d48d62cd7bd908f9a19cb00c4ec524a9.ppt
- Количество слайдов: 33
 Human Genome Project Chapter 9
Human Genome Project Chapter 9
 Central Points (1) § Large, international project analyzing human genome § Gene mapping to locate human genes § Number of surprises as human genome analyzed
Central Points (1) § Large, international project analyzing human genome § Gene mapping to locate human genes § Number of surprises as human genome analyzed
 Central Points (2) § Scientists apply information from Human Genome Project (HGP) to medical diagnosis and treatment § Gene therapy is a future application of the HGP § Ethical and legal aspects of the HGP discussed
Central Points (2) § Scientists apply information from Human Genome Project (HGP) to medical diagnosis and treatment § Gene therapy is a future application of the HGP § Ethical and legal aspects of the HGP discussed
 Case A: The Future Tells All § Natalie had fetal cells collected from her blood § DNA of fetus analyzed § Fetal sequencing helped them “know” their baby before meeting it
Case A: The Future Tells All § Natalie had fetal cells collected from her blood § DNA of fetus analyzed § Fetal sequencing helped them “know” their baby before meeting it
 9. 1 Goals of HGP (1) 1. Create maps of the human and other creatures’ genomes 2. Find location of all genes 3. Compile lists of expressed genes and nonexpressed sequences 4. Discover function of all genes
9. 1 Goals of HGP (1) 1. Create maps of the human and other creatures’ genomes 2. Find location of all genes 3. Compile lists of expressed genes and nonexpressed sequences 4. Discover function of all genes
 9. 1 Goals of HGP (2) 5. ID proteins encoded by genes and their functions 6. Compare genes and proteins between species 7. Analyze DNA differences between genomes 8. Set up and manage databases based on genomes discovered
9. 1 Goals of HGP (2) 5. ID proteins encoded by genes and their functions 6. Compare genes and proteins between species 7. Analyze DNA differences between genomes 8. Set up and manage databases based on genomes discovered
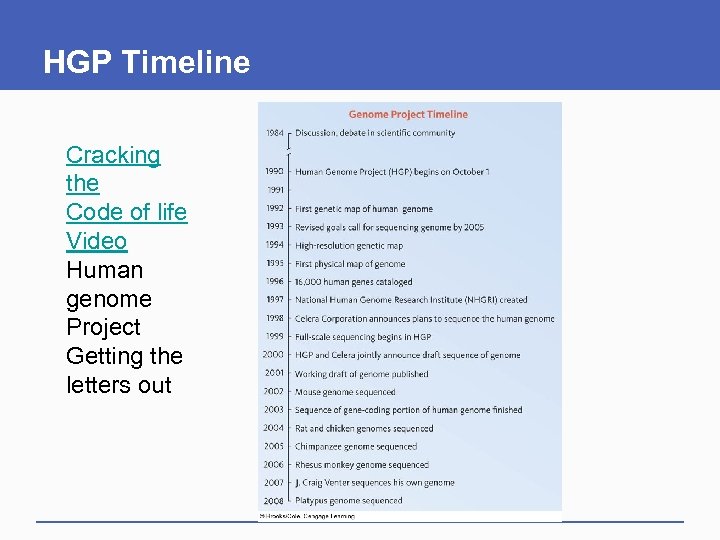 HGP Timeline Cracking the Code of life Video Human genome Project Getting the letters out
HGP Timeline Cracking the Code of life Video Human genome Project Getting the letters out
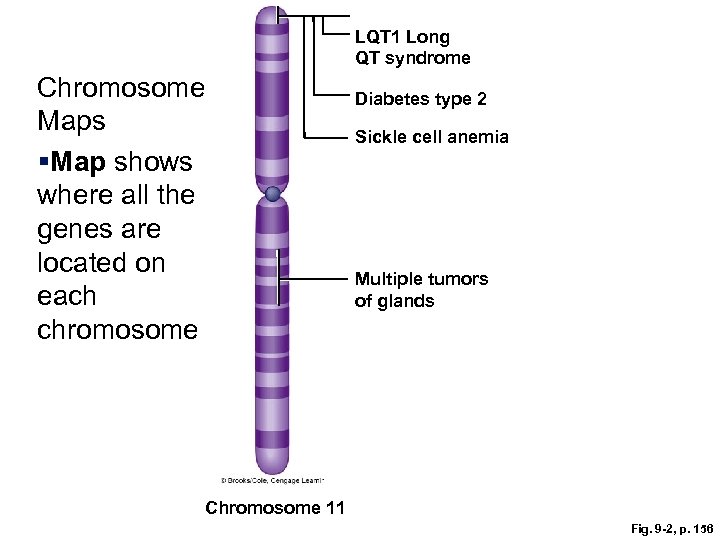 LQT 1 Long QT syndrome Chromosome Maps §Map shows where all the genes are located on each chromosome Diabetes type 2 Sickle cell anemia Multiple tumors of glands Chromosome 11 Fig. 9 -2, p. 156
LQT 1 Long QT syndrome Chromosome Maps §Map shows where all the genes are located on each chromosome Diabetes type 2 Sickle cell anemia Multiple tumors of glands Chromosome 11 Fig. 9 -2, p. 156
 Methods § Began in 1990, human genome ~3. 2 billion nucleotides § Required development of automated methods § Bioinformatics created software, web-based databases, and research tools to collect, analyze, and store information § Genomics: study of genomes
Methods § Began in 1990, human genome ~3. 2 billion nucleotides § Required development of automated methods § Bioinformatics created software, web-based databases, and research tools to collect, analyze, and store information § Genomics: study of genomes
 HGP Now § Portion that carries genes was sequenced in 2003 § Function of remaining 15% unknown and currently being sequenced § Sequenced portion studied to ID genes and assign functions § Proteomics: study of protein structure and function
HGP Now § Portion that carries genes was sequenced in 2003 § Function of remaining 15% unknown and currently being sequenced § Sequenced portion studied to ID genes and assign functions § Proteomics: study of protein structure and function
 9. 2 Gene Mapping § Genes close together on same chromosome tend to be inherited together and show linkage § In 1936, hemophilia and color blindness found to be linked, both on X chromosome § Difficult to map genes on autosomes, requires very large families with two specific genetic traits
9. 2 Gene Mapping § Genes close together on same chromosome tend to be inherited together and show linkage § In 1936, hemophilia and color blindness found to be linked, both on X chromosome § Difficult to map genes on autosomes, requires very large families with two specific genetic traits
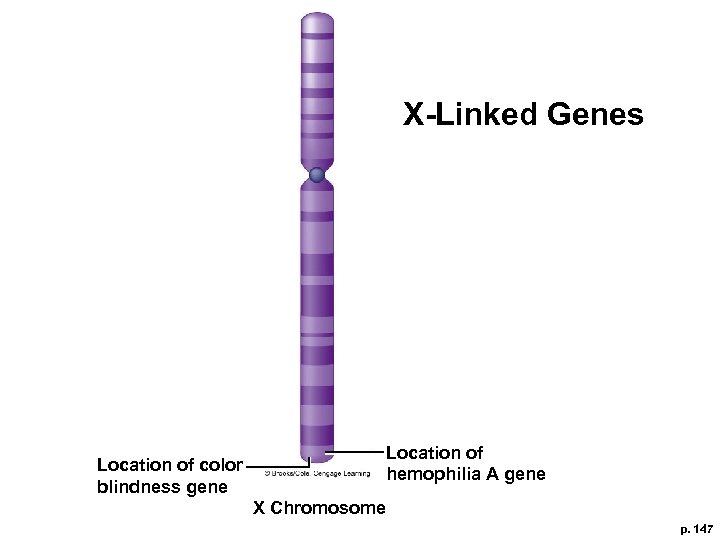 X-Linked Genes Location of hemophilia A gene Location of color blindness gene X Chromosome p. 147
X-Linked Genes Location of hemophilia A gene Location of color blindness gene X Chromosome p. 147
 Linked Genes Separate by Crossing Over § Separation of the two alleles is result of crossing over between two genes § Occurs randomly in meiosis § Frequency of crossing over related to distance between two genes § Linkage map of a chromosome can be constructed
Linked Genes Separate by Crossing Over § Separation of the two alleles is result of crossing over between two genes § Occurs randomly in meiosis § Frequency of crossing over related to distance between two genes § Linkage map of a chromosome can be constructed
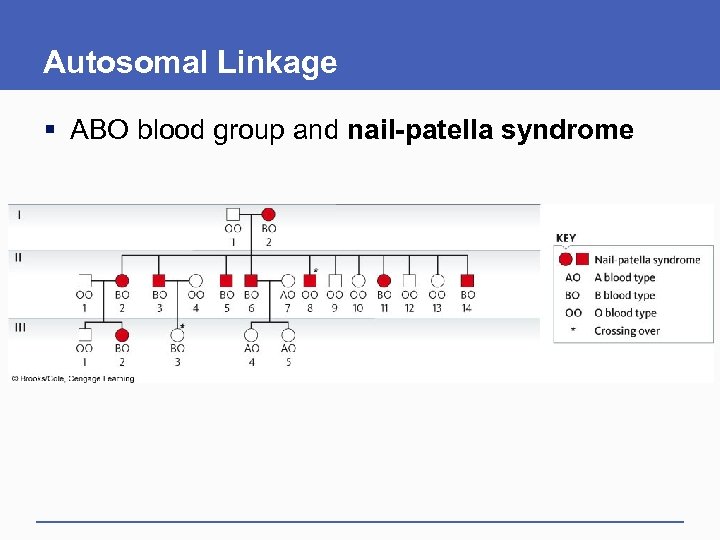 Autosomal Linkage § ABO blood group and nail-patella syndrome
Autosomal Linkage § ABO blood group and nail-patella syndrome
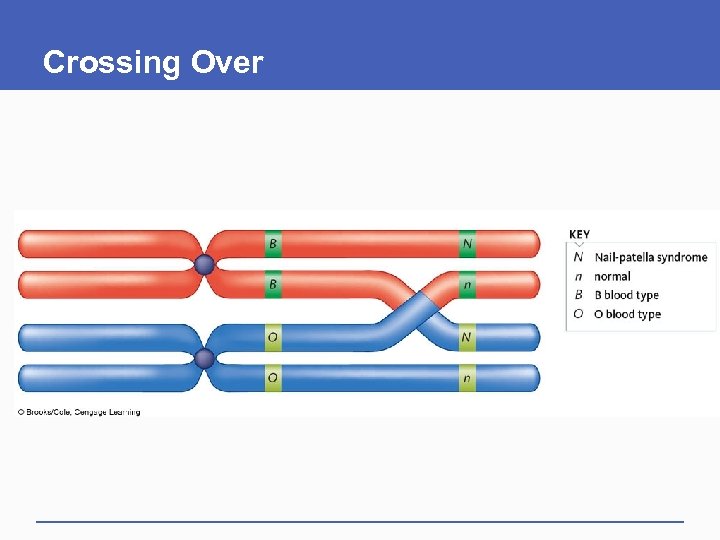 Crossing Over
Crossing Over
 Linkage or Genetic Map § Order of genes on a chromosome and distance between them § Expressed as percentage of crossing-over events § 10% = 10 map units or centimorgans (c. M) apart § From this pedigree, frequency of crossing over: 2/16 = 12. 5%. or 12. 5 c. M (actual value ~10 c. M)
Linkage or Genetic Map § Order of genes on a chromosome and distance between them § Expressed as percentage of crossing-over events § 10% = 10 map units or centimorgans (c. M) apart § From this pedigree, frequency of crossing over: 2/16 = 12. 5%. or 12. 5 c. M (actual value ~10 c. M)
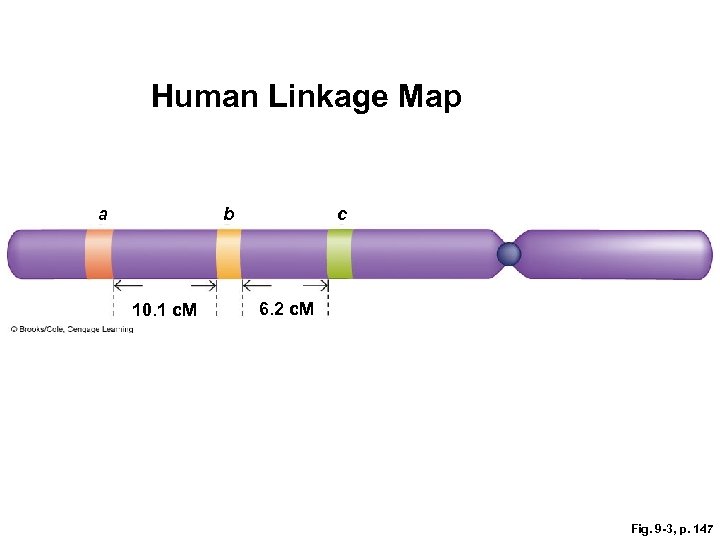 Human Linkage Map a b 10. 1 c. M c 6. 2 c. M Fig. 9 -3, p. 147
Human Linkage Map a b 10. 1 c. M c 6. 2 c. M Fig. 9 -3, p. 147
 Positional Cloning § Markers identified that show differences in: • Restriction enzyme cutting sites • Number of repeated DNA sequences (i. e. , STRs) § Markers assigned to chromosomes § Used to follow genetic disorder in pedigrees § Map one gene at a time, and by late 1980 s, more than 3, 500 genes and markers
Positional Cloning § Markers identified that show differences in: • Restriction enzyme cutting sites • Number of repeated DNA sequences (i. e. , STRs) § Markers assigned to chromosomes § Used to follow genetic disorder in pedigrees § Map one gene at a time, and by late 1980 s, more than 3, 500 genes and markers
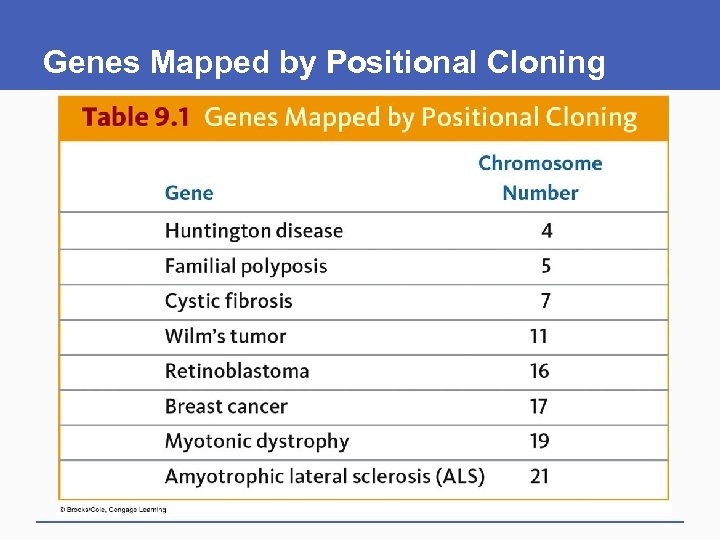 Genes Mapped by Positional Cloning
Genes Mapped by Positional Cloning
 DNA Sequencing Today § Can rapidly sequence DNA with computer programs § Sequence entire DNA sequence in genome § Uses high-speed sequencers and computers § Allowed HGP to succeed
DNA Sequencing Today § Can rapidly sequence DNA with computer programs § Sequence entire DNA sequence in genome § Uses high-speed sequencers and computers § Allowed HGP to succeed
 9. 3 Whole Genome Sequencing § Construct a genomic library that contains all sequences in a genome § Fragments of DNA placed in a DNA sequencer § Generates nucleotide sequence (As, Cs, Gs, and Ts) § Assemblers (specialized software) produce sequence of genome
9. 3 Whole Genome Sequencing § Construct a genomic library that contains all sequences in a genome § Fragments of DNA placed in a DNA sequencer § Generates nucleotide sequence (As, Cs, Gs, and Ts) § Assemblers (specialized software) produce sequence of genome
 Finding Genes from Sequence § Software programs scan sequences, searching for promoter sequence § Sequences that follow promoter are genes § AA sequence determined by matching the nucleotide triplets to corresponding AA § ID protein encoded by this gene
Finding Genes from Sequence § Software programs scan sequences, searching for promoter sequence § Sequences that follow promoter are genes § AA sequence determined by matching the nucleotide triplets to corresponding AA § ID protein encoded by this gene
 9. 4 What Have We Learned? § > 3 billion nucleotides of DNA § ~5% genes code for proteins § Many remnants of genome’s evolutionary history § > Half the genome is repetitive DNA
9. 4 What Have We Learned? § > 3 billion nucleotides of DNA § ~5% genes code for proteins § Many remnants of genome’s evolutionary history § > Half the genome is repetitive DNA
 Types Repetitive DNA § 45%: transposons • New copies can move (or transpose) • Most not functional • Do not replicate and move around § 17%: LINE 1 sequence § 10%: Alu sequences § Others including short tandem repeats (STRs)
Types Repetitive DNA § 45%: transposons • New copies can move (or transpose) • Most not functional • Do not replicate and move around § 17%: LINE 1 sequence § 10%: Alu sequences § Others including short tandem repeats (STRs)
 Other Surprises from HGP § 20, 000– 25, 000 genes but > 500, 000 known proteins (possibly exceed 2 million) § Possible mechanisms • During processing m. RNA, the coding segments can be rearranged • Proteins modified after synthesis § Human Proteome Project (HUPO): ID proteins, functions, and interactions
Other Surprises from HGP § 20, 000– 25, 000 genes but > 500, 000 known proteins (possibly exceed 2 million) § Possible mechanisms • During processing m. RNA, the coding segments can be rearranged • Proteins modified after synthesis § Human Proteome Project (HUPO): ID proteins, functions, and interactions
 9. 5 How Can Information Be Used? 1. ID location of gene 2. Function of the protein encoded by this gene 3. How the mutated gene or its protein product results in a disorder § Allow development of treatments and medications
9. 5 How Can Information Be Used? 1. ID location of gene 2. Function of the protein encoded by this gene 3. How the mutated gene or its protein product results in a disorder § Allow development of treatments and medications
 Cystic Fibrosis (CF) Gene § Positional cloning ID gene, long arm of chromosome 7 § Isolated nucleotide sequence, ID AA sequence of CF protein § Compared to databases of other organisms, protein in plasma membrane § Now developing medications
Cystic Fibrosis (CF) Gene § Positional cloning ID gene, long arm of chromosome 7 § Isolated nucleotide sequence, ID AA sequence of CF protein § Compared to databases of other organisms, protein in plasma membrane § Now developing medications
 9. 6 Future of Genome Sequencing § New technologies to reduce cost and time § Make sequencing routine in medical care § Possible for doctors to monitor your health § Provide: • Information to reduce risks for certain diseases • Early diagnosis of conditions
9. 6 Future of Genome Sequencing § New technologies to reduce cost and time § Make sequencing routine in medical care § Possible for doctors to monitor your health § Provide: • Information to reduce risks for certain diseases • Early diagnosis of conditions
 9. 7 Gene Therapy § Recombinant DNA technology to treat genetic disorders § Transfer copies of normal genes into cells (or people) with defective copies of these genes § Normal genes directs synthesis of the normal protein
9. 7 Gene Therapy § Recombinant DNA technology to treat genetic disorders § Transfer copies of normal genes into cells (or people) with defective copies of these genes § Normal genes directs synthesis of the normal protein
 How Are Genes Transferred? § Cells removed from the body § Normal copies inserted using virus, or vector § Cells grown in the laboratory § Checked that normal gene actively making protein § Cells transferred back into the body
How Are Genes Transferred? § Cells removed from the body § Normal copies inserted using virus, or vector § Cells grown in the laboratory § Checked that normal gene actively making protein § Cells transferred back into the body
 First Gene Therapy Patient (1990) § Ashanti De. Silva had severe combined immunodeficiency disorder (SCID) § No functional immune system, die from infection § Inserted gene for adenosine deaminase (ADA) into her white blood cells § Treated cells injected into her, allowed her to develop an immune system Ongoing dilemma: Parkinsons My father, my brother and me video
First Gene Therapy Patient (1990) § Ashanti De. Silva had severe combined immunodeficiency disorder (SCID) § No functional immune system, die from infection § Inserted gene for adenosine deaminase (ADA) into her white blood cells § Treated cells injected into her, allowed her to develop an immune system Ongoing dilemma: Parkinsons My father, my brother and me video
 Problems with Gene Therapy § In many cases, gene therapy has not worked § Few patients developed leukemia § At least two people died § Scientists working to correct problems § Need to develop new approaches to use genes to treatment genetic diseases § Gene patent video start 27: 33 - end 36: 33
Problems with Gene Therapy § In many cases, gene therapy has not worked § Few patients developed leukemia § At least two people died § Scientists working to correct problems § Need to develop new approaches to use genes to treatment genetic diseases § Gene patent video start 27: 33 - end 36: 33
 9. 8 Legal and Ethical Issues
9. 8 Legal and Ethical Issues


