cd9fbc424270856e0786784397d9c9b6.ppt
- Количество слайдов: 29

Identifying Applicability Domains for Quantitative Structure Property Relationships Mordechai Shachama, Neima Braunerb Georgi St. Cholakovc and Roumiana P. Statevad, a. Dept. Chem. Eng. , Ben-Gurion University Beer-Sheva, Israel b. School of Engineering, Tel-Aviv University Tel-Aviv, Israel c. Dept. Org. Synth. and Fuels, University of Chemical Technology and Metallurgy, Sofia, Bulgaria d. Institute of Chemical Engineering, Bulgarian Academy of Sciences, Sofia 1113, Bulgaria
The Needs ØPhysicochemical and biological properties are needed for risk assessment, environmental impact assessment and process design, analysis and optimization ØThe number of the compounds used at present by the industry or those of its immediate interest ~100, 000. Those theoretically possible and may be of future interest several tens of millions. ØDIPPR 801 database contains 2101 compounds (33 constant properties, 15 temperature dependent properties)
Presentation Outline ØReview of Structure-Property Relationships (QSPR) based on Molecular Descriptors ØThe “Targeted” and “Homologous Series” QSPR Methods ØRepresentation of Liquid and Gas Properties by Molecular Descriptors Ø Representation of Normal Melting Temperature by Molecular Descriptors ØLong Range Extrapolation from small Training Sets

References for the New Techniques "A Structurally "Targeted" QSPR Method for Property Prediction". Ind. Eng. Chem. Res. , 45, 8430 -8437 (2006 ) Molecular descriptors database- 1280 (non-constant) descriptors for 324 compounds (hydrocarbons and oxygen containing organic compounds). The descriptors are calculated using the Dragon program (version 5. 4, DRAGON http: //www. talete. mi. it ) Physical properties databases: DIPPR (http: //dippr. byu. edu ) NIST (http: //webbook. nist. gov/chemistry/ )
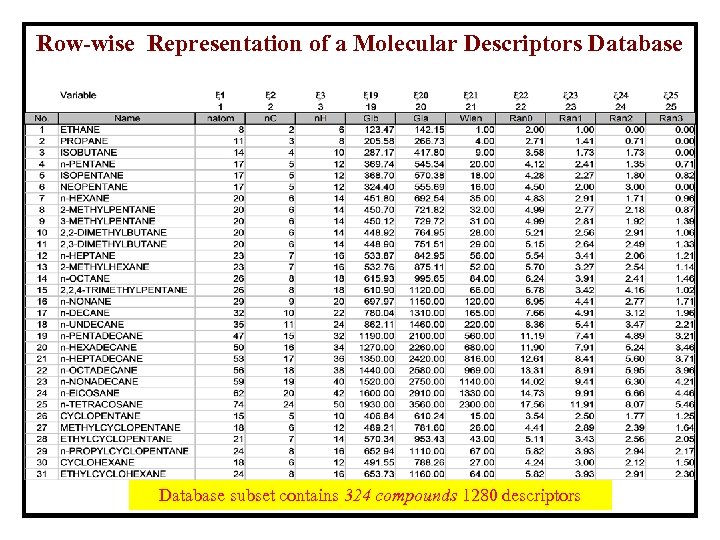
Row-wise Representation of a Molecular Descriptors Database subset contains 324 compounds 1280 descriptors
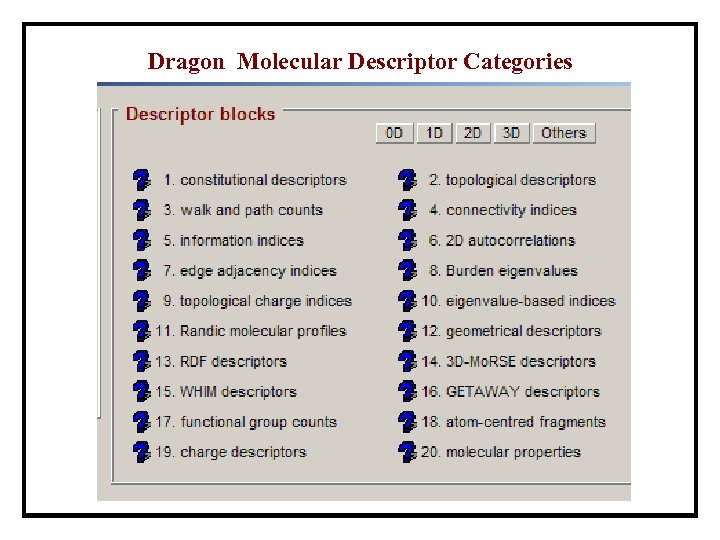
Dragon Molecular Descriptor Categories
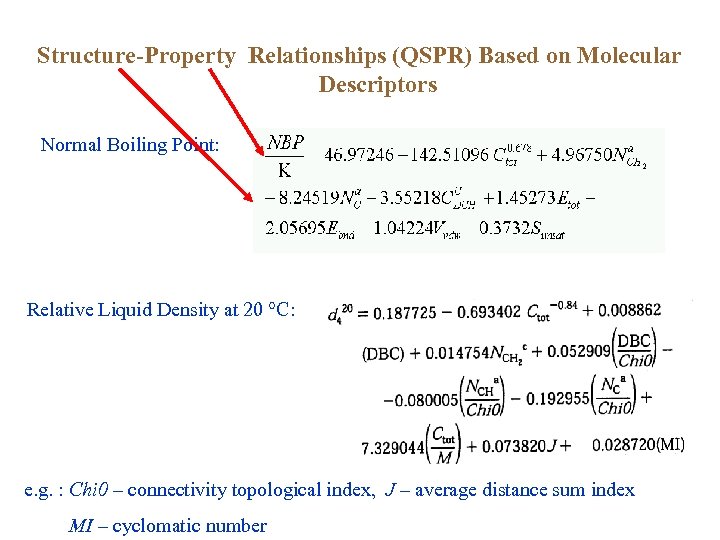
Structure-Property Relationships (QSPR) Based on Molecular Descriptors Normal Boiling Point: Relative Liquid Density at 20 °C: e. g. : Chi 0 – connectivity topological index, J – average distance sum index MI – cyclomatic number
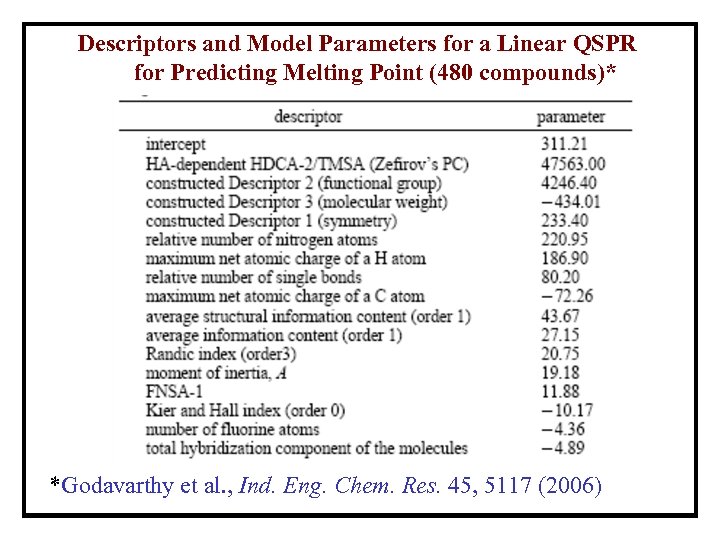
Descriptors and Model Parameters for a Linear QSPR for Predicting Melting Point (480 compounds)* *Godavarthy et al. , Ind. Eng. Chem. Res. 45, 5117 (2006)
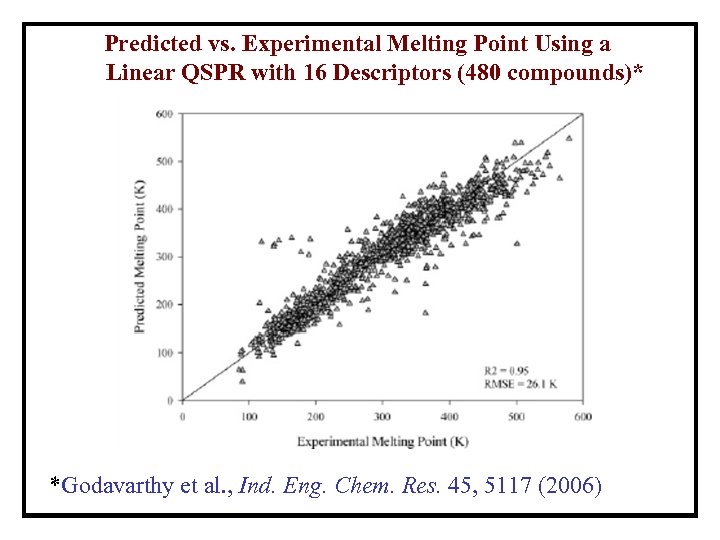
Predicted vs. Experimental Melting Point Using a Linear QSPR with 16 Descriptors (480 compounds)* *Godavarthy et al. , Ind. Eng. Chem. Res. 45, 5117 (2006)
Limitations of the QSPR Techniques with Unrestricted Applicability Domains ØComplex , often nonlinear QSPRs are needed in order to match the great variability of property values caused by the many structural differences between the various compounds. ØPrediction errors are very large especially for properties which are highly sensitive to structural differences (i. e. solid properties) ØThe accuracy of the property prediction will be much higher for compounds which are well represented in the “training set" than for compounds which are sparsely represented. No systematic way is offered to categorize a particular target compound. ØFor a target compound of unmeasured properties it is impossible to assess the prediction accuracy.
The “Targeted” and “Homologous Series” QSPR Methods ØIn the TQSPR method, a similarity group of compounds for a target compound is first identified, using correlation coefficients between vectors of descriptors as measures of “similarity”. ØIn the HS-QSPR method the members of the homologous series are assigned into the “similarity group”. ØIn the second step a linear QSPR is tailored to a particular property of the target compound. ØRow-wise representation (a row of descriptors for each compound) of a subset of the database, which contains only the members of the similarity group is used to derive the QSPR. ØOnly the HS-QSPR method is discussed here
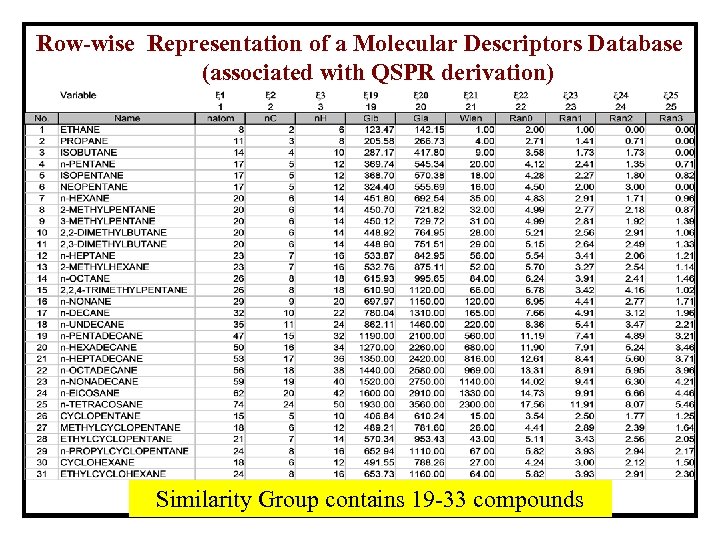
Row-wise Representation of a Molecular Descriptors Database (associated with QSPR derivation) Similarity Group contains 19 -33 compounds
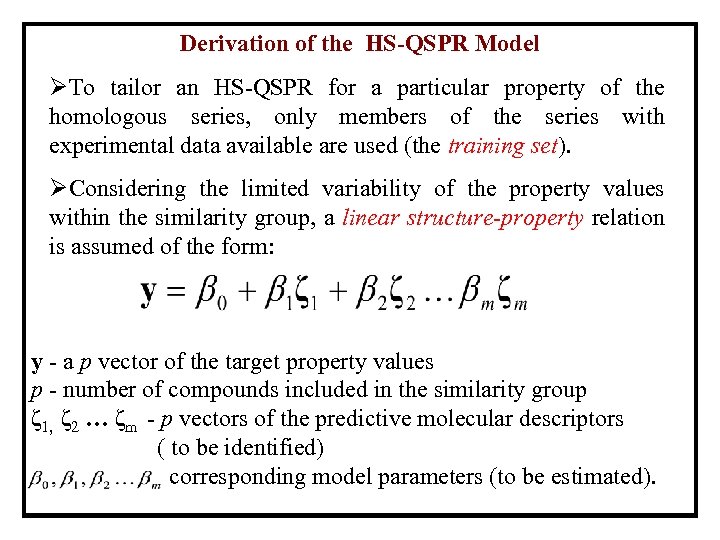
Derivation of the HS-QSPR Model ØTo tailor an HS-QSPR for a particular property of the homologous series, only members of the series with experimental data available are used (the training set). ØConsidering the limited variability of the property values within the similarity group, a linear structure-property relation is assumed of the form: y - a p vector of the target property values p - number of compounds included in the similarity group ζ 1, ζ 2 … ζm - p vectors of the predictive molecular descriptors ( to be identified) corresponding model parameters (to be estimated).
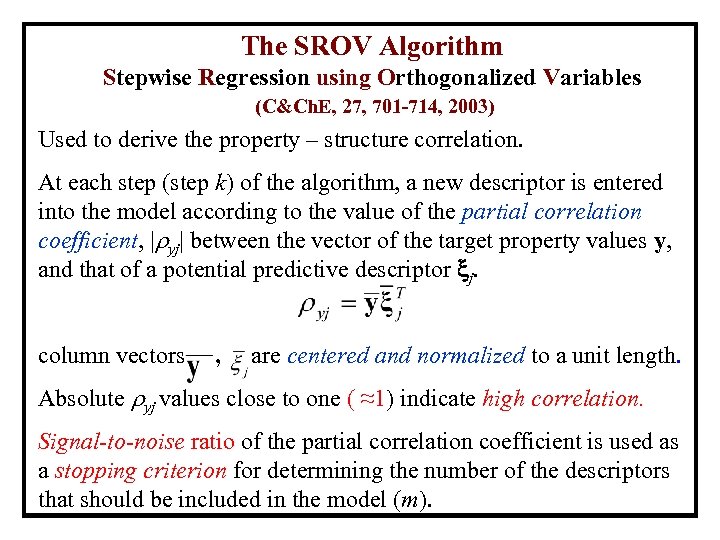
The SROV Algorithm Stepwise Regression using Orthogonalized Variables (C&Ch. E, 27, 701 -714, 2003) Used to derive the property – structure correlation. At each step (step k) of the algorithm, a new descriptor is entered into the model according to the value of the partial correlation coefficient, | yj| between the vector of the target property values y, and that of a potential predictive descriptor j. column vectors , are centered and normalized to a unit length. Absolute yj values close to one ( ≈1) indicate high correlation. Signal-to-noise ratio of the partial correlation coefficient is used as a stopping criterion for determining the number of the descriptors that should be included in the model (m).
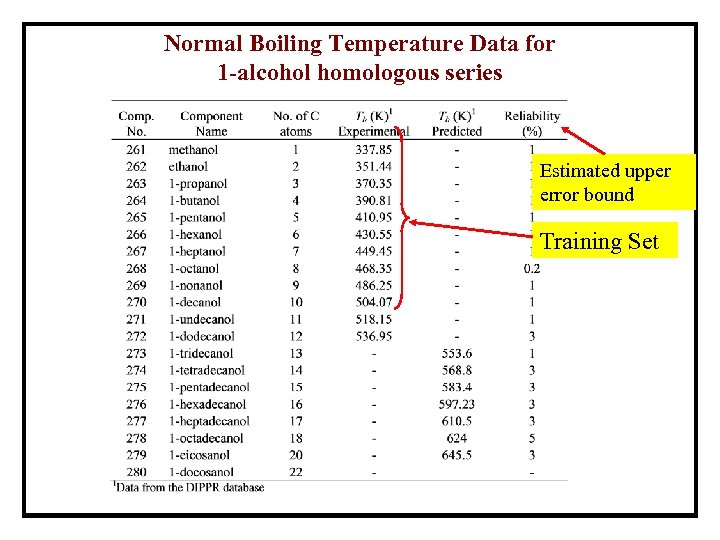
Normal Boiling Temperature Data for 1 -alcohol homologous series Estimated upper error bound Training Set
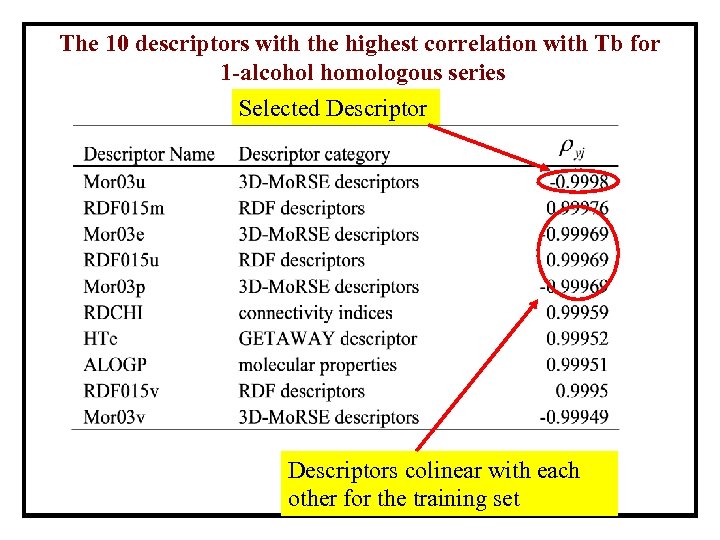
The 10 descriptors with the highest correlation with Tb for 1 -alcohol homologous series Selected Descriptors colinear with each other for the training set
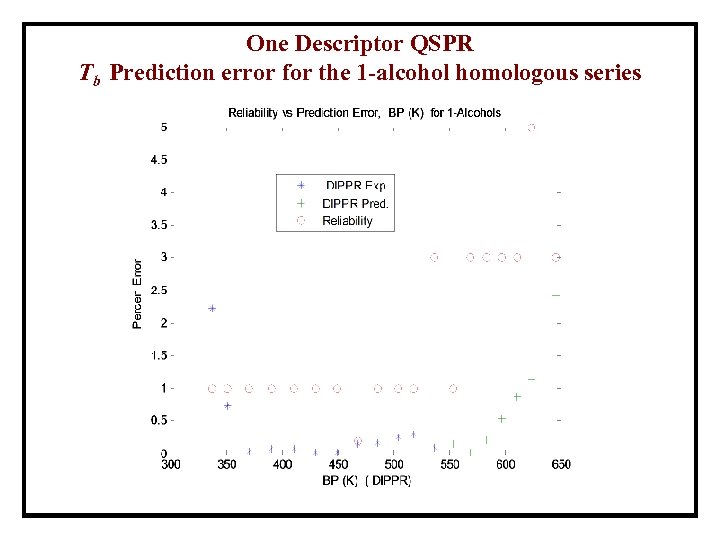
One Descriptor QSPR Tb Prediction error for the 1 -alcohol homologous series
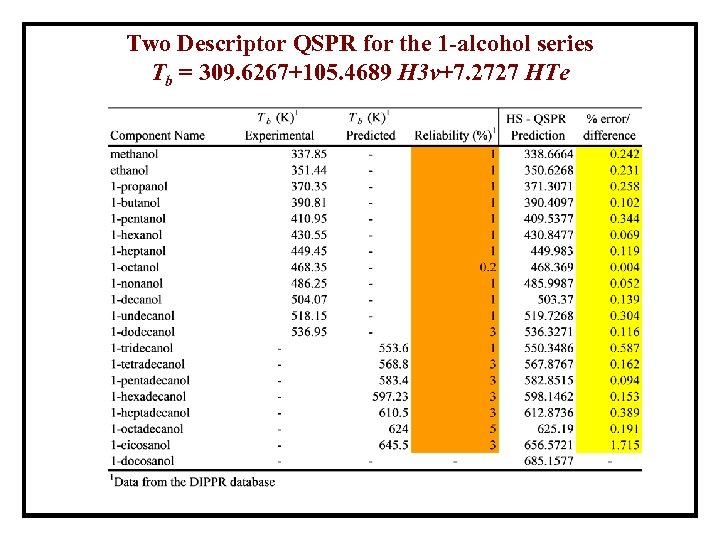
Two Descriptor QSPR for the 1 -alcohol series Tb = 309. 6267+105. 4689 H 3 v+7. 2727 HTe
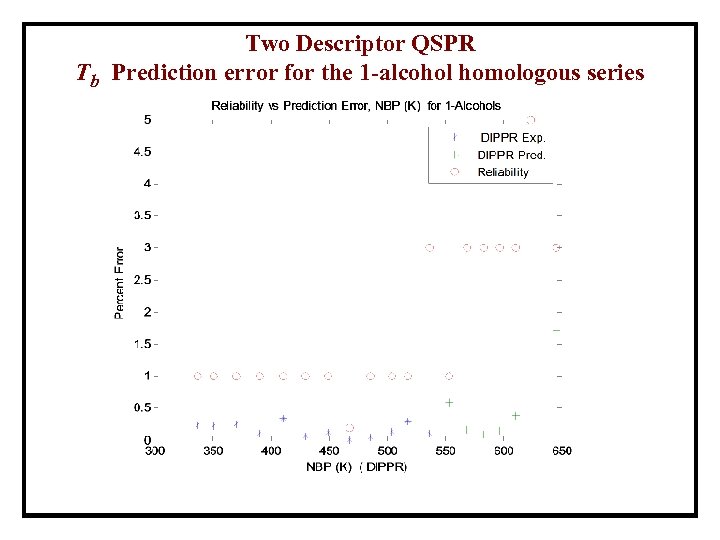
Two Descriptor QSPR Tb Prediction error for the 1 -alcohol homologous series
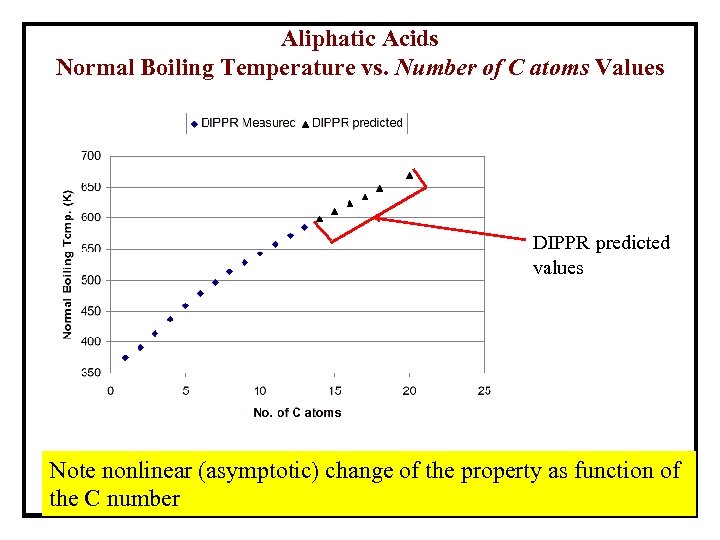
Aliphatic Acids Normal Boiling Temperature vs. Number of C atoms Values DIPPR predicted values Note nonlinear (asymptotic) change of the property as function of the C number
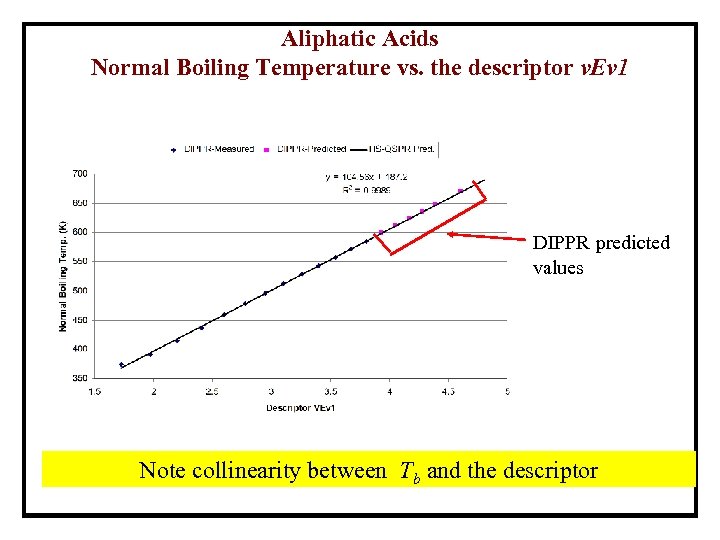
Aliphatic Acids Normal Boiling Temperature vs. the descriptor v. Ev 1 DIPPR predicted values Note collinearity between Tb and the descriptor
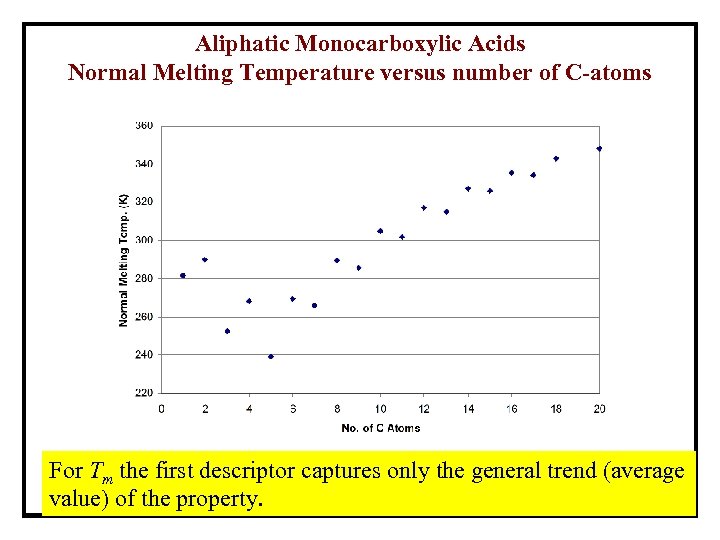
Aliphatic Monocarboxylic Acids Normal Melting Temperature versus number of C-atoms For Tm the first descriptor captures only the general trend (average value) of the property.
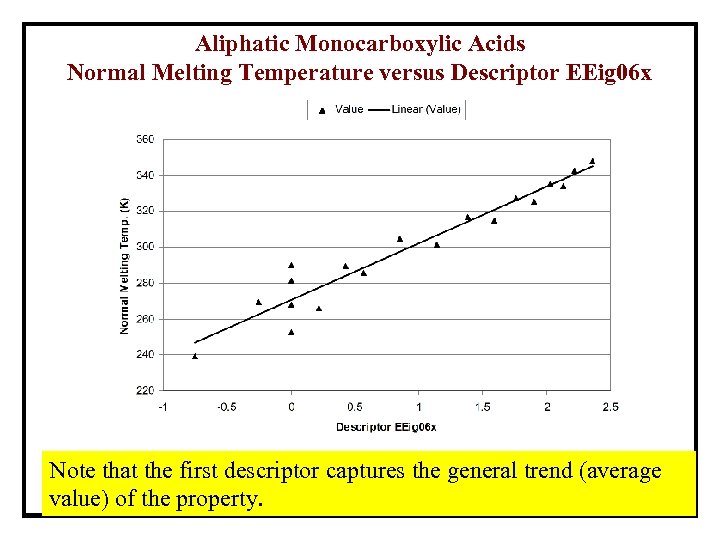
Aliphatic Monocarboxylic Acids Normal Melting Temperature versus Descriptor EEig 06 x Note that the first descriptor captures the general trend (average value) of the property.
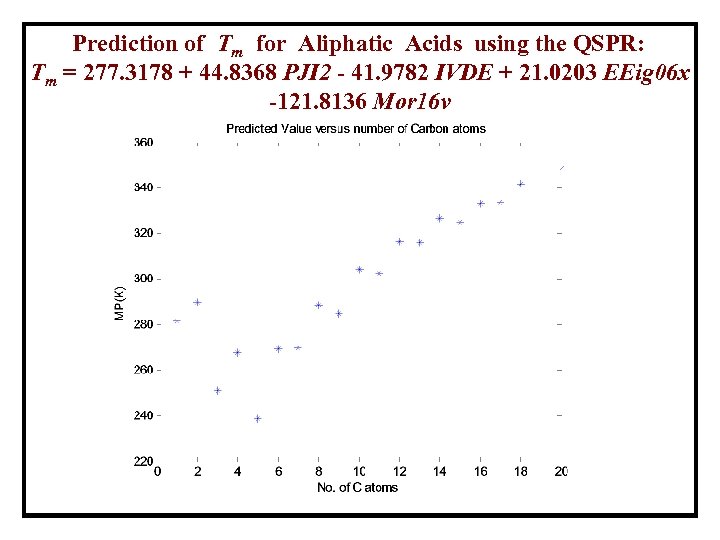
Prediction of Tm for Aliphatic Acids using the QSPR: Tm = 277. 3178 + 44. 8368 PJI 2 - 41. 9782 IVDE + 21. 0203 EEig 06 x -121. 8136 Mor 16 v
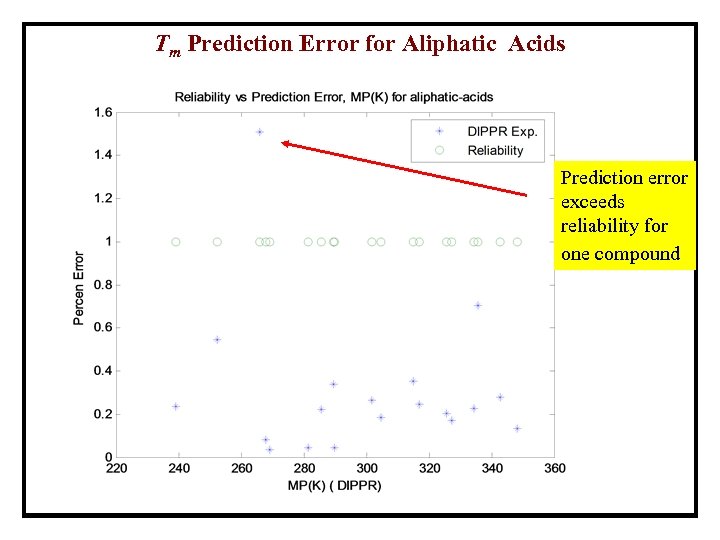
Tm Prediction Error for Aliphatic Acids Prediction error exceeds reliability for one compound
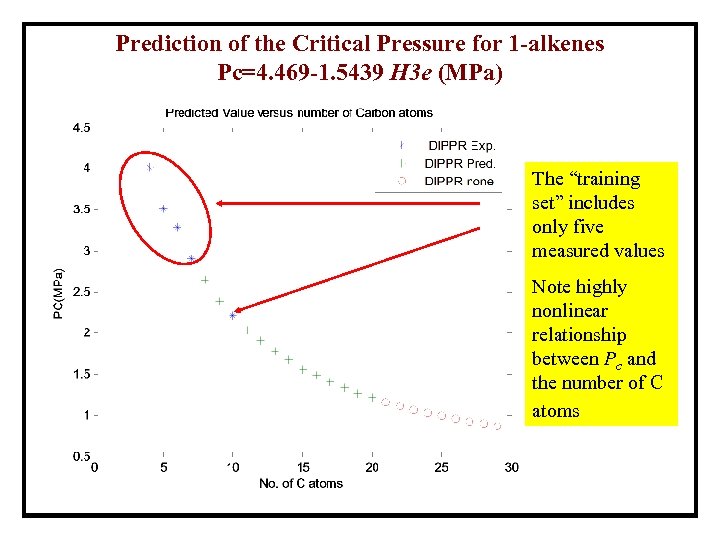
Prediction of the Critical Pressure for 1 -alkenes Pc=4. 469 -1. 5439 H 3 e (MPa) The “training set” includes only five measured values Note highly nonlinear relationship between Pc and the number of C atoms
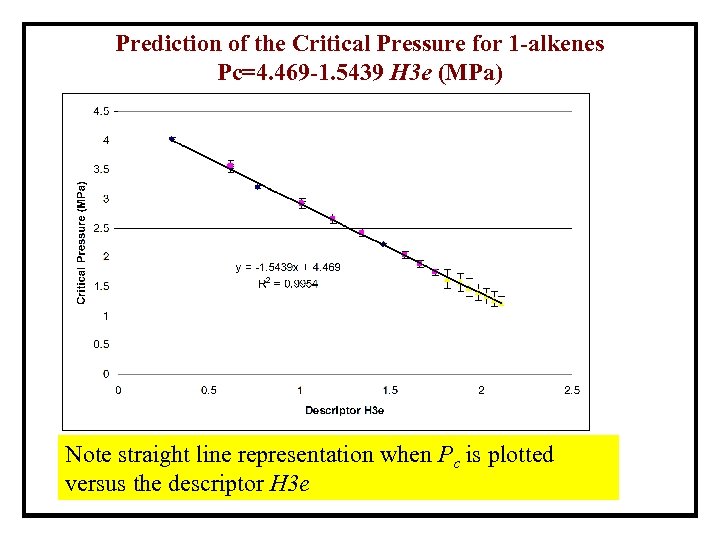
Prediction of the Critical Pressure for 1 -alkenes Pc=4. 469 -1. 5439 H 3 e (MPa) Note straight line representation when Pc is plotted versus the descriptor H 3 e
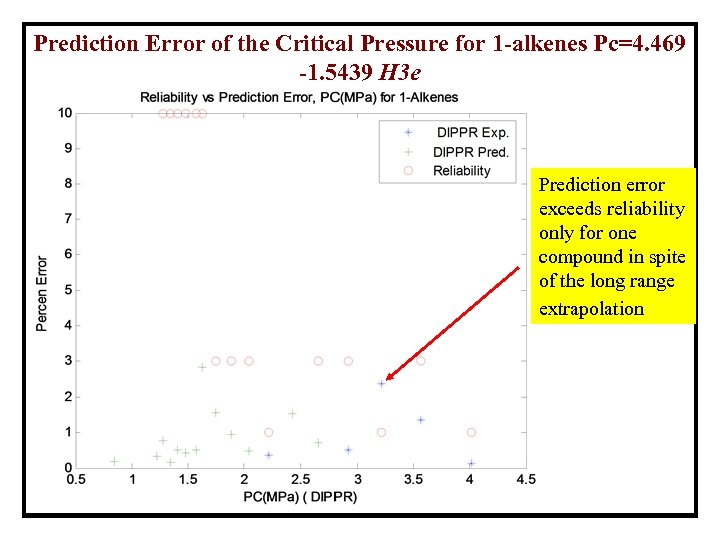
Prediction Error of the Critical Pressure for 1 -alkenes Pc=4. 469 -1. 5439 H 3 e Prediction error exceeds reliability only for one compound in spite of the long range extrapolation
Conclusions Selecting the molecular descriptors that exhibit the highest level of collinearity with a particular property from a very large pool of descriptors enables developing simple linear QSPRs for prediction of properties of homologous series with the characteristics: 1. Prediction of constant properties (including solid properties) within experimental error (reliability) level. 2. Long range extrapolation from small training sets of 3 -5 compounds for which experimental data is available. 3. Use of linear QSPRs that include one to four descriptors. 4. The maximal prediction error of the melting point temperature is 3 K. This is smaller by at least an order of magnitude than the errors reported in the literature.
cd9fbc424270856e0786784397d9c9b6.ppt