e8ede19de05b76279f9b7a3297bae5bb.ppt
- Количество слайдов: 38
 II. GENOMICS: ANALYIS OF MULTIPLE MACROMOLECULES AT THE SAME TIME
II. GENOMICS: ANALYIS OF MULTIPLE MACROMOLECULES AT THE SAME TIME
 Genomics Structural Functional Integrative
Genomics Structural Functional Integrative
 Structural genomics ü Genome libraries ü DNA sequencing ü Genome projects ü Polymorphisms ü RFLP
Structural genomics ü Genome libraries ü DNA sequencing ü Genome projects ü Polymorphisms ü RFLP
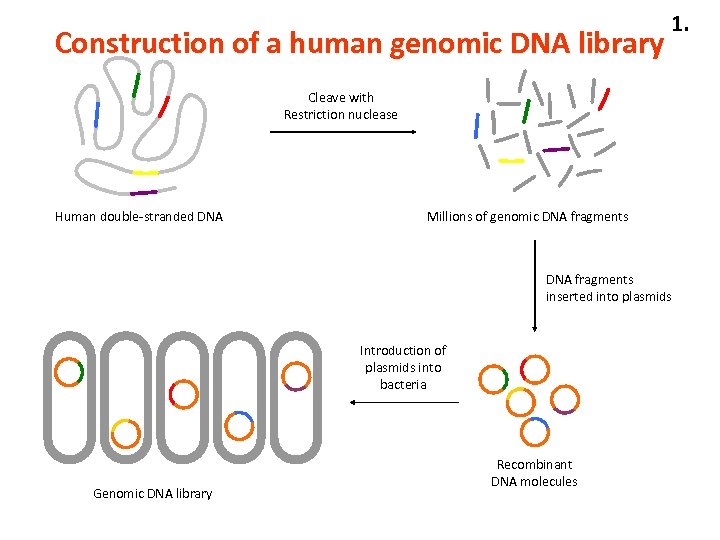 Construction of a human genomic DNA library 1. Cleave with Restriction nuclease Human double-stranded DNA Millions of genomic DNA fragments inserted into plasmids Introduction of plasmids into bacteria Genomic DNA library Recombinant DNA molecules
Construction of a human genomic DNA library 1. Cleave with Restriction nuclease Human double-stranded DNA Millions of genomic DNA fragments inserted into plasmids Introduction of plasmids into bacteria Genomic DNA library Recombinant DNA molecules
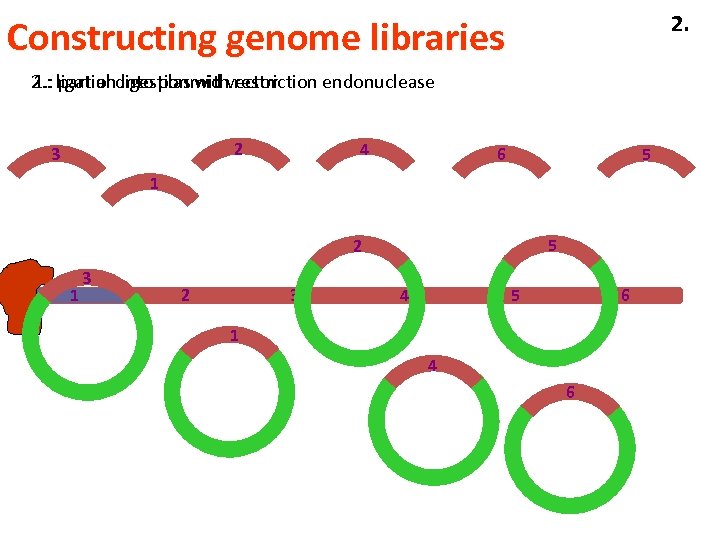 2. Constructing genome libraries 2. : ligationdigestion withvector 1. : partial into plasmid restriction endonuclease 4 2 3 6 5 1 5 2 1 3 2 3 4 5 6 1 4 6
2. Constructing genome libraries 2. : ligationdigestion withvector 1. : partial into plasmid restriction endonuclease 4 2 3 6 5 1 5 2 1 3 2 3 4 5 6 1 4 6
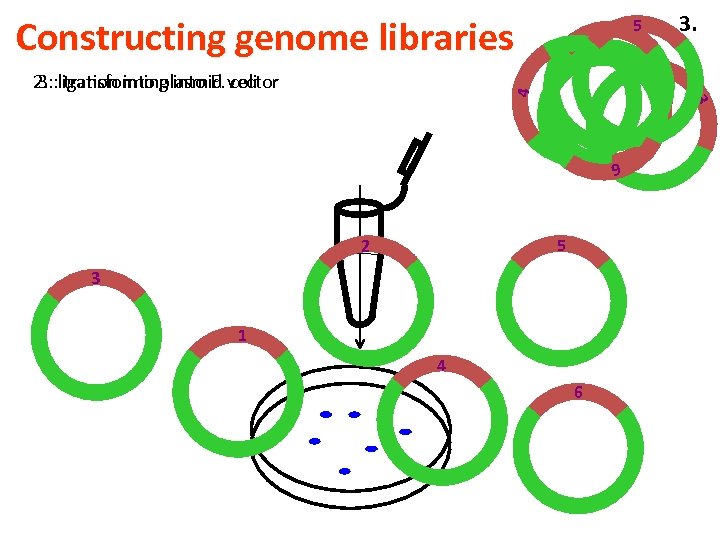 Constructing genome libraries 3. 5 3 4 2. : ligation into plasmid vector 3. : transforming into E. coli 2 1 6 5 2 3 1 4 6
Constructing genome libraries 3. 5 3 4 2. : ligation into plasmid vector 3. : transforming into E. coli 2 1 6 5 2 3 1 4 6
 Genome Libraries Genome library: collection of clones, in wich every pieces of the genome of a particular organism can be found. Usage: sequencing (genome projects), isolation of genes. c. DNA library: The c. DNA library contains a c. DNA copy of each m. RNA of an organism (tissue or cell type). It represents the transcriptome. Usage: gene structure determination, isolation of c. DNSs (intronles gene). 4.
Genome Libraries Genome library: collection of clones, in wich every pieces of the genome of a particular organism can be found. Usage: sequencing (genome projects), isolation of genes. c. DNA library: The c. DNA library contains a c. DNA copy of each m. RNA of an organism (tissue or cell type). It represents the transcriptome. Usage: gene structure determination, isolation of c. DNSs (intronles gene). 4.
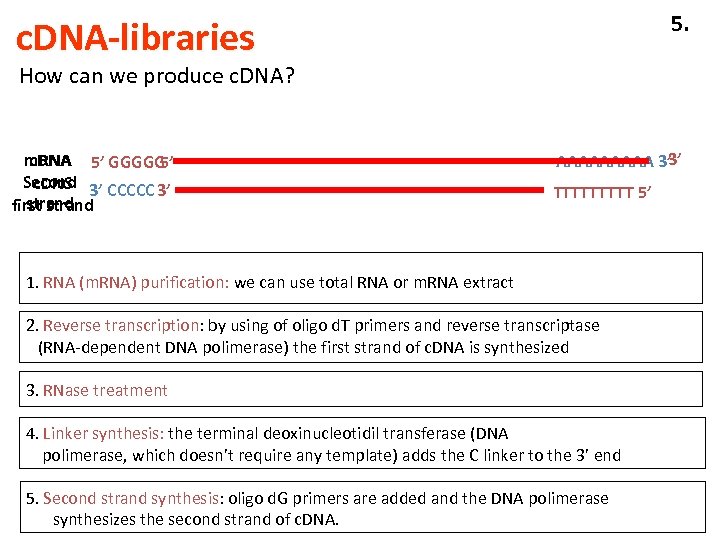 5. c. DNA-libraries How can we produce c. DNA? c. DNA m. RNA 5’ GGGGG 5’ Second 3’ CCCCC 3’ c. DNS strand first strand 3’ AAAAA 3’ TTTTT 5’ 1. RNA (m. RNA) purification: we can use total RNA or m. RNA extract 2. Reverse transcription: by using of oligo d. T primers and reverse transcriptase (RNA-dependent DNA polimerase) the first strand of c. DNA is synthesized 3. RNase treatment 4. Linker synthesis: the terminal deoxinucleotidil transferase (DNA polimerase, which doesn’t require any template) adds the C linker to the 3’ end 5. Second strand synthesis: oligo d. G primers are added and the DNA polimerase synthesizes the second strand of c. DNA.
5. c. DNA-libraries How can we produce c. DNA? c. DNA m. RNA 5’ GGGGG 5’ Second 3’ CCCCC 3’ c. DNS strand first strand 3’ AAAAA 3’ TTTTT 5’ 1. RNA (m. RNA) purification: we can use total RNA or m. RNA extract 2. Reverse transcription: by using of oligo d. T primers and reverse transcriptase (RNA-dependent DNA polimerase) the first strand of c. DNA is synthesized 3. RNase treatment 4. Linker synthesis: the terminal deoxinucleotidil transferase (DNA polimerase, which doesn’t require any template) adds the C linker to the 3’ end 5. Second strand synthesis: oligo d. G primers are added and the DNA polimerase synthesizes the second strand of c. DNA.
 DNA sequencing – Sanger method v Dideoxynucleotide chain termination or Stop-nucleotide-method or Chain termination method anger Frederic S 6.
DNA sequencing – Sanger method v Dideoxynucleotide chain termination or Stop-nucleotide-method or Chain termination method anger Frederic S 6.
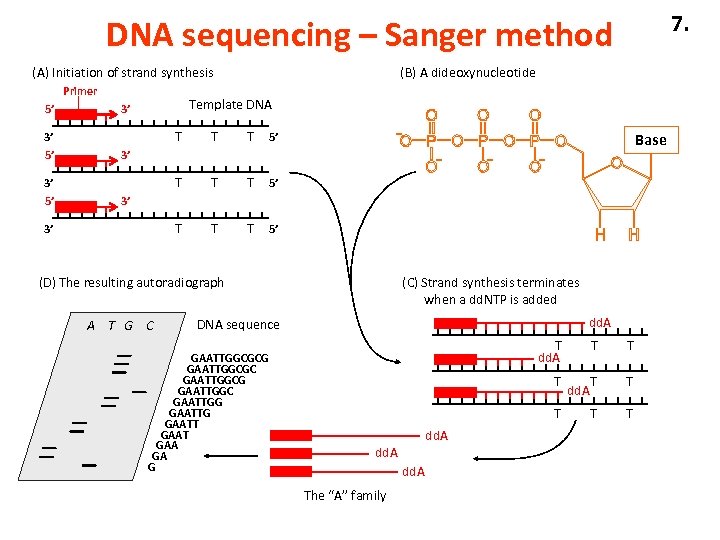 7. DNA sequencing – Sanger method (A) Initiation of strand synthesis Primer 5’ Template DNA 3’ T 3’ 5’ T T T 5’ T 3’ 5’ (B) A dideoxynucleotide T T 5’ Base 3’ 3’ 3’ (D) The resulting autoradiograph A T G C (C) Strand synthesis terminates when a dd. NTP is added dd. A DNA sequence GAATTGGCGCG GAATTGGCGC GAATTGGCG GAATTGGC GAATTGG GAATT GAAT GAA GA G T dd. A T T dd. A The “A” family T dd. A T T T
7. DNA sequencing – Sanger method (A) Initiation of strand synthesis Primer 5’ Template DNA 3’ T 3’ 5’ T T T 5’ T 3’ 5’ (B) A dideoxynucleotide T T 5’ Base 3’ 3’ 3’ (D) The resulting autoradiograph A T G C (C) Strand synthesis terminates when a dd. NTP is added dd. A DNA sequence GAATTGGCGCG GAATTGGCGC GAATTGGCG GAATTGGC GAATTGG GAATT GAAT GAA GA G T dd. A T T dd. A The “A” family T dd. A T T T
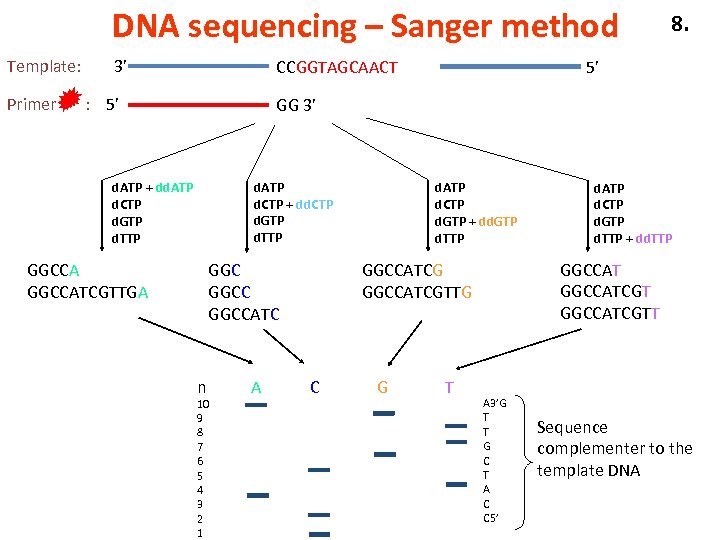 DNA sequencing – Sanger method Template: Primer 3’ CCGGTAGCAACT : 5’ 8. 5’ GG 3’ d. ATP d. CTP + dd. CTP d. GTP d. TTP d. ATP + dd. ATP d. CTP d. GTP d. TTP GGCCATCGTTGA GGCCATC n 10 9 8 7 6 5 4 3 2 1 A d. ATP d. CTP d. GTP + dd. GTP d. TTP GGCCATCGTT GGCCATCGTTG C G T d. ATP d. CTP d. GTP d. TTP + dd. TTP A 3’G T T G C T A C C 5’ Sequence complementer to the template DNA
DNA sequencing – Sanger method Template: Primer 3’ CCGGTAGCAACT : 5’ 8. 5’ GG 3’ d. ATP d. CTP + dd. CTP d. GTP d. TTP d. ATP + dd. ATP d. CTP d. GTP d. TTP GGCCATCGTTGA GGCCATC n 10 9 8 7 6 5 4 3 2 1 A d. ATP d. CTP d. GTP + dd. GTP d. TTP GGCCATCGTT GGCCATCGTTG C G T d. ATP d. CTP d. GTP d. TTP + dd. TTP A 3’G T T G C T A C C 5’ Sequence complementer to the template DNA
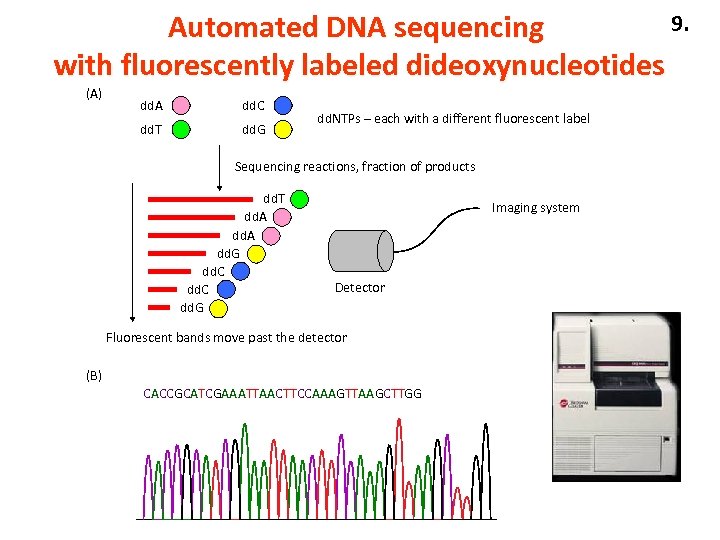 9. Automated DNA sequencing with fluorescently labeled dideoxynucleotides (A) dd. A dd. C dd. T dd. G dd. NTPs – each with a different fluorescent label Sequencing reactions, fraction of products dd. T dd. A dd. G dd. C dd. G Imaging system Detector Fluorescent bands move past the detector (B) CACCGCATCGAAATTAACTTCCAAAGTTAAGCTTGG
9. Automated DNA sequencing with fluorescently labeled dideoxynucleotides (A) dd. A dd. C dd. T dd. G dd. NTPs – each with a different fluorescent label Sequencing reactions, fraction of products dd. T dd. A dd. G dd. C dd. G Imaging system Detector Fluorescent bands move past the detector (B) CACCGCATCGAAATTAACTTCCAAAGTTAAGCTTGG
 The Human Genome Project Craig Venter Francis Collins 10.
The Human Genome Project Craig Venter Francis Collins 10.
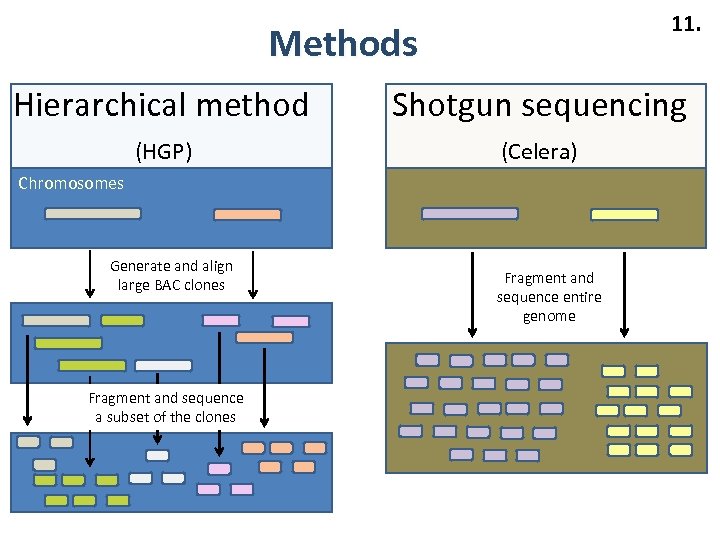 11. Methods Hierarchical method Shotgun sequencing (HGP) (Celera) Chromosomes Generate and align large BAC clones Fragment and sequence a subset of the clones Fragment and sequence entire genome
11. Methods Hierarchical method Shotgun sequencing (HGP) (Celera) Chromosomes Generate and align large BAC clones Fragment and sequence a subset of the clones Fragment and sequence entire genome
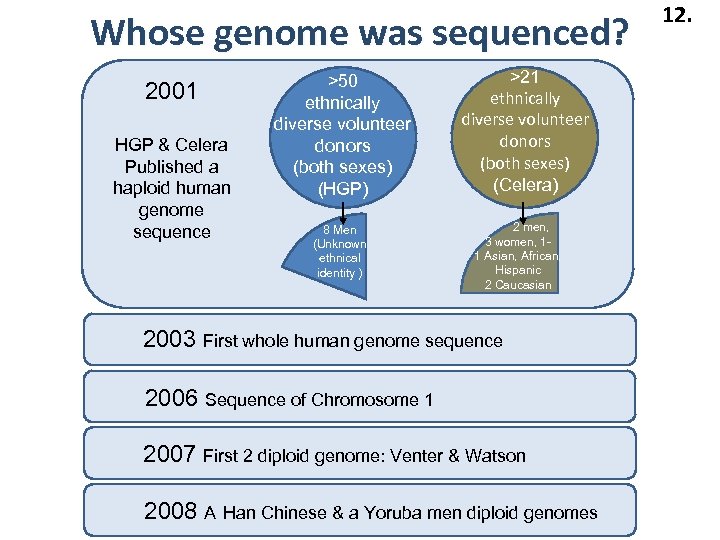 Whose genome was sequenced? 2001 HGP & Celera Published a haploid human genome sequence >50 ethnically diverse volunteer donors (both sexes) (HGP) 8 Men (Unknown ethnical identity ) >21 ethnically diverse volunteer donors (both sexes) (Celera) 2 men, 3 women, 11 Asian, African, Hispanic 2 Caucasian 2003 First whole human genome sequence 2006 Sequence of Chromosome 1 2007 First 2 diploid genome: Venter & Watson 2008 A Han Chinese & a Yoruba men diploid genomes 12.
Whose genome was sequenced? 2001 HGP & Celera Published a haploid human genome sequence >50 ethnically diverse volunteer donors (both sexes) (HGP) 8 Men (Unknown ethnical identity ) >21 ethnically diverse volunteer donors (both sexes) (Celera) 2 men, 3 women, 11 Asian, African, Hispanic 2 Caucasian 2003 First whole human genome sequence 2006 Sequence of Chromosome 1 2007 First 2 diploid genome: Venter & Watson 2008 A Han Chinese & a Yoruba men diploid genomes 12.
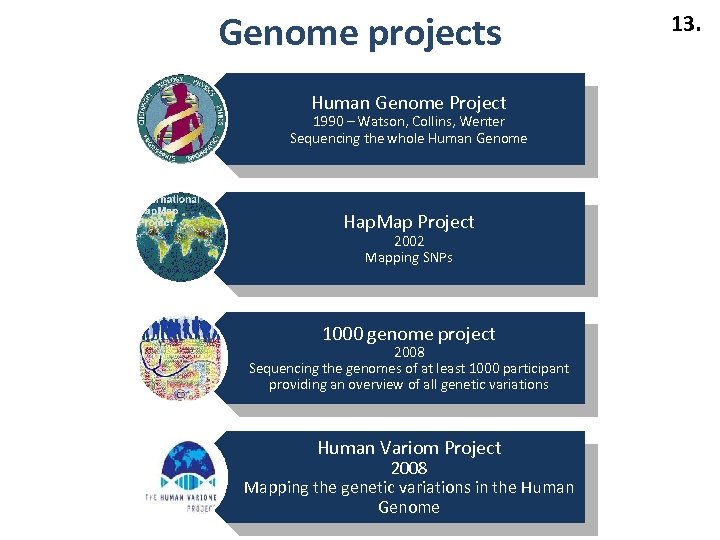 Genome projects Human Genome Project 1990 – Watson, Collins, Wenter Sequencing the whole Human Genome Hap. Map Project 2002 Mapping SNPs 1000 genome project 2008 Sequencing the genomes of at least 1000 participant providing an overview of all genetic variations Human Variom Project 2008 Mapping the genetic variations in the Human Genome 13.
Genome projects Human Genome Project 1990 – Watson, Collins, Wenter Sequencing the whole Human Genome Hap. Map Project 2002 Mapping SNPs 1000 genome project 2008 Sequencing the genomes of at least 1000 participant providing an overview of all genetic variations Human Variom Project 2008 Mapping the genetic variations in the Human Genome 13.
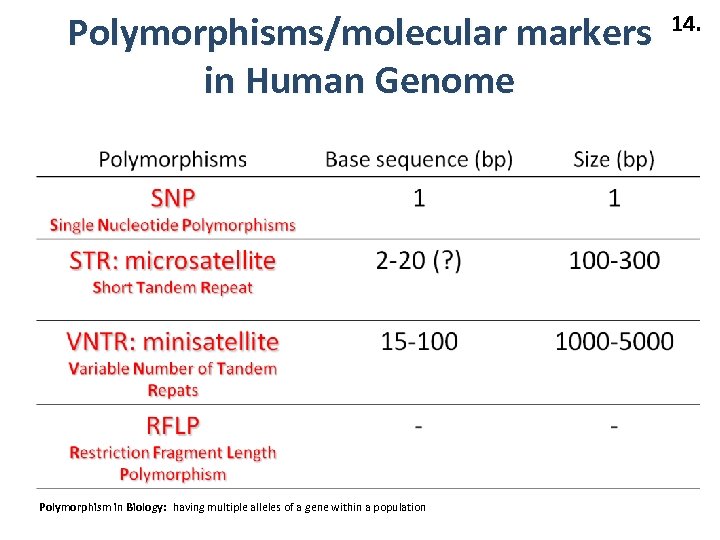 Polymorphisms/molecular markers in Human Genome Polymorphism in Biology: having multiple alleles of a gene within a population 14.
Polymorphisms/molecular markers in Human Genome Polymorphism in Biology: having multiple alleles of a gene within a population 14.
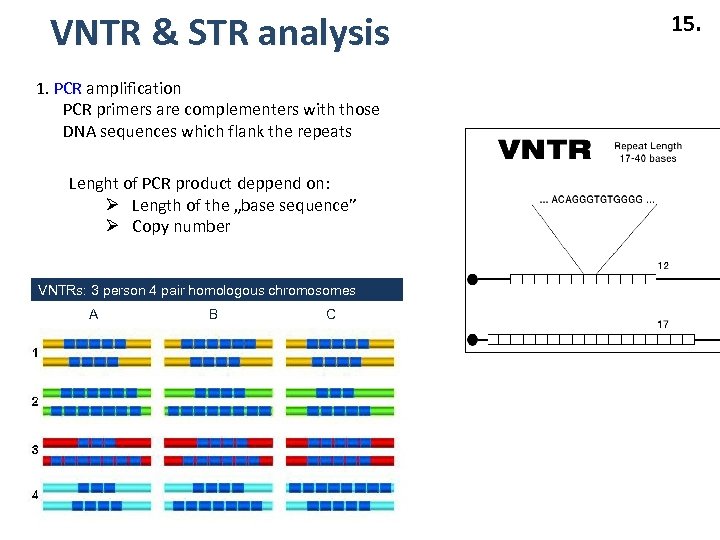 VNTR & STR analysis 1. PCR amplification PCR primers are complementers with those DNA sequences which flank the repeats Lenght of PCR product deppend on: Ø Length of the „base sequence” Ø Copy number VNTRs: 3 person 4 pair homologous chromosomes A B C 15.
VNTR & STR analysis 1. PCR amplification PCR primers are complementers with those DNA sequences which flank the repeats Lenght of PCR product deppend on: Ø Length of the „base sequence” Ø Copy number VNTRs: 3 person 4 pair homologous chromosomes A B C 15.
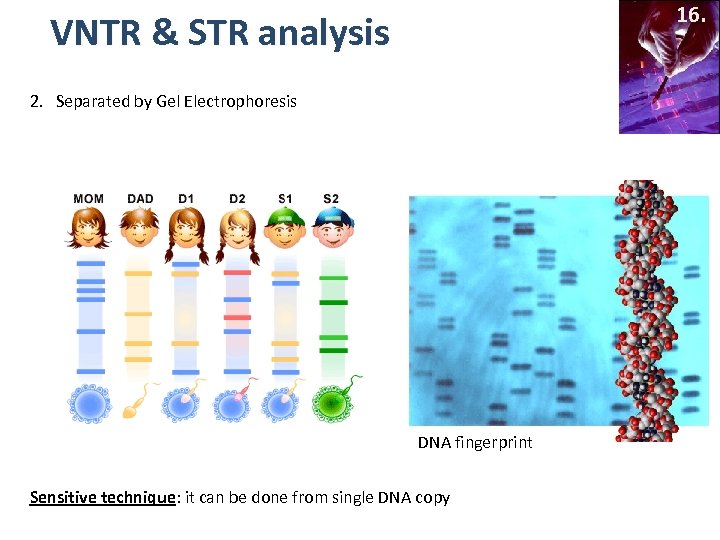 16. VNTR & STR analysis 2. Separated by Gel Electrophoresis DNA fingerprint Sensitive technique: it can be done from single DNA copy
16. VNTR & STR analysis 2. Separated by Gel Electrophoresis DNA fingerprint Sensitive technique: it can be done from single DNA copy
 SNPs A variation in the base sequence occuring at any given single position in the genome (for example C instead of T). ACGGCTAA It is found in more than 1% of the population. 17.
SNPs A variation in the base sequence occuring at any given single position in the genome (for example C instead of T). ACGGCTAA It is found in more than 1% of the population. 17.
 SNPs A variation in the base sequence occuring at any given single position in the genome (for example C instead of T). ATGGCTAA It is found in more than 1% of the population. 18.
SNPs A variation in the base sequence occuring at any given single position in the genome (for example C instead of T). ATGGCTAA It is found in more than 1% of the population. 18.
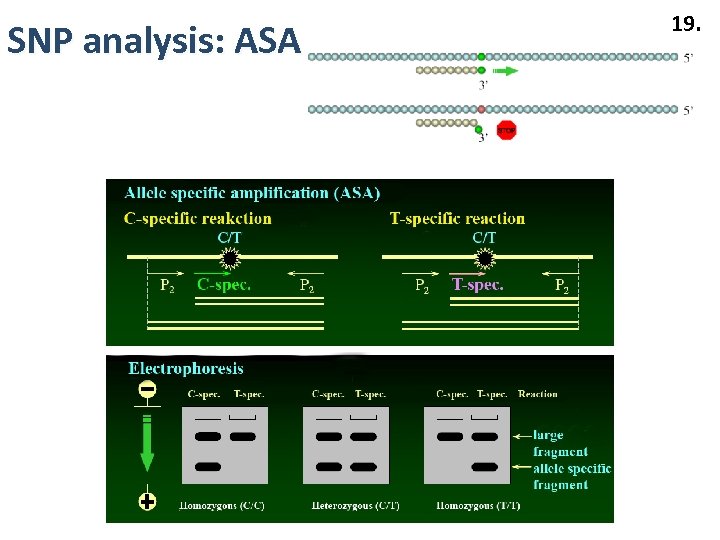 SNP analysis: ASA 19.
SNP analysis: ASA 19.
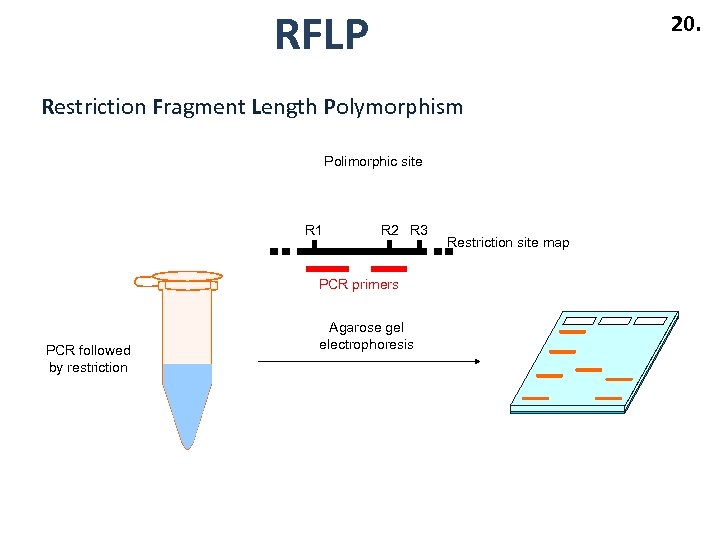 RFLP 20. Restriction Fragment Length Polymorphism Polimorphic site R 1 R 2 R 3 PCR primers PCR followed by restriction Agarose gel electrophoresis Restriction site map
RFLP 20. Restriction Fragment Length Polymorphism Polimorphic site R 1 R 2 R 3 PCR primers PCR followed by restriction Agarose gel electrophoresis Restriction site map
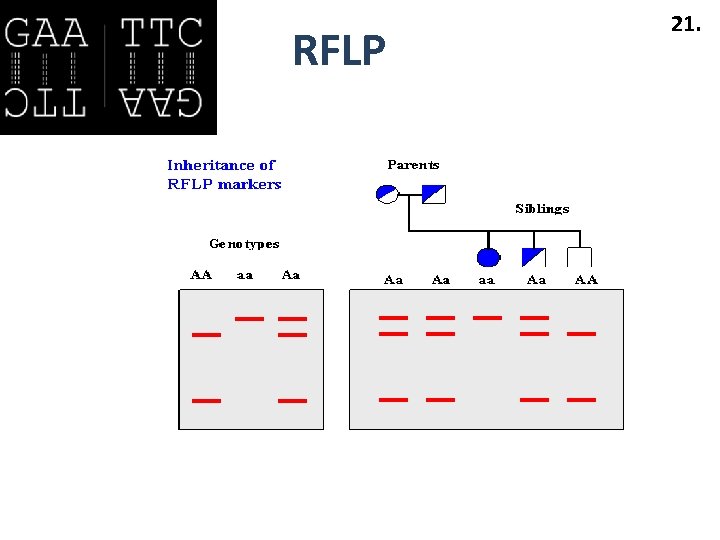 RFLP 21.
RFLP 21.
 Functional Genomics Microchip Microarray scanner Real-Time PCR cycler 22.
Functional Genomics Microchip Microarray scanner Real-Time PCR cycler 22.
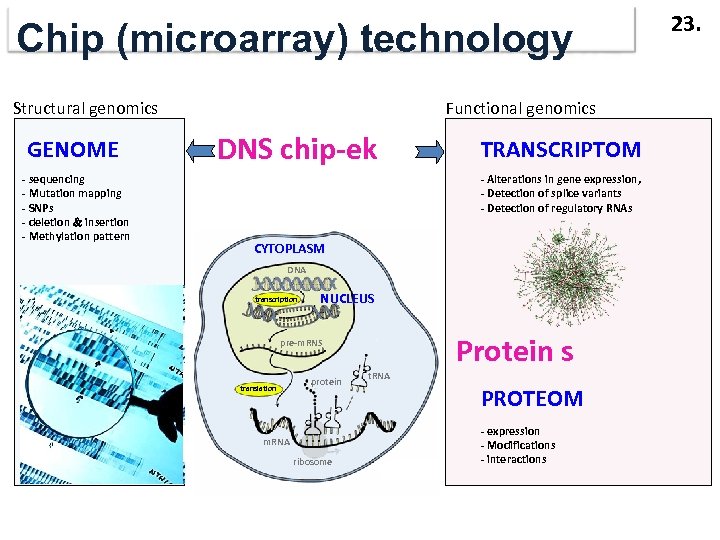 Chip (microarray) technology Structural genomics GENOME - sequencing - Mutation mapping - SNPs - deletion insertion - Methylation pattern Functional genomics DNS chip-ek TRANSCRIPTOM - Alterations in gene expression, - Detection of splice variants - Detection of regulatory RNAs CYTOPLASM DNA transcription NUCLEUS Protein s pre-m. RNS translation protein m. RNA ribosome t. RNA PROTEOM - expression - Modifications - interactions 23.
Chip (microarray) technology Structural genomics GENOME - sequencing - Mutation mapping - SNPs - deletion insertion - Methylation pattern Functional genomics DNS chip-ek TRANSCRIPTOM - Alterations in gene expression, - Detection of splice variants - Detection of regulatory RNAs CYTOPLASM DNA transcription NUCLEUS Protein s pre-m. RNS translation protein m. RNA ribosome t. RNA PROTEOM - expression - Modifications - interactions 23.
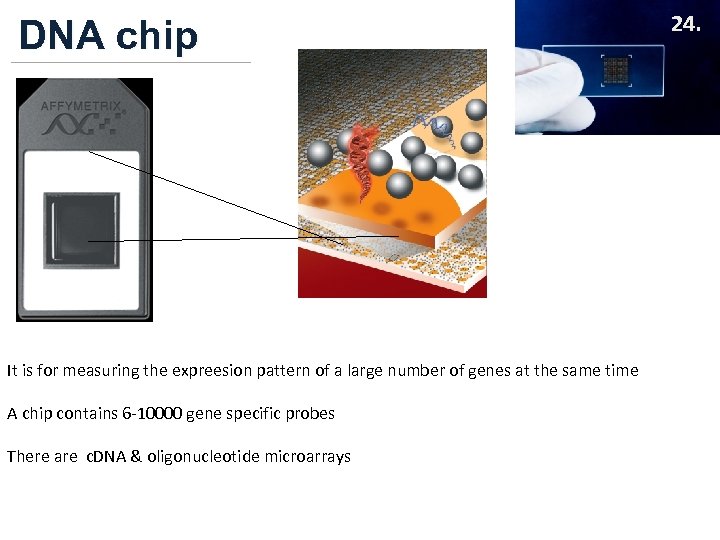 DNA chip It is for measuring the expreesion pattern of a large number of genes at the same time A chip contains 6 -10000 gene specific probes There are c. DNA & oligonucleotide microarrays 24.
DNA chip It is for measuring the expreesion pattern of a large number of genes at the same time A chip contains 6 -10000 gene specific probes There are c. DNA & oligonucleotide microarrays 24.
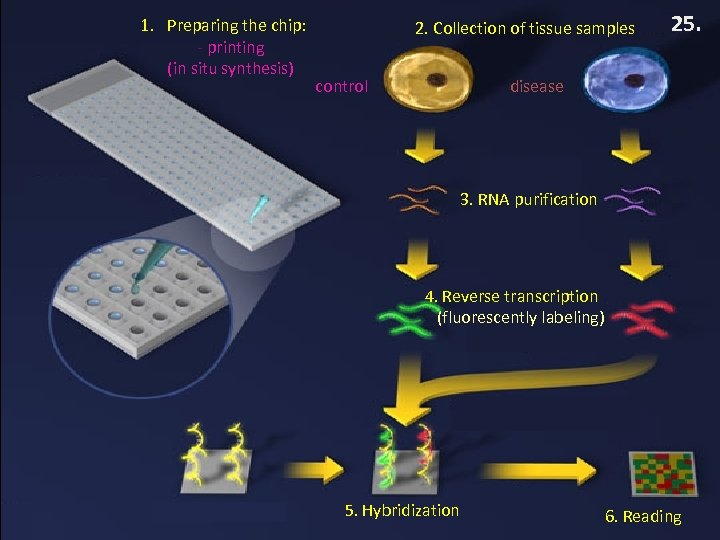 1. Preparing the chip: - printing (in situ synthesis) 2. Collection of tissue samples control 25. disease 3. RNA purification 4. Reverse transcription (fluorescently labeling) 5. Hybridization 6. Reading
1. Preparing the chip: - printing (in situ synthesis) 2. Collection of tissue samples control 25. disease 3. RNA purification 4. Reverse transcription (fluorescently labeling) 5. Hybridization 6. Reading
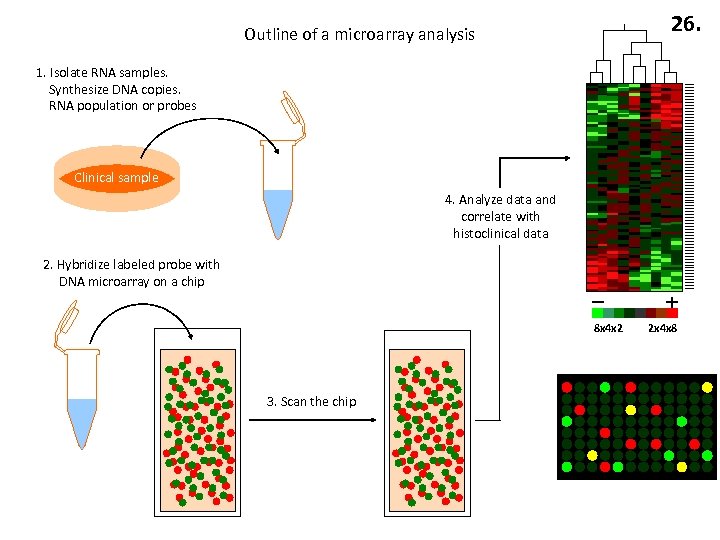 26. Outline of a microarray analysis 1. Isolate RNA samples. Synthesize DNA copies. RNA population or probes Clinical sample 4. Analyze data and correlate with histoclinical data 2. Hybridize labeled probe with DNA microarray on a chip 8 x 4 x 2 3. Scan the chip 2 x 4 x 8
26. Outline of a microarray analysis 1. Isolate RNA samples. Synthesize DNA copies. RNA population or probes Clinical sample 4. Analyze data and correlate with histoclinical data 2. Hybridize labeled probe with DNA microarray on a chip 8 x 4 x 2 3. Scan the chip 2 x 4 x 8
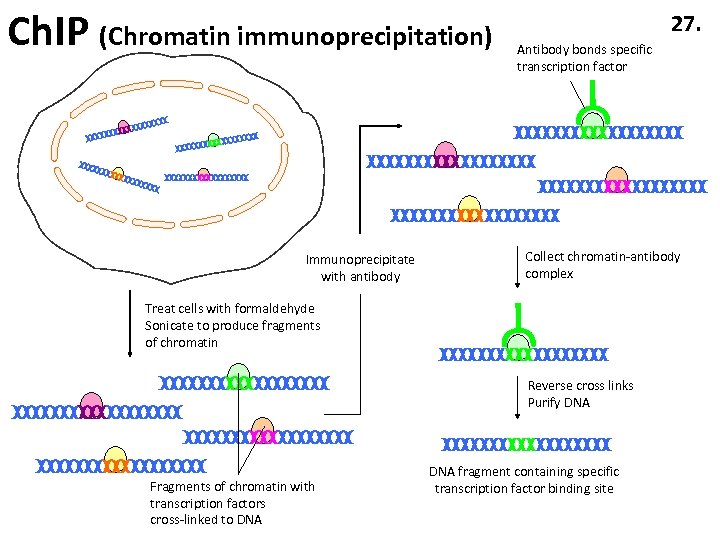 Ch. IP (Chromatin immunoprecipitation) Immunoprecipitate with antibody 27. Antibody bonds specific transcription factor Collect chromatin-antibody complex Treat cells with formaldehyde Sonicate to produce fragments of chromatin Reverse cross links Purify DNA Fragments of chromatin with transcription factors cross-linked to DNA fragment containing specific transcription factor binding site
Ch. IP (Chromatin immunoprecipitation) Immunoprecipitate with antibody 27. Antibody bonds specific transcription factor Collect chromatin-antibody complex Treat cells with formaldehyde Sonicate to produce fragments of chromatin Reverse cross links Purify DNA Fragments of chromatin with transcription factors cross-linked to DNA fragment containing specific transcription factor binding site
 Real-Time-PCR q Used to amplify and simultaneously quantify a targeted DNA molecule q. Detection of fluoresce at each cycle during PCR reaction → Real-Time q. No gel-based analysis at the end of the PCR reaction q. Computer based analysis of the cycle fluorescence time course Real-Time PCR cycler 28.
Real-Time-PCR q Used to amplify and simultaneously quantify a targeted DNA molecule q. Detection of fluoresce at each cycle during PCR reaction → Real-Time q. No gel-based analysis at the end of the PCR reaction q. Computer based analysis of the cycle fluorescence time course Real-Time PCR cycler 28.
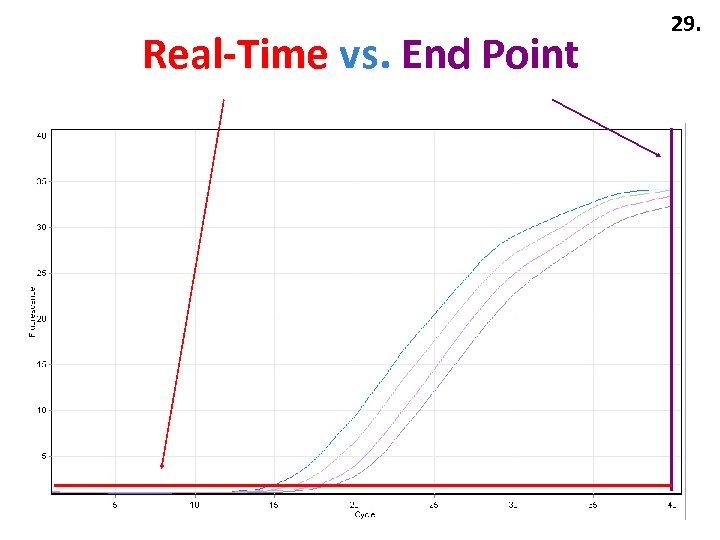 Real-Time vs. End Point 29.
Real-Time vs. End Point 29.
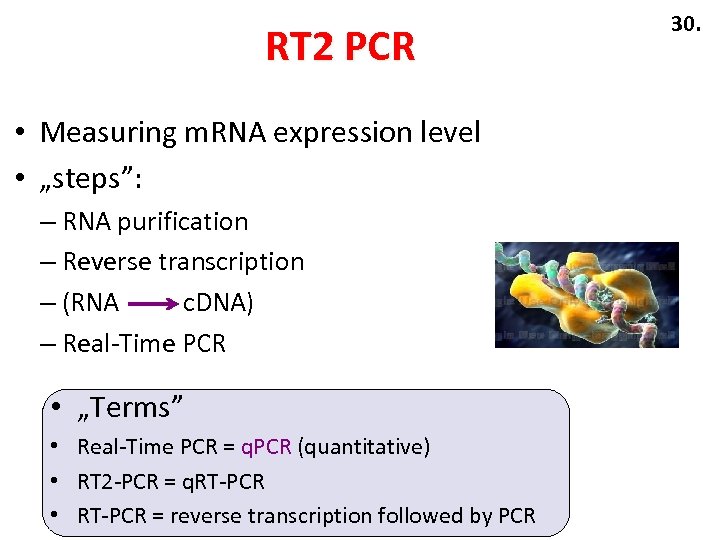 RT 2 PCR • Measuring m. RNA expression level • „steps”: – RNA purification – Reverse transcription – (RNA c. DNA) – Real-Time PCR • „Terms” • Real-Time PCR = q. PCR (quantitative) • RT 2 -PCR = q. RT-PCR • RT-PCR = reverse transcription followed by PCR 30.
RT 2 PCR • Measuring m. RNA expression level • „steps”: – RNA purification – Reverse transcription – (RNA c. DNA) – Real-Time PCR • „Terms” • Real-Time PCR = q. PCR (quantitative) • RT 2 -PCR = q. RT-PCR • RT-PCR = reverse transcription followed by PCR 30.
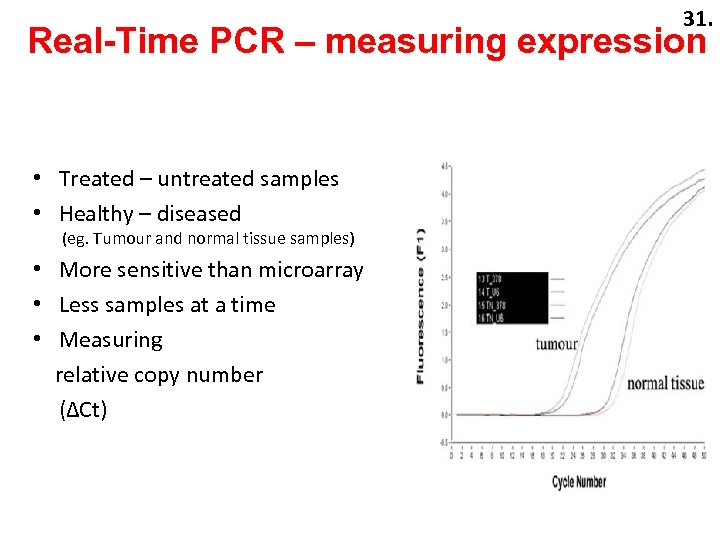 31. 33. Real-Time PCR – measuring expression • Treated – untreated samples • Healthy – diseased (eg. Tumour and normal tissue samples) • More sensitive than microarray • Less samples at a time • Measuring relative copy number (∆Ct)
31. 33. Real-Time PCR – measuring expression • Treated – untreated samples • Healthy – diseased (eg. Tumour and normal tissue samples) • More sensitive than microarray • Less samples at a time • Measuring relative copy number (∆Ct)
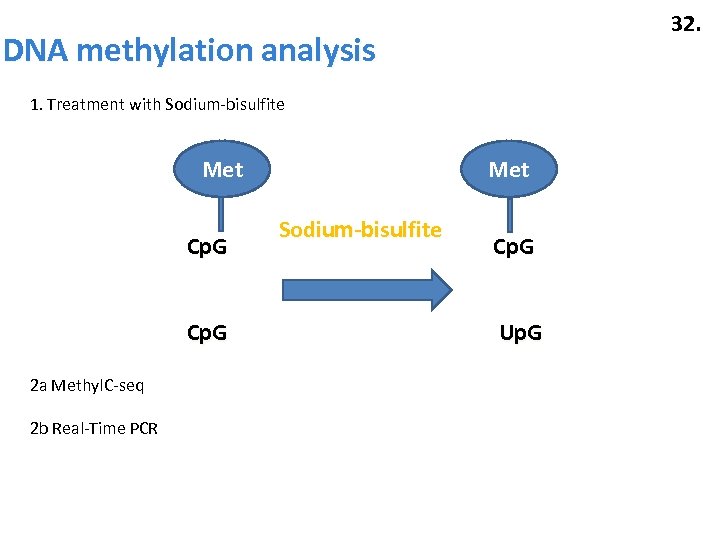 32. DNA methylation analysis 1. Treatment with Sodium-bisulfite Met Cp. G 2 a Methyl. C-seq 2 b Real-Time PCR Met Sodium-bisulfite Cp. G Up. G
32. DNA methylation analysis 1. Treatment with Sodium-bisulfite Met Cp. G 2 a Methyl. C-seq 2 b Real-Time PCR Met Sodium-bisulfite Cp. G Up. G
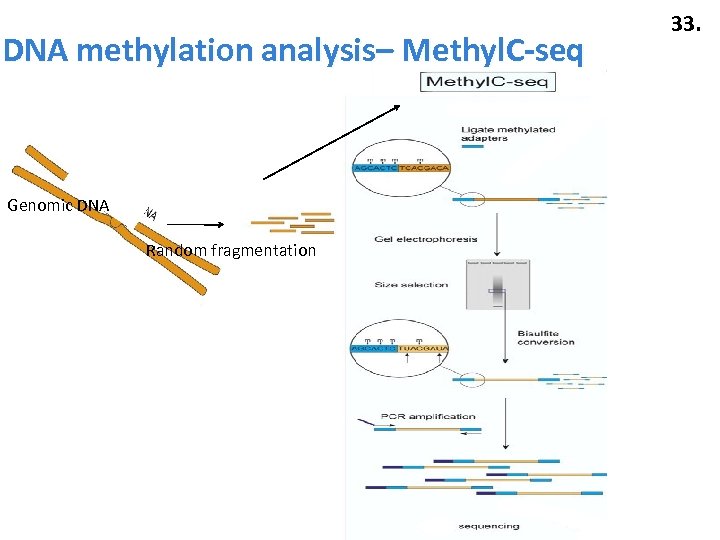 DNA methylation analysis– Methyl. C-seq Genomic DNA Random fragmentation 33.
DNA methylation analysis– Methyl. C-seq Genomic DNA Random fragmentation 33.
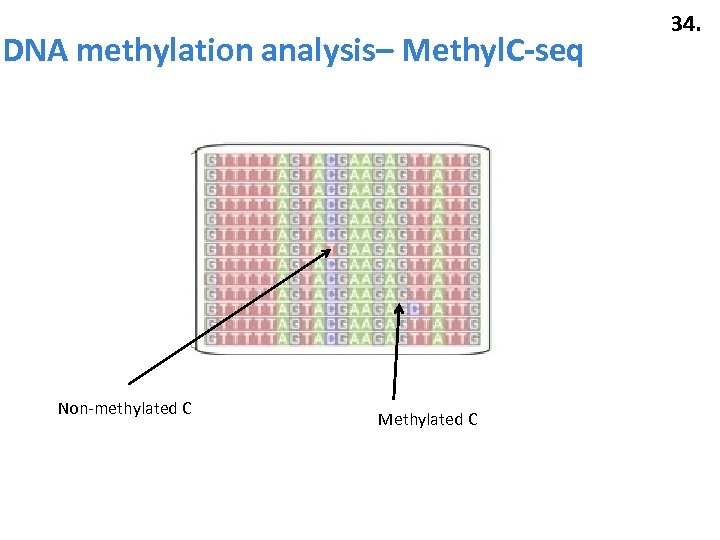 DNA methylation analysis– Methyl. C-seq Non-methylated C Methylated C 34.
DNA methylation analysis– Methyl. C-seq Non-methylated C Methylated C 34.
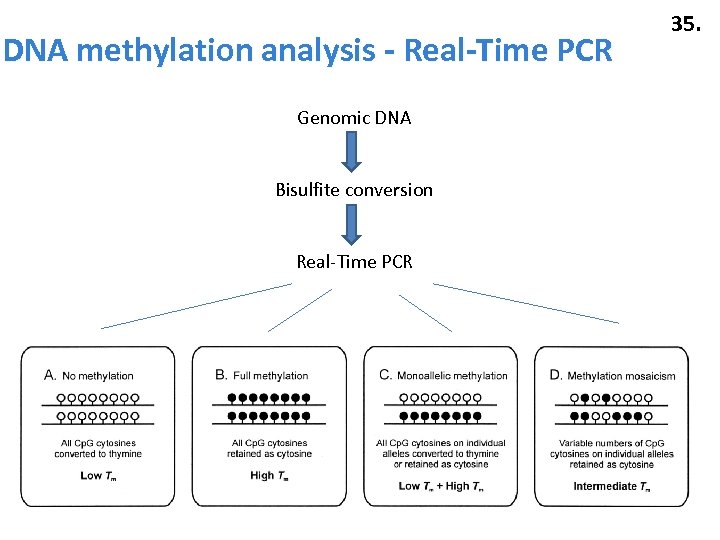 DNA methylation analysis - Real-Time PCR Genomic DNA Bisulfite conversion Real-Time PCR 35.
DNA methylation analysis - Real-Time PCR Genomic DNA Bisulfite conversion Real-Time PCR 35.


