, MLM Permeability: High No HERG liability at 10 u. M No Cyp liabilities @ 10 u. M Stable in HLM, MLM Permeability: High No HERG liability at 10 u. M In vivo PK Bioavailability ≥ 30%, Half life ≥ 3 hrs (mice, rat & dog) In vivo Murine tumor models 40– 50 mg/kg body weight-treats effectively without side effects Murine model-tumor regression Safety No cardiac/Hepatotoxicity IP NCE’s Patentable NCE’s MW <500 (<500 )
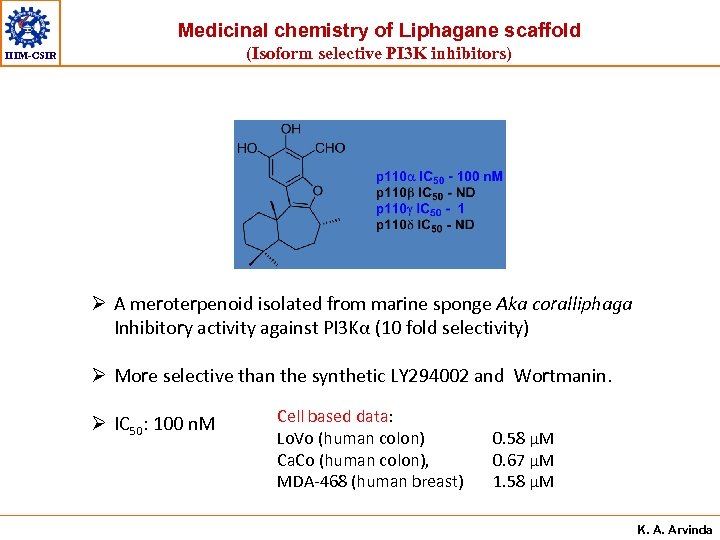 Medicinal chemistry of Liphagane scaffold (Isoform selective PI 3 K inhibitors) IIIM-CSIR Ø A meroterpenoid isolated from marine sponge Aka coralliphaga Inhibitory activity against PI 3 Kα (10 fold selectivity) Ø More selective than the synthetic LY 294002 and Wortmanin. Ø IC 50: 100 n. M Cell based data: Lo. Vo (human colon) Ca. Co (human colon), MDA-468 (human breast) 0. 58 µM 0. 67 µM 1. 58 µM K. A. Arvinda
Medicinal chemistry of Liphagane scaffold (Isoform selective PI 3 K inhibitors) IIIM-CSIR Ø A meroterpenoid isolated from marine sponge Aka coralliphaga Inhibitory activity against PI 3 Kα (10 fold selectivity) Ø More selective than the synthetic LY 294002 and Wortmanin. Ø IC 50: 100 n. M Cell based data: Lo. Vo (human colon) Ca. Co (human colon), MDA-468 (human breast) 0. 58 µM 0. 67 µM 1. 58 µM K. A. Arvinda
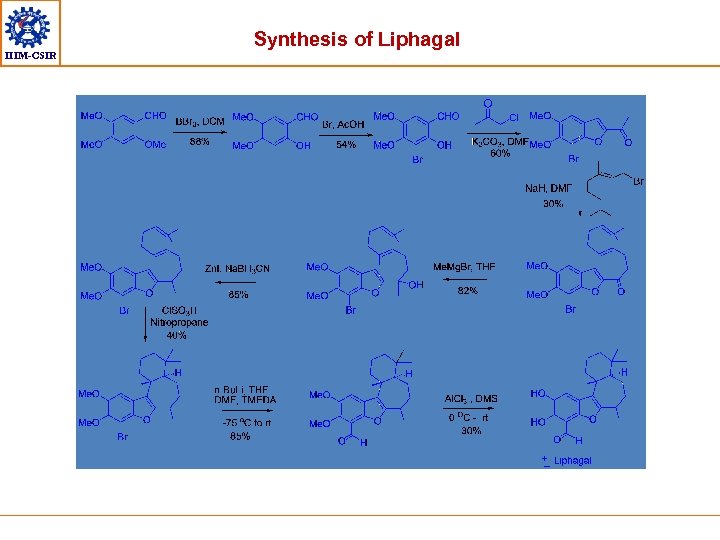 IIIM-CSIR Synthesis of Liphagal
IIIM-CSIR Synthesis of Liphagal
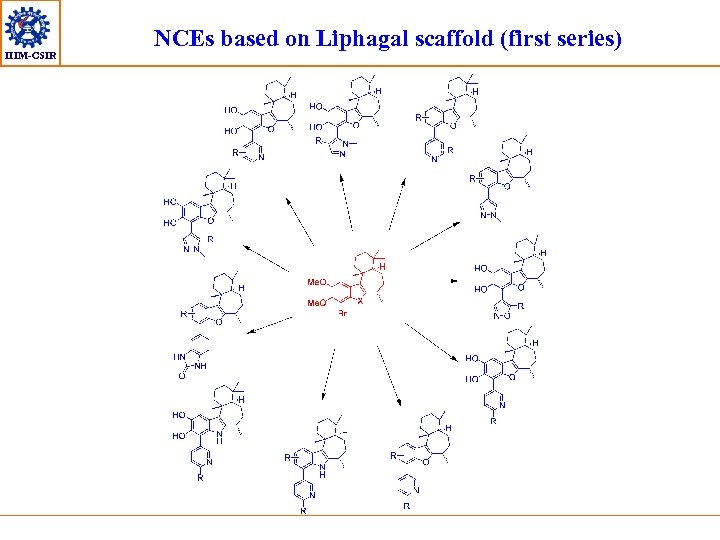 IIIM-CSIR NCEs based on Liphagal scaffold (first series)
IIIM-CSIR NCEs based on Liphagal scaffold (first series)
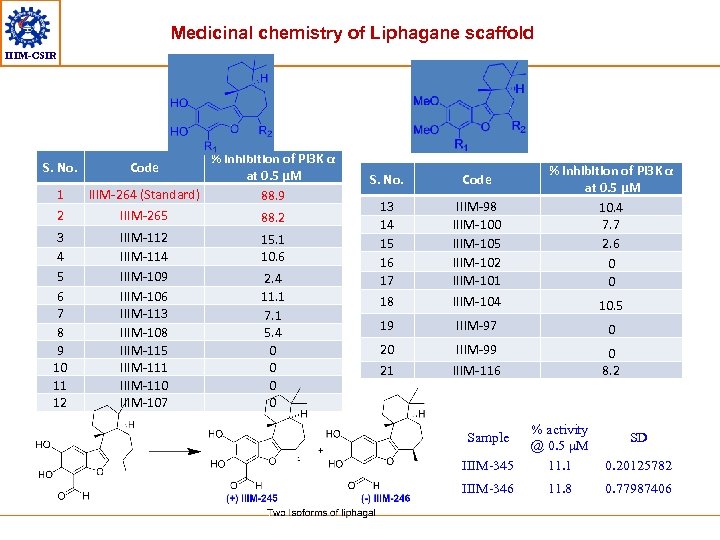 Medicinal chemistry of Liphagane scaffold IIIM-CSIR S. No. 1 2 3 4 5 6 7 8 9 10 11 12 % inhibition of Pi 3 K α at 0. 5 μM IIIM-264 (Standard) 88. 9 IIIM-265 88. 2 Code IIIM-112 IIIM-114 IIIM-109 IIIM-106 IIIM-113 IIIM-108 IIIM-115 IIIM-111 IIIM-110 IIIM-107 15. 1 10. 6 2. 4 11. 1 7. 1 5. 4 0 0 % inhibition of Pi 3 K α at 0. 5 μM 10. 4 7. 7 2. 6 0 0 S. No. Code 13 14 15 16 17 18 IIIM-98 IIIM-100 IIIM-105 IIIM-102 IIIM-101 IIIM-104 19 IIIM-97 0 20 IIIM-99 21 IIIM-116 0 8. 2 10. 5 IIIM-345 % activity @ 0. 5 μM 11. 1 0. 20125782 IIIM-346 11. 8 0. 77987406 Sample SD
Medicinal chemistry of Liphagane scaffold IIIM-CSIR S. No. 1 2 3 4 5 6 7 8 9 10 11 12 % inhibition of Pi 3 K α at 0. 5 μM IIIM-264 (Standard) 88. 9 IIIM-265 88. 2 Code IIIM-112 IIIM-114 IIIM-109 IIIM-106 IIIM-113 IIIM-108 IIIM-115 IIIM-111 IIIM-110 IIIM-107 15. 1 10. 6 2. 4 11. 1 7. 1 5. 4 0 0 % inhibition of Pi 3 K α at 0. 5 μM 10. 4 7. 7 2. 6 0 0 S. No. Code 13 14 15 16 17 18 IIIM-98 IIIM-100 IIIM-105 IIIM-102 IIIM-101 IIIM-104 19 IIIM-97 0 20 IIIM-99 21 IIIM-116 0 8. 2 10. 5 IIIM-345 % activity @ 0. 5 μM 11. 1 0. 20125782 IIIM-346 11. 8 0. 77987406 Sample SD
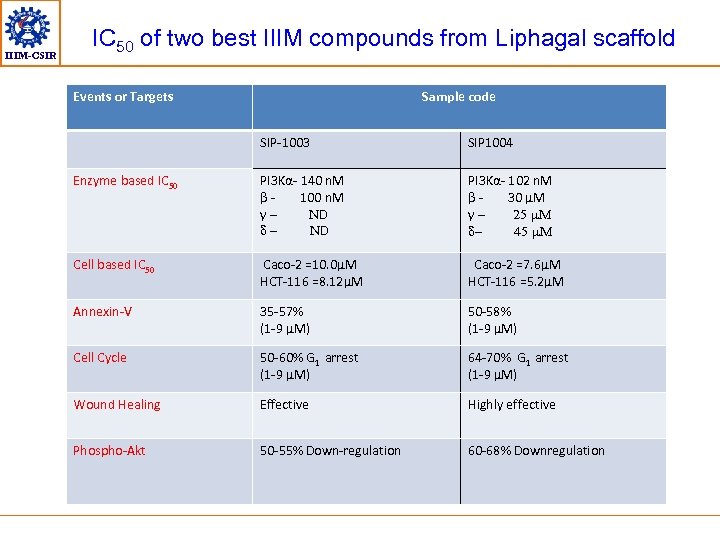 IIIM-CSIR IC 50 of two best IIIM compounds from Liphagal scaffold Events or Targets Sample code SIP-1003 SIP 1004 Enzyme based IC 50 PI 3 Kα- 140 n. M β 100 n. M γND ND PI 3 Kα- 102 n. M β 30 µM γ 25 m. M 45 m. M Cell based IC 50 Caco-2 =10. 0µM HCT-116 =8. 12µM Caco-2 =7. 6µM HCT-116 =5. 2µM Annexin-V 35 -57% (1 -9 µM) 50 -58% (1 -9 µM) Cell Cycle 50 -60% G 1 arrest (1 -9 µM) 64 -70% G 1 arrest (1 -9 µM) Wound Healing Effective Highly effective Phospho-Akt 50 -55% Down-regulation 60 -68% Downregulation
IIIM-CSIR IC 50 of two best IIIM compounds from Liphagal scaffold Events or Targets Sample code SIP-1003 SIP 1004 Enzyme based IC 50 PI 3 Kα- 140 n. M β 100 n. M γND ND PI 3 Kα- 102 n. M β 30 µM γ 25 m. M 45 m. M Cell based IC 50 Caco-2 =10. 0µM HCT-116 =8. 12µM Caco-2 =7. 6µM HCT-116 =5. 2µM Annexin-V 35 -57% (1 -9 µM) 50 -58% (1 -9 µM) Cell Cycle 50 -60% G 1 arrest (1 -9 µM) 64 -70% G 1 arrest (1 -9 µM) Wound Healing Effective Highly effective Phospho-Akt 50 -55% Down-regulation 60 -68% Downregulation
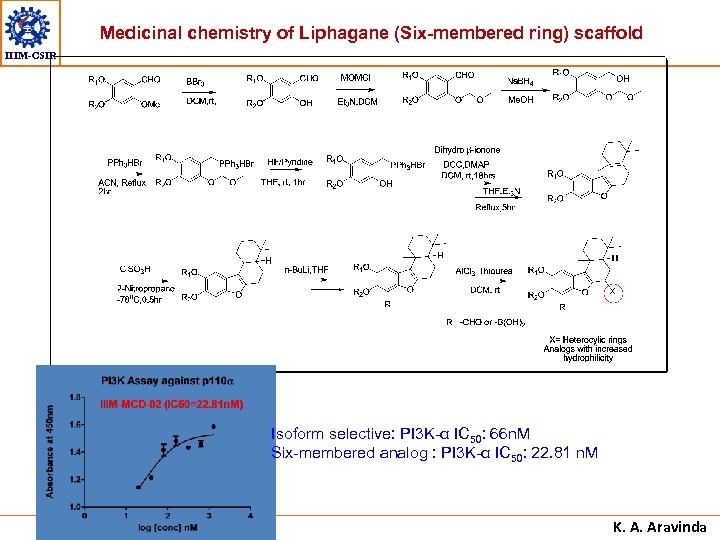 Medicinal chemistry of Liphagane (Six-membered ring) scaffold IIIM-CSIR Isoform selective: PI 3 K-α IC 50: 66 n. M Six-membered analog : PI 3 K-α IC 50: 22. 81 n. M K. A. Aravinda
Medicinal chemistry of Liphagane (Six-membered ring) scaffold IIIM-CSIR Isoform selective: PI 3 K-α IC 50: 66 n. M Six-membered analog : PI 3 K-α IC 50: 22. 81 n. M K. A. Aravinda
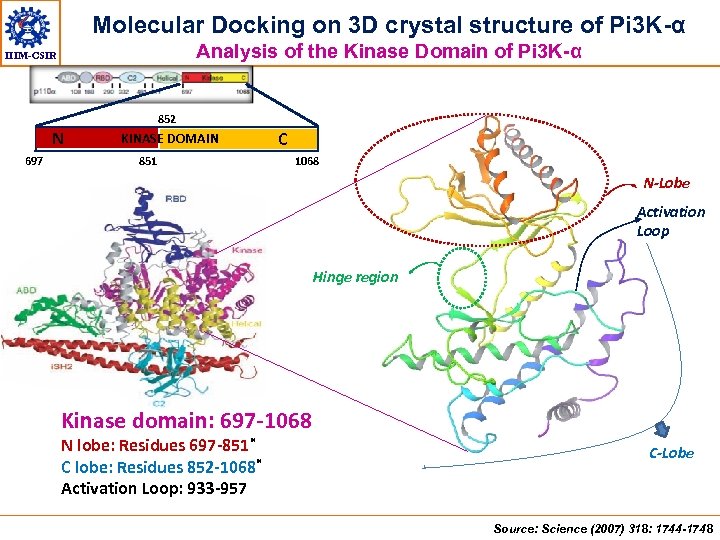 Molecular Docking on 3 D crystal structure of Pi 3 K-α Analysis of the Kinase Domain of Pi 3 K-α IIIM-CSIR 852 N 697 KINASE DOMAIN 851 C 1068 N-Lobe Activation Loop Hinge region Kinase domain: 697 -1068 N lobe: Residues 697 -851* C lobe: Residues 852 -1068* Activation Loop: 933 -957 C-Lobe Source: Science (2007) 318: 1744 -1748
Molecular Docking on 3 D crystal structure of Pi 3 K-α Analysis of the Kinase Domain of Pi 3 K-α IIIM-CSIR 852 N 697 KINASE DOMAIN 851 C 1068 N-Lobe Activation Loop Hinge region Kinase domain: 697 -1068 N lobe: Residues 697 -851* C lobe: Residues 852 -1068* Activation Loop: 933 -957 C-Lobe Source: Science (2007) 318: 1744 -1748
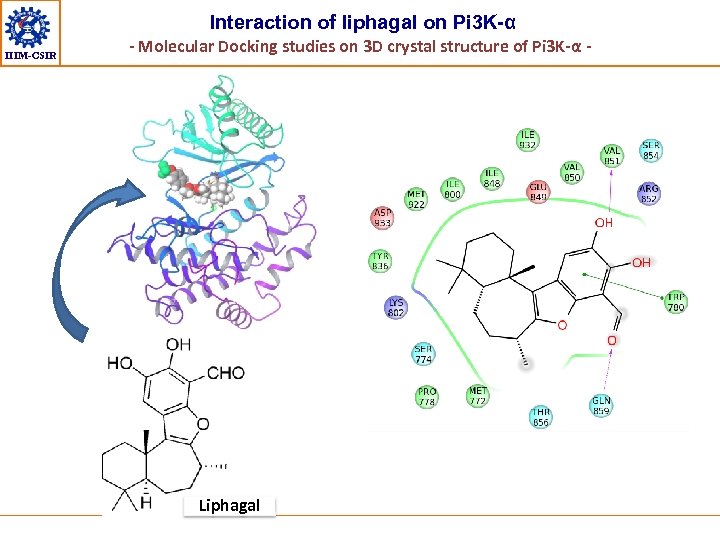 Interaction of liphagal on Pi 3 K-α IIIM-CSIR - Molecular Docking studies on 3 D crystal structure of Pi 3 K-α - Liphagal
Interaction of liphagal on Pi 3 K-α IIIM-CSIR - Molecular Docking studies on 3 D crystal structure of Pi 3 K-α - Liphagal
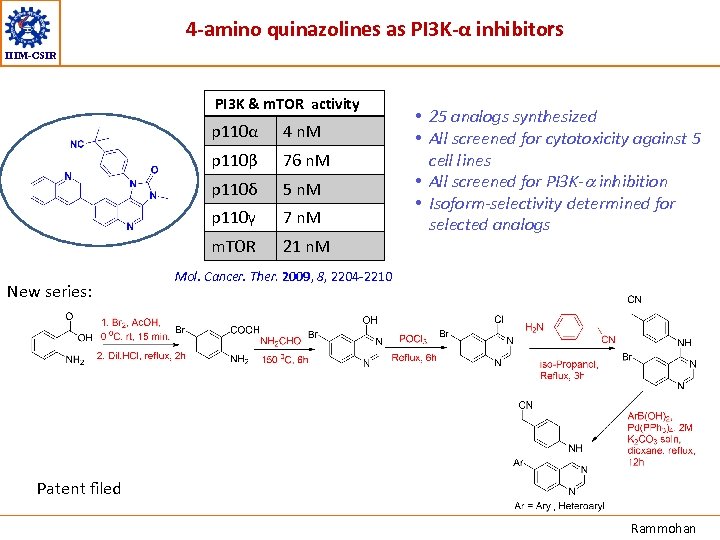 4 -amino quinazolines as PI 3 K-α inhibitors IIIM-CSIR PI 3 K & m. TOR activity p 110α p 110β 76 n. M p 110δ 5 n. M p 110γ 7 n. M m. TOR New series: 4 n. M • 25 analogs synthesized • All screened for cytotoxicity against 5 cell lines • All screened for PI 3 K-a inhibition • Isoform-selectivity determined for selected analogs 21 n. M Mol. Cancer. Ther. 2009, 8, 2204 -2210 Patent filed Rammohan
4 -amino quinazolines as PI 3 K-α inhibitors IIIM-CSIR PI 3 K & m. TOR activity p 110α p 110β 76 n. M p 110δ 5 n. M p 110γ 7 n. M m. TOR New series: 4 n. M • 25 analogs synthesized • All screened for cytotoxicity against 5 cell lines • All screened for PI 3 K-a inhibition • Isoform-selectivity determined for selected analogs 21 n. M Mol. Cancer. Ther. 2009, 8, 2204 -2210 Patent filed Rammohan
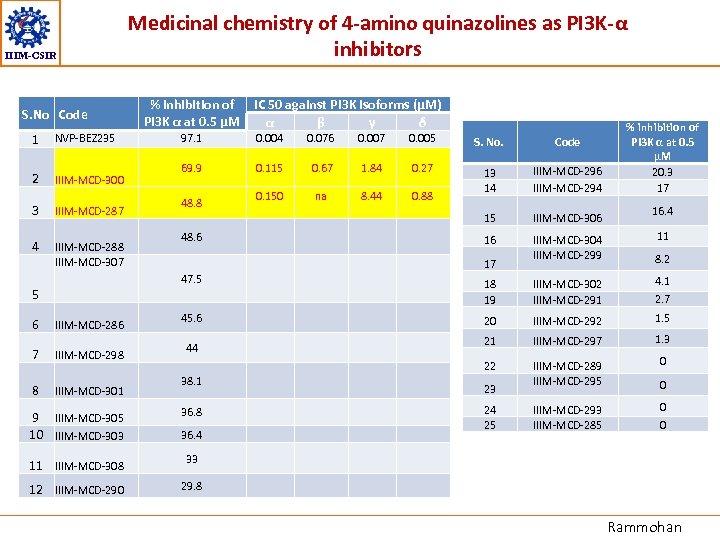 IIIM-CSIR S. No Code 1 NVP-BEZ 235 2 IIIM-MCD-300 3 IIIM-MCD-287 4 IIIM-MCD-288 IIIM-MCD-307 % inhibition of IC 50 against PI 3 K isoforms (µM) PI 3 K α at 0. 5 µM α β γ δ 97. 1 0. 004 0. 076 0. 007 0. 005 S. No. Code 69. 9 0. 115 0. 67 1. 84 0. 27 0. 150 na 8. 44 0. 88 13 14 IIIM-MCD-296 IIIM-MCD-294 15 IIIM-MCD-306 16 IIIM-MCD-304 IIIM-MCD-299 48. 8 48. 6 17 47. 5 5 6 Medicinal chemistry of 4 -amino quinazolines as PI 3 K-α inhibitors IIIM-MCD-286 7 IIIM-MCD-298 8 IIIM-MCD-301 % inhibition of PI 3 K α at 0. 5 µM 20. 3 17 16. 4 11 8. 2 44 38. 1 9 IIIM-MCD-305 10 IIIM-MCD-303 36. 8 11 IIIM-MCD-308 4. 1 20 IIIM-MCD-292 1. 5 21 IIIM-MCD-297 1. 3 IIIM-MCD-289 IIIM-MCD-295 0 IIIM-MCD-293 IIIM-MCD-285 0 23 24 25 2. 7 0 33 12 IIIM-MCD-290 IIIM-MCD-302 IIIM-MCD-291 22 45. 6 18 19 29. 8 36. 4 0 Rammohan
IIIM-CSIR S. No Code 1 NVP-BEZ 235 2 IIIM-MCD-300 3 IIIM-MCD-287 4 IIIM-MCD-288 IIIM-MCD-307 % inhibition of IC 50 against PI 3 K isoforms (µM) PI 3 K α at 0. 5 µM α β γ δ 97. 1 0. 004 0. 076 0. 007 0. 005 S. No. Code 69. 9 0. 115 0. 67 1. 84 0. 27 0. 150 na 8. 44 0. 88 13 14 IIIM-MCD-296 IIIM-MCD-294 15 IIIM-MCD-306 16 IIIM-MCD-304 IIIM-MCD-299 48. 8 48. 6 17 47. 5 5 6 Medicinal chemistry of 4 -amino quinazolines as PI 3 K-α inhibitors IIIM-MCD-286 7 IIIM-MCD-298 8 IIIM-MCD-301 % inhibition of PI 3 K α at 0. 5 µM 20. 3 17 16. 4 11 8. 2 44 38. 1 9 IIIM-MCD-305 10 IIIM-MCD-303 36. 8 11 IIIM-MCD-308 4. 1 20 IIIM-MCD-292 1. 5 21 IIIM-MCD-297 1. 3 IIIM-MCD-289 IIIM-MCD-295 0 IIIM-MCD-293 IIIM-MCD-285 0 23 24 25 2. 7 0 33 12 IIIM-MCD-290 IIIM-MCD-302 IIIM-MCD-291 22 45. 6 18 19 29. 8 36. 4 0 Rammohan
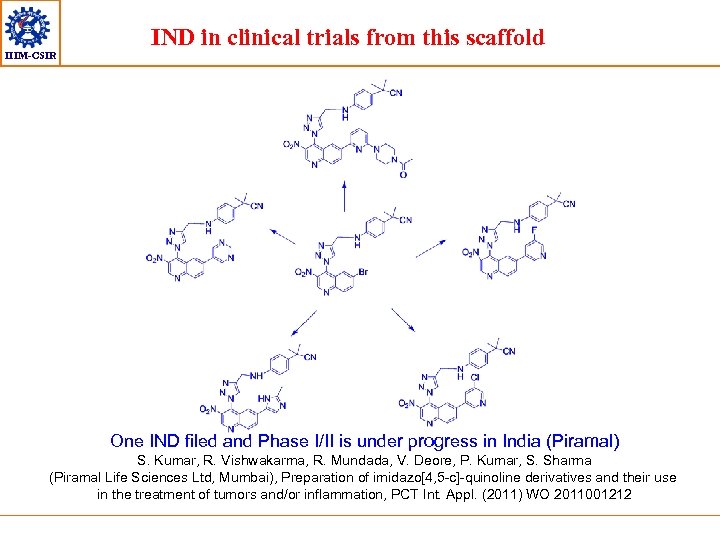 IIIM-CSIR IND in clinical trials from this scaffold One IND filed and Phase I/II is under progress in India (Piramal) S. Kumar, R. Vishwakarma, R. Mundada, V. Deore, P. Kumar, S. Sharma (Piramal Life Sciences Ltd, Mumbai), Preparation of imidazo[4, 5 -c]-quinoline derivatives and their use in the treatment of tumors and/or inflammation, PCT Int. Appl. (2011) WO 2011001212
IIIM-CSIR IND in clinical trials from this scaffold One IND filed and Phase I/II is under progress in India (Piramal) S. Kumar, R. Vishwakarma, R. Mundada, V. Deore, P. Kumar, S. Sharma (Piramal Life Sciences Ltd, Mumbai), Preparation of imidazo[4, 5 -c]-quinoline derivatives and their use in the treatment of tumors and/or inflammation, PCT Int. Appl. (2011) WO 2011001212
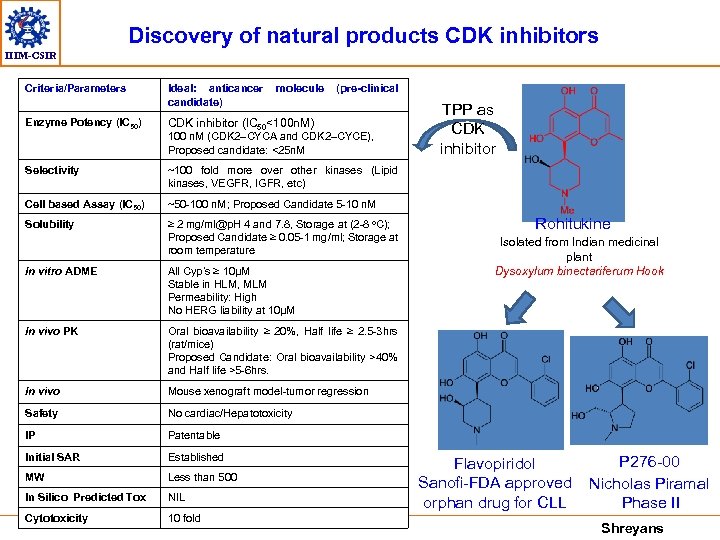 Discovery of natural products CDK inhibitors IIIM-CSIR Criteria/Parameters Ideal: anticancer candidate) molecule Enzyme Potency (IC 50) (pre-clinical CDK inhibitor (IC 50<100 n. M) 100 n. M (CDK 2–CYCA and CDK 2–CYCE), Proposed candidate: <25 n. M Selectivity ~100 fold more over other kinases (Lipid kinases, VEGFR, IGFR, etc) Cell based Assay (IC 50) ~50 -100 n. M; Proposed Candidate 5 -10 n. M Solubility ≥ 2 mg/ml@p. H 4 and 7. 8, Storage at (2 -8 o. C); Proposed Candidate ≥ 0. 05 -1 mg/ml; Storage at room temperature TPP as CDK inhibitor In vitro ADME All Cyp’s ≥ 10µM Stable in HLM, MLM Permeability: High No HERG liability at 10µM In vivo PK Mouse xenograft model-tumor regression Safety No cardiac/Hepatotoxicity IP Patentable Initial SAR Established MW Less than 500 In Silico Predicted Tox NIL Cytotoxicity 10 fold Isolated from Indian medicinal plant Dysoxylum binectariferum Hook Oral bioavailability ≥ 20%, Half life ≥ 2. 5 -3 hrs (rat/mice) Proposed Candidate: Oral bioavailability >40% and Half life >5 -6 hrs. In vivo Rohitukine Flavopiridol Sanofi-FDA approved orphan drug for CLL P 276 -00 Nicholas Piramal Phase II Shreyans
Discovery of natural products CDK inhibitors IIIM-CSIR Criteria/Parameters Ideal: anticancer candidate) molecule Enzyme Potency (IC 50) (pre-clinical CDK inhibitor (IC 50<100 n. M) 100 n. M (CDK 2–CYCA and CDK 2–CYCE), Proposed candidate: <25 n. M Selectivity ~100 fold more over other kinases (Lipid kinases, VEGFR, IGFR, etc) Cell based Assay (IC 50) ~50 -100 n. M; Proposed Candidate 5 -10 n. M Solubility ≥ 2 mg/ml@p. H 4 and 7. 8, Storage at (2 -8 o. C); Proposed Candidate ≥ 0. 05 -1 mg/ml; Storage at room temperature TPP as CDK inhibitor In vitro ADME All Cyp’s ≥ 10µM Stable in HLM, MLM Permeability: High No HERG liability at 10µM In vivo PK Mouse xenograft model-tumor regression Safety No cardiac/Hepatotoxicity IP Patentable Initial SAR Established MW Less than 500 In Silico Predicted Tox NIL Cytotoxicity 10 fold Isolated from Indian medicinal plant Dysoxylum binectariferum Hook Oral bioavailability ≥ 20%, Half life ≥ 2. 5 -3 hrs (rat/mice) Proposed Candidate: Oral bioavailability >40% and Half life >5 -6 hrs. In vivo Rohitukine Flavopiridol Sanofi-FDA approved orphan drug for CLL P 276 -00 Nicholas Piramal Phase II Shreyans
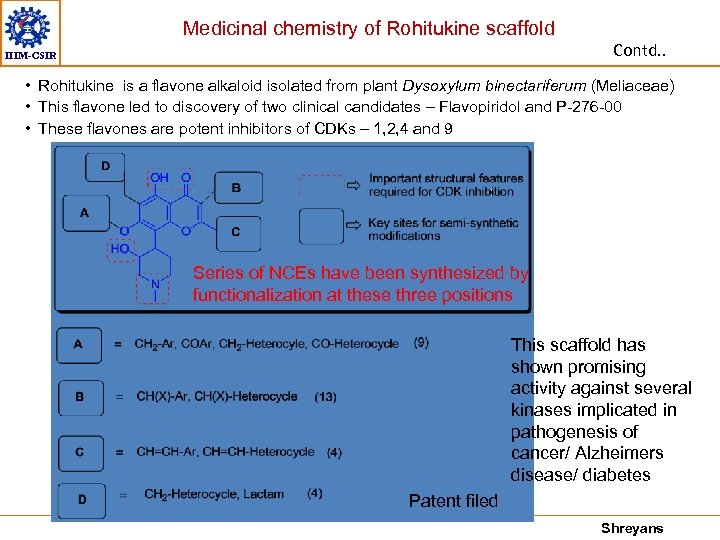 Medicinal chemistry of Rohitukine scaffold Contd. . IIIM-CSIR • Rohitukine is a flavone alkaloid isolated from plant Dysoxylum binectariferum (Meliaceae) • This flavone led to discovery of two clinical candidates – Flavopiridol and P-276 -00 • These flavones are potent inhibitors of CDKs – 1, 2, 4 and 9 Series of NCEs have been synthesized by functionalization at these three positions This scaffold has shown promising activity against several kinases implicated in pathogenesis of cancer/ Alzheimers disease/ diabetes Patent filed Shreyans
Medicinal chemistry of Rohitukine scaffold Contd. . IIIM-CSIR • Rohitukine is a flavone alkaloid isolated from plant Dysoxylum binectariferum (Meliaceae) • This flavone led to discovery of two clinical candidates – Flavopiridol and P-276 -00 • These flavones are potent inhibitors of CDKs – 1, 2, 4 and 9 Series of NCEs have been synthesized by functionalization at these three positions This scaffold has shown promising activity against several kinases implicated in pathogenesis of cancer/ Alzheimers disease/ diabetes Patent filed Shreyans
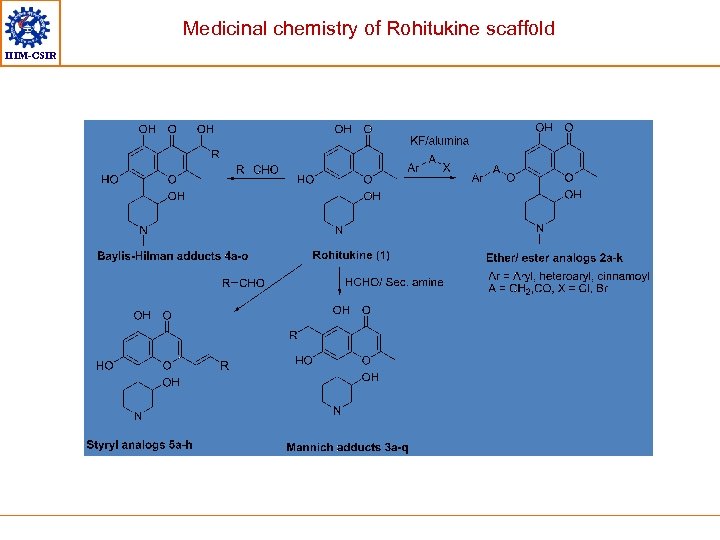 Medicinal chemistry of Rohitukine scaffold IIIM-CSIR
Medicinal chemistry of Rohitukine scaffold IIIM-CSIR
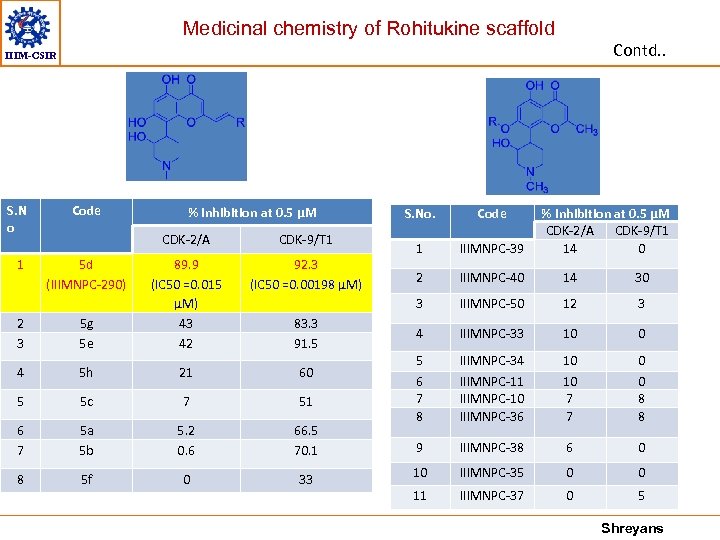 Medicinal chemistry of Rohitukine scaffold Contd. . IIIM-CSIR S. N o 1 Code % Inhibition at 0. 5 μM 5 d (IIIMNPC-290) CDK-9/T 1 92. 3 (IC 50 =0. 00198 μM) 2 3 5 g 5 e 89. 9 (IC 50 =0. 015 μM) 43 42 4 5 h 21 60 5 5 c 7 51 6 7 5 a 5 b 5. 2 0. 6 66. 5 70. 1 8 5 f 0 33 83. 3 91. 5 Code 1 IIIMNPC-39 2 IIIMNPC-40 14 30 3 CDK-2/A S. No. % Inhibition at 0. 5 μM CDK-2/A CDK-9/T 1 14 0 IIIMNPC-50 12 3 4 IIIMNPC-33 10 0 5 6 7 8 IIIMNPC-34 IIIMNPC-11 IIIMNPC-10 IIIMNPC-36 10 10 7 7 0 0 8 8 9 IIIMNPC-38 6 0 10 IIIMNPC-35 0 0 11 IIIMNPC-37 0 5 Shreyans
Medicinal chemistry of Rohitukine scaffold Contd. . IIIM-CSIR S. N o 1 Code % Inhibition at 0. 5 μM 5 d (IIIMNPC-290) CDK-9/T 1 92. 3 (IC 50 =0. 00198 μM) 2 3 5 g 5 e 89. 9 (IC 50 =0. 015 μM) 43 42 4 5 h 21 60 5 5 c 7 51 6 7 5 a 5 b 5. 2 0. 6 66. 5 70. 1 8 5 f 0 33 83. 3 91. 5 Code 1 IIIMNPC-39 2 IIIMNPC-40 14 30 3 CDK-2/A S. No. % Inhibition at 0. 5 μM CDK-2/A CDK-9/T 1 14 0 IIIMNPC-50 12 3 4 IIIMNPC-33 10 0 5 6 7 8 IIIMNPC-34 IIIMNPC-11 IIIMNPC-10 IIIMNPC-36 10 10 7 7 0 0 8 8 9 IIIMNPC-38 6 0 10 IIIMNPC-35 0 0 11 IIIMNPC-37 0 5 Shreyans
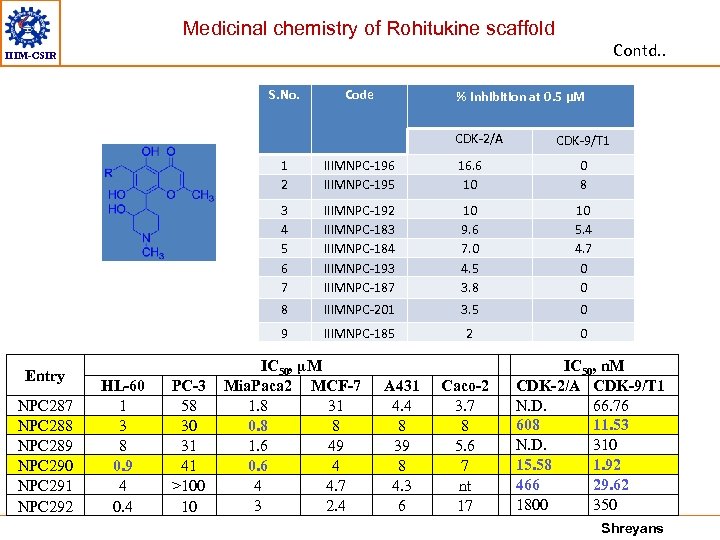 Medicinal chemistry of Rohitukine scaffold Contd. . IIIM-CSIR S. No. Code % Inhibition at 0. 5 μM CDK-2/A CDK-9/T 1 1 2 NPC 287 NPC 288 NPC 289 NPC 290 NPC 291 NPC 292 IIIMNPC-183 IIIMNPC-184 IIIMNPC-193 IIIMNPC-187 10 9. 6 7. 0 4. 5 3. 8 10 5. 4 4. 7 0 0 IIIMNPC-201 3. 5 0 9 PC-3 58 30 31 41 >100 10 0 8 8 HL-60 1 3 8 0. 9 4 0. 4 16. 6 10 3 4 5 6 7 Entry IIIMNPC-196 IIIMNPC-195 IIIMNPC-185 2 0 IC 50, µM Mia. Paca 2 MCF-7 1. 8 31 0. 8 8 1. 6 49 0. 6 4 4 4. 7 3 2. 4 A 431 4. 4 8 39 8 4. 3 6 Caco-2 3. 7 8 5. 6 7 nt 17 IC 50, n. M CDK-2/A CDK-9/T 1 N. D. 66. 76 608 11. 53 N. D. 310 15. 58 1. 92 466 29. 62 1800 350 Shreyans
Medicinal chemistry of Rohitukine scaffold Contd. . IIIM-CSIR S. No. Code % Inhibition at 0. 5 μM CDK-2/A CDK-9/T 1 1 2 NPC 287 NPC 288 NPC 289 NPC 290 NPC 291 NPC 292 IIIMNPC-183 IIIMNPC-184 IIIMNPC-193 IIIMNPC-187 10 9. 6 7. 0 4. 5 3. 8 10 5. 4 4. 7 0 0 IIIMNPC-201 3. 5 0 9 PC-3 58 30 31 41 >100 10 0 8 8 HL-60 1 3 8 0. 9 4 0. 4 16. 6 10 3 4 5 6 7 Entry IIIMNPC-196 IIIMNPC-195 IIIMNPC-185 2 0 IC 50, µM Mia. Paca 2 MCF-7 1. 8 31 0. 8 8 1. 6 49 0. 6 4 4 4. 7 3 2. 4 A 431 4. 4 8 39 8 4. 3 6 Caco-2 3. 7 8 5. 6 7 nt 17 IC 50, n. M CDK-2/A CDK-9/T 1 N. D. 66. 76 608 11. 53 N. D. 310 15. 58 1. 92 466 29. 62 1800 350 Shreyans
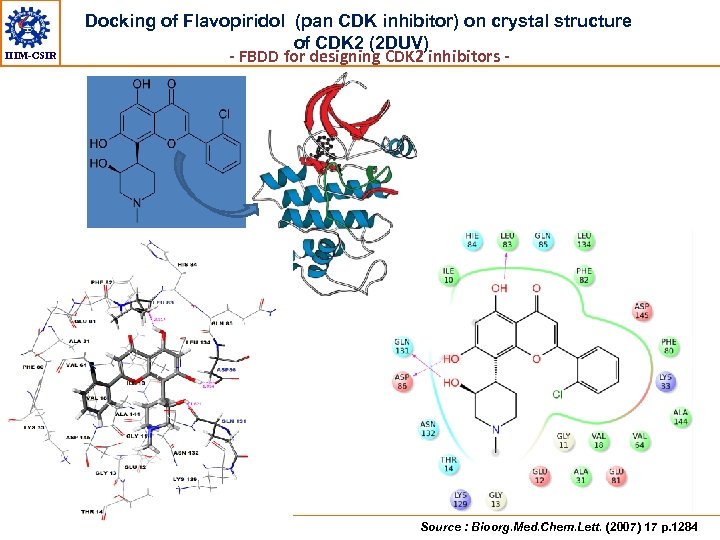 IIIM-CSIR Docking of Flavopiridol (pan CDK inhibitor) on crystal structure of CDK 2 (2 DUV) - FBDD for designing CDK 2 inhibitors - Source : Bioorg. Med. Chem. Lett. (2007) 17 p. 1284
IIIM-CSIR Docking of Flavopiridol (pan CDK inhibitor) on crystal structure of CDK 2 (2 DUV) - FBDD for designing CDK 2 inhibitors - Source : Bioorg. Med. Chem. Lett. (2007) 17 p. 1284
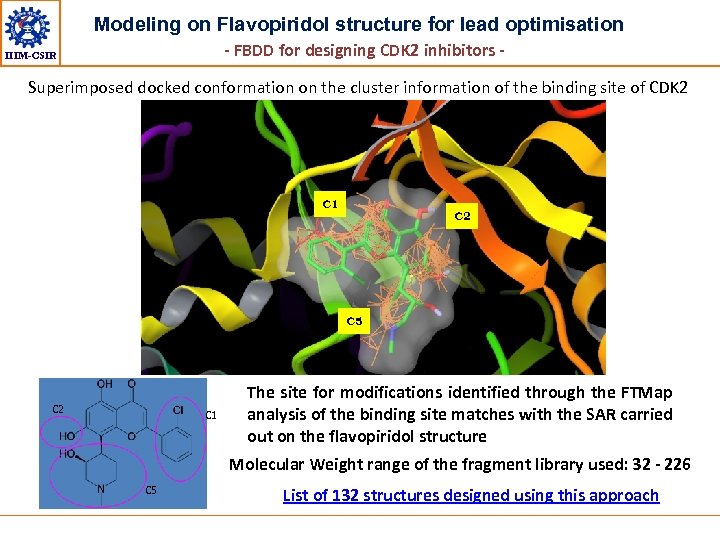 Modeling on Flavopiridol structure for lead optimisation - FBDD for designing CDK 2 inhibitors - IIIM-CSIR Superimposed docked conformation on the cluster information of the binding site of CDK 2 C 1 C 2 C 5 C 2 C 1 The site for modifications identified through the FTMap analysis of the binding site matches with the SAR carried out on the flavopiridol structure Molecular Weight range of the fragment library used: 32 - 226 C 5 List of 132 structures designed using this approach
Modeling on Flavopiridol structure for lead optimisation - FBDD for designing CDK 2 inhibitors - IIIM-CSIR Superimposed docked conformation on the cluster information of the binding site of CDK 2 C 1 C 2 C 5 C 2 C 1 The site for modifications identified through the FTMap analysis of the binding site matches with the SAR carried out on the flavopiridol structure Molecular Weight range of the fragment library used: 32 - 226 C 5 List of 132 structures designed using this approach
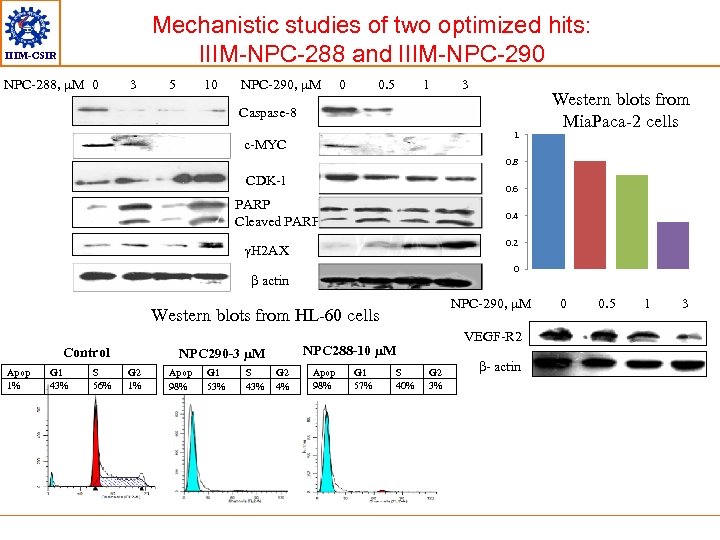 Mechanistic studies of two optimized hits: IIIM-NPC-288 and IIIM-NPC-290 IIIM-CSIR NPC-288, µM 0 3 5 10 NPC-290, µM 0 0. 5 1 3 Caspase-8 1 c-MYC Western blots from Mia. Paca-2 cells 0. 8 CDK-1 0. 6 PARP Cleaved PARP 0. 4 0. 2 γH 2 AX 0 β actin NPC-290, µM Western blots from HL-60 cells Control Apop 1% G 1 43% S 56% NPC 290 -3 µM G 2 1% Apop 98% G 1 53% S 43% VEGF-R 2 NPC 288 -10 µM G 2 4% Apop 98% G 1 57% S 40% G 2 3% β- actin 0 0. 5 1 3
Mechanistic studies of two optimized hits: IIIM-NPC-288 and IIIM-NPC-290 IIIM-CSIR NPC-288, µM 0 3 5 10 NPC-290, µM 0 0. 5 1 3 Caspase-8 1 c-MYC Western blots from Mia. Paca-2 cells 0. 8 CDK-1 0. 6 PARP Cleaved PARP 0. 4 0. 2 γH 2 AX 0 β actin NPC-290, µM Western blots from HL-60 cells Control Apop 1% G 1 43% S 56% NPC 290 -3 µM G 2 1% Apop 98% G 1 53% S 43% VEGF-R 2 NPC 288 -10 µM G 2 4% Apop 98% G 1 57% S 40% G 2 3% β- actin 0 0. 5 1 3
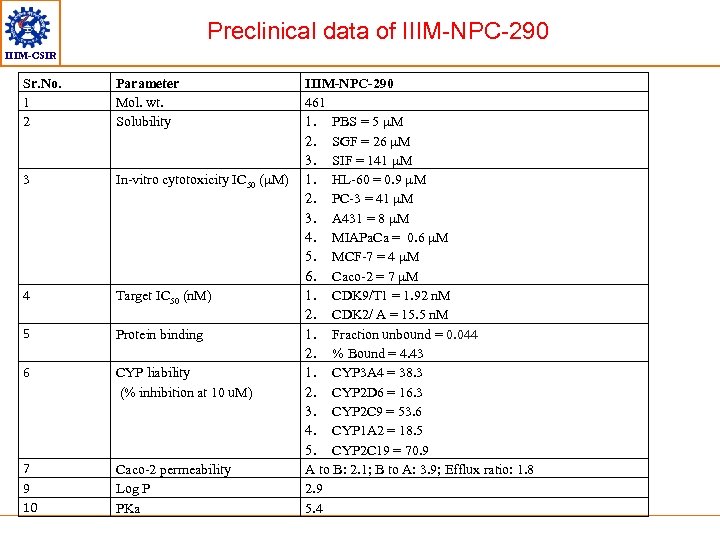 Preclinical data of IIIM-NPC-290 IIIM-CSIR Sr. No. 1 2 Parameter Mol. wt. Solubility 3 In-vitro cytotoxicity IC 50 (µM) 4 Target IC 50 (n. M) 5 Protein binding 6 CYP liability (% inhibition at 10 u. M) 7 9 10 Caco-2 permeability Log P PKa IIIM-NPC-290 461 1. PBS = 5 µM 2. SGF = 26 µM 3. SIF = 141 µM 1. HL-60 = 0. 9 µM 2. PC-3 = 41 µM 3. A 431 = 8 µM 4. MIAPa. Ca = 0. 6 µM 5. MCF-7 = 4 µM 6. Caco-2 = 7 µM 1. CDK 9/T 1 = 1. 92 n. M 2. CDK 2/ A = 15. 5 n. M 1. Fraction unbound = 0. 044 2. % Bound = 4. 43 1. CYP 3 A 4 = 38. 3 2. CYP 2 D 6 = 16. 3 3. CYP 2 C 9 = 53. 6 4. CYP 1 A 2 = 18. 5 5. CYP 2 C 19 = 70. 9 A to B: 2. 1; B to A: 3. 9; Efflux ratio: 1. 8 2. 9 5. 4
Preclinical data of IIIM-NPC-290 IIIM-CSIR Sr. No. 1 2 Parameter Mol. wt. Solubility 3 In-vitro cytotoxicity IC 50 (µM) 4 Target IC 50 (n. M) 5 Protein binding 6 CYP liability (% inhibition at 10 u. M) 7 9 10 Caco-2 permeability Log P PKa IIIM-NPC-290 461 1. PBS = 5 µM 2. SGF = 26 µM 3. SIF = 141 µM 1. HL-60 = 0. 9 µM 2. PC-3 = 41 µM 3. A 431 = 8 µM 4. MIAPa. Ca = 0. 6 µM 5. MCF-7 = 4 µM 6. Caco-2 = 7 µM 1. CDK 9/T 1 = 1. 92 n. M 2. CDK 2/ A = 15. 5 n. M 1. Fraction unbound = 0. 044 2. % Bound = 4. 43 1. CYP 3 A 4 = 38. 3 2. CYP 2 D 6 = 16. 3 3. CYP 2 C 9 = 53. 6 4. CYP 1 A 2 = 18. 5 5. CYP 2 C 19 = 70. 9 A to B: 2. 1; B to A: 3. 9; Efflux ratio: 1. 8 2. 9 5. 4
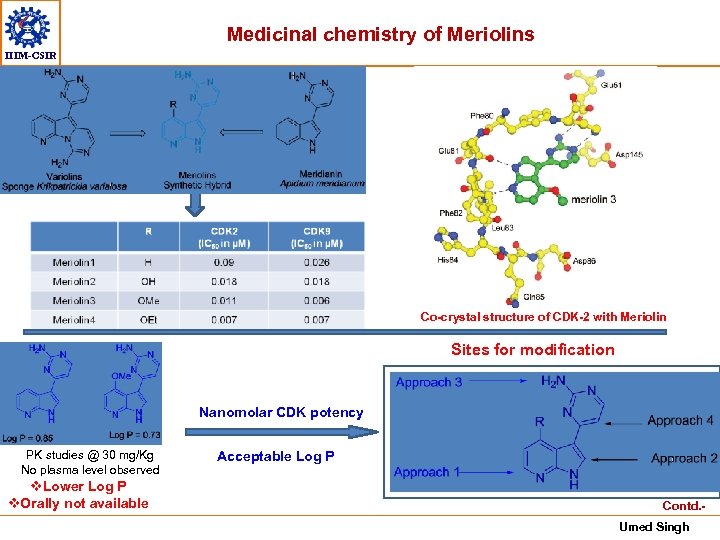 Medicinal chemistry of Meriolins IIIM-CSIR Co-crystal structure of CDK-2 with Meriolin Sites for modification Nanomolar CDK potency PK studies @ 30 mg/Kg No plasma level observed v. Lower Log P v. Orally not available Acceptable Log P Contd. Umed Singh
Medicinal chemistry of Meriolins IIIM-CSIR Co-crystal structure of CDK-2 with Meriolin Sites for modification Nanomolar CDK potency PK studies @ 30 mg/Kg No plasma level observed v. Lower Log P v. Orally not available Acceptable Log P Contd. Umed Singh
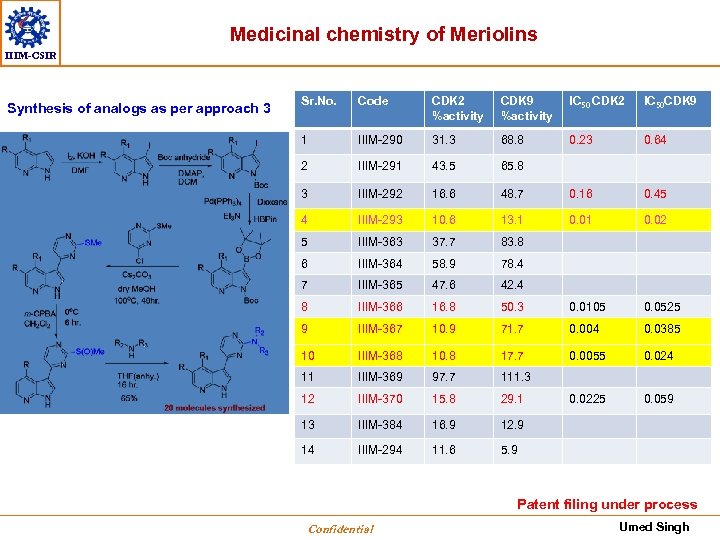 Medicinal chemistry of Meriolins IIIM-CSIR Synthesis of analogs as per approach 3 Sr. No. Code CDK 2 %activity CDK 9 %activity IC 50 CDK 2 IC 50 CDK 9 1 IIIM-290 31. 3 68. 8 0. 23 0. 64 2 IIIM-291 43. 5 65. 8 3 IIIM-292 16. 6 48. 7 0. 16 0. 45 4 IIIM-293 10. 6 13. 1 0. 02 5 IIIM-363 37. 7 83. 8 6 IIIM-364 58. 9 78. 4 7 IIIM-365 47. 6 42. 4 8 IIIM-366 16. 8 50. 3 0. 0105 0. 0525 9 IIIM-367 10. 9 71. 7 0. 004 0. 0385 10 IIIM-368 10. 8 17. 7 0. 0055 0. 024 11 IIIM-369 97. 7 111. 3 12 IIIM-370 15. 8 29. 1 0. 0225 0. 059 13 IIIM-384 16. 9 12. 9 14 IIIM-294 11. 6 5. 9 Patent filing under process Confidential Umed Singh
Medicinal chemistry of Meriolins IIIM-CSIR Synthesis of analogs as per approach 3 Sr. No. Code CDK 2 %activity CDK 9 %activity IC 50 CDK 2 IC 50 CDK 9 1 IIIM-290 31. 3 68. 8 0. 23 0. 64 2 IIIM-291 43. 5 65. 8 3 IIIM-292 16. 6 48. 7 0. 16 0. 45 4 IIIM-293 10. 6 13. 1 0. 02 5 IIIM-363 37. 7 83. 8 6 IIIM-364 58. 9 78. 4 7 IIIM-365 47. 6 42. 4 8 IIIM-366 16. 8 50. 3 0. 0105 0. 0525 9 IIIM-367 10. 9 71. 7 0. 004 0. 0385 10 IIIM-368 10. 8 17. 7 0. 0055 0. 024 11 IIIM-369 97. 7 111. 3 12 IIIM-370 15. 8 29. 1 0. 0225 0. 059 13 IIIM-384 16. 9 12. 9 14 IIIM-294 11. 6 5. 9 Patent filing under process Confidential Umed Singh
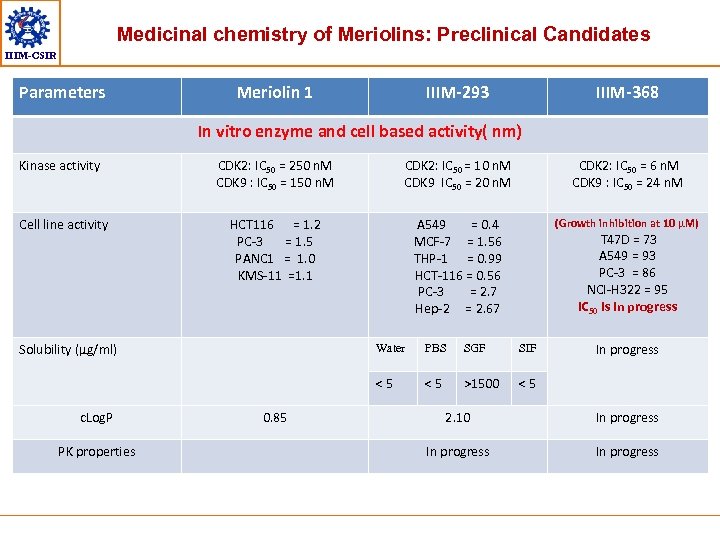 Medicinal chemistry of Meriolins: Preclinical Candidates IIIM-CSIR Parameters Meriolin 1 IIIM-293 IIIM-368 In vitro enzyme and cell based activity( nm) Kinase activity CDK 2: IC 50 = 250 n. M CDK 9 : IC 50 = 150 n. M CDK 2: IC 50 = 10 n. M CDK 9 IC 50 = 20 n. M CDK 2: IC 50 = 6 n. M CDK 9 : IC 50 = 24 n. M Cell line activity HCT 116 = 1. 2 PC-3 = 1. 5 PANC 1 = 1. 0 KMS-11 =1. 1 A 549 = 0. 4 MCF-7 = 1. 56 THP-1 = 0. 99 HCT-116 = 0. 56 PC-3 = 2. 7 Hep-2 = 2. 67 (Growth inhibition at 10 µM) Solubility (µg/ml) T 47 D = 73 A 549 = 93 PC-3 = 86 NCI-H 322 = 95 IC 50 is in progress PK properties 0. 85 PBS SGF SIF <5 c. Log. P Water <5 >1500 In progress <5 2. 10 In progress
Medicinal chemistry of Meriolins: Preclinical Candidates IIIM-CSIR Parameters Meriolin 1 IIIM-293 IIIM-368 In vitro enzyme and cell based activity( nm) Kinase activity CDK 2: IC 50 = 250 n. M CDK 9 : IC 50 = 150 n. M CDK 2: IC 50 = 10 n. M CDK 9 IC 50 = 20 n. M CDK 2: IC 50 = 6 n. M CDK 9 : IC 50 = 24 n. M Cell line activity HCT 116 = 1. 2 PC-3 = 1. 5 PANC 1 = 1. 0 KMS-11 =1. 1 A 549 = 0. 4 MCF-7 = 1. 56 THP-1 = 0. 99 HCT-116 = 0. 56 PC-3 = 2. 7 Hep-2 = 2. 67 (Growth inhibition at 10 µM) Solubility (µg/ml) T 47 D = 73 A 549 = 93 PC-3 = 86 NCI-H 322 = 95 IC 50 is in progress PK properties 0. 85 PBS SGF SIF <5 c. Log. P Water <5 >1500 In progress <5 2. 10 In progress
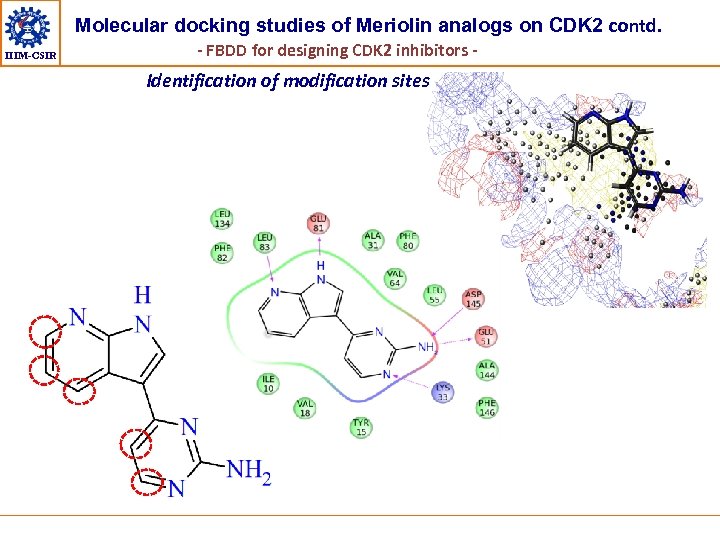 Molecular docking studies of Meriolin analogs on CDK 2 contd. IIIM-CSIR - FBDD for designing CDK 2 inhibitors - Identification of modification sites
Molecular docking studies of Meriolin analogs on CDK 2 contd. IIIM-CSIR - FBDD for designing CDK 2 inhibitors - Identification of modification sites
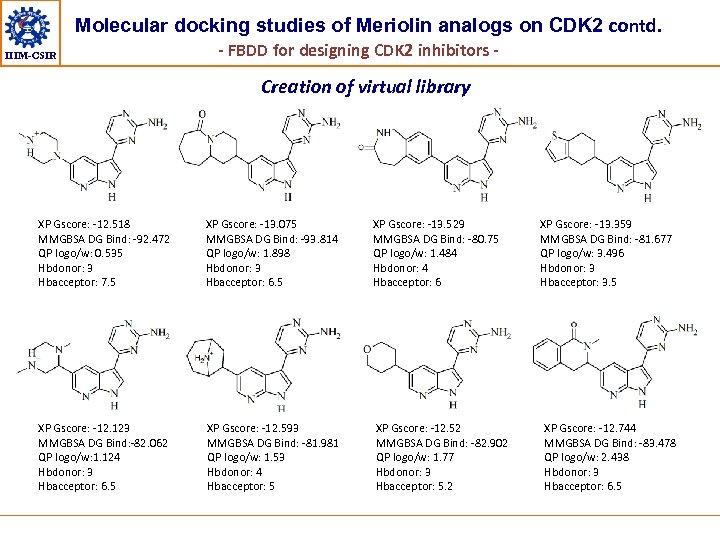 Molecular docking studies of Meriolin analogs on CDK 2 contd. IIIM-CSIR - FBDD for designing CDK 2 inhibitors - Creation of virtual library XP Gscore: -12. 518 MMGBSA DG Bind: -92. 472 QP logo/w: 0. 535 Hbdonor: 3 Hbacceptor: 7. 5 XP Gscore: -13. 075 MMGBSA DG Bind: -93. 814 QP logo/w: 1. 898 Hbdonor: 3 Hbacceptor: 6. 5 XP Gscore: -13. 529 MMGBSA DG Bind: -80. 75 QP logo/w: 1. 484 Hbdonor: 4 Hbacceptor: 6 XP Gscore: -12. 123 MMGBSA DG Bind: -82. 062 QP logo/w: 1. 124 Hbdonor: 3 Hbacceptor: 6. 5 XP Gscore: -12. 593 MMGBSA DG Bind: -81. 981 QP logo/w: 1. 53 Hbdonor: 4 Hbacceptor: 5 XP Gscore: -12. 52 MMGBSA DG Bind: -82. 902 QP logo/w: 1. 77 Hbdonor: 3 Hbacceptor: 5. 2 XP Gscore: -13. 359 MMGBSA DG Bind: -81. 677 QP logo/w: 3. 496 Hbdonor: 3 Hbacceptor: 3. 5 XP Gscore: -12. 744 MMGBSA DG Bind: -83. 478 QP logo/w: 2. 438 Hbdonor: 3 Hbacceptor: 6. 5
Molecular docking studies of Meriolin analogs on CDK 2 contd. IIIM-CSIR - FBDD for designing CDK 2 inhibitors - Creation of virtual library XP Gscore: -12. 518 MMGBSA DG Bind: -92. 472 QP logo/w: 0. 535 Hbdonor: 3 Hbacceptor: 7. 5 XP Gscore: -13. 075 MMGBSA DG Bind: -93. 814 QP logo/w: 1. 898 Hbdonor: 3 Hbacceptor: 6. 5 XP Gscore: -13. 529 MMGBSA DG Bind: -80. 75 QP logo/w: 1. 484 Hbdonor: 4 Hbacceptor: 6 XP Gscore: -12. 123 MMGBSA DG Bind: -82. 062 QP logo/w: 1. 124 Hbdonor: 3 Hbacceptor: 6. 5 XP Gscore: -12. 593 MMGBSA DG Bind: -81. 981 QP logo/w: 1. 53 Hbdonor: 4 Hbacceptor: 5 XP Gscore: -12. 52 MMGBSA DG Bind: -82. 902 QP logo/w: 1. 77 Hbdonor: 3 Hbacceptor: 5. 2 XP Gscore: -13. 359 MMGBSA DG Bind: -81. 677 QP logo/w: 3. 496 Hbdonor: 3 Hbacceptor: 3. 5 XP Gscore: -12. 744 MMGBSA DG Bind: -83. 478 QP logo/w: 2. 438 Hbdonor: 3 Hbacceptor: 6. 5
 IIIM-CSIR NCEs in preclinical development Hits identified (4 for PI 3 K and 3 for CDK) taken forward for lead optimization and preclinical development Ongoing studies • Optimization of lead • PK profile, in vivo animal studies • Cyp liabilities, Pre-formulation studies • Kinase profiling
IIIM-CSIR NCEs in preclinical development Hits identified (4 for PI 3 K and 3 for CDK) taken forward for lead optimization and preclinical development Ongoing studies • Optimization of lead • PK profile, in vivo animal studies • Cyp liabilities, Pre-formulation studies • Kinase profiling
 IIIM-CSIR Kinase Inhibitors of Marine Origin Chem. Rev. 2013, 113, 6761− 6815
IIIM-CSIR Kinase Inhibitors of Marine Origin Chem. Rev. 2013, 113, 6761− 6815
 Our Patents (2012 -2013) IIIM-CSIR 1. R. A. Vishwakarma, S. D. Sawant, P. P. Singh, A. H. Dar, P. R. Sharma, A. K. Saxena, A. Nargotra, A. A. K. Kollaru, R. Mudududdla, A. K. Qazi et. al. Bororic acid bearing liphagane compounds as inhibitors of PI 3 Ka and/or b PCT Int. Appl. (2012) WO 2013/140417 A 1 2. R. A. Vishwakarma, S. D. Sawant, P. P. Singh, A. H. Dar, A. Nargotra, P. R. Sharma, D. M. Mondhe; Isoform selective C-ring substituted purinyl, tetrazolyl, isatinyl-quinazolinone analogs as anticancer agents and inhibitors of PI 3 K-a/b; PCT Application: 150 NF/2012 3. S. K. Jain, T. Sidiq, S. Meena, A. Khajuria, R. A. Vishwakarma, S. Bharate Bibishan. Tetrahydro-2 H-Pyrano [3, 2 -C] Isochromene-6 -Ones for the treatment of inflammatory Disorders; PCT Appl. Filed, 1565 DEL 2013 4. D. M. Mondhe, S. C. Taneja, S. Koul, J. K. Dhar, A. K. Saxena, R. K. Johri, Z. A. Wani, S. A. Andotra, S. C. Sharma, S. Singh, P. N. Gupta, R. A. Vishwakarma; A novel formulation useful in Cancer chemotherapy; PCT Appl. Filed. 2554/DEL/2012 5. R. A. Vishwakarma, S. B. Bharate, S. Bhushan, S. K. Jain, S. Meena, S. K Guru, A. S. Pathania, S. Kumar; Cyclin-Dependent Kinase Inhibition by 5, 7 -Dihydroxy-8 -(3 -Hydroxy-1 -Methyl. Piperidin-4 -Yl)-2 -Methyl-4 H-Chromen-4 -One Analogs; PCT Appl. Filed 1142 DEL 2013 6. P. P. Singh, R. A. Vishwakarma; 6 -Nitro-2, 3 -Dihydroimidazo [2, 1 -b] oxazoles for the treatment of M. tuberculosis and a process for the preparation thereof ; PCT Appl. Filed 0225 NF 2012 7. R. A. Vishwakarma, S. B. Bharate, S. Bhushan, S. K. Jain, S. Meena, A. Khajuria, S. K. Bhola et. al. New Chromone alkaloid dysoline for the treatment of cancer and inflammatory disorders. ; PCT Appl. Filed 1077 DEL 2013 8. Deepika Singh, Jai Parkash Sharma, Sundeep Jaglan, Abid Hamid Dar, Varun Partap Singh, Ram A. Vishwakarma; Brachiatins: Novel anticancer agents from an endophytic fungus Trichoderma longibrachiatum, process for their production and use thereof; PCT Appl. Filed 2563 DEL 2013 9. S. D. Sawant, G. Lakshma Reddy, M. Srinivas, S. S. Hussain, D M Ishaq, A. Nargotra, P. Mahajan, R. A. Vishwakarma; Novel Pyrazolopyrimidies for treatment of impotence and process for the preparation thereof; PCT Appl. Filed 0106 NF 2013 10. R. A. Vishwakarma, S. B. Bharate, S. Bhushan, RR Yadav, S. K. Guru, P. Joshi, 6 -Aryl-3 -phenylamino-quinazoline analogs as phosphoinositide 3 -kinase inhibitors; PCT Appl. Filed 0117/NF/2013 Patent filed on 23 -May-2013 11. S. Balachandran, C. J. Dinsmore, A. Roychowdhury, R. Sharma, R. A. Vishwakarma. Preparation of morpholinosulfonyl indole compounds as modulators of insulin-like growth factor I receptors and insulin receptors for treating cancer, PCT Int. Appl. (2012), WO 2012145471 A 1
Our Patents (2012 -2013) IIIM-CSIR 1. R. A. Vishwakarma, S. D. Sawant, P. P. Singh, A. H. Dar, P. R. Sharma, A. K. Saxena, A. Nargotra, A. A. K. Kollaru, R. Mudududdla, A. K. Qazi et. al. Bororic acid bearing liphagane compounds as inhibitors of PI 3 Ka and/or b PCT Int. Appl. (2012) WO 2013/140417 A 1 2. R. A. Vishwakarma, S. D. Sawant, P. P. Singh, A. H. Dar, A. Nargotra, P. R. Sharma, D. M. Mondhe; Isoform selective C-ring substituted purinyl, tetrazolyl, isatinyl-quinazolinone analogs as anticancer agents and inhibitors of PI 3 K-a/b; PCT Application: 150 NF/2012 3. S. K. Jain, T. Sidiq, S. Meena, A. Khajuria, R. A. Vishwakarma, S. Bharate Bibishan. Tetrahydro-2 H-Pyrano [3, 2 -C] Isochromene-6 -Ones for the treatment of inflammatory Disorders; PCT Appl. Filed, 1565 DEL 2013 4. D. M. Mondhe, S. C. Taneja, S. Koul, J. K. Dhar, A. K. Saxena, R. K. Johri, Z. A. Wani, S. A. Andotra, S. C. Sharma, S. Singh, P. N. Gupta, R. A. Vishwakarma; A novel formulation useful in Cancer chemotherapy; PCT Appl. Filed. 2554/DEL/2012 5. R. A. Vishwakarma, S. B. Bharate, S. Bhushan, S. K. Jain, S. Meena, S. K Guru, A. S. Pathania, S. Kumar; Cyclin-Dependent Kinase Inhibition by 5, 7 -Dihydroxy-8 -(3 -Hydroxy-1 -Methyl. Piperidin-4 -Yl)-2 -Methyl-4 H-Chromen-4 -One Analogs; PCT Appl. Filed 1142 DEL 2013 6. P. P. Singh, R. A. Vishwakarma; 6 -Nitro-2, 3 -Dihydroimidazo [2, 1 -b] oxazoles for the treatment of M. tuberculosis and a process for the preparation thereof ; PCT Appl. Filed 0225 NF 2012 7. R. A. Vishwakarma, S. B. Bharate, S. Bhushan, S. K. Jain, S. Meena, A. Khajuria, S. K. Bhola et. al. New Chromone alkaloid dysoline for the treatment of cancer and inflammatory disorders. ; PCT Appl. Filed 1077 DEL 2013 8. Deepika Singh, Jai Parkash Sharma, Sundeep Jaglan, Abid Hamid Dar, Varun Partap Singh, Ram A. Vishwakarma; Brachiatins: Novel anticancer agents from an endophytic fungus Trichoderma longibrachiatum, process for their production and use thereof; PCT Appl. Filed 2563 DEL 2013 9. S. D. Sawant, G. Lakshma Reddy, M. Srinivas, S. S. Hussain, D M Ishaq, A. Nargotra, P. Mahajan, R. A. Vishwakarma; Novel Pyrazolopyrimidies for treatment of impotence and process for the preparation thereof; PCT Appl. Filed 0106 NF 2013 10. R. A. Vishwakarma, S. B. Bharate, S. Bhushan, RR Yadav, S. K. Guru, P. Joshi, 6 -Aryl-3 -phenylamino-quinazoline analogs as phosphoinositide 3 -kinase inhibitors; PCT Appl. Filed 0117/NF/2013 Patent filed on 23 -May-2013 11. S. Balachandran, C. J. Dinsmore, A. Roychowdhury, R. Sharma, R. A. Vishwakarma. Preparation of morpholinosulfonyl indole compounds as modulators of insulin-like growth factor I receptors and insulin receptors for treating cancer, PCT Int. Appl. (2012), WO 2012145471 A 1
 Byproducts of discovery projects: Publications (2012 -13) IIIM-CSIR 1. V. Venkateswarlu, KA Arvinda Kumar, S. Balgotra, G. L. Reddy, M. Srinivas, R. A. Vishwakarma, S. D. Sawant, Chemistry, Eur. J. , 20, 1 -6, (2014) 2. N. Mupparapu, S. Khan, S. Battula, M. Kushwaha, A. P. Gupta, Q. N. Ahmed, R. A. Vishwakarma, Organic Letters, 16, 1152 -1155 (2014) 3. P. Kannaboina, K. Anilkumar, S. Aravinda, R. A. Vishwakarma, P. Das, Organic Letters, 15, 5718 -5721 (2013) 4. S. B. Bharate, S. D. Sawant, P. P. Singh and R. A. Vishwakarma, Chemical Reviews, 113, 6761 -6815 (2013). 5. Singh, P. P. ; Aithagani, S. K. ; Yadav, M. ; Singh, V. P. ; Vishwakarma, R. A. . J. Org. Chem. , 2013, 78, 2639– 2648 6. R. Mudududdla, S. K. Jain, J. B. Bharate, A. P. Gupta, B. Singh, R. A. Vishwakarma, S. B. Bharate, J. Org. Chem. 77, 8821 -8827 (2012) 7. P. P. Singh, T. Thatikonda, K. A. Kumar, Aravinda; S. D. Sawant, B. Singh, A. K. Sharma, P. R. Sharma, D. Singh, R. A. Vishwakarma, J. Org. Chem. , 77, 5823 -5828 (2012) 8. S. B. Bharate, R. Mudududdla, J. B. Bharate, N. Battini, S. Battula, R. R. Yadav, B. Singh, R. A. Vishwakarma, Org. Biomol. Chem. 10, 5143 -5150 (2012) 9. P. P. Singh, S. Gudup, H. Ayuri, S. Ambala, M. Yadav, S. D. Sawant and R. A. Vishwakarma, Org. Biomol. Chem. , 10, 1587 -1597 (2012) 10. P. P. Singh, S. Gudup, S. Ambala, U. Singh, S. Dadhwal, B. Singh, S. D. Sawant and R. A. Vishwakarma, Chem. Commun. , 47, 5852 -5854 (2011) 11. S. Manda, B, Singh, S. B. Bharate, R. A. Vishwakarma, Med. Chem. Commun. , 3, 1098 -1103 (2012) 12. Bharate, S. B. ; Yadav, R. R. ; Khan, S. I. ; Tekwani, B. L. ; Jacob, M. R. ; Khan, I. A. ; . Med. Chem. Commun. 2013, 4, 1042 -1048.
Byproducts of discovery projects: Publications (2012 -13) IIIM-CSIR 1. V. Venkateswarlu, KA Arvinda Kumar, S. Balgotra, G. L. Reddy, M. Srinivas, R. A. Vishwakarma, S. D. Sawant, Chemistry, Eur. J. , 20, 1 -6, (2014) 2. N. Mupparapu, S. Khan, S. Battula, M. Kushwaha, A. P. Gupta, Q. N. Ahmed, R. A. Vishwakarma, Organic Letters, 16, 1152 -1155 (2014) 3. P. Kannaboina, K. Anilkumar, S. Aravinda, R. A. Vishwakarma, P. Das, Organic Letters, 15, 5718 -5721 (2013) 4. S. B. Bharate, S. D. Sawant, P. P. Singh and R. A. Vishwakarma, Chemical Reviews, 113, 6761 -6815 (2013). 5. Singh, P. P. ; Aithagani, S. K. ; Yadav, M. ; Singh, V. P. ; Vishwakarma, R. A. . J. Org. Chem. , 2013, 78, 2639– 2648 6. R. Mudududdla, S. K. Jain, J. B. Bharate, A. P. Gupta, B. Singh, R. A. Vishwakarma, S. B. Bharate, J. Org. Chem. 77, 8821 -8827 (2012) 7. P. P. Singh, T. Thatikonda, K. A. Kumar, Aravinda; S. D. Sawant, B. Singh, A. K. Sharma, P. R. Sharma, D. Singh, R. A. Vishwakarma, J. Org. Chem. , 77, 5823 -5828 (2012) 8. S. B. Bharate, R. Mudududdla, J. B. Bharate, N. Battini, S. Battula, R. R. Yadav, B. Singh, R. A. Vishwakarma, Org. Biomol. Chem. 10, 5143 -5150 (2012) 9. P. P. Singh, S. Gudup, H. Ayuri, S. Ambala, M. Yadav, S. D. Sawant and R. A. Vishwakarma, Org. Biomol. Chem. , 10, 1587 -1597 (2012) 10. P. P. Singh, S. Gudup, S. Ambala, U. Singh, S. Dadhwal, B. Singh, S. D. Sawant and R. A. Vishwakarma, Chem. Commun. , 47, 5852 -5854 (2011) 11. S. Manda, B, Singh, S. B. Bharate, R. A. Vishwakarma, Med. Chem. Commun. , 3, 1098 -1103 (2012) 12. Bharate, S. B. ; Yadav, R. R. ; Khan, S. I. ; Tekwani, B. L. ; Jacob, M. R. ; Khan, I. A. ; . Med. Chem. Commun. 2013, 4, 1042 -1048.
 IIIM-CSIR Publications (2012 -13) 13. S. B. Bharate, A. K. Padala, B. A. Dar, R. R. Yadav, B. Singh and R. A. Vishwakarma, Tetrahedron Lett. , 54, 3558 -3561 (2013) 14. S. B. Bharate, R. Mudududdla, R. Sharma and R. A. Vishwakarma, Tetrahedron Lett. , 54, 2913 -2915 (2013) 15. S. K. Jain, S. Meenaa, b, A. K. Qazi, A. Hussain, S. K. Bhola, R. Kshirsagar, K. Pari, A. Khajuria, A. Hamid, R. Uma Shaanker, S. B. Bharate and R. A. Vishwakarma, Tetrahedron Lett. , In Press 2013 16. B. Singh, R. Parshad, R. K. Khajuria, S. K. Guru, A. S. Pathania, R. Sharma, R. Chib, S. Aravinda, V. K. Gupta, I. A. Khan, S. Bhushan, Sandip B. Bharate, R. A. Vishwakarma, Tetrahedron Lett. In Press: 2013 17. S. D. Sawant, M. Srinivas, K. A. Aravinda Kumar, G. Lakshma Reddy, P. P. Singh, B. Singh, A. K. Sharma, P. R. Sharma and R. A. Vishwakarma, Tetrahedron Lett. , 54, 5351 -5354 (2013) 18. R. M. Yadav, R. A. Vishwakarma and S. B. Bharate, Tetrahedron Lett, 53, 5958 -5960 (2012) 19. S. D. Sawant, M. Srinivas, G. Lakshma Reddy, P. P. Singh, R. A. Vishwakarma, Tetrahedron Lett. , 53, 61956198 (2012) 20. R. R. Yadav, N. Battini, R. Mudududdla, J. B. Bharate, N. Muparappu, S. B. Bharate and R. A. Vishwakarma, Tetrahedron Lett. , 53, 2222 -2225 (2012) 21. S. Mohammed, A. K. Padala, B. A. Dar, B. Singh, B. Sreedhar, R. A. Vishwakarma and S. B. Bharate, Tetrahedron 68, 8156 -8162 (2012) 22. S. K. Jain, A. S. Pathania, S. Meena, R. Sharma, A. Sharma, B. Singh, B. D. Gupta, S. Bhushan, S. B. Bharate, and R. A. Vishwakarma, J. Nat. Prod. In Press, 2013: DOI: 10. 1021/np 400433 g 23. M. K. Zilla, M. Qadri, A. S. Pathania, G. A. Strobel, Y. Nalli, S. Kumar, S. K. Guru, S. Bhushan, S. K. Singh, R. A. Vishwakarma et al; Phytochemistry, In Press, 2013; DOI: 10. 1016/j. phytochem. 2013. 06. 021 24. M. Sen, B. Shah, S. Rakshit, V. Singh, B. Padmanabhan, M. Pomusamy, K. Pari, R. A. Vishwakarma, D. Nandi and P. P. Sadhale, Plo. S Pathogens, 7, e 1002384 (2011)
IIIM-CSIR Publications (2012 -13) 13. S. B. Bharate, A. K. Padala, B. A. Dar, R. R. Yadav, B. Singh and R. A. Vishwakarma, Tetrahedron Lett. , 54, 3558 -3561 (2013) 14. S. B. Bharate, R. Mudududdla, R. Sharma and R. A. Vishwakarma, Tetrahedron Lett. , 54, 2913 -2915 (2013) 15. S. K. Jain, S. Meenaa, b, A. K. Qazi, A. Hussain, S. K. Bhola, R. Kshirsagar, K. Pari, A. Khajuria, A. Hamid, R. Uma Shaanker, S. B. Bharate and R. A. Vishwakarma, Tetrahedron Lett. , In Press 2013 16. B. Singh, R. Parshad, R. K. Khajuria, S. K. Guru, A. S. Pathania, R. Sharma, R. Chib, S. Aravinda, V. K. Gupta, I. A. Khan, S. Bhushan, Sandip B. Bharate, R. A. Vishwakarma, Tetrahedron Lett. In Press: 2013 17. S. D. Sawant, M. Srinivas, K. A. Aravinda Kumar, G. Lakshma Reddy, P. P. Singh, B. Singh, A. K. Sharma, P. R. Sharma and R. A. Vishwakarma, Tetrahedron Lett. , 54, 5351 -5354 (2013) 18. R. M. Yadav, R. A. Vishwakarma and S. B. Bharate, Tetrahedron Lett, 53, 5958 -5960 (2012) 19. S. D. Sawant, M. Srinivas, G. Lakshma Reddy, P. P. Singh, R. A. Vishwakarma, Tetrahedron Lett. , 53, 61956198 (2012) 20. R. R. Yadav, N. Battini, R. Mudududdla, J. B. Bharate, N. Muparappu, S. B. Bharate and R. A. Vishwakarma, Tetrahedron Lett. , 53, 2222 -2225 (2012) 21. S. Mohammed, A. K. Padala, B. A. Dar, B. Singh, B. Sreedhar, R. A. Vishwakarma and S. B. Bharate, Tetrahedron 68, 8156 -8162 (2012) 22. S. K. Jain, A. S. Pathania, S. Meena, R. Sharma, A. Sharma, B. Singh, B. D. Gupta, S. Bhushan, S. B. Bharate, and R. A. Vishwakarma, J. Nat. Prod. In Press, 2013: DOI: 10. 1021/np 400433 g 23. M. K. Zilla, M. Qadri, A. S. Pathania, G. A. Strobel, Y. Nalli, S. Kumar, S. K. Guru, S. Bhushan, S. K. Singh, R. A. Vishwakarma et al; Phytochemistry, In Press, 2013; DOI: 10. 1016/j. phytochem. 2013. 06. 021 24. M. Sen, B. Shah, S. Rakshit, V. Singh, B. Padmanabhan, M. Pomusamy, K. Pari, R. A. Vishwakarma, D. Nandi and P. P. Sadhale, Plo. S Pathogens, 7, e 1002384 (2011)
 IIIM-CSIR Acknowledgements Parvinder Pal Singh K. A. Aravind Kumar M. Nagaraju S. D. Sawant M. Srinivas Sudhakar Manda Sandip B. Bharate M. Ramesh Mohammed Shabbir V. Venkateshwaralu Shreyans K. Jain Rammohan R. Yadav Baljinder Singh T. Thanusha Abubakar Wani Parthasarathi Das Naveed Qazi Ajay Kumar S. C. Sharma Shashi Bhushan Amit Nargotra Umed Singh Prashant Joshi D. Saidulu Collaborators: • Jubilant Biosys, Bangalore • Vimta Labs, Hyderabad • International Center for Kinase Profiling (ICKP), University of Dundee, UK • Dr. Raj Hirwani and Team, URDIP, Pune (for patentability search)
IIIM-CSIR Acknowledgements Parvinder Pal Singh K. A. Aravind Kumar M. Nagaraju S. D. Sawant M. Srinivas Sudhakar Manda Sandip B. Bharate M. Ramesh Mohammed Shabbir V. Venkateshwaralu Shreyans K. Jain Rammohan R. Yadav Baljinder Singh T. Thanusha Abubakar Wani Parthasarathi Das Naveed Qazi Ajay Kumar S. C. Sharma Shashi Bhushan Amit Nargotra Umed Singh Prashant Joshi D. Saidulu Collaborators: • Jubilant Biosys, Bangalore • Vimta Labs, Hyderabad • International Center for Kinase Profiling (ICKP), University of Dundee, UK • Dr. Raj Hirwani and Team, URDIP, Pune (for patentability search)


