6- RNA Editing 2013.ppt
- Количество слайдов: 97

 И я сжег все, чему поклонялся, Поклонился всему, что сжигал. .
И я сжег все, чему поклонялся, Поклонился всему, что сжигал. .

 RNA Editing: Against the central dogma Unexpextedly researches encounter a gene with a sequence of nucleotides that does not match exactly that in its RNA product Редактирование РНК В. В. Вельков 2013
RNA Editing: Against the central dogma Unexpextedly researches encounter a gene with a sequence of nucleotides that does not match exactly that in its RNA product Редактирование РНК В. В. Вельков 2013
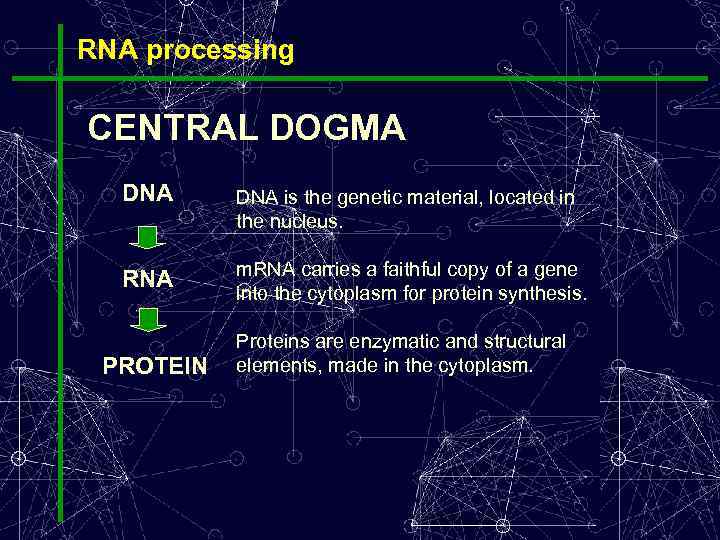 RNA processing CENTRAL DOGMA DNA is the genetic material, located in the nucleus. RNA m. RNA carries a faithful copy of a gene into the cytoplasm for protein synthesis. PROTEIN Proteins are enzymatic and structural elements, made in the cytoplasm.
RNA processing CENTRAL DOGMA DNA is the genetic material, located in the nucleus. RNA m. RNA carries a faithful copy of a gene into the cytoplasm for protein synthesis. PROTEIN Proteins are enzymatic and structural elements, made in the cytoplasm.
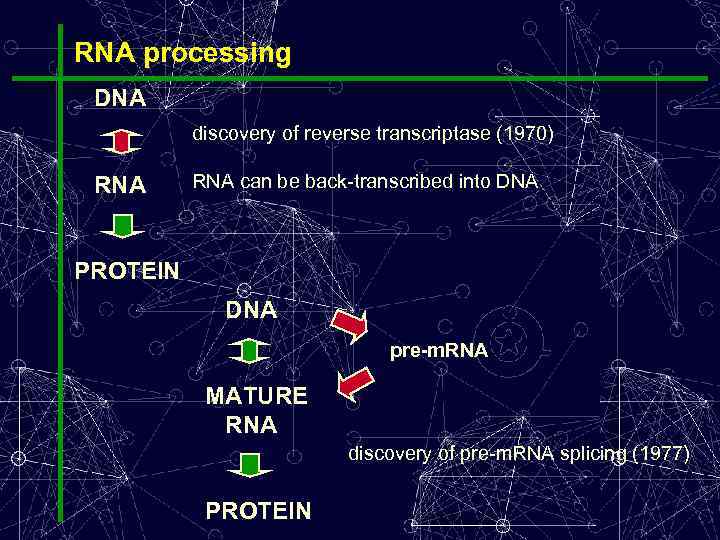 RNA processing DNA discovery of reverse transcriptase (1970) RNA can be back-transcribed into DNA PROTEIN DNA pre-m. RNA MATURE RNA discovery of pre-m. RNA splicing (1977) PROTEIN
RNA processing DNA discovery of reverse transcriptase (1970) RNA can be back-transcribed into DNA PROTEIN DNA pre-m. RNA MATURE RNA discovery of pre-m. RNA splicing (1977) PROTEIN
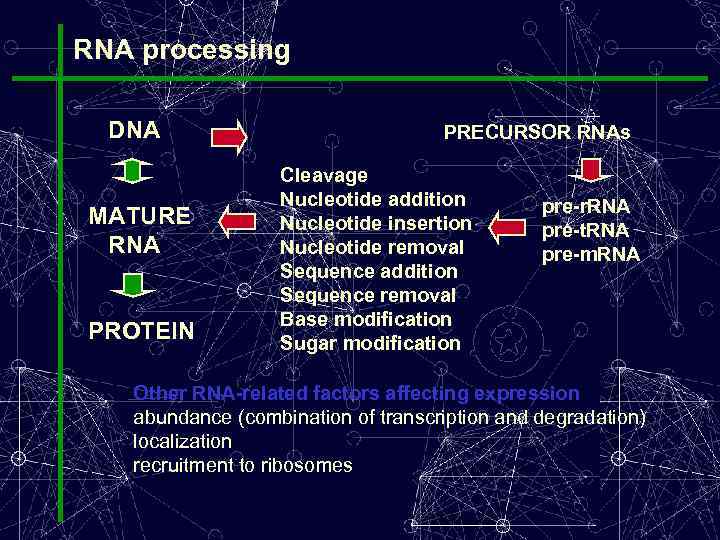 RNA processing DNA MATURE RNA PROTEIN PRECURSOR RNAs Cleavage Nucleotide addition Nucleotide insertion Nucleotide removal Sequence addition Sequence removal Base modification Sugar modification pre-r. RNA pre-t. RNA pre-m. RNA Other RNA-related factors affecting expression abundance (combination of transcription and degradation) localization recruitment to ribosomes
RNA processing DNA MATURE RNA PROTEIN PRECURSOR RNAs Cleavage Nucleotide addition Nucleotide insertion Nucleotide removal Sequence addition Sequence removal Base modification Sugar modification pre-r. RNA pre-t. RNA pre-m. RNA Other RNA-related factors affecting expression abundance (combination of transcription and degradation) localization recruitment to ribosomes
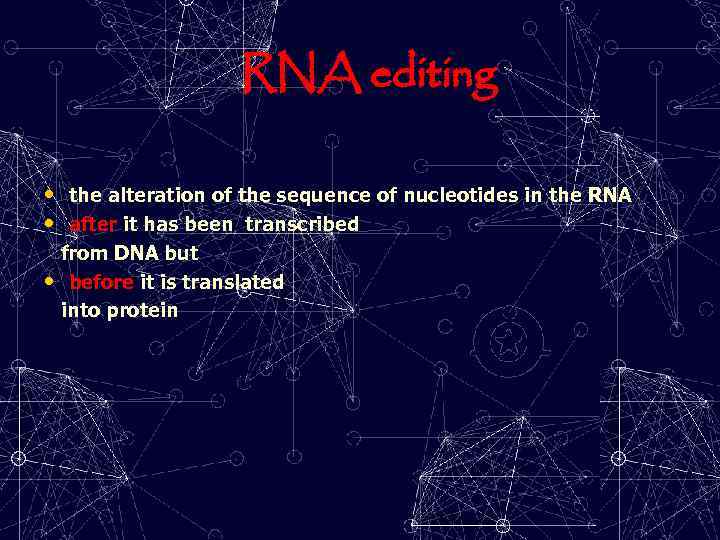 RNA editing • the alteration of the sequence of nucleotides in the RNA • after it has been transcribed from DNA but • before it is translated into protein
RNA editing • the alteration of the sequence of nucleotides in the RNA • after it has been transcribed from DNA but • before it is translated into protein
 РНК редактирование • • Изменение информационного содержания в пре-м. РНК Обнаружено у эукариотных м. РНК, р. РНК, т. РНК. Происходит в ядрах Митохондриях пластидах Не найдено у прокариот
РНК редактирование • • Изменение информационного содержания в пре-м. РНК Обнаружено у эукариотных м. РНК, р. РНК, т. РНК. Происходит в ядрах Митохондриях пластидах Не найдено у прокариот
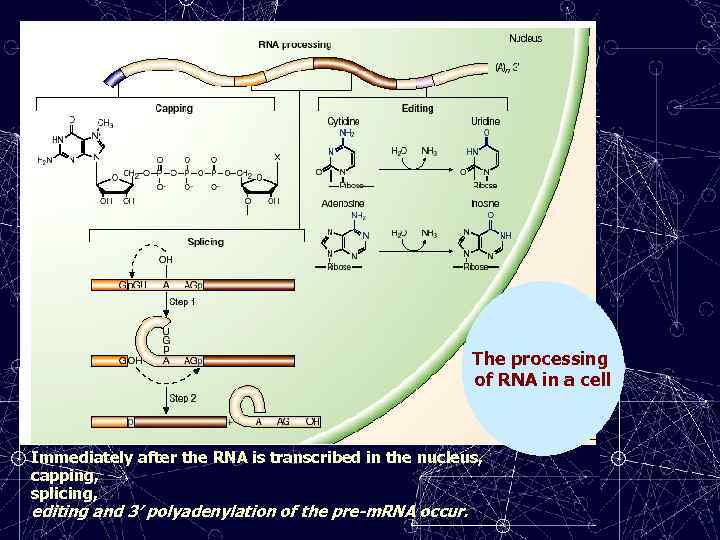 The processing of RNA in a cell Immediately after the RNA is transcribed in the nucleus, capping, splicing, editing and 3′ polyadenylation of the pre-m. RNA occur.
The processing of RNA in a cell Immediately after the RNA is transcribed in the nucleus, capping, splicing, editing and 3′ polyadenylation of the pre-m. RNA occur.
 Молекулярные механизмы редактирования РНК • Модификация (конверсия) C в U A в I – путем дезаминирования • Матричное инсерционное добавление нуклеотидов • Делетирование нуклеотидов
Молекулярные механизмы редактирования РНК • Модификация (конверсия) C в U A в I – путем дезаминирования • Матричное инсерционное добавление нуклеотидов • Делетирование нуклеотидов
 В результате редактирования РНК • Возникает непрерывная рамка считывания (ранее ее не было), • Прерывается рамка считывания, • Изменяется аминокислотная последовательность кодируемого белка
В результате редактирования РНК • Возникает непрерывная рамка считывания (ранее ее не было), • Прерывается рамка считывания, • Изменяется аминокислотная последовательность кодируемого белка
 AGACGGUC AUG CGU AAU UCU GUA GGC UGC CAA UGGC M R N S V G C Q *
AGACGGUC AUG CGU AAU UCU GUA GGC UGC CAA UGGC M R N S V G C Q *
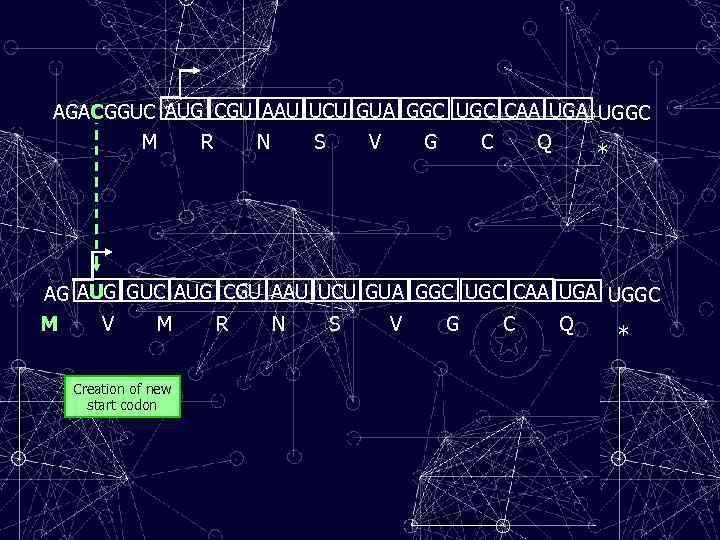 AGACGGUC AUG CGU AAU UCU GUA GGC UGC CAA UGGC M R N S V G C Q * AG AUG GUC AUG CGU AAU UCU GUA GGC UGC CAA UGGC M V M R N S V G C Q * Creation of new start codon
AGACGGUC AUG CGU AAU UCU GUA GGC UGC CAA UGGC M R N S V G C Q * AG AUG GUC AUG CGU AAU UCU GUA GGC UGC CAA UGGC M V M R N S V G C Q * Creation of new start codon
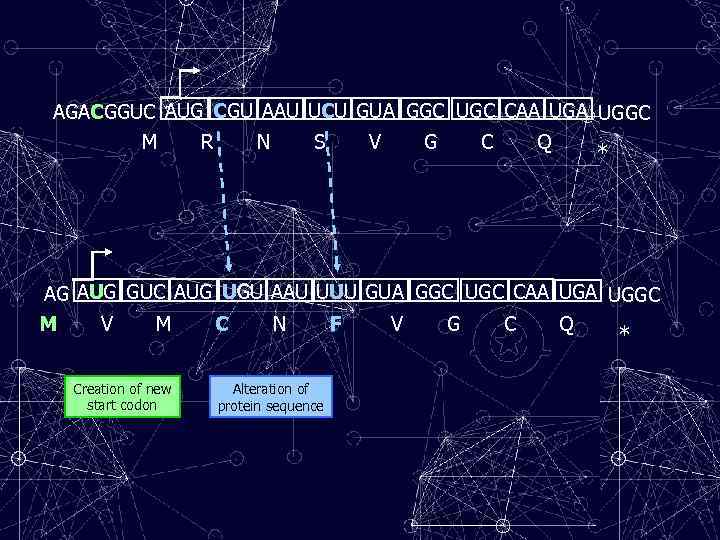 AGACGGUC AUG CGU AAU UCU GUA GGC UGC CAA UGGC M R N S V G C Q * AG AUG GUC AUG UGU AAU UUU GUA GGC UGC CAA UGGC M V M C N F V G C Q * Creation of new start codon Alteration of protein sequence
AGACGGUC AUG CGU AAU UCU GUA GGC UGC CAA UGGC M R N S V G C Q * AG AUG GUC AUG UGU AAU UUU GUA GGC UGC CAA UGGC M V M C N F V G C Q * Creation of new start codon Alteration of protein sequence
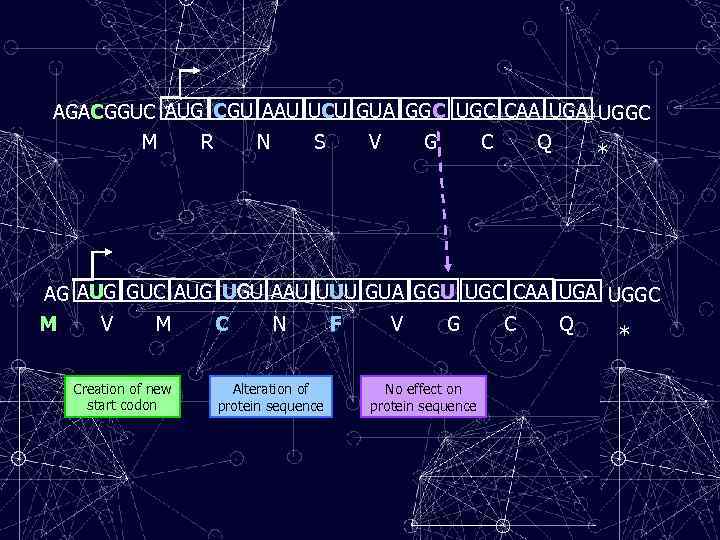 AGACGGUC AUG CGU AAU UCU GUA GGC UGC CAA UGGC M R N S V G C Q * AG AUG GUC AUG UGU AAU UUU GUA GGU UGC CAA UGGC M V M C N F V G C Q * Creation of new start codon Alteration of protein sequence No effect on protein sequence
AGACGGUC AUG CGU AAU UCU GUA GGC UGC CAA UGGC M R N S V G C Q * AG AUG GUC AUG UGU AAU UUU GUA GGU UGC CAA UGGC M V M C N F V G C Q * Creation of new start codon Alteration of protein sequence No effect on protein sequence
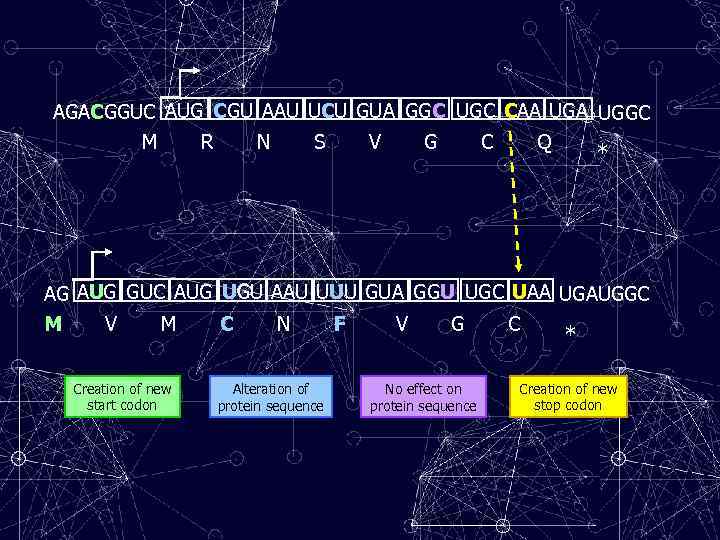 AGACGGUC AUG CGU AAU UCU GUA GGC UGC CAA UGGC M R N S V G C Q * AG AUG GUC AUG UGU AAU UUU GUA GGU UGC UAA UGAUGGC M V M Creation of new start codon C N Alteration of protein sequence F V G No effect on protein sequence C * Creation of new stop codon
AGACGGUC AUG CGU AAU UCU GUA GGC UGC CAA UGGC M R N S V G C Q * AG AUG GUC AUG UGU AAU UUU GUA GGU UGC UAA UGAUGGC M V M Creation of new start codon C N Alteration of protein sequence F V G No effect on protein sequence C * Creation of new stop codon
 Модификационное редактирование Substitution Editing
Модификационное редактирование Substitution Editing
 Модификационное редактирование • Дезаминирование C → U • Менее часто U - > C, G - > A, U - > A, ( у человека и мыши)
Модификационное редактирование • Дезаминирование C → U • Менее часто U - > C, G - > A, U - > A, ( у человека и мыши)
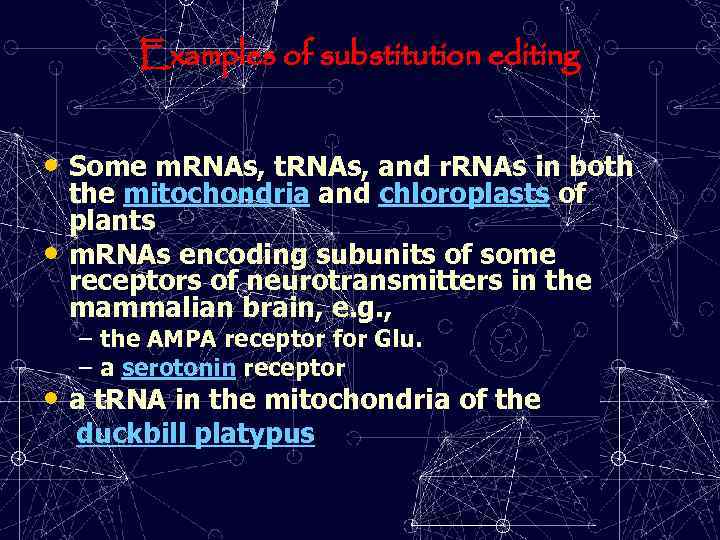 Examples of substitution editing • Some m. RNAs, t. RNAs, and r. RNAs in both • the mitochondria and chloroplasts of plants m. RNAs encoding subunits of some receptors of neurotransmitters in the mammalian brain, e. g. , – the AMPA receptor for Glu. – a serotonin receptor • a t. RNA in the mitochondria of the duckbill platypus
Examples of substitution editing • Some m. RNAs, t. RNAs, and r. RNAs in both • the mitochondria and chloroplasts of plants m. RNAs encoding subunits of some receptors of neurotransmitters in the mammalian brain, e. g. , – the AMPA receptor for Glu. – a serotonin receptor • a t. RNA in the mitochondria of the duckbill platypus
 Duckbill platypus
Duckbill platypus
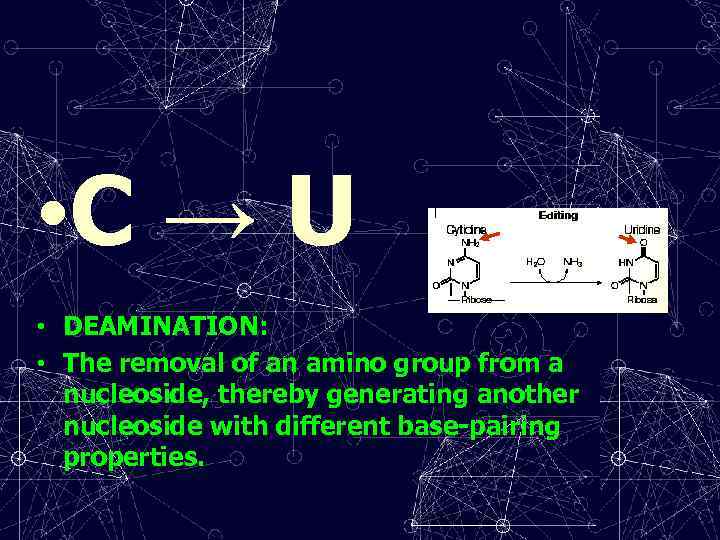 • C → U • DEAMINATION: • The removal of an amino group from a nucleoside, thereby generating another nucleoside with different base-pairing properties.
• C → U • DEAMINATION: • The removal of an amino group from a nucleoside, thereby generating another nucleoside with different base-pairing properties.
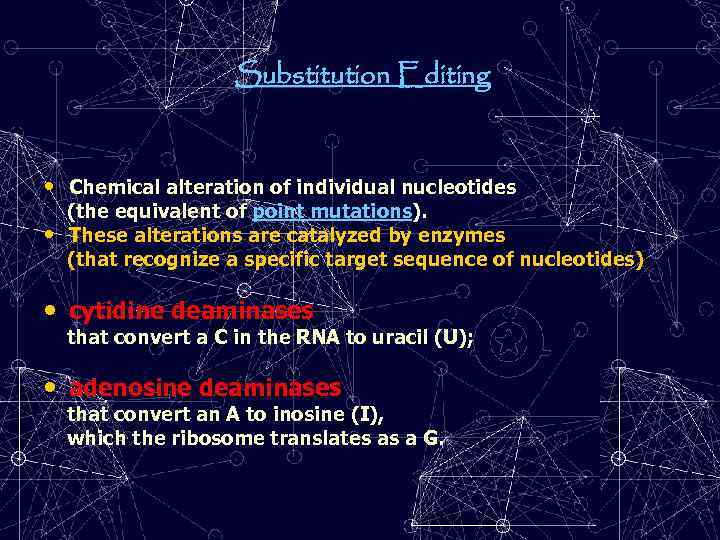 Substitution Editing • Chemical alteration of individual nucleotides (the equivalent of point mutations). • These alterations are catalyzed by enzymes (that recognize a specific target sequence of nucleotides) • cytidine deaminases that convert a C in the RNA to uracil (U); • adenosine deaminases that convert an A to inosine (I), which the ribosome translates as a G.
Substitution Editing • Chemical alteration of individual nucleotides (the equivalent of point mutations). • These alterations are catalyzed by enzymes (that recognize a specific target sequence of nucleotides) • cytidine deaminases that convert a C in the RNA to uracil (U); • adenosine deaminases that convert an A to inosine (I), which the ribosome translates as a G.
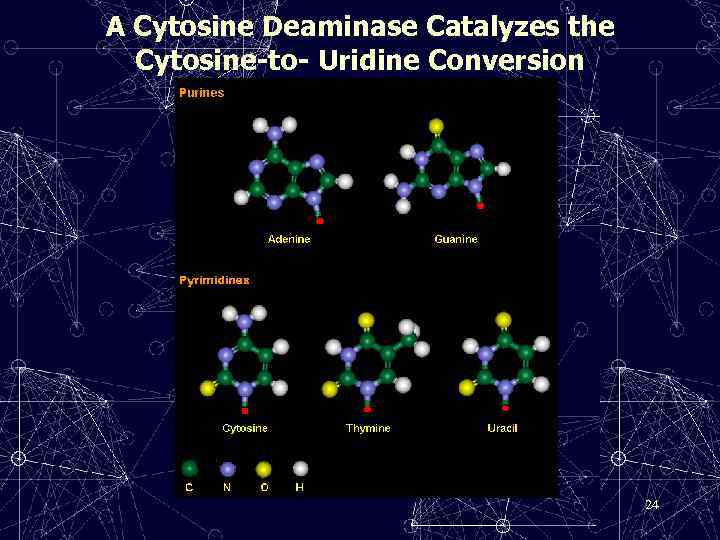 A Cytosine Deaminase Catalyzes the Cytosine-to- Uridine Conversion 24
A Cytosine Deaminase Catalyzes the Cytosine-to- Uridine Conversion 24
 APOB is a component of the plasma lipoproteins and is crucial for the transport of cholesterol and of triglycerides in the plasma. There are two forms of APOB: APOB 100 and the shorter APOB 48 isoform, which results from the DEAMINATION of C → U at nucleotide position 6666 (C 6666) in the APOB m. RNA, which causes the change of a glutamine to a translational stop codon. .
APOB is a component of the plasma lipoproteins and is crucial for the transport of cholesterol and of triglycerides in the plasma. There are two forms of APOB: APOB 100 and the shorter APOB 48 isoform, which results from the DEAMINATION of C → U at nucleotide position 6666 (C 6666) in the APOB m. RNA, which causes the change of a glutamine to a translational stop codon. .
 C -> U RNA Editing: APOB The APOB 100 isoform is synthesized only in the liver and is used to assemble the very-low-density lipoprotein (VLDL) that is necessary for the transport of endogenously synthesized TRIGLYCERIDES and cholesterol. VLDL is metabolized to intermediate-density lipoprotein(IDL) and subsequently to LOW-DENSITY LIPOPROTEIN (LDL). The carboxyl terminus of APOB 100 interacts with the LDL receptor, and LDL is removed from the circulation. Such a functional interaction is medically important, as high levels of LDL cholesterol is one of the main risk factors for coronary heart disease. Conversely, APOB 48, which lacks the carboxyl terminus of APOB 100, is generated in the small intestine and is necessary for the synthesis and secretion of CHYLOMICRONS In humans, this editing event occurs in the small intestine b ut not in the liver.
C -> U RNA Editing: APOB The APOB 100 isoform is synthesized only in the liver and is used to assemble the very-low-density lipoprotein (VLDL) that is necessary for the transport of endogenously synthesized TRIGLYCERIDES and cholesterol. VLDL is metabolized to intermediate-density lipoprotein(IDL) and subsequently to LOW-DENSITY LIPOPROTEIN (LDL). The carboxyl terminus of APOB 100 interacts with the LDL receptor, and LDL is removed from the circulation. Such a functional interaction is medically important, as high levels of LDL cholesterol is one of the main risk factors for coronary heart disease. Conversely, APOB 48, which lacks the carboxyl terminus of APOB 100, is generated in the small intestine and is necessary for the synthesis and secretion of CHYLOMICRONS In humans, this editing event occurs in the small intestine b ut not in the liver.
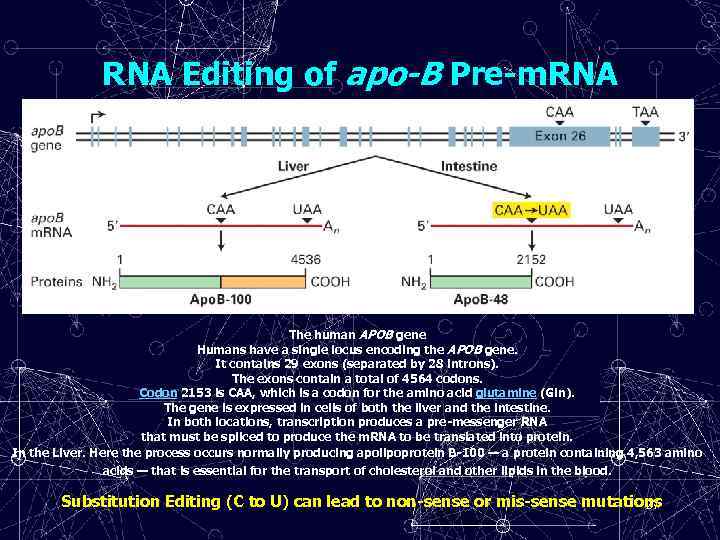 RNA Editing of apo-B Pre-m. RNA The human APOB gene Humans have a single locus encoding the APOB gene. It contains 29 exons (separated by 28 introns). The exons contain a total of 4564 codons. Codon 2153 is CAA, which is a codon for the amino acid glutamine (Gln). The gene is expressed in cells of both the liver and the intestine. In both locations, transcription produces a pre-messenger RNA that must be spliced to produce the m. RNA to be translated into protein. In the Liver. Here the process occurs normally producing apolipoprotein B-100 — a protein containing 4, 563 amino acids — that is essential for the transport of cholesterol and other lipids in the blood. Substitution Editing (C to U) can lead to non-sense or mis-sense mutations 27
RNA Editing of apo-B Pre-m. RNA The human APOB gene Humans have a single locus encoding the APOB gene. It contains 29 exons (separated by 28 introns). The exons contain a total of 4564 codons. Codon 2153 is CAA, which is a codon for the amino acid glutamine (Gln). The gene is expressed in cells of both the liver and the intestine. In both locations, transcription produces a pre-messenger RNA that must be spliced to produce the m. RNA to be translated into protein. In the Liver. Here the process occurs normally producing apolipoprotein B-100 — a protein containing 4, 563 amino acids — that is essential for the transport of cholesterol and other lipids in the blood. Substitution Editing (C to U) can lead to non-sense or mis-sense mutations 27
 Editosome
Editosome
 APOBEC-1 • Фермент, дезаминирующий C в U • Sequence specific subunit of Editosome
APOBEC-1 • Фермент, дезаминирующий C в U • Sequence specific subunit of Editosome
 Editing of Apolipoprotein B The Editosome A cytidine deaminase activity – apobec (apo. B m. RNA) editing enzyme catalytic subunit) A cytidine deaminase activity is involved – apobec (apo. B m. RNA editing enzyme catalytic subunit) Both recognize sequences flanking the C to be edited
Editing of Apolipoprotein B The Editosome A cytidine deaminase activity – apobec (apo. B m. RNA) editing enzyme catalytic subunit) A cytidine deaminase activity is involved – apobec (apo. B m. RNA editing enzyme catalytic subunit) Both recognize sequences flanking the C to be edited
 APOBEC-1 • A family of enzymes known as APOBEC-1 • • • related proteins (ARPs) whose function is to change the nucleotide sequence of RNA or DNA. Such 'power' would need to be precisely 'tuned' so that it only affected select nucleic acids sequences, in certain cells at vary particular times. The enzyme expression and activity are highly regulated
APOBEC-1 • A family of enzymes known as APOBEC-1 • • • related proteins (ARPs) whose function is to change the nucleotide sequence of RNA or DNA. Such 'power' would need to be precisely 'tuned' so that it only affected select nucleic acids sequences, in certain cells at vary particular times. The enzyme expression and activity are highly regulated
 Cytidine deamination in vitro and in vivo “Editosome” Editosome assembly in cells or in vitro requires a complex protein composition with an aggregate size of 27 S. In vitro editing activity requires a - homodimer of APOBEC-1 (catalytic subunit for cytidine-to-uridine editing of apolipoprotein B m. RNA-1), and a - 65 k. Da RNA binding protein, ACF (APOBEC-1 complementation factor). APOBEC-1 demonstrates a weak and non-specific RNA binding activity to AU-rich apo. B sequences and alone could not edit apo. B m. RNA. Editing site specificity and RNA editing activity are imparted upon APOBEC-1 through its interactions with ACF and potentially other auxiliary proteins that bind to APOBEC-1 and/or apo. B m. RNA and modulate the efficiency of the editing reaction.
Cytidine deamination in vitro and in vivo “Editosome” Editosome assembly in cells or in vitro requires a complex protein composition with an aggregate size of 27 S. In vitro editing activity requires a - homodimer of APOBEC-1 (catalytic subunit for cytidine-to-uridine editing of apolipoprotein B m. RNA-1), and a - 65 k. Da RNA binding protein, ACF (APOBEC-1 complementation factor). APOBEC-1 demonstrates a weak and non-specific RNA binding activity to AU-rich apo. B sequences and alone could not edit apo. B m. RNA. Editing site specificity and RNA editing activity are imparted upon APOBEC-1 through its interactions with ACF and potentially other auxiliary proteins that bind to APOBEC-1 and/or apo. B m. RNA and modulate the efficiency of the editing reaction.

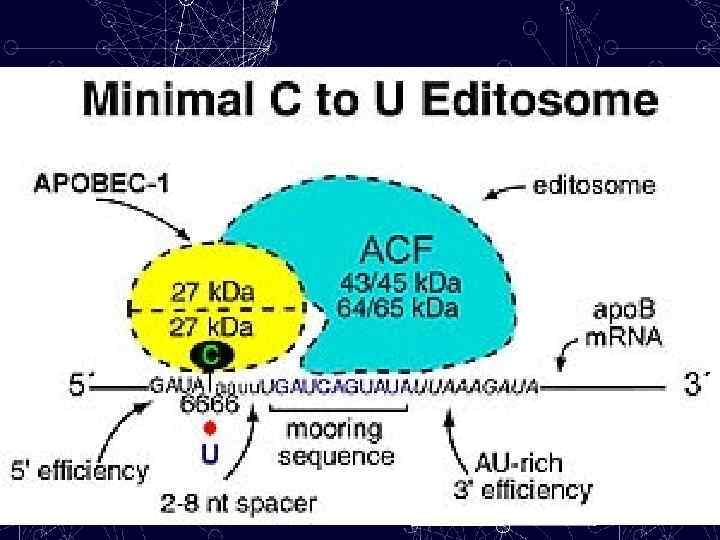
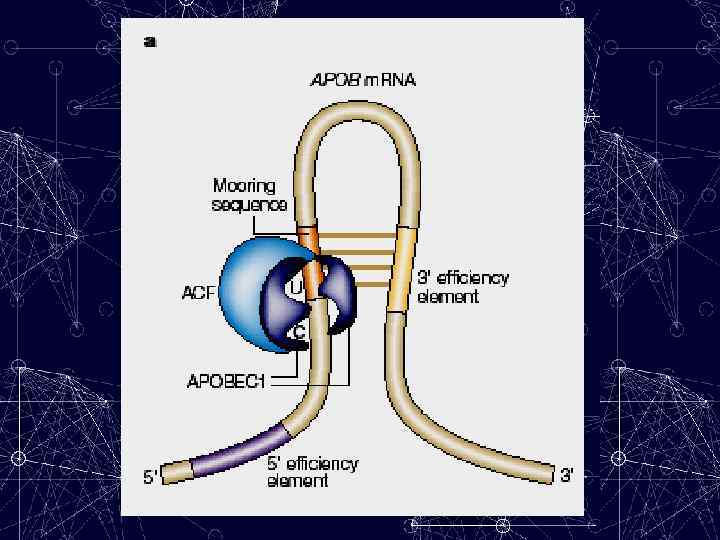
 ACF APOBEC-1 MOORING SEQUENCE An 11 -nucleotide motif, which is located 3′ of the cytidine that is edited in APOB m. RNA. Both APOBEC-1 and ACF bind to this motif.
ACF APOBEC-1 MOORING SEQUENCE An 11 -nucleotide motif, which is located 3′ of the cytidine that is edited in APOB m. RNA. Both APOBEC-1 and ACF bind to this motif.
 Mechanism of Mooring: C → U editing The human APOB 100 locus spans 43 kb, has 29 exons and encodes one of the largest known proteins (4, 536 amino acids). The editing site lies in exon 26, which, at 7, 572 nucleotides, is one of the largest known exons. Although the m. RNA is 14 kb, editing occurs with exact precision at C 6666 and requires both C 6666 - trans-acting factors and - cis-acting sequence elements that surround the cytosine that is edited. A mooring sequence of 11 nucleotides that is situated downstream of the editing site and is separated from the target cytosine by a spacer element that is usually four nucleotides long. Mooring- швартовка (англ)
Mechanism of Mooring: C → U editing The human APOB 100 locus spans 43 kb, has 29 exons and encodes one of the largest known proteins (4, 536 amino acids). The editing site lies in exon 26, which, at 7, 572 nucleotides, is one of the largest known exons. Although the m. RNA is 14 kb, editing occurs with exact precision at C 6666 and requires both C 6666 - trans-acting factors and - cis-acting sequence elements that surround the cytosine that is edited. A mooring sequence of 11 nucleotides that is situated downstream of the editing site and is separated from the target cytosine by a spacer element that is usually four nucleotides long. Mooring- швартовка (англ)
 A mooring sequence The site is flanked by - 3′ and 5′ “efficiency elements” (sequences that have been identified that are required for efficient editing), and there is an additional requirement for A+U-rich bulk RNA. The mooring sequence and the 3′ efficiency element form a double-stranded (ds) stem that is predicted to position the edited cytosine in a favorable configuration for deamination.
A mooring sequence The site is flanked by - 3′ and 5′ “efficiency elements” (sequences that have been identified that are required for efficient editing), and there is an additional requirement for A+U-rich bulk RNA. The mooring sequence and the 3′ efficiency element form a double-stranded (ds) stem that is predicted to position the edited cytosine in a favorable configuration for deamination.
 A -> I
A -> I
 A -> I The first example of A to I editing in an m. RNA was found in the mammalian brain, in transcripts of the gene encoding the ionotropic glutamate receptor subunit, Glu. R -B. Other examples have appeared in numerous signaling components of the nervous systems of vertebrates and invertebrates.
A -> I The first example of A to I editing in an m. RNA was found in the mammalian brain, in transcripts of the gene encoding the ionotropic glutamate receptor subunit, Glu. R -B. Other examples have appeared in numerous signaling components of the nervous systems of vertebrates and invertebrates.
 A → I RNA editing - ADAR. The enzymes that deaminate adenosine to inosine in ds. RNA are members of a family of Adenosine Deaminases that Act on RNA-ADAR.
A → I RNA editing - ADAR. The enzymes that deaminate adenosine to inosine in ds. RNA are members of a family of Adenosine Deaminases that Act on RNA-ADAR.
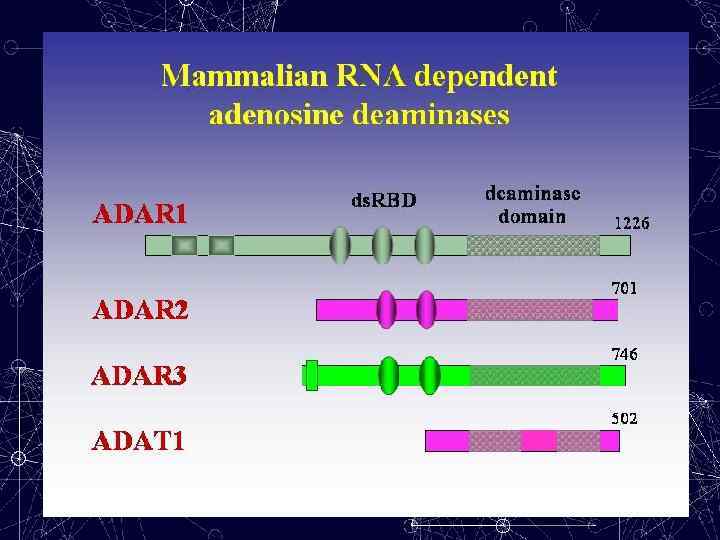
 A → I RNA editing In humans, there are three members of this family -ADAR 1, ADAR 2 and ADAR 3, the names reflecting the order in which they were identified. ADAR 1 and ADAR 2, which are almost ubiquitously expressed, can convert adenosine to inosine, both in long ds. RNA duplexes and in specific pre-m. RNAs. Several isoforms of these two enzymes exist that vary in their editing activity.
A → I RNA editing In humans, there are three members of this family -ADAR 1, ADAR 2 and ADAR 3, the names reflecting the order in which they were identified. ADAR 1 and ADAR 2, which are almost ubiquitously expressed, can convert adenosine to inosine, both in long ds. RNA duplexes and in specific pre-m. RNAs. Several isoforms of these two enzymes exist that vary in their editing activity.
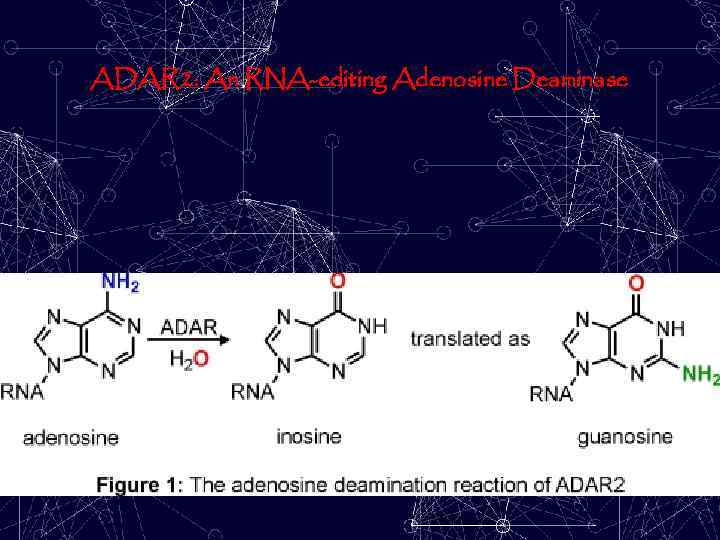 ADAR 2: An RNA-editing Adenosine Deaminase
ADAR 2: An RNA-editing Adenosine Deaminase
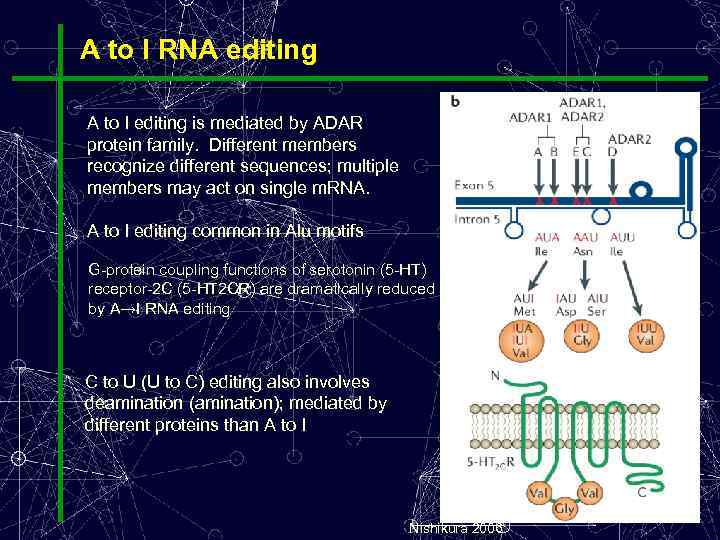 A to I RNA editing A to I editing is mediated by ADAR protein family. Different members recognize different sequences; multiple members may act on single m. RNA. A to I editing common in Alu motifs G-protein coupling functions of serotonin (5 -HT) receptor-2 C (5 -HT 2 CR) are dramatically reduced by A→I RNA editing C to U (U to C) editing also involves deamination (amination); mediated by different proteins than A to I Nishikura 2006
A to I RNA editing A to I editing is mediated by ADAR protein family. Different members recognize different sequences; multiple members may act on single m. RNA. A to I editing common in Alu motifs G-protein coupling functions of serotonin (5 -HT) receptor-2 C (5 -HT 2 CR) are dramatically reduced by A→I RNA editing C to U (U to C) editing also involves deamination (amination); mediated by different proteins than A to I Nishikura 2006
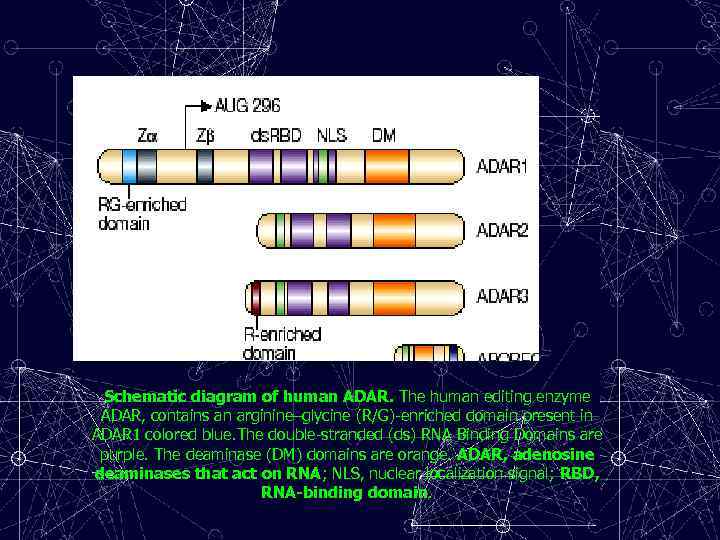 Schematic diagram of human ADAR. The human editing enzyme ADAR, contains an arginine–glycine (R/G)-enriched domain present in ADAR 1 colored blue. The double-stranded (ds) RNA Binding Domains are purple. The deaminase (DM) domains are orange. ADAR, adenosine deaminases that act on RNA; NLS, nuclear localization signal; RBD, RNA-binding domain.
Schematic diagram of human ADAR. The human editing enzyme ADAR, contains an arginine–glycine (R/G)-enriched domain present in ADAR 1 colored blue. The double-stranded (ds) RNA Binding Domains are purple. The deaminase (DM) domains are orange. ADAR, adenosine deaminases that act on RNA; NLS, nuclear localization signal; RBD, RNA-binding domain.
 Mechanism of ADAR editing ADAR proteins do not require cofactors to deaminate adenosine to inosine through HYDROLYTIC DEAMINATION. The ADARs are unique in that they recognize the adenosine to be edited not by a surrounding consensus sequence but by the structure of the duplex that is formed between the editing site and an editing site complementary sequence (ECS) that is usually located in a downstream intron. The ds. RNA-binding domains (ds. RBDs) found in ADARs mediate the binding to the duplex.
Mechanism of ADAR editing ADAR proteins do not require cofactors to deaminate adenosine to inosine through HYDROLYTIC DEAMINATION. The ADARs are unique in that they recognize the adenosine to be edited not by a surrounding consensus sequence but by the structure of the duplex that is formed between the editing site and an editing site complementary sequence (ECS) that is usually located in a downstream intron. The ds. RNA-binding domains (ds. RBDs) found in ADARs mediate the binding to the duplex.
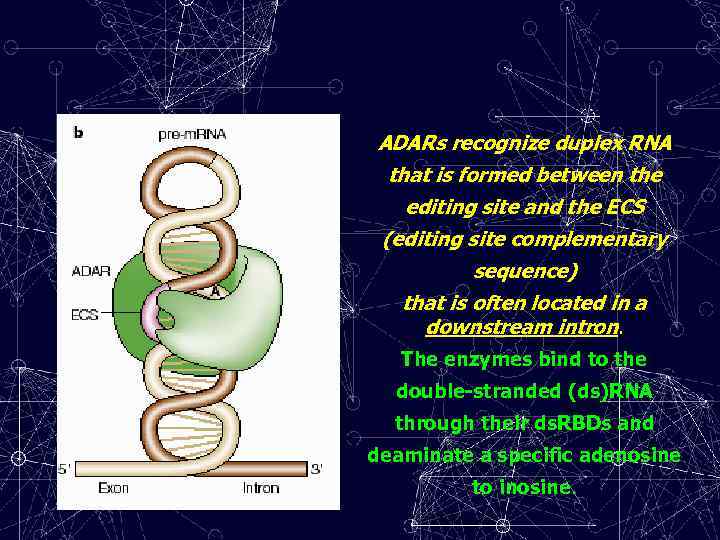 ADARs recognize duplex RNA that is formed between the editing site and the ECS (editing site complementary sequence) that is often located in a downstream intron. The enzymes bind to the double-stranded (ds)RNA through their ds. RBDs and deaminate a specific adenosine to inosine.
ADARs recognize duplex RNA that is formed between the editing site and the ECS (editing site complementary sequence) that is often located in a downstream intron. The enzymes bind to the double-stranded (ds)RNA through their ds. RBDs and deaminate a specific adenosine to inosine.
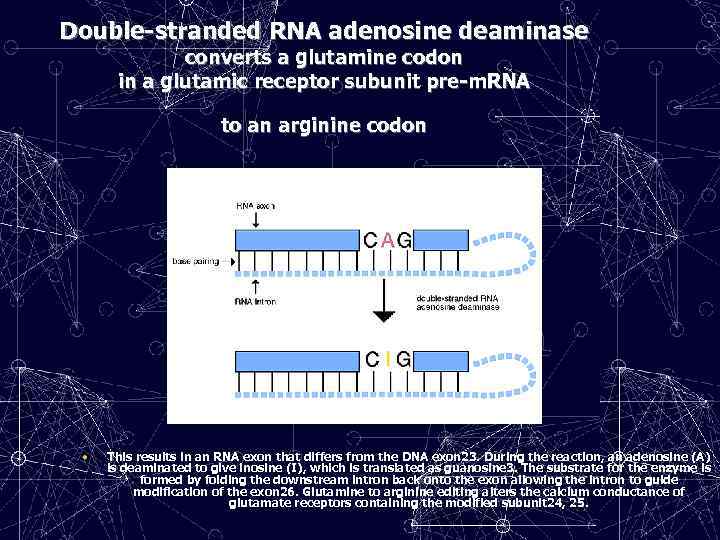 Double-stranded RNA adenosine deaminase converts a glutamine codon in a glutamic receptor subunit pre-m. RNA to an arginine codon • • T This results in an RNA exon that differs from the DNA exon 23. During the reaction, an adenosine (A) is deaminated to give inosine (I), which is translated as guanosine 3. The substrate for the enzyme is formed by folding the downstream intron back onto the exon allowing the intron to guide modification of the exon 26. Glutamine to arginine editing alters the calcium conductance of glutamate receptors containing the modified subunit 24, 25.
Double-stranded RNA adenosine deaminase converts a glutamine codon in a glutamic receptor subunit pre-m. RNA to an arginine codon • • T This results in an RNA exon that differs from the DNA exon 23. During the reaction, an adenosine (A) is deaminated to give inosine (I), which is translated as guanosine 3. The substrate for the enzyme is formed by folding the downstream intron back onto the exon allowing the intron to guide modification of the exon 26. Glutamine to arginine editing alters the calcium conductance of glutamate receptors containing the modified subunit 24, 25.
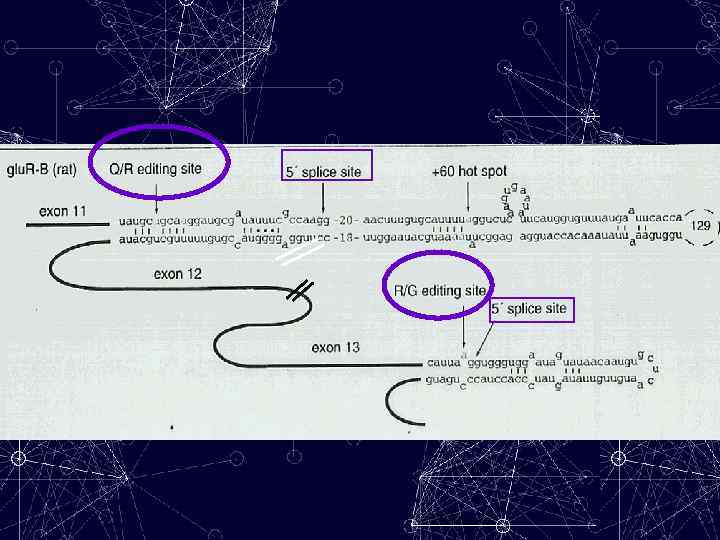
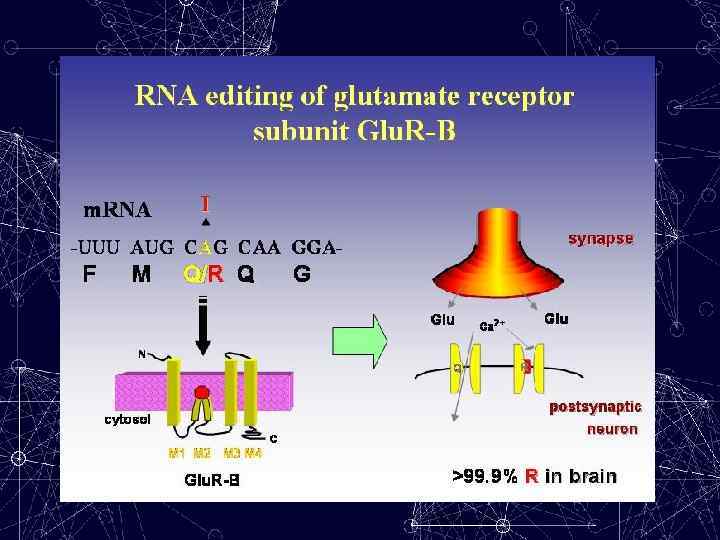
 Editing of a specific adenosine in a pre-m. RNA is usually not 100% efficient, one exception being the glutamine/arginine (Q/R) site in Glu. R-B. Deamination of a specific adenosine at this site changes the codon Q →R — a change that is crucial for the correct functioning of the receptor. Editing at this position in the Glu. R-B transcript controls the Ca 2+ permeability of heteromeric α-amino-3 hydroxy-5 methylisoxazole-4 -propionate (AMPA) receptors, which mediate fast-excitatory-synaptic transmission in the CNS. Transgenic mice that were unable to edit only at this position had epileptic seizures and died within three weeks of birth, presumably due to increased Ca 2+ permeability of the AMPA receptors.
Editing of a specific adenosine in a pre-m. RNA is usually not 100% efficient, one exception being the glutamine/arginine (Q/R) site in Glu. R-B. Deamination of a specific adenosine at this site changes the codon Q →R — a change that is crucial for the correct functioning of the receptor. Editing at this position in the Glu. R-B transcript controls the Ca 2+ permeability of heteromeric α-amino-3 hydroxy-5 methylisoxazole-4 -propionate (AMPA) receptors, which mediate fast-excitatory-synaptic transmission in the CNS. Transgenic mice that were unable to edit only at this position had epileptic seizures and died within three weeks of birth, presumably due to increased Ca 2+ permeability of the AMPA receptors.
 Functional Importance of EDITING Editing of the Glu. R-B Q/R site is a major determinant of the Ca 2+ permeability of multimeric Glu. R channels that incorporate the Glu. R -B subunit. Editing of the Glu. R-B, -C and -D R/G sites affects the rate of recovery from desensitization of Glu. R channels. Editing of the serotonin receptor 5 -HT 2 C results in greatly reduced efficiency of G-protein coupling in certain edited forms.
Functional Importance of EDITING Editing of the Glu. R-B Q/R site is a major determinant of the Ca 2+ permeability of multimeric Glu. R channels that incorporate the Glu. R -B subunit. Editing of the Glu. R-B, -C and -D R/G sites affects the rate of recovery from desensitization of Glu. R channels. Editing of the serotonin receptor 5 -HT 2 C results in greatly reduced efficiency of G-protein coupling in certain edited forms.
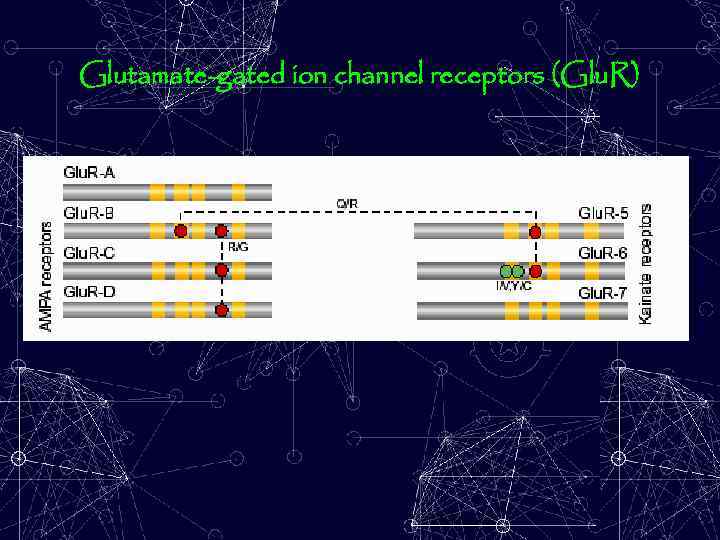 Glutamate-gated ion channel receptors (Glu. R)
Glutamate-gated ion channel receptors (Glu. R)
 Editing modifies the GABAA receptor subunit 3 Adenosine to inosine (A-to-I) pre-m. RNA editing by the ADAR enzyme family has the potential to increase the variety of the proteome. This editing by adenosine deamination is essential in mammals for a functional brain. J. OHLSON, J. S. PEDERSEN, D. HAUSSLER, and M. OHMAN RNA (2007), 13: 698– 703.
Editing modifies the GABAA receptor subunit 3 Adenosine to inosine (A-to-I) pre-m. RNA editing by the ADAR enzyme family has the potential to increase the variety of the proteome. This editing by adenosine deamination is essential in mammals for a functional brain. J. OHLSON, J. S. PEDERSEN, D. HAUSSLER, and M. OHMAN RNA (2007), 13: 698– 703.
 In mammals, RNA editing by site-selective adenosine deamination regulates key functional properties of neurotransmitter receptors in the CNS. Glutamate receptor subunit B is nearly 100% edited at one position (the QR-site), which is essential for normal receptor function. In mouse models, a slightly reduced rate of QR-site editing is associated with early onset epilepsy and premature death. It was found that in tissues from malignant human brain tumors, this editing position of glutamate receptor subunit B is substantially underedited compared with control tissues. Alterations in editing and alternative splicing of serotonin receptor 5 -HT 2 C transcripts were also seen. These changes correlate with a decrease in enzymatic activity of the editing enzyme ADAR 2, as deduced from analysis of ADAR 2 self-editing. This suggests a role for RNA editing in tumor progression and may provide a molecular model explaining the occurrence of epileptic seizures in association with malignant gliomas.
In mammals, RNA editing by site-selective adenosine deamination regulates key functional properties of neurotransmitter receptors in the CNS. Glutamate receptor subunit B is nearly 100% edited at one position (the QR-site), which is essential for normal receptor function. In mouse models, a slightly reduced rate of QR-site editing is associated with early onset epilepsy and premature death. It was found that in tissues from malignant human brain tumors, this editing position of glutamate receptor subunit B is substantially underedited compared with control tissues. Alterations in editing and alternative splicing of serotonin receptor 5 -HT 2 C transcripts were also seen. These changes correlate with a decrease in enzymatic activity of the editing enzyme ADAR 2, as deduced from analysis of ADAR 2 self-editing. This suggests a role for RNA editing in tumor progression and may provide a molecular model explaining the occurrence of epileptic seizures in association with malignant gliomas.
 Five adenosines within the coding sequence of the serotonin 2 C receptor (5 -HT 2 C) pre-m. RNA are converted to inosines by RNA editing (named A, B, C (E), C, and D sites). In human prefrontal cortex (PFC), the most abundant 5 HT 2 C m. RNA sequences result from editing at the A site, or from the editing combinations ACC, ABCD, and ABD. In suicide victims with a history of major depression, C site editing is significantly increased, D site editing is significantly decreased. Treatment of mice with the antidepressant drug fluoxetine (Prozac) causes changes in C and D site editing that are exactly opposite to those seen in suicide victims. Thus, one outcome of fluoxetine treatment may be to reverse the abnormalities in 5 -HT 2 C pre-m. RNA editing seen in depressed suicide victims.
Five adenosines within the coding sequence of the serotonin 2 C receptor (5 -HT 2 C) pre-m. RNA are converted to inosines by RNA editing (named A, B, C (E), C, and D sites). In human prefrontal cortex (PFC), the most abundant 5 HT 2 C m. RNA sequences result from editing at the A site, or from the editing combinations ACC, ABCD, and ABD. In suicide victims with a history of major depression, C site editing is significantly increased, D site editing is significantly decreased. Treatment of mice with the antidepressant drug fluoxetine (Prozac) causes changes in C and D site editing that are exactly opposite to those seen in suicide victims. Thus, one outcome of fluoxetine treatment may be to reverse the abnormalities in 5 -HT 2 C pre-m. RNA editing seen in depressed suicide victims.
 Among the edited m. RNAs are those that encode: - the glutamate-gated ion channel receptors (Glu. R); - the serotonin (5 -HT 2 C) receptor; - the voltage-gated calcium and sodium channels, and - glutamate-gated chloride channel, and the - voltage-gated potassium channel pre-m. RNA. Editing in these transcripts changes the coding potential of the RNA so that different protein isoforms are generated.
Among the edited m. RNAs are those that encode: - the glutamate-gated ion channel receptors (Glu. R); - the serotonin (5 -HT 2 C) receptor; - the voltage-gated calcium and sodium channels, and - glutamate-gated chloride channel, and the - voltage-gated potassium channel pre-m. RNA. Editing in these transcripts changes the coding potential of the RNA so that different protein isoforms are generated.
 In A to I Editing— Splicing occurs AFTER Editing because the double-stranded RNA must be formed between the upstream Exon and the downstream Intron. RNA editing may even influence splicing
In A to I Editing— Splicing occurs AFTER Editing because the double-stranded RNA must be formed between the upstream Exon and the downstream Intron. RNA editing may even influence splicing
 RNA editing can regulate Alternative splicing
RNA editing can regulate Alternative splicing
 RNA editing can regulate splicing by targeting the adenosines involved in splicing. Rat ADAR 2 edits own pre-m. RNA such that a 3′ splice site is generated, the use of which adds 47 nucleotides to rat ADAR 2 and changes the predicted open reading frame. Internal translation initiation then leads to the production of an active enzyme, but lower protein levels are expressed, because the internal translation initiation is relatively inefficient. Self-editing results in a lower ADAR 2 concentration, so this process can be thought of as a negative autoregulatory mechanism whereby rat ADAR 2 can regulate protein expression by changing a downstream splice site. AUG
RNA editing can regulate splicing by targeting the adenosines involved in splicing. Rat ADAR 2 edits own pre-m. RNA such that a 3′ splice site is generated, the use of which adds 47 nucleotides to rat ADAR 2 and changes the predicted open reading frame. Internal translation initiation then leads to the production of an active enzyme, but lower protein levels are expressed, because the internal translation initiation is relatively inefficient. Self-editing results in a lower ADAR 2 concentration, so this process can be thought of as a negative autoregulatory mechanism whereby rat ADAR 2 can regulate protein expression by changing a downstream splice site. AUG
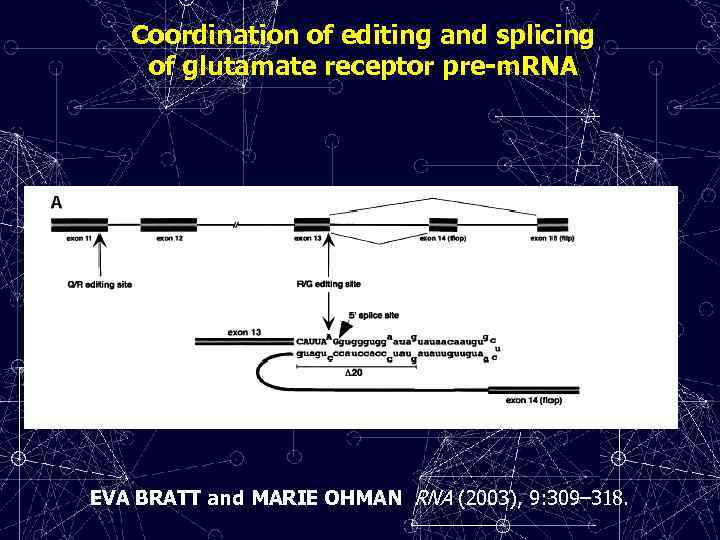 Coordination of editing and splicing of glutamate receptor pre-m. RNA EVA BRATT and MARIE OHMAN RNA (2003), 9: 309– 318.
Coordination of editing and splicing of glutamate receptor pre-m. RNA EVA BRATT and MARIE OHMAN RNA (2003), 9: 309– 318.
 Why bother with RNA editing? Why not just change a nucleotide through mutation? - One intrinsic advantage of editing a nucleotide over having change ‘hard-wired’ into the genome through mutation is the regulation of the degree to which a coding position is modified within m. RNAs. - Certain pre-m. RNA editing sites vary greatly in the frequency with which their editing is detected in vivo, ranging from a few percent to nearly 100%. - Editing introduces levels of expression intermediate to the usual genetic variation (i. e. 0, 1 or 2 copies), possibly conferring selective advantage. In addition, because the known ADAR target m. RNAs are commonly edited at multiple positions independently the combinatorial effect of editing greatly increases the number of protein products that can be generated from an edited gene.
Why bother with RNA editing? Why not just change a nucleotide through mutation? - One intrinsic advantage of editing a nucleotide over having change ‘hard-wired’ into the genome through mutation is the regulation of the degree to which a coding position is modified within m. RNAs. - Certain pre-m. RNA editing sites vary greatly in the frequency with which their editing is detected in vivo, ranging from a few percent to nearly 100%. - Editing introduces levels of expression intermediate to the usual genetic variation (i. e. 0, 1 or 2 copies), possibly conferring selective advantage. In addition, because the known ADAR target m. RNAs are commonly edited at multiple positions independently the combinatorial effect of editing greatly increases the number of protein products that can be generated from an edited gene.
 Stephen William Hawking Стивен Уильям Хокинг один из наиболее влиятельных и известных физиков-теоретиков нашего времени
Stephen William Hawking Стивен Уильям Хокинг один из наиболее влиятельных и известных физиков-теоретиков нашего времени




 Боковой (латеральный) амиотрофический склероз Болезнь моторных нейронов, болезнь Шарко , болезнь Лу Ге рига — медленно прогрессирующее, неизлечимое дегенеративное заболевание центральной нервной системы неизвестной этиологии, при котором происходит поражение как верхних (моторная кора головного мозга) так и нижних (передние рога спинного мозга и ядра черепно-мозговых нервов) двигательных нейронов, что приводит к параличам и последующей атрофии мышц. Характеризуется прогрессирующим поражением двигательных нейронов, сопровождаемым параличом (парезом) конечностей и атрофией мышц. Смерть наступает от инфекций дыхательных путей или отказа дыхательной мускулатуры.
Боковой (латеральный) амиотрофический склероз Болезнь моторных нейронов, болезнь Шарко , болезнь Лу Ге рига — медленно прогрессирующее, неизлечимое дегенеративное заболевание центральной нервной системы неизвестной этиологии, при котором происходит поражение как верхних (моторная кора головного мозга) так и нижних (передние рога спинного мозга и ядра черепно-мозговых нервов) двигательных нейронов, что приводит к параличам и последующей атрофии мышц. Характеризуется прогрессирующим поражением двигательных нейронов, сопровождаемым параличом (парезом) конечностей и атрофией мышц. Смерть наступает от инфекций дыхательных путей или отказа дыхательной мускулатуры.
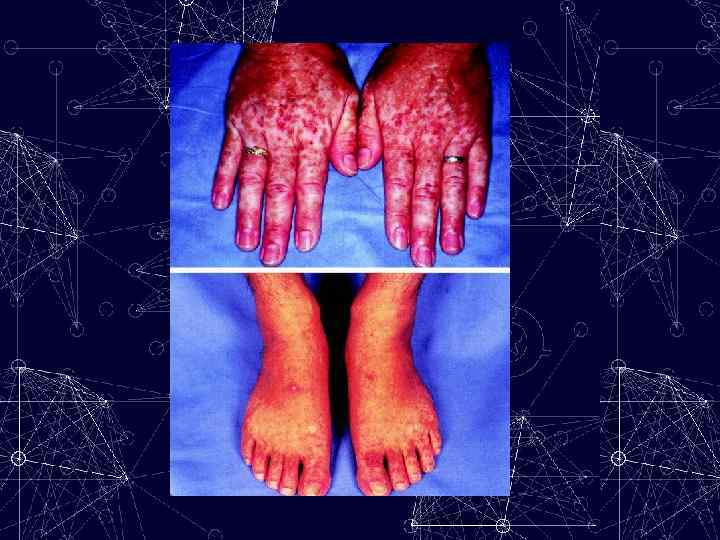
 Defects in RNA editing Are associated with some human cancers as well as with Amylotrophic lateral sclerosis Kwak S, Kawahara Y. Deficient RNA editing of Glu. R 2 and neuronal death in amyotropic lateral sclerosis. J Mol Med 2005; 83(2): 110 -20. Morabito MV, Emeson RB. RNA editing as a therapeutic target for CNS disorders. Neuropsychopharmacology Reviews (2009) 34, 246;
Defects in RNA editing Are associated with some human cancers as well as with Amylotrophic lateral sclerosis Kwak S, Kawahara Y. Deficient RNA editing of Glu. R 2 and neuronal death in amyotropic lateral sclerosis. J Mol Med 2005; 83(2): 110 -20. Morabito MV, Emeson RB. RNA editing as a therapeutic target for CNS disorders. Neuropsychopharmacology Reviews (2009) 34, 246;
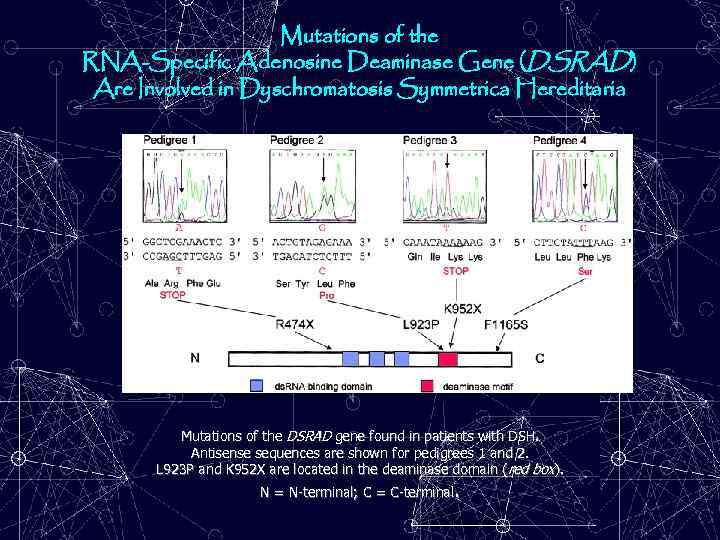 Mutations of the RNA-Specific Adenosine Deaminase Gene (DSRAD) Are Involved in Dyschromatosis Symmetrica Hereditaria Mutations of the DSRAD gene found in patients with DSH. Antisense sequences are shown for pedigrees 1 and 2. L 923 P and K 952 X are located in the deaminase domain (red box). N = N-terminal; C = C-terminal.
Mutations of the RNA-Specific Adenosine Deaminase Gene (DSRAD) Are Involved in Dyschromatosis Symmetrica Hereditaria Mutations of the DSRAD gene found in patients with DSH. Antisense sequences are shown for pedigrees 1 and 2. L 923 P and K 952 X are located in the deaminase domain (red box). N = N-terminal; C = C-terminal.
 Инсерционное редактирование РНК
Инсерционное редактирование РНК
 • Вставка (или удаление) U в однонитевые РНК (Physarium и трипаносомы) • РНК становится длиннее, чем транскрипт • В трипаносомах м. РНК становится в 2 раза длиннее транскрипта
• Вставка (или удаление) U в однонитевые РНК (Physarium и трипаносомы) • РНК становится длиннее, чем транскрипт • В трипаносомах м. РНК становится в 2 раза длиннее транскрипта
 Insertion/Deletion Editing • Several genes encoded in the mitochondrial • • • DNA of Trypanosoma brucei (the cause of sleeping sickness in humans) encode transcripts that must be edited to make the m. RNA molecules that will be translated into protein. • Example: the gene for one • of the subunits of cytochrome c oxidase
Insertion/Deletion Editing • Several genes encoded in the mitochondrial • • • DNA of Trypanosoma brucei (the cause of sleeping sickness in humans) encode transcripts that must be edited to make the m. RNA molecules that will be translated into protein. • Example: the gene for one • of the subunits of cytochrome c oxidase
 RNA editing is required for creation of translatable m. RNAs in mitochondria of all kinetoplastids * * * * ** * * ** **** * ** ** ** * * * ** * * ** ** *** T. brucei COIII edited m. RNA sequence Black = encoded nucleotides; u = U’s added by editing; * = encoded U’s deleted by editing
RNA editing is required for creation of translatable m. RNAs in mitochondria of all kinetoplastids * * * * ** * * ** **** * ** ** ** * * * ** * * ** ** *** T. brucei COIII edited m. RNA sequence Black = encoded nucleotides; u = U’s added by editing; * = encoded U’s deleted by editing

 Editing Mechanism • Post-transcriptional • Guide RNAs (g. RNAs) direct editing – g. RNAs are small and complementary to portions of the edited m. RNA – Base-pairing of g. RNA with unedited RNA gives mismatched regions, which are recognized by the editing machinery – Machinery includes an Endonuclease, a Terminal Uridylyl Transferase (TUTase), and a RNA ligase • Editing is directional, from 3’ to 5’
Editing Mechanism • Post-transcriptional • Guide RNAs (g. RNAs) direct editing – g. RNAs are small and complementary to portions of the edited m. RNA – Base-pairing of g. RNA with unedited RNA gives mismatched regions, which are recognized by the editing machinery – Machinery includes an Endonuclease, a Terminal Uridylyl Transferase (TUTase), and a RNA ligase • Editing is directional, from 3’ to 5’
 g. RNA • Guide RNA`s are tiny RNA`s (50 -70 nt) that guide the Editosomes through base-pairing with the m. RNA`s in editing regions. • They are complimentary to small segments of the m. RNA and function as templates.
g. RNA • Guide RNA`s are tiny RNA`s (50 -70 nt) that guide the Editosomes through base-pairing with the m. RNA`s in editing regions. • They are complimentary to small segments of the m. RNA and function as templates.
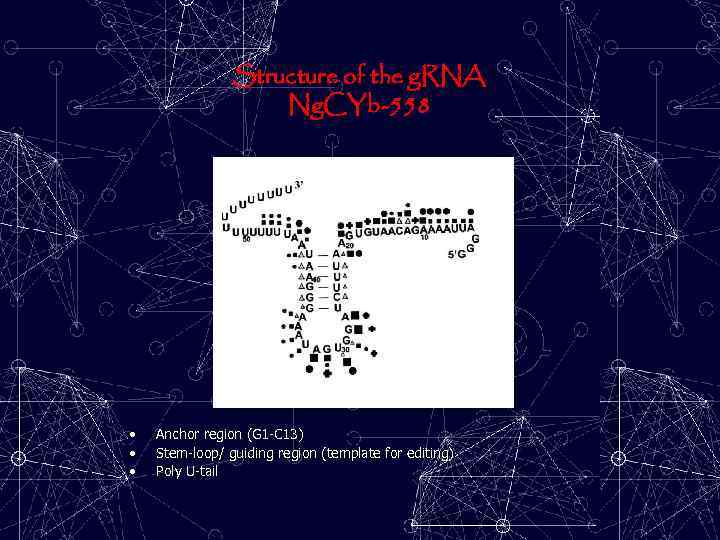 Structure of the g. RNA Ng. CYb-558 • • • Anchor region (G 1 -C 13) Stem-loop/ guiding region (template for editing) Poly U-tail
Structure of the g. RNA Ng. CYb-558 • • • Anchor region (G 1 -C 13) Stem-loop/ guiding region (template for editing) Poly U-tail
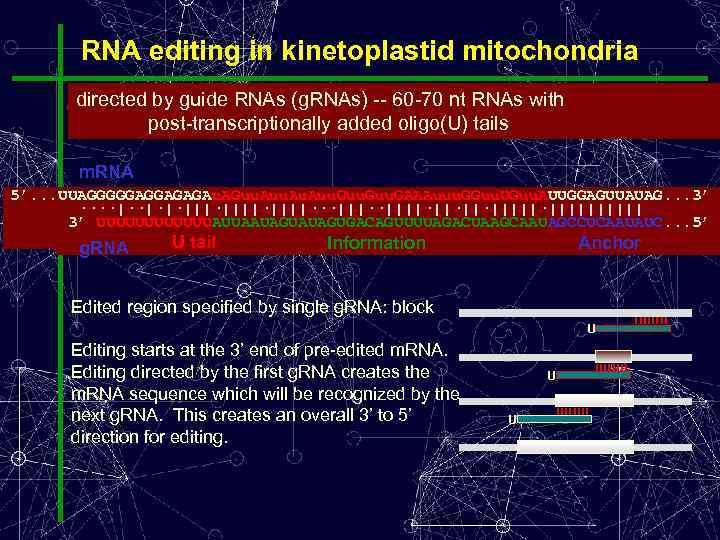 RNA editing in kinetoplastid mitochondria directed by guide RNAs (g. RNAs) -- 60 -70 nt RNAs with post-transcriptionally added oligo(U) tails m. RNA 5’. . . UUAGGGGGAGGAGAGAu. AGuu. Au. Auu. Guu. GAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . 3’. . . GUUAGGGGGAGGAGAGAAGuu. Au. Auu. Guu. GAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . AGGUUAGGGGGAGGAGAGAAGAuu. Guu. GAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . AAAGGUUAGGGGGAGGAGAGAAGAAu. Auu. Guu. GAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . GAAAGGUUAGGGGGAGGAGAGAAGAAAuu. Guu. GAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . AGGAAAGGUUAGGGGGAGGAGAGAAGAAAGuu. GAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . GCAGGAAAGGUUAGGGGGAGGAGAGAAGAAAGGuu. GAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . GAGCAGGAAAGGUUAGGGGGAGGAGAGAAGAAAGGGAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . AAAGAGCAGGAAAGGUUAGGGGGAGGAGAGAAGAAAGGuu. UGuu. AUUGGAGUUAUAG. . . AGAGCAGGAAAGGUUAGGGGGAGGAGAGAAGAAAGGGAAAGUUGuu. AUUGGAGUUAUAG. . . AAAGAGCAGGAAAGGUUAGGGGGAGGAGAGAAGAAAGGGAAAGUUGUGuu. AUUGGAGUUAUAG. . . AAAGAGCAGGAAAGGUUAGGGGGAGGAGAGAAGAAAGGGAAAGUUGUGAUUGGAGUUAUAG. . . ····|··|·|·||||·||||···||||·||·||·|||||||||| ||·||||···|||··||||·||·||·|||||·|||||||||| ||··||||·||·||·|||||·|||||||||| ·|||||||||| ||·|||||||||| 3’ UUUUUUAUUAAUAGUGACAGUUUUAGACUAAGCAAUAGCCUCAAUAUC. . . 5’ UUAAUAGUGACAGUUUUAGACUAAGCAAUAGCCUCAAUAUC. . . UAGUAUAGUGACAGUUUUAGACUAAGCAAUAGCCUCAAUAUC. . . GCAAUAGCCUCAAUAUC. . . UUUUUUUUUUUUAUUAAUAG Information UUUUUUUUUUUUAUUAAUAGUAUAGUGACAG UUUUUUUUUUUUAUUAAUAGUAUAGUGACAGUUUUAGACUAAGCAA UUUUUUAUUAAUAGUGACAGUUUUAGACUAAGCAAU Anchor U tail g. RNA Edited region specified by single g. RNA: block U Editing starts at the 3’ end of pre-edited m. RNA. Editing directed by the first g. RNA creates the m. RNA sequence which will be recognized by the next g. RNA. This creates an overall 3’ to 5’ direction for editing. U U
RNA editing in kinetoplastid mitochondria directed by guide RNAs (g. RNAs) -- 60 -70 nt RNAs with post-transcriptionally added oligo(U) tails m. RNA 5’. . . UUAGGGGGAGGAGAGAu. AGuu. Au. Auu. Guu. GAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . 3’. . . GUUAGGGGGAGGAGAGAAGuu. Au. Auu. Guu. GAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . AGGUUAGGGGGAGGAGAGAAGAuu. Guu. GAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . AAAGGUUAGGGGGAGGAGAGAAGAAu. Auu. Guu. GAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . GAAAGGUUAGGGGGAGGAGAGAAGAAAuu. Guu. GAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . AGGAAAGGUUAGGGGGAGGAGAGAAGAAAGuu. GAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . GCAGGAAAGGUUAGGGGGAGGAGAGAAGAAAGGuu. GAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . GAGCAGGAAAGGUUAGGGGGAGGAGAGAAGAAAGGGAAAuuu. GGuu. UGuu. AUUGGAGUUAUAG. . . AAAGAGCAGGAAAGGUUAGGGGGAGGAGAGAAGAAAGGuu. UGuu. AUUGGAGUUAUAG. . . AGAGCAGGAAAGGUUAGGGGGAGGAGAGAAGAAAGGGAAAGUUGuu. AUUGGAGUUAUAG. . . AAAGAGCAGGAAAGGUUAGGGGGAGGAGAGAAGAAAGGGAAAGUUGUGuu. AUUGGAGUUAUAG. . . AAAGAGCAGGAAAGGUUAGGGGGAGGAGAGAAGAAAGGGAAAGUUGUGAUUGGAGUUAUAG. . . ····|··|·|·||||·||||···||||·||·||·|||||||||| ||·||||···|||··||||·||·||·|||||·|||||||||| ||··||||·||·||·|||||·|||||||||| ·|||||||||| ||·|||||||||| 3’ UUUUUUAUUAAUAGUGACAGUUUUAGACUAAGCAAUAGCCUCAAUAUC. . . 5’ UUAAUAGUGACAGUUUUAGACUAAGCAAUAGCCUCAAUAUC. . . UAGUAUAGUGACAGUUUUAGACUAAGCAAUAGCCUCAAUAUC. . . GCAAUAGCCUCAAUAUC. . . UUUUUUUUUUUUAUUAAUAG Information UUUUUUUUUUUUAUUAAUAGUAUAGUGACAG UUUUUUUUUUUUAUUAAUAGUAUAGUGACAGUUUUAGACUAAGCAA UUUUUUAUUAAUAGUGACAGUUUUAGACUAAGCAAU Anchor U tail g. RNA Edited region specified by single g. RNA: block U Editing starts at the 3’ end of pre-edited m. RNA. Editing directed by the first g. RNA creates the m. RNA sequence which will be recognized by the next g. RNA. This creates an overall 3’ to 5’ direction for editing. U U
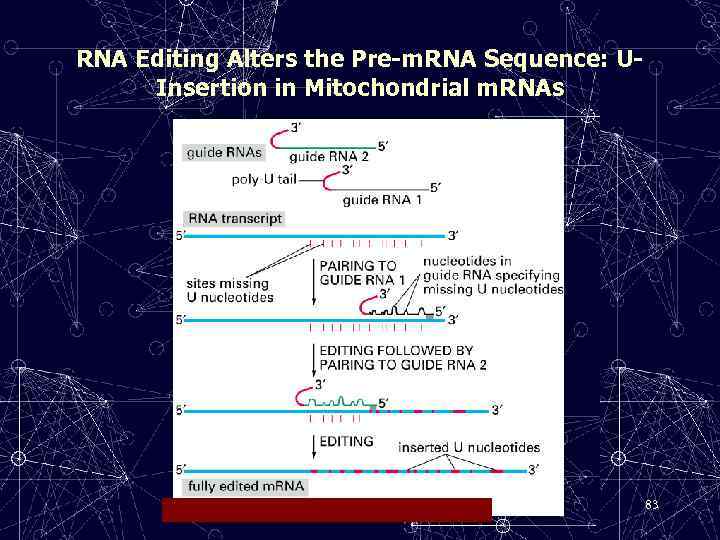 RNA Editing Alters the Pre-m. RNA Sequence: UInsertion in Mitochondrial m. RNAs 83
RNA Editing Alters the Pre-m. RNA Sequence: UInsertion in Mitochondrial m. RNAs 83
 Механизм редактирования г. РНК
Механизм редактирования г. РНК
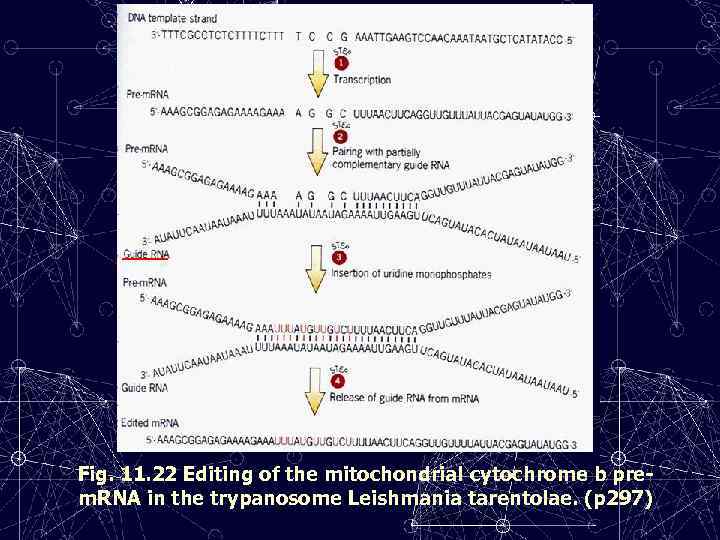 Fig. 11. 22 Editing of the mitochondrial cytochrome b prem. RNA in the trypanosome Leishmania tarentolae. (p 297)
Fig. 11. 22 Editing of the mitochondrial cytochrome b prem. RNA in the trypanosome Leishmania tarentolae. (p 297)
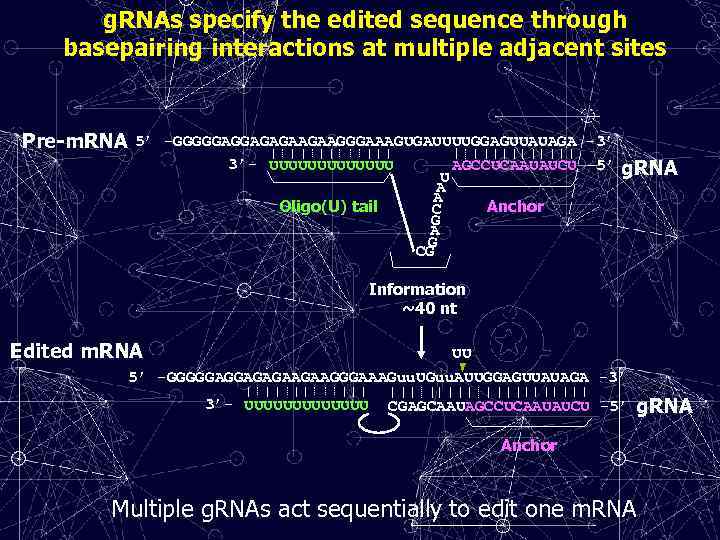 g. RNAs specify the edited sequence through basepairing interactions at multiple adjacent sites Pre-m. RNA 5’ -GGGGGAGGAGAGAAGAAGGGAAAGUGAUUUUGGAGUUAUAGA -3’ 3’- UUUUUUU AGCCUCAAUAUCU -5’ U A A Oligo(U) tail Anchor C G A G CG g. RNA Information ~40 nt Edited m. RNA UU 5’ -GGGGGAGGAGAGAAGAAGGGAAAGuu. UGuu. AUUGGAGUUAUAGA -3’ 3’- UUUUUUU CGAGCAAUAGCCUCAAUAUCU -5’ g. RNA Anchor Multiple g. RNAs act sequentially to edit one m. RNA
g. RNAs specify the edited sequence through basepairing interactions at multiple adjacent sites Pre-m. RNA 5’ -GGGGGAGGAGAGAAGAAGGGAAAGUGAUUUUGGAGUUAUAGA -3’ 3’- UUUUUUU AGCCUCAAUAUCU -5’ U A A Oligo(U) tail Anchor C G A G CG g. RNA Information ~40 nt Edited m. RNA UU 5’ -GGGGGAGGAGAGAAGAAGGGAAAGuu. UGuu. AUUGGAGUUAUAGA -3’ 3’- UUUUUUU CGAGCAAUAGCCUCAAUAUCU -5’ g. RNA Anchor Multiple g. RNAs act sequentially to edit one m. RNA
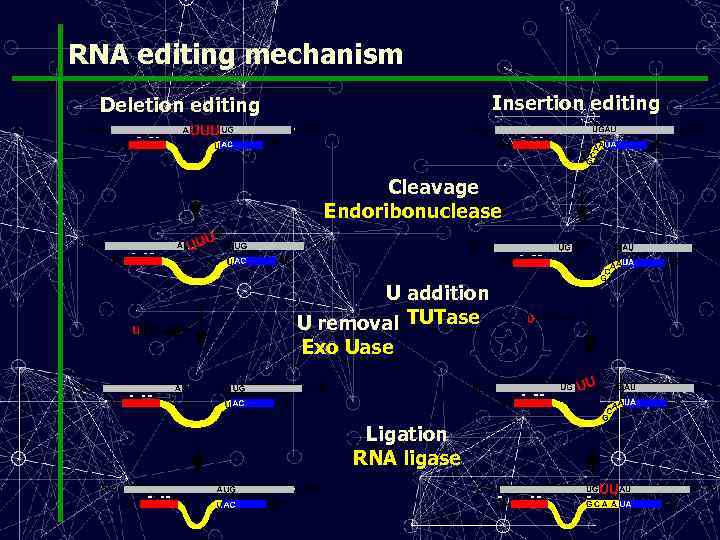 RNA editing mechanism Insertion editing Deletion editing Cleavage Endoribonuclease U addition U removal TUTase Exo Uase Ligation RNA ligase
RNA editing mechanism Insertion editing Deletion editing Cleavage Endoribonuclease U addition U removal TUTase Exo Uase Ligation RNA ligase
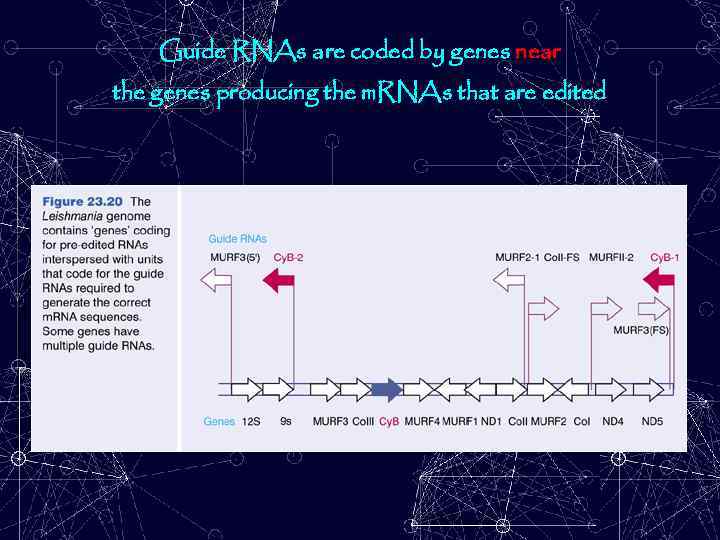 Guide RNAs are coded by genes near the genes producing the m. RNAs that are edited
Guide RNAs are coded by genes near the genes producing the m. RNAs that are edited
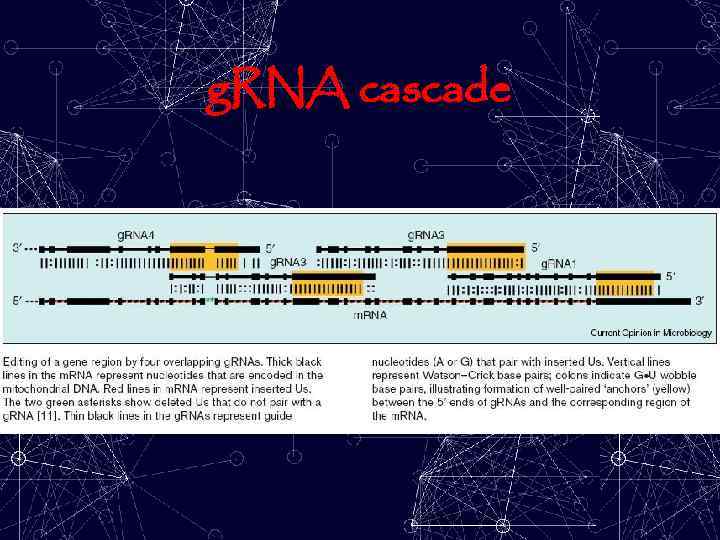 g. RNA cascade
g. RNA cascade
 Why RNA Editing?
Why RNA Editing?
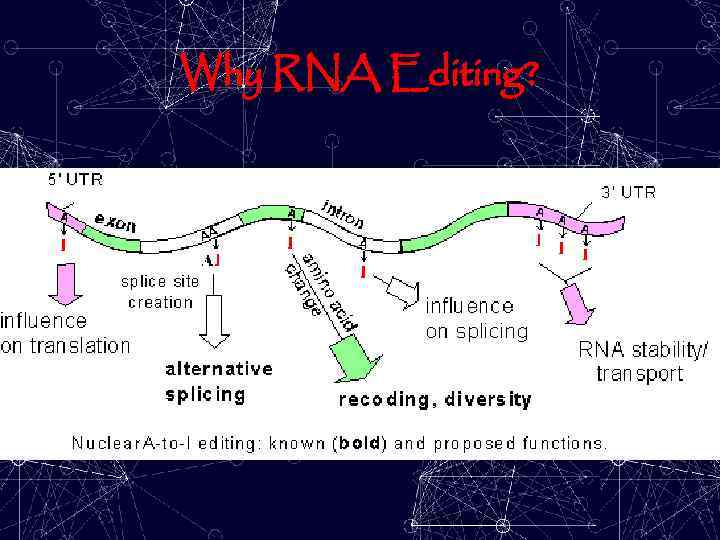 Why RNA Editing?
Why RNA Editing?
 Why RNA Editing? • Like alternative splicing — it is a mechanism • - to increase the number of different proteins available without the need to increase the number of genes in the genome. • So it can create proteins with slightly different functions to use in specialized circumstances. • There is evidence that Drosophila (and humans) • use editing to create subtle differences in the properties of – some voltage-gated ion channels and – some receptors of neurotransmitters in different regions of the brain.
Why RNA Editing? • Like alternative splicing — it is a mechanism • - to increase the number of different proteins available without the need to increase the number of genes in the genome. • So it can create proteins with slightly different functions to use in specialized circumstances. • There is evidence that Drosophila (and humans) • use editing to create subtle differences in the properties of – some voltage-gated ion channels and – some receptors of neurotransmitters in different regions of the brain.
 Редактирование РНК может изменять • • • Кодоновый состав РНК Рамку считывания Путь сплайсинга Сайт инциации трансляции Сайт терминации трансляции Стабильность РНК
Редактирование РНК может изменять • • • Кодоновый состав РНК Рамку считывания Путь сплайсинга Сайт инциации трансляции Сайт терминации трансляции Стабильность РНК
 RNA editing changes the sequence of an RNA from that encoded by DNA, producing a functional transcript. First considered a bizarre relic; now recognized as widespread RNA editing has been reported in: protozoa, plants and mammals, not yet fungi or prokaryotes nuclear, mitochondrial, chloroplast, and viral RNAs m. RNA, t. RNA, r. RNA Two general types Base modification (deaminase) A to I double-stranded mechanism, seen in viruses, human genes C to U, U to C seen in chloroplasts, plant mitochondria, human genes Insertion/deletion U insertion/deletion, seen in kinetoplastid protozoa mono/di nucleotide insertion, seen in Physarum nucleotide replacement, seen in Acanthamoeba t. RNAs
RNA editing changes the sequence of an RNA from that encoded by DNA, producing a functional transcript. First considered a bizarre relic; now recognized as widespread RNA editing has been reported in: protozoa, plants and mammals, not yet fungi or prokaryotes nuclear, mitochondrial, chloroplast, and viral RNAs m. RNA, t. RNA, r. RNA Two general types Base modification (deaminase) A to I double-stranded mechanism, seen in viruses, human genes C to U, U to C seen in chloroplasts, plant mitochondria, human genes Insertion/deletion U insertion/deletion, seen in kinetoplastid protozoa mono/di nucleotide insertion, seen in Physarum nucleotide replacement, seen in Acanthamoeba t. RNAs
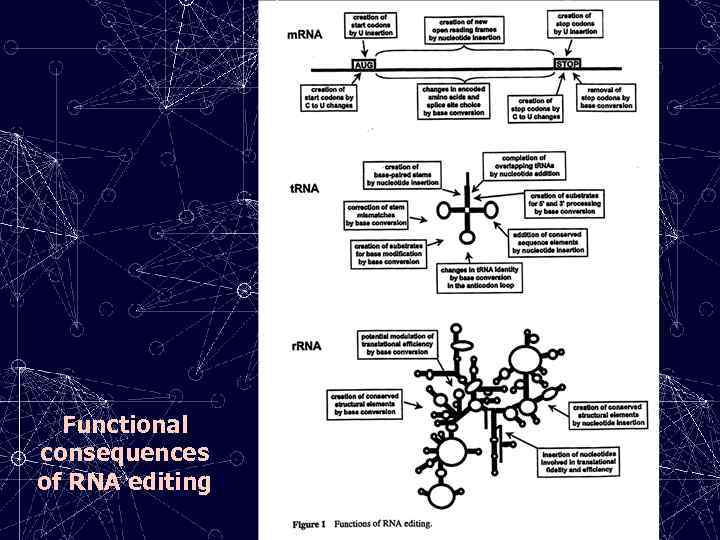 Functional consequences of RNA editing
Functional consequences of RNA editing
 Редактирование РНК и будущая эволюция?
Редактирование РНК и будущая эволюция?
 Спасибо за внимание
Спасибо за внимание


