d8e920a3ab4ff32dfac66d6b13378c20.ppt
- Количество слайдов: 32
 IEEE International Conference on Computer Design Tsung-Wei Huang and Tsung-Yi Ho http: //eda. csie. ncku. edu. tw Department of Computer Science and Information Engineering National Cheng Kung University Tainan, Taiwan NCKU CSIE EDALAB
IEEE International Conference on Computer Design Tsung-Wei Huang and Tsung-Yi Ho http: //eda. csie. ncku. edu. tw Department of Computer Science and Information Engineering National Cheng Kung University Tainan, Taiwan NCKU CSIE EDALAB
 Outline Introduction Problem Formulation Algorithms Experimental Results Conclusion NCKU CSIE EDALAB
Outline Introduction Problem Formulation Algorithms Experimental Results Conclusion NCKU CSIE EDALAB
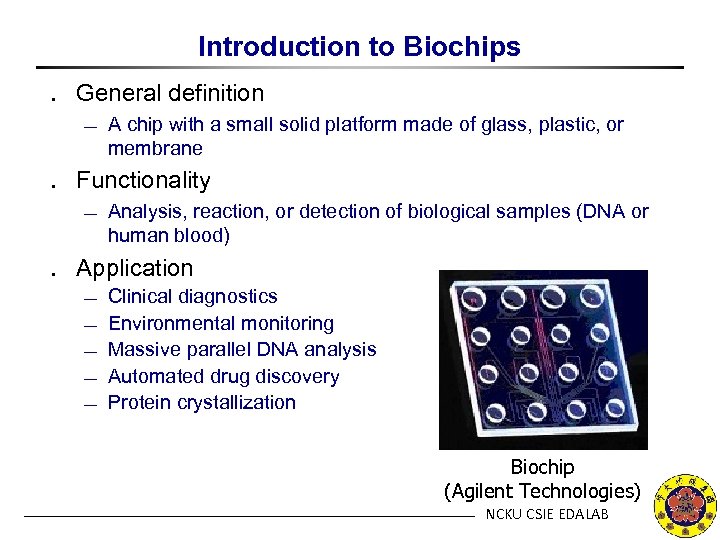 Introduction to Biochips ․ General definition ¾ A chip with a small solid platform made of glass, plastic, or membrane ․ Functionality ¾ Analysis, reaction, or detection of biological samples (DNA or human blood) ․ Application ¾ ¾ ¾ Clinical diagnostics Environmental monitoring Massive parallel DNA analysis Automated drug discovery Protein crystallization Biochip (Agilent Technologies) NCKU CSIE EDALAB
Introduction to Biochips ․ General definition ¾ A chip with a small solid platform made of glass, plastic, or membrane ․ Functionality ¾ Analysis, reaction, or detection of biological samples (DNA or human blood) ․ Application ¾ ¾ ¾ Clinical diagnostics Environmental monitoring Massive parallel DNA analysis Automated drug discovery Protein crystallization Biochip (Agilent Technologies) NCKU CSIE EDALAB
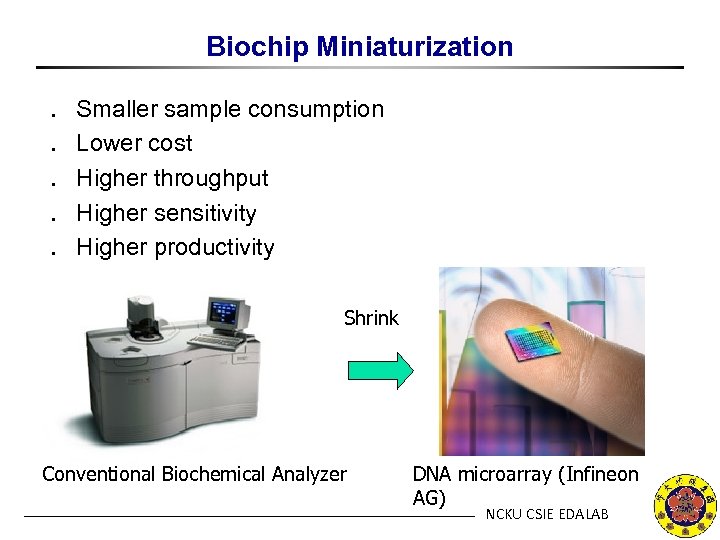 Biochip Miniaturization ․ ․ ․ Smaller sample consumption Lower cost Higher throughput Higher sensitivity Higher productivity Shrink Conventional Biochemical Analyzer DNA microarray (Infineon AG) NCKU CSIE EDALAB
Biochip Miniaturization ․ ․ ․ Smaller sample consumption Lower cost Higher throughput Higher sensitivity Higher productivity Shrink Conventional Biochemical Analyzer DNA microarray (Infineon AG) NCKU CSIE EDALAB
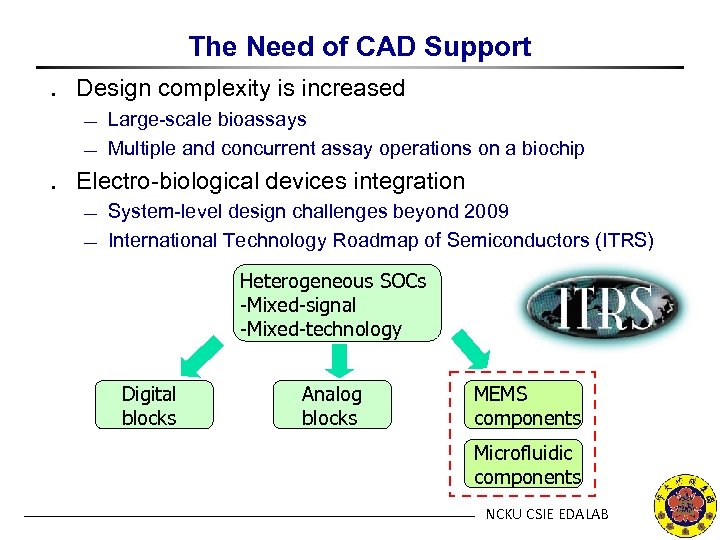 The Need of CAD Support ․ Design complexity is increased ¾ ¾ Large-scale bioassays Multiple and concurrent assay operations on a biochip ․ Electro-biological devices integration ¾ ¾ System-level design challenges beyond 2009 International Technology Roadmap of Semiconductors (ITRS) Heterogeneous SOCs -Mixed-signal -Mixed-technology Digital blocks Analog blocks MEMS components Microfluidic components NCKU CSIE EDALAB
The Need of CAD Support ․ Design complexity is increased ¾ ¾ Large-scale bioassays Multiple and concurrent assay operations on a biochip ․ Electro-biological devices integration ¾ ¾ System-level design challenges beyond 2009 International Technology Roadmap of Semiconductors (ITRS) Heterogeneous SOCs -Mixed-signal -Mixed-technology Digital blocks Analog blocks MEMS components Microfluidic components NCKU CSIE EDALAB
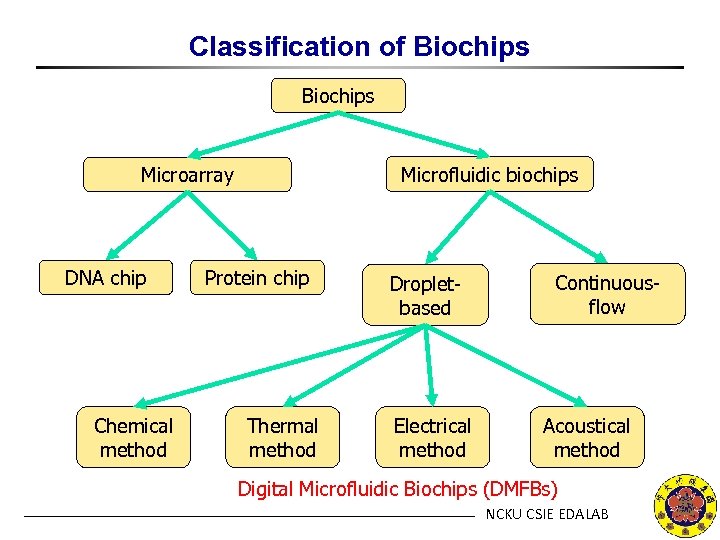 Classification of Biochips Microfluidic biochips Microarray DNA chip Chemical method Protein chip Thermal method Dropletbased Electrical method Continuousflow Acoustical method Digital Microfluidic Biochips (DMFBs) NCKU CSIE EDALAB
Classification of Biochips Microfluidic biochips Microarray DNA chip Chemical method Protein chip Thermal method Dropletbased Electrical method Continuousflow Acoustical method Digital Microfluidic Biochips (DMFBs) NCKU CSIE EDALAB
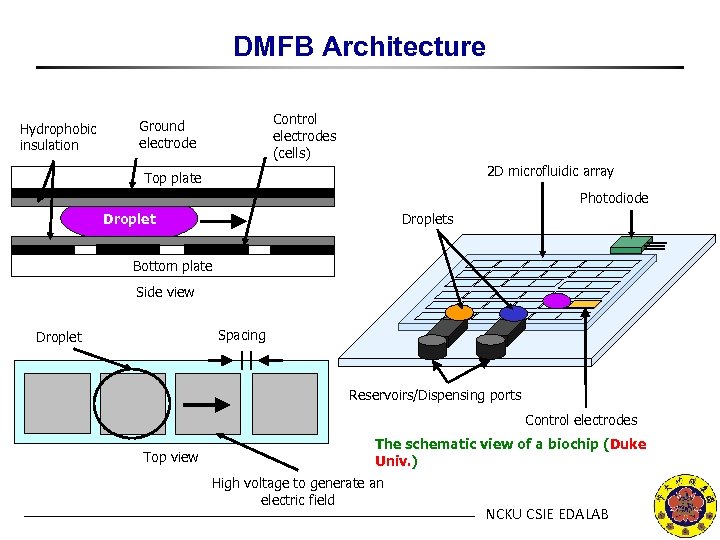 DMFB Architecture Hydrophobic insulation Control electrodes (cells) Ground electrode 2 D microfluidic array Top plate Photodiode Droplets Bottom plate Side view Spacing Droplet Reservoirs/Dispensing ports Control electrodes Top view The schematic view of a biochip (Duke Univ. ) High voltage to generate an electric field NCKU CSIE EDALAB
DMFB Architecture Hydrophobic insulation Control electrodes (cells) Ground electrode 2 D microfluidic array Top plate Photodiode Droplets Bottom plate Side view Spacing Droplet Reservoirs/Dispensing ports Control electrodes Top view The schematic view of a biochip (Duke Univ. ) High voltage to generate an electric field NCKU CSIE EDALAB
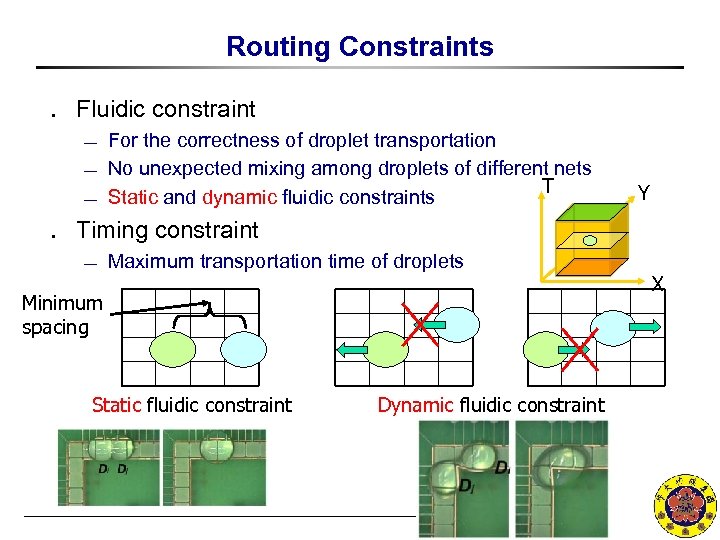 Routing Constraints ․ Fluidic constraint ¾ ¾ ¾ For the correctness of droplet transportation No unexpected mixing among droplets of different nets T Static and dynamic fluidic constraints Y ․ Timing constraint ¾ Maximum transportation time of droplets X Minimum spacing Static fluidic constraint Dynamic fluidic constraint NCKU CSIE EDALAB
Routing Constraints ․ Fluidic constraint ¾ ¾ ¾ For the correctness of droplet transportation No unexpected mixing among droplets of different nets T Static and dynamic fluidic constraints Y ․ Timing constraint ¾ Maximum transportation time of droplets X Minimum spacing Static fluidic constraint Dynamic fluidic constraint NCKU CSIE EDALAB
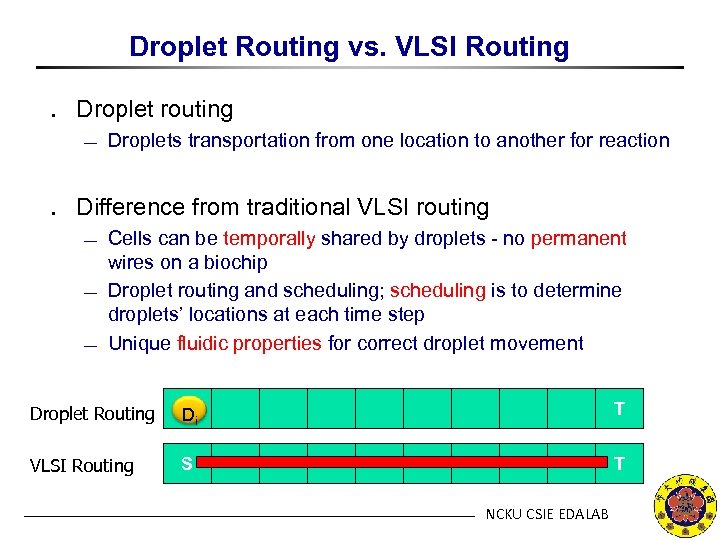 Droplet Routing vs. VLSI Routing ․ Droplet routing ¾ Droplets transportation from one location to another for reaction ․ Difference from traditional VLSI routing ¾ ¾ ¾ Cells can be temporally shared by droplets - no permanent wires on a biochip Droplet routing and scheduling; scheduling is to determine droplets’ locations at each time step Unique fluidic properties for correct droplet movement Droplet Routing S Di T VLSI Routing S T NCKU CSIE EDALAB
Droplet Routing vs. VLSI Routing ․ Droplet routing ¾ Droplets transportation from one location to another for reaction ․ Difference from traditional VLSI routing ¾ ¾ ¾ Cells can be temporally shared by droplets - no permanent wires on a biochip Droplet routing and scheduling; scheduling is to determine droplets’ locations at each time step Unique fluidic properties for correct droplet movement Droplet Routing S Di T VLSI Routing S T NCKU CSIE EDALAB
 Outline Introduction Problem Formulation Algorithms Experimental Results Conclusion NCKU CSIE EDALAB
Outline Introduction Problem Formulation Algorithms Experimental Results Conclusion NCKU CSIE EDALAB
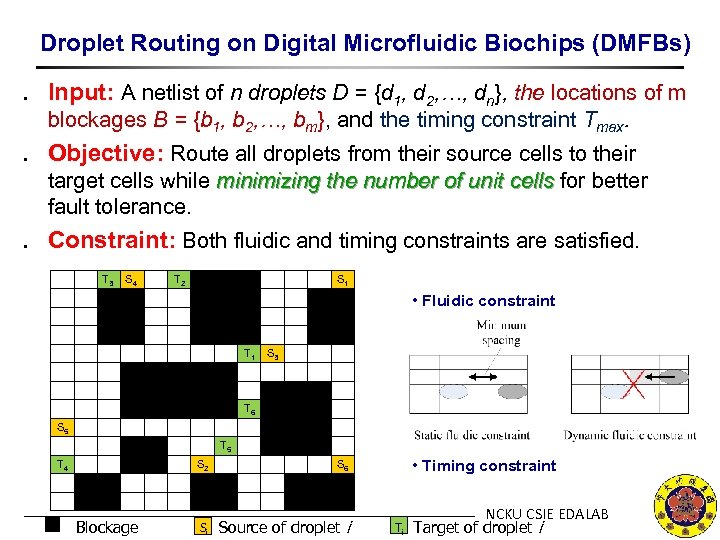 Droplet Routing on Digital Microfluidic Biochips (DMFBs) ․ Input: A netlist of n droplets D = {d 1, d 2, …, dn}, the locations of m blockages B = {b 1, b 2, …, bm}, and the timing constraint Tmax. ․ Objective: Route all droplets from their source cells to their target cells while minimizing the number of unit cells for better fault tolerance. ․ Constraint: Both fluidic and timing constraints are satisfied. T 3 S 4 T 2 S 1 • Fluidic constraint T 1 S 3 T 6 S 5 T 4 S 2 Blockage Si Source of droplet i • Timing constraint S 6 Ti NCKU CSIE EDALAB Target of droplet i
Droplet Routing on Digital Microfluidic Biochips (DMFBs) ․ Input: A netlist of n droplets D = {d 1, d 2, …, dn}, the locations of m blockages B = {b 1, b 2, …, bm}, and the timing constraint Tmax. ․ Objective: Route all droplets from their source cells to their target cells while minimizing the number of unit cells for better fault tolerance. ․ Constraint: Both fluidic and timing constraints are satisfied. T 3 S 4 T 2 S 1 • Fluidic constraint T 1 S 3 T 6 S 5 T 4 S 2 Blockage Si Source of droplet i • Timing constraint S 6 Ti NCKU CSIE EDALAB Target of droplet i
![Related Work l Prioritized A*-search algorithm [K. Böhringer, TCAD’ 06] l l l Open Related Work l Prioritized A*-search algorithm [K. Böhringer, TCAD’ 06] l l l Open](https://present5.com/presentation/d8e920a3ab4ff32dfac66d6b13378c20/image-12.jpg) Related Work l Prioritized A*-search algorithm [K. Böhringer, TCAD’ 06] l l l Open shortest path first algorithm [Griffith et al, TCAD’ 06] l l Alternative routing path generation and droplet scheduling Random selection Network flow-based approach [Yuh et al, ICCAD’ 07] l l l Layout patterns with routing table No dynamic reconfiguration Two-stage algorithm [Su et al, DATE’ 06] l l A*-search for each droplet based on its priority High-priority droplets may block low-priority droplets Maximize the number of nets routed Min-cost Max-flow formulation + prioritized A* search High-performance approach [Cho and Pan, ISPD’ 08] l l Capable of handing routing obstacles Routing order decided by bypassibility of targets NCKU CSIE EDALAB
Related Work l Prioritized A*-search algorithm [K. Böhringer, TCAD’ 06] l l l Open shortest path first algorithm [Griffith et al, TCAD’ 06] l l Alternative routing path generation and droplet scheduling Random selection Network flow-based approach [Yuh et al, ICCAD’ 07] l l l Layout patterns with routing table No dynamic reconfiguration Two-stage algorithm [Su et al, DATE’ 06] l l A*-search for each droplet based on its priority High-priority droplets may block low-priority droplets Maximize the number of nets routed Min-cost Max-flow formulation + prioritized A* search High-performance approach [Cho and Pan, ISPD’ 08] l l Capable of handing routing obstacles Routing order decided by bypassibility of targets NCKU CSIE EDALAB
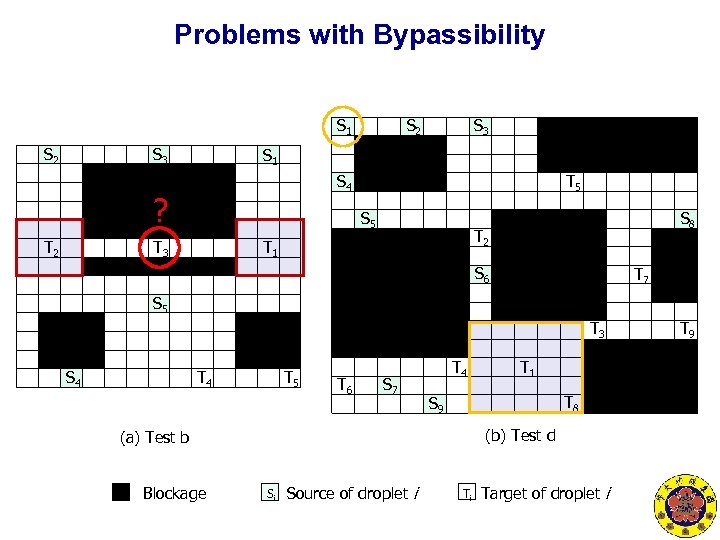 Problems with Bypassibility S 1 S 2 S 3 S 1 S 4 ? T 2 S 2 T 5 S 5 T 2 T 1 T 3 S 8 S 6 T 7 S 5 T 3 S 4 T 5 T 6 S 7 T 4 T 8 S 9 (b) Test d (a) Test b Blockage T 1 Si Source of droplet i Ti Target of droplet i T 9
Problems with Bypassibility S 1 S 2 S 3 S 1 S 4 ? T 2 S 2 T 5 S 5 T 2 T 1 T 3 S 8 S 6 T 7 S 5 T 3 S 4 T 5 T 6 S 7 T 4 T 8 S 9 (b) Test d (a) Test b Blockage T 1 Si Source of droplet i Ti Target of droplet i T 9
 Outline Introduction Problem Formulation Preferred Routing Track Construction Algorithms Routing Ordering by Entropy Equation Experimental Results Routing Compaction by Dynamic Programming Conclusion NCKU CSIE EDALAB
Outline Introduction Problem Formulation Preferred Routing Track Construction Algorithms Routing Ordering by Entropy Equation Experimental Results Routing Compaction by Dynamic Programming Conclusion NCKU CSIE EDALAB
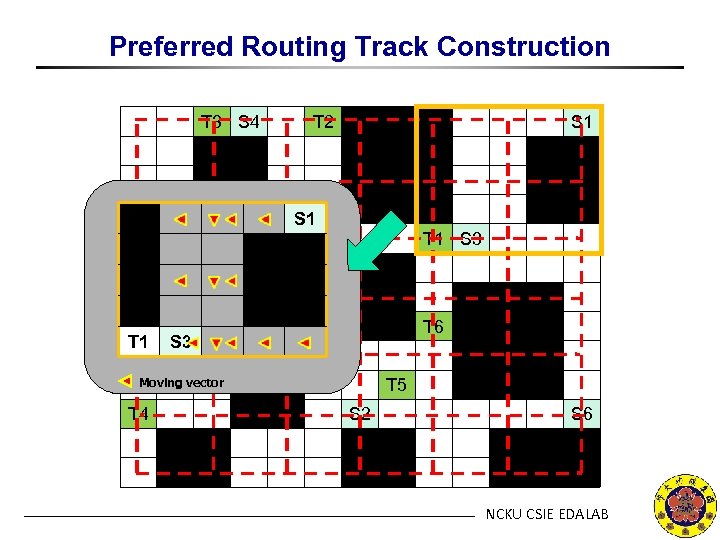 Preferred Routing Track Construction T 3 S 4 T 2 S 1 T 1 S 3 T 1 S 5 T 6 S 3 Moving vector T 4 T 5 S 2 S 6 NCKU CSIE EDALAB
Preferred Routing Track Construction T 3 S 4 T 2 S 1 T 1 S 3 T 1 S 5 T 6 S 3 Moving vector T 4 T 5 S 2 S 6 NCKU CSIE EDALAB
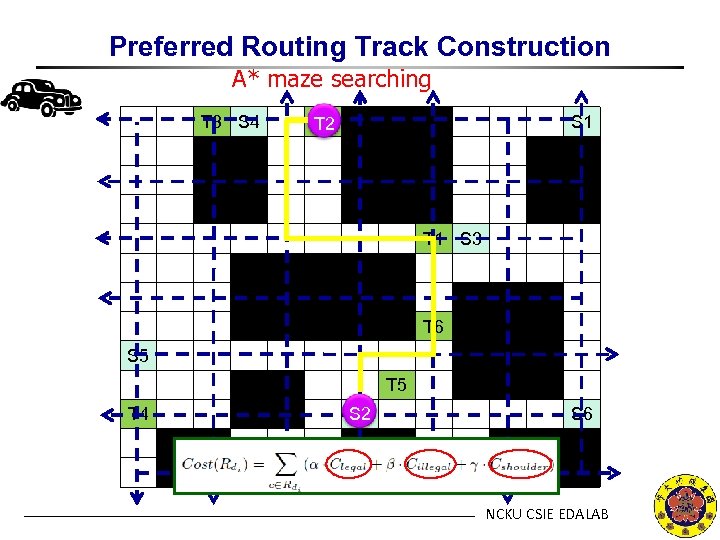 Preferred Routing Track Construction A* maze searching T 3 S 4 T 2 S 1 T 1 S 3 T 6 S 5 T 4 S 2 S 6 NCKU CSIE EDALAB
Preferred Routing Track Construction A* maze searching T 3 S 4 T 2 S 1 T 1 S 3 T 6 S 5 T 4 S 2 S 6 NCKU CSIE EDALAB
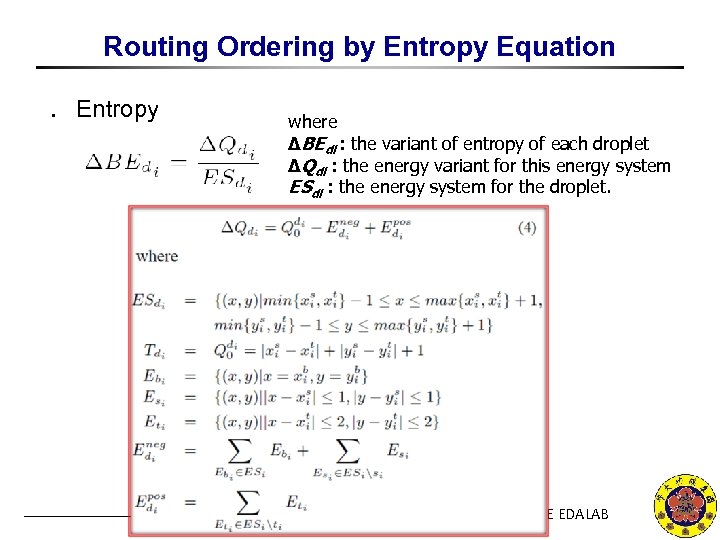 Routing Ordering by Entropy Equation ․ Entropy where ΔBEdi : the variant of entropy of each droplet ΔQdi : the energy variant for this energy system ESdi : the energy system for the droplet. NCKU CSIE EDALAB
Routing Ordering by Entropy Equation ․ Entropy where ΔBEdi : the variant of entropy of each droplet ΔQdi : the energy variant for this energy system ESdi : the energy system for the droplet. NCKU CSIE EDALAB
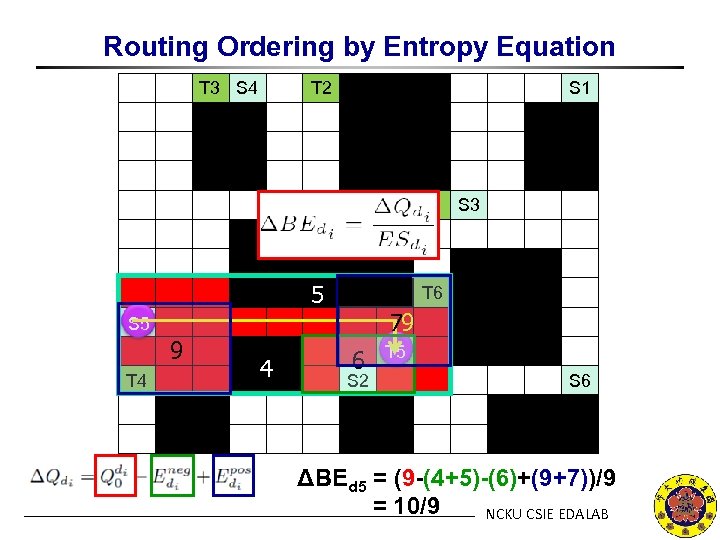 Routing Ordering by Entropy Equation T 3 S 4 T 2 S 1 T 1 S 3 5 T 6 79 S 5 9 T 4 4 6 S 2 T 5 S 6 ΔBEd 5 = (9 -(4+5)-(6)+(9+7))/9 = 10/9 NCKU CSIE EDALAB
Routing Ordering by Entropy Equation T 3 S 4 T 2 S 1 T 1 S 3 5 T 6 79 S 5 9 T 4 4 6 S 2 T 5 S 6 ΔBEd 5 = (9 -(4+5)-(6)+(9+7))/9 = 10/9 NCKU CSIE EDALAB
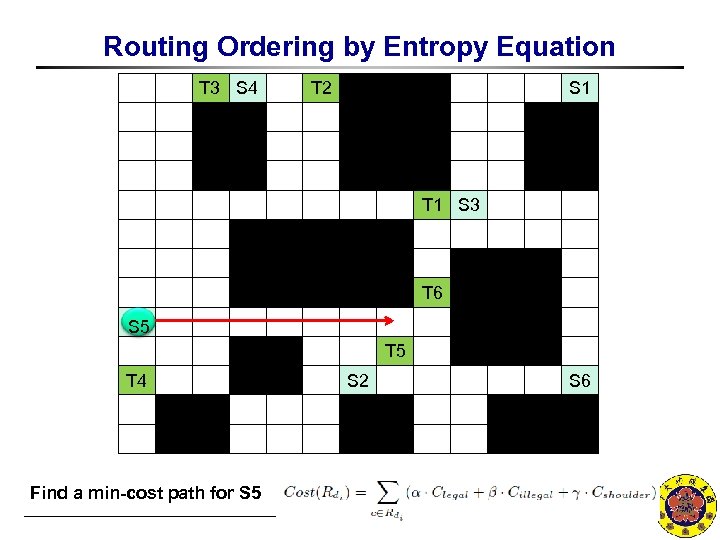 Routing Ordering by Entropy Equation T 3 S 4 T 2 S 1 T 1 S 3 T 6 S 5 T 5 T 4 S 2 S 6 Find a min-cost path for S 5 NCKU CSIE EDALAB
Routing Ordering by Entropy Equation T 3 S 4 T 2 S 1 T 1 S 3 T 6 S 5 T 5 T 4 S 2 S 6 Find a min-cost path for S 5 NCKU CSIE EDALAB
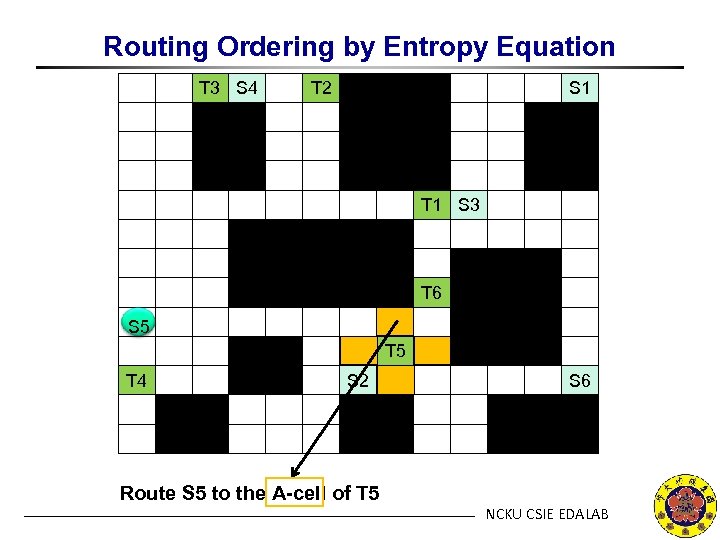 Routing Ordering by Entropy Equation T 3 S 4 T 2 S 1 T 1 S 3 T 6 S 5 T 4 S 2 Route S 5 to the A-cell of T 5 S 6 NCKU CSIE EDALAB
Routing Ordering by Entropy Equation T 3 S 4 T 2 S 1 T 1 S 3 T 6 S 5 T 4 S 2 Route S 5 to the A-cell of T 5 S 6 NCKU CSIE EDALAB
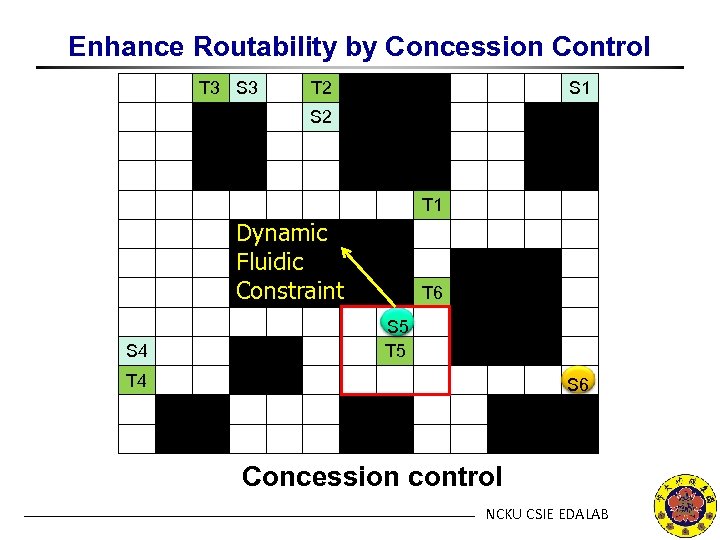 Enhance Routability by Concession Control T 3 S 3 T 2 S 1 S 2 T 1 Dynamic Fluidic Constraint S 4 T 6 S 5 T 4 S 6 Concession control NCKU CSIE EDALAB
Enhance Routability by Concession Control T 3 S 3 T 2 S 1 S 2 T 1 Dynamic Fluidic Constraint S 4 T 6 S 5 T 4 S 6 Concession control NCKU CSIE EDALAB
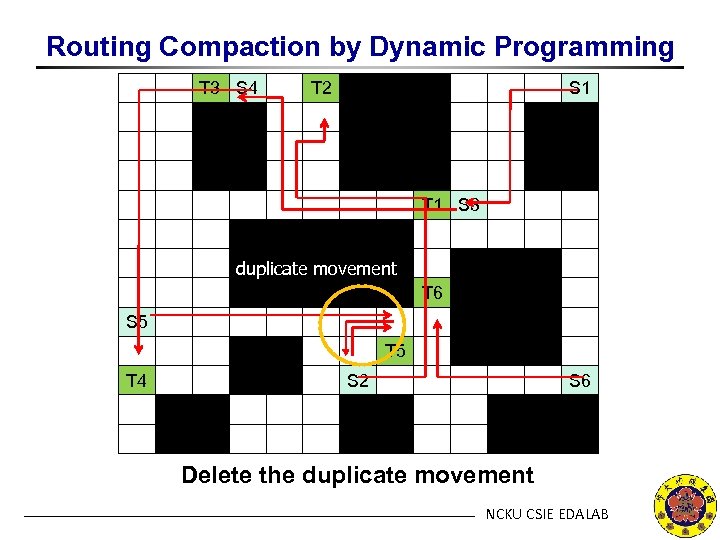 Routing Compaction by Dynamic Programming T 3 S 4 T 2 S 1 T 1 S 3 duplicate movement T 6 S 5 T 4 S 2 S 6 Delete the duplicate movement NCKU CSIE EDALAB
Routing Compaction by Dynamic Programming T 3 S 4 T 2 S 1 T 1 S 3 duplicate movement T 6 S 5 T 4 S 2 S 6 Delete the duplicate movement NCKU CSIE EDALAB
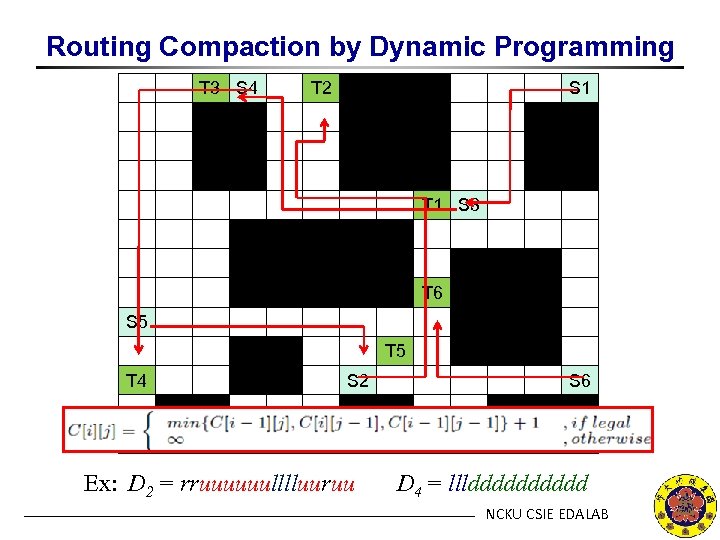 Routing Compaction by Dynamic Programming T 3 S 4 T 2 S 1 T 1 S 3 T 6 S 5 T 4 S 2 Ex: D 2 = rruuuuuulllluuruu S 6 D 4 = lllddddd NCKU CSIE EDALAB
Routing Compaction by Dynamic Programming T 3 S 4 T 2 S 1 T 1 S 3 T 6 S 5 T 4 S 2 Ex: D 2 = rruuuuuulllluuruu S 6 D 4 = lllddddd NCKU CSIE EDALAB
 Outline Introduction Problem Formulation Algorithms Experimental Results Conclusion NCKU CSIE EDALAB
Outline Introduction Problem Formulation Algorithms Experimental Results Conclusion NCKU CSIE EDALAB
 Experimental Settings ․ Implemented our algorithm in C++ language on a 2 GHz 64 -bit Linux machine w/ 8 GB memory ․ Compared with three state-of-the-art algorithms ¾ ¾ ¾ Prioritized A* search [K. Böhringer, TCAD’ 06] Network-flow algorithm [Yuh et al, ICCAD’ 07] High-performance algorithm [Cho and Pan, ISPD’ 08] ․ Tested on three benchmark suites ¾ ¾ ¾ Benchmark I [30] [Cho and Pan, ISPD’ 08] Benchmark II [10] [Self generated] Benchmark III [4] [Su and Chakrabarty, DAC’ 05] ※Benchmark II: (1) bounding boxes of droplets are overlapped; (2) nx 1 or 1 xn narrow routing regions are used for routing; (3) the density of blockage area is over 30%. NCKU CSIE EDALAB
Experimental Settings ․ Implemented our algorithm in C++ language on a 2 GHz 64 -bit Linux machine w/ 8 GB memory ․ Compared with three state-of-the-art algorithms ¾ ¾ ¾ Prioritized A* search [K. Böhringer, TCAD’ 06] Network-flow algorithm [Yuh et al, ICCAD’ 07] High-performance algorithm [Cho and Pan, ISPD’ 08] ․ Tested on three benchmark suites ¾ ¾ ¾ Benchmark I [30] [Cho and Pan, ISPD’ 08] Benchmark II [10] [Self generated] Benchmark III [4] [Su and Chakrabarty, DAC’ 05] ※Benchmark II: (1) bounding boxes of droplets are overlapped; (2) nx 1 or 1 xn narrow routing regions are used for routing; (3) the density of blockage area is over 30%. NCKU CSIE EDALAB
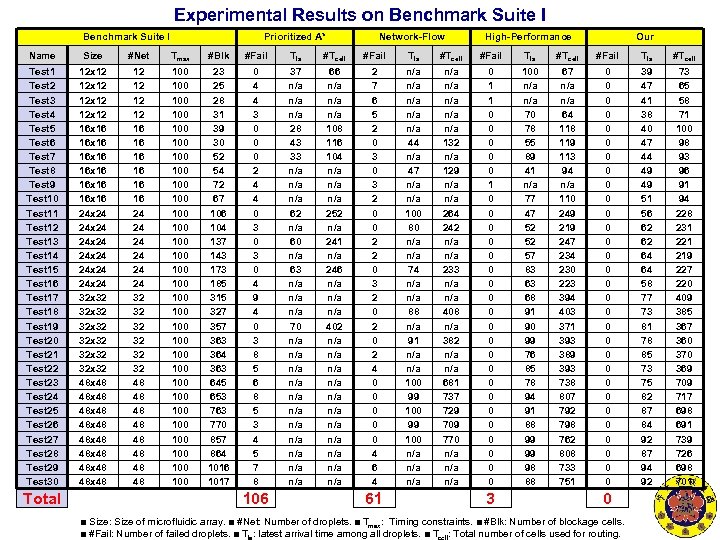 Experimental Results on Benchmark Suite I Prioritized A* Network-Flow High-Performance Our Name Size #Net Tmax #Blk #Fail Tla #Tcell Test 1 Test 2 Test 3 Test 4 Test 5 Test 6 Test 7 Test 8 Test 9 Test 10 Test 11 Test 12 Test 13 Test 14 Test 15 Test 16 Test 17 Test 18 Test 19 Test 20 Test 21 Test 22 Test 23 Test 24 Test 25 Test 26 Test 27 Test 28 Test 29 Test 30 12 x 12 16 x 16 16 x 16 24 x 24 24 x 24 32 x 32 32 x 32 48 x 48 48 x 48 12 12 16 16 16 24 24 24 32 32 32 48 48 100 100 100 100 100 100 100 100 23 25 28 31 39 30 52 54 72 67 106 104 137 143 173 185 315 327 357 363 364 363 645 653 763 770 857 864 1016 1017 0 4 4 3 0 0 0 2 4 4 0 3 0 4 9 4 0 3 8 5 6 8 5 3 4 5 7 8 37 n/a n/a 28 43 33 n/a n/a 62 n/a 60 n/a 63 n/a n/a 70 n/a n/a n/a 66 n/a n/a 108 116 104 n/a n/a 252 n/a 241 n/a 246 n/a n/a 402 n/a n/a n/a 2 7 6 5 2 0 3 2 0 0 2 2 0 3 2 0 2 4 0 0 0 4 6 4 n/a n/a n/a 44 n/a 47 n/a 100 80 n/a 74 n/a 88 n/a 91 n/a 100 99 100 n/a n/a 132 n/a 129 n/a 264 242 n/a 233 n/a 408 n/a 382 n/a 681 737 729 709 770 n/a n/a 0 1 1 0 0 0 0 0 0 0 100 n/a 70 78 55 89 41 n/a 77 47 52 52 57 83 63 68 91 90 99 76 85 78 94 91 88 99 99 98 88 67 n/a 64 118 119 113 94 n/a 110 249 219 247 234 230 223 394 403 371 393 389 393 738 807 792 798 762 808 733 751 0 0 0 0 0 0 0 0 39 47 41 38 40 47 44 49 49 51 56 62 62 64 64 58 77 73 81 78 85 73 75 82 87 84 92 87 94 92 73 65 58 71 100 98 93 96 91 94 228 231 221 219 227 220 409 385 367 360 370 369 709 717 698 691 739 726 698 701 Total 106 61 3 0 ■ Size: Size of microfluidic array. ■ #Net: Number of droplets. ■ Tmax: Timing constraints. ■ #Blk: Number of blockage cells. ■ #Fail: Number of failed droplets. ■ Tla: latest arrival time among all droplets. ■ Tcell: Total number of cells used for routing.
Experimental Results on Benchmark Suite I Prioritized A* Network-Flow High-Performance Our Name Size #Net Tmax #Blk #Fail Tla #Tcell Test 1 Test 2 Test 3 Test 4 Test 5 Test 6 Test 7 Test 8 Test 9 Test 10 Test 11 Test 12 Test 13 Test 14 Test 15 Test 16 Test 17 Test 18 Test 19 Test 20 Test 21 Test 22 Test 23 Test 24 Test 25 Test 26 Test 27 Test 28 Test 29 Test 30 12 x 12 16 x 16 16 x 16 24 x 24 24 x 24 32 x 32 32 x 32 48 x 48 48 x 48 12 12 16 16 16 24 24 24 32 32 32 48 48 100 100 100 100 100 100 100 100 23 25 28 31 39 30 52 54 72 67 106 104 137 143 173 185 315 327 357 363 364 363 645 653 763 770 857 864 1016 1017 0 4 4 3 0 0 0 2 4 4 0 3 0 4 9 4 0 3 8 5 6 8 5 3 4 5 7 8 37 n/a n/a 28 43 33 n/a n/a 62 n/a 60 n/a 63 n/a n/a 70 n/a n/a n/a 66 n/a n/a 108 116 104 n/a n/a 252 n/a 241 n/a 246 n/a n/a 402 n/a n/a n/a 2 7 6 5 2 0 3 2 0 0 2 2 0 3 2 0 2 4 0 0 0 4 6 4 n/a n/a n/a 44 n/a 47 n/a 100 80 n/a 74 n/a 88 n/a 91 n/a 100 99 100 n/a n/a 132 n/a 129 n/a 264 242 n/a 233 n/a 408 n/a 382 n/a 681 737 729 709 770 n/a n/a 0 1 1 0 0 0 0 0 0 0 100 n/a 70 78 55 89 41 n/a 77 47 52 52 57 83 63 68 91 90 99 76 85 78 94 91 88 99 99 98 88 67 n/a 64 118 119 113 94 n/a 110 249 219 247 234 230 223 394 403 371 393 389 393 738 807 792 798 762 808 733 751 0 0 0 0 0 0 0 0 39 47 41 38 40 47 44 49 49 51 56 62 62 64 64 58 77 73 81 78 85 73 75 82 87 84 92 87 94 92 73 65 58 71 100 98 93 96 91 94 228 231 221 219 227 220 409 385 367 360 370 369 709 717 698 691 739 726 698 701 Total 106 61 3 0 ■ Size: Size of microfluidic array. ■ #Net: Number of droplets. ■ Tmax: Timing constraints. ■ #Blk: Number of blockage cells. ■ #Fail: Number of failed droplets. ■ Tla: latest arrival time among all droplets. ■ Tcell: Total number of cells used for routing.
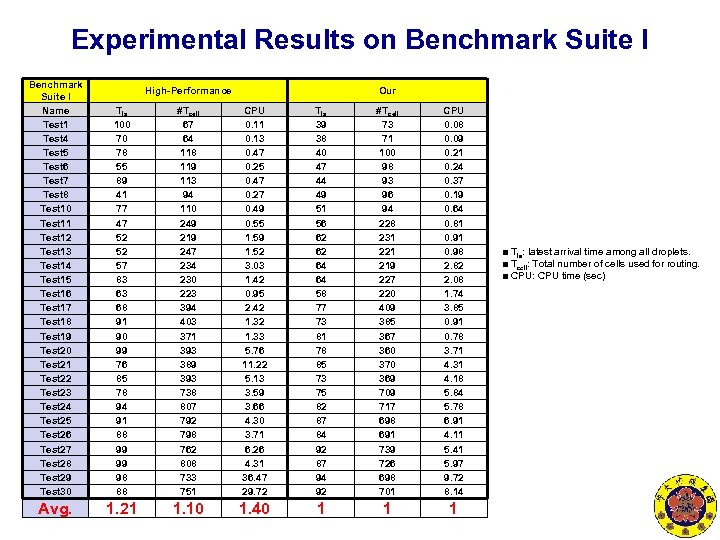 Experimental Results on Benchmark Suite I Name Test 1 Test 4 Test 5 Test 6 Test 7 Test 8 Test 10 Test 11 Test 12 Test 13 Test 14 Test 15 Test 16 Test 17 Test 18 Test 19 Test 20 Test 21 Test 22 Test 23 Test 24 Test 25 Test 26 Test 27 Test 28 Test 29 Test 30 Tla 100 70 78 55 89 41 77 47 52 52 57 83 63 68 91 90 99 76 85 78 94 91 88 99 99 98 88 #Tcell 67 64 118 119 113 94 110 249 219 247 234 230 223 394 403 371 393 389 393 738 807 792 798 762 808 733 751 CPU 0. 11 0. 13 0. 47 0. 25 0. 47 0. 27 0. 49 0. 55 1. 59 1. 52 3. 03 1. 42 0. 95 2. 42 1. 33 5. 76 11. 22 5. 13 3. 59 3. 66 4. 30 3. 71 6. 26 4. 31 36. 47 29. 72 Tla 39 38 40 47 44 49 51 56 62 62 64 64 58 77 73 81 78 85 73 75 82 87 84 92 87 94 92 #Tcell 73 71 100 98 93 96 94 228 231 221 219 227 220 409 385 367 360 370 369 709 717 698 691 739 726 698 701 CPU 0. 08 0. 09 0. 21 0. 24 0. 37 0. 19 0. 64 0. 81 0. 98 2. 82 2. 08 1. 74 3. 85 0. 91 0. 78 3. 71 4. 31 4. 18 5. 84 5. 78 6. 91 4. 11 5. 41 5. 97 9. 72 8. 14 Avg. 1. 21 1. 10 1. 40 1 1 1 High-Performance Our ■ Tla: latest arrival time among all droplets. ■ Tcell: Total number of cells used for routing. ■ CPU: CPU time (sec)
Experimental Results on Benchmark Suite I Name Test 1 Test 4 Test 5 Test 6 Test 7 Test 8 Test 10 Test 11 Test 12 Test 13 Test 14 Test 15 Test 16 Test 17 Test 18 Test 19 Test 20 Test 21 Test 22 Test 23 Test 24 Test 25 Test 26 Test 27 Test 28 Test 29 Test 30 Tla 100 70 78 55 89 41 77 47 52 52 57 83 63 68 91 90 99 76 85 78 94 91 88 99 99 98 88 #Tcell 67 64 118 119 113 94 110 249 219 247 234 230 223 394 403 371 393 389 393 738 807 792 798 762 808 733 751 CPU 0. 11 0. 13 0. 47 0. 25 0. 47 0. 27 0. 49 0. 55 1. 59 1. 52 3. 03 1. 42 0. 95 2. 42 1. 33 5. 76 11. 22 5. 13 3. 59 3. 66 4. 30 3. 71 6. 26 4. 31 36. 47 29. 72 Tla 39 38 40 47 44 49 51 56 62 62 64 64 58 77 73 81 78 85 73 75 82 87 84 92 87 94 92 #Tcell 73 71 100 98 93 96 94 228 231 221 219 227 220 409 385 367 360 370 369 709 717 698 691 739 726 698 701 CPU 0. 08 0. 09 0. 21 0. 24 0. 37 0. 19 0. 64 0. 81 0. 98 2. 82 2. 08 1. 74 3. 85 0. 91 0. 78 3. 71 4. 31 4. 18 5. 84 5. 78 6. 91 4. 11 5. 41 5. 97 9. 72 8. 14 Avg. 1. 21 1. 10 1. 40 1 1 1 High-Performance Our ■ Tla: latest arrival time among all droplets. ■ Tcell: Total number of cells used for routing. ■ CPU: CPU time (sec)
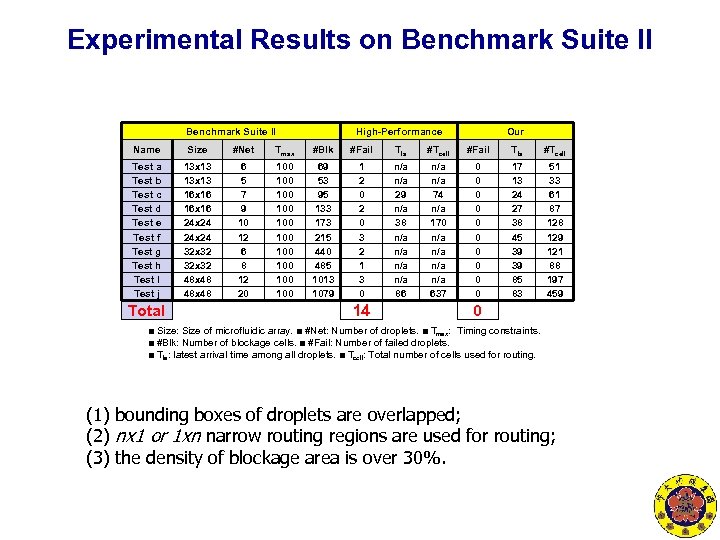 Experimental Results on Benchmark Suite II High-Performance Our Name Size #Net Tmax #Blk #Fail Tla #Tcell Test a Test b Test c Test d Test e Test f Test g Test h Test I Test j 13 x 13 16 x 16 24 x 24 32 x 32 48 x 48 6 5 7 9 10 12 6 8 12 20 100 100 100 69 53 95 133 173 215 440 485 1013 1079 1 2 0 3 2 1 3 0 n/a 29 n/a 38 n/a n/a 86 n/a 74 n/a 170 n/a n/a 637 0 0 0 0 0 17 13 24 27 38 45 39 39 85 83 51 33 61 87 128 129 121 88 197 459 Total 14 0 ■ Size: Size of microfluidic array. ■ #Net: Number of droplets. ■ Tmax: Timing constraints. ■ #Blk: Number of blockage cells. ■ #Fail: Number of failed droplets. ■ Tla: latest arrival time among all droplets. ■ Tcell: Total number of cells used for routing. (1) bounding boxes of droplets are overlapped; (2) nx 1 or 1 xn narrow routing regions are used for routing; (3) the density of blockage area is over 30%.
Experimental Results on Benchmark Suite II High-Performance Our Name Size #Net Tmax #Blk #Fail Tla #Tcell Test a Test b Test c Test d Test e Test f Test g Test h Test I Test j 13 x 13 16 x 16 24 x 24 32 x 32 48 x 48 6 5 7 9 10 12 6 8 12 20 100 100 100 69 53 95 133 173 215 440 485 1013 1079 1 2 0 3 2 1 3 0 n/a 29 n/a 38 n/a n/a 86 n/a 74 n/a 170 n/a n/a 637 0 0 0 0 0 17 13 24 27 38 45 39 39 85 83 51 33 61 87 128 129 121 88 197 459 Total 14 0 ■ Size: Size of microfluidic array. ■ #Net: Number of droplets. ■ Tmax: Timing constraints. ■ #Blk: Number of blockage cells. ■ #Fail: Number of failed droplets. ■ Tla: latest arrival time among all droplets. ■ Tcell: Total number of cells used for routing. (1) bounding boxes of droplets are overlapped; (2) nx 1 or 1 xn narrow routing regions are used for routing; (3) the density of blockage area is over 30%.
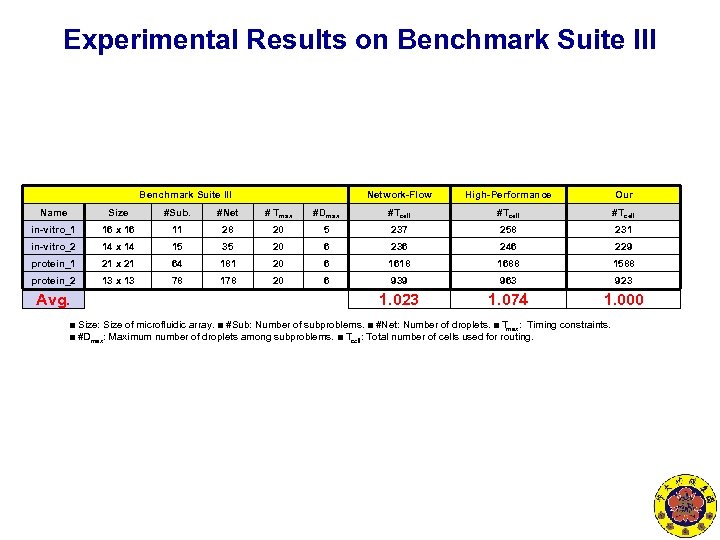 Experimental Results on Benchmark Suite III Network-Flow High-Performance Our Name Size #Sub. #Net # Tmax #Dmax #Tcell in-vitro_1 16 x 16 11 28 20 5 237 258 231 in-vitro_2 14 x 14 15 35 20 6 236 246 229 protein_1 21 x 21 64 181 20 6 1618 1688 1588 protein_2 13 x 13 78 178 20 6 939 963 923 1. 074 1. 000 Avg. ■ Size: Size of microfluidic array. ■ #Sub: Number of subproblems. ■ #Net: Number of droplets. ■ Tmax: Timing constraints. ■ #Dmax: Maximum number of droplets among subproblems. ■ Tcell: Total number of cells used for routing.
Experimental Results on Benchmark Suite III Network-Flow High-Performance Our Name Size #Sub. #Net # Tmax #Dmax #Tcell in-vitro_1 16 x 16 11 28 20 5 237 258 231 in-vitro_2 14 x 14 15 35 20 6 236 246 229 protein_1 21 x 21 64 181 20 6 1618 1688 1588 protein_2 13 x 13 78 178 20 6 939 963 923 1. 074 1. 000 Avg. ■ Size: Size of microfluidic array. ■ #Sub: Number of subproblems. ■ #Net: Number of droplets. ■ Tmax: Timing constraints. ■ #Dmax: Maximum number of droplets among subproblems. ■ Tcell: Total number of cells used for routing.
 Outline Introduction Problem Formulation Algorithms Experimental Results Conclusion NCKU CSIE EDALAB
Outline Introduction Problem Formulation Algorithms Experimental Results Conclusion NCKU CSIE EDALAB
 Conclusion ․ We proposed a fast routability- and performance-driven ․ ․ droplet router for DMFBs. Experimental results demonstrated that our algorithm achieves 100% routing completion for all test cases in three Benchmark Suites while the previous algorithms are not. Furthermore, the experimental results shown that our algorithm can achieve better timing result (Tla) and fault tolerance (Tcell) and faster runtime (CPU) with the best known results. NCKU CSIE EDALAB
Conclusion ․ We proposed a fast routability- and performance-driven ․ ․ droplet router for DMFBs. Experimental results demonstrated that our algorithm achieves 100% routing completion for all test cases in three Benchmark Suites while the previous algorithms are not. Furthermore, the experimental results shown that our algorithm can achieve better timing result (Tla) and fault tolerance (Tcell) and faster runtime (CPU) with the best known results. NCKU CSIE EDALAB
 NCKU CSIE EDALAB
NCKU CSIE EDALAB


