2e3b8708297311269ac544ba6aebde68.ppt
- Количество слайдов: 94
 Human Anatomy and Physiology I CELLS: THE LIVING UNITS Chapter 3
Human Anatomy and Physiology I CELLS: THE LIVING UNITS Chapter 3
 Chapter 3 Outline § Overview of Cells § The Plasma Membrane Phospholipid bilayer, membrane junctions, cell-environment interactions § Membrane Transport Active and Passive Processes § The Cytoplasm and Organelles § The Nucleus § The Cell Life Cycle (Mitosis)
Chapter 3 Outline § Overview of Cells § The Plasma Membrane Phospholipid bilayer, membrane junctions, cell-environment interactions § Membrane Transport Active and Passive Processes § The Cytoplasm and Organelles § The Nucleus § The Cell Life Cycle (Mitosis)
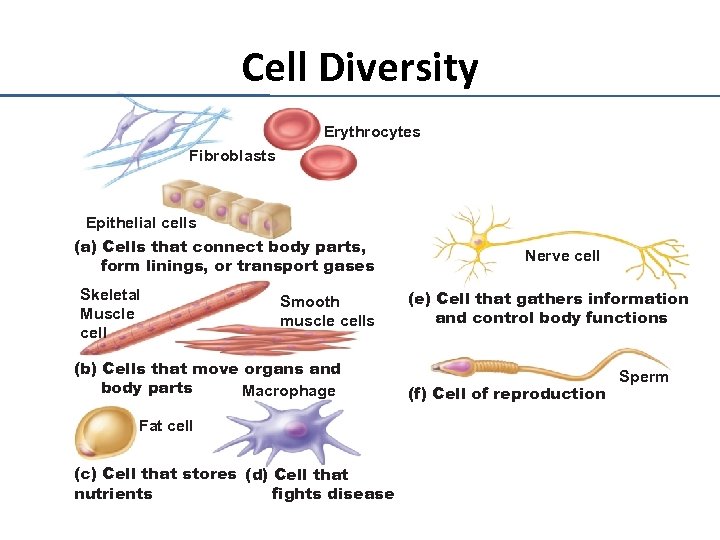 Cell Diversity Erythrocytes Fibroblasts Epithelial cells (a) Cells that connect body parts, form linings, or transport gases Skeletal Muscle cell Smooth muscle cells (b) Cells that move organs and body parts Macrophage Fat cell (c) Cell that stores (d) Cell that nutrients fights disease Nerve cell (e) Cell that gathers information and control body functions (f) Cell of reproduction Sperm
Cell Diversity Erythrocytes Fibroblasts Epithelial cells (a) Cells that connect body parts, form linings, or transport gases Skeletal Muscle cell Smooth muscle cells (b) Cells that move organs and body parts Macrophage Fat cell (c) Cell that stores (d) Cell that nutrients fights disease Nerve cell (e) Cell that gathers information and control body functions (f) Cell of reproduction Sperm
 Generalized Eukaryotic Cell • All cells have some common structures and functions • Human cells have three basic parts – Plasma membrane—flexible outer boundary – Cytoplasm—intracellular fluid containing organelles – Nucleus—control center
Generalized Eukaryotic Cell • All cells have some common structures and functions • Human cells have three basic parts – Plasma membrane—flexible outer boundary – Cytoplasm—intracellular fluid containing organelles – Nucleus—control center
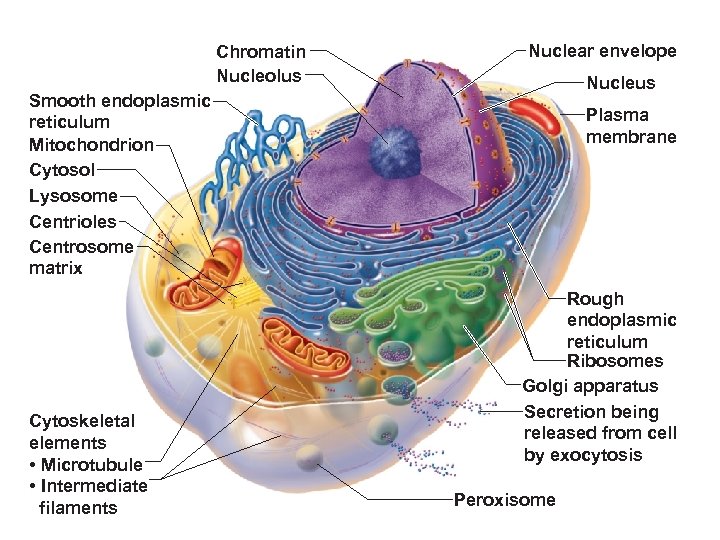 Chromatin Nucleolus Nuclear envelope Nucleus Smooth endoplasmic reticulum Mitochondrion Cytosol Lysosome Centrioles Centrosome matrix Cytoskeletal elements • Microtubule • Intermediate filaments Plasma membrane Rough endoplasmic reticulum Ribosomes Golgi apparatus Secretion being released from cell by exocytosis Peroxisome
Chromatin Nucleolus Nuclear envelope Nucleus Smooth endoplasmic reticulum Mitochondrion Cytosol Lysosome Centrioles Centrosome matrix Cytoskeletal elements • Microtubule • Intermediate filaments Plasma membrane Rough endoplasmic reticulum Ribosomes Golgi apparatus Secretion being released from cell by exocytosis Peroxisome
 Plasma Membrane § Bimolecular layer of lipids and proteins in a constantly changing fluid mosaic § Plays a dynamic role in cellular activity § Separates intracellular fluid (ICF) from extracellular fluid (ECF) – Interstitial fluid (IF) = ECF that surrounds cells
Plasma Membrane § Bimolecular layer of lipids and proteins in a constantly changing fluid mosaic § Plays a dynamic role in cellular activity § Separates intracellular fluid (ICF) from extracellular fluid (ECF) – Interstitial fluid (IF) = ECF that surrounds cells
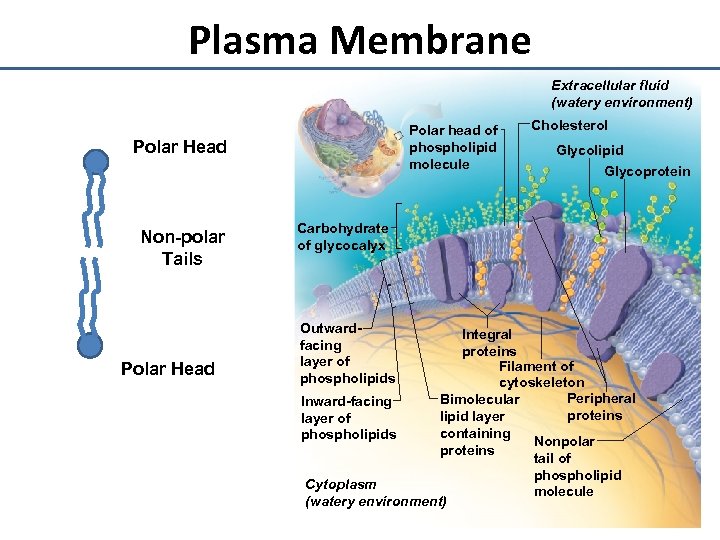 Plasma Membrane Extracellular fluid (watery environment) Polar head of phospholipid molecule Polar Head Non-polar Tails Polar Head Cholesterol Glycolipid Glycoprotein Carbohydrate of glycocalyx Outwardfacing layer of phospholipids Integral proteins Filament of cytoskeleton Peripheral Bimolecular Inward-facing proteins lipid layer of containing phospholipids Nonpolar proteins tail of phospholipid Cytoplasm molecule (watery environment)
Plasma Membrane Extracellular fluid (watery environment) Polar head of phospholipid molecule Polar Head Non-polar Tails Polar Head Cholesterol Glycolipid Glycoprotein Carbohydrate of glycocalyx Outwardfacing layer of phospholipids Integral proteins Filament of cytoskeleton Peripheral Bimolecular Inward-facing proteins lipid layer of containing phospholipids Nonpolar proteins tail of phospholipid Cytoplasm molecule (watery environment)
 Membrane Lipids § 75% Phospholipids (lipid bilayer) – Phosphate heads: polar and hydrophilic – Fatty acid tails: nonpolar and hydrophobic § 5% Glycolipids – Lipids with polar sugar groups on outer membrane surface § 20% Cholesterol – Increases membrane stability and fluidity
Membrane Lipids § 75% Phospholipids (lipid bilayer) – Phosphate heads: polar and hydrophilic – Fatty acid tails: nonpolar and hydrophobic § 5% Glycolipids – Lipids with polar sugar groups on outer membrane surface § 20% Cholesterol – Increases membrane stability and fluidity
 Membrane Proteins § Integral proteins – Firmly inserted into the membrane (most are transmembrane) – Functions • Transport proteins (channels and carriers) • Enzymes • Receptors
Membrane Proteins § Integral proteins – Firmly inserted into the membrane (most are transmembrane) – Functions • Transport proteins (channels and carriers) • Enzymes • Receptors
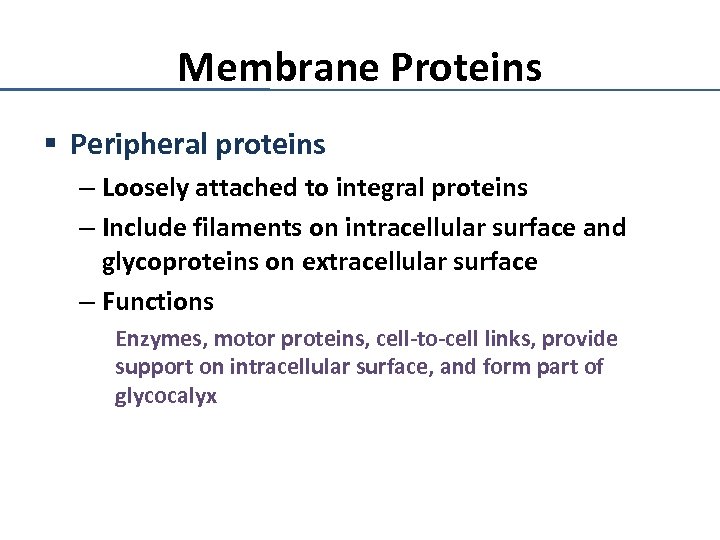 Membrane Proteins § Peripheral proteins – Loosely attached to integral proteins – Include filaments on intracellular surface and glycoproteins on extracellular surface – Functions Enzymes, motor proteins, cell-to-cell links, provide support on intracellular surface, and form part of glycocalyx
Membrane Proteins § Peripheral proteins – Loosely attached to integral proteins – Include filaments on intracellular surface and glycoproteins on extracellular surface – Functions Enzymes, motor proteins, cell-to-cell links, provide support on intracellular surface, and form part of glycocalyx
 What is the outer covering of the cell called? A. Nucleus B. Cytoplasm C. Plasma Membrane
What is the outer covering of the cell called? A. Nucleus B. Cytoplasm C. Plasma Membrane
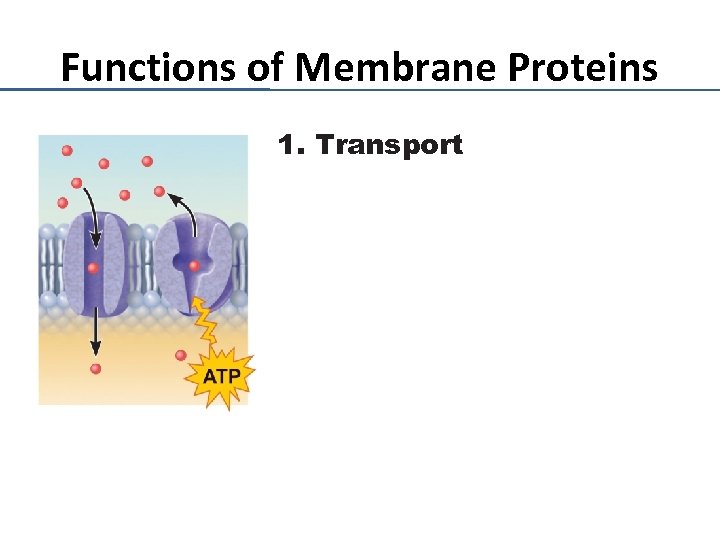 Functions of Membrane Proteins 1. Transport
Functions of Membrane Proteins 1. Transport
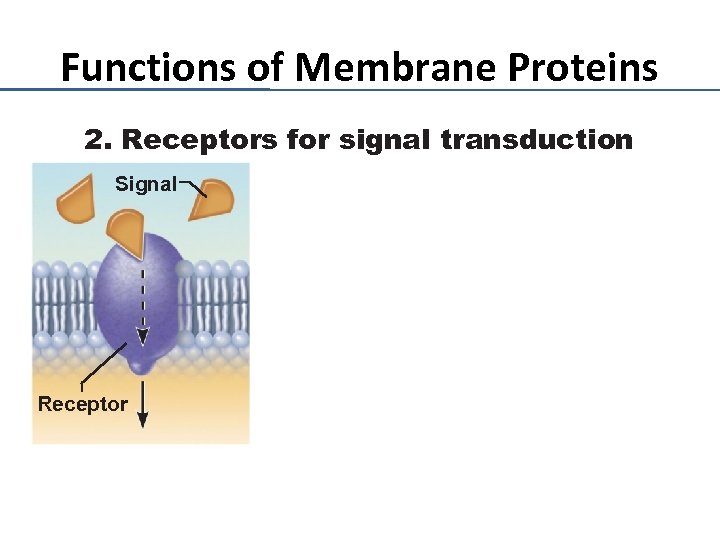 Functions of Membrane Proteins 2. Receptors for signal transduction Signal Receptor
Functions of Membrane Proteins 2. Receptors for signal transduction Signal Receptor
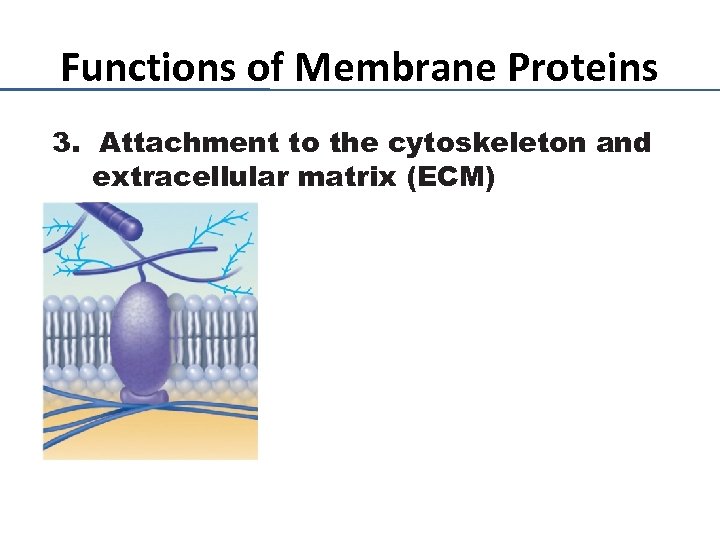 Functions of Membrane Proteins 3. Attachment to the cytoskeleton and extracellular matrix (ECM)
Functions of Membrane Proteins 3. Attachment to the cytoskeleton and extracellular matrix (ECM)
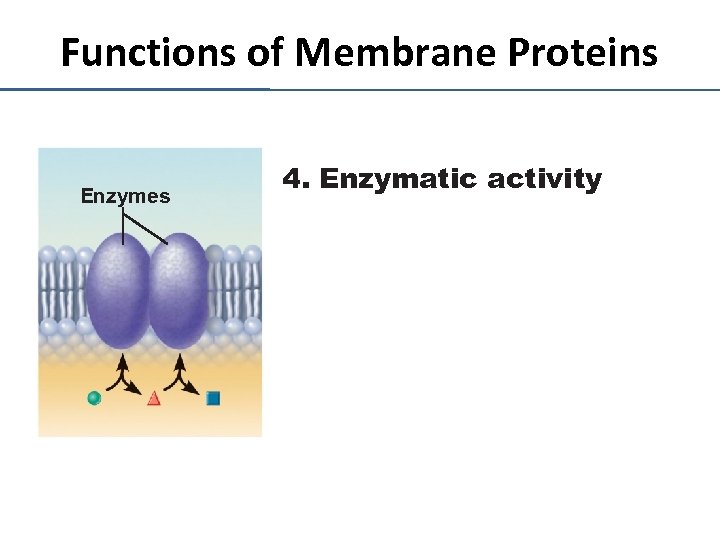 Functions of Membrane Proteins Enzymes 4. Enzymatic activity
Functions of Membrane Proteins Enzymes 4. Enzymatic activity
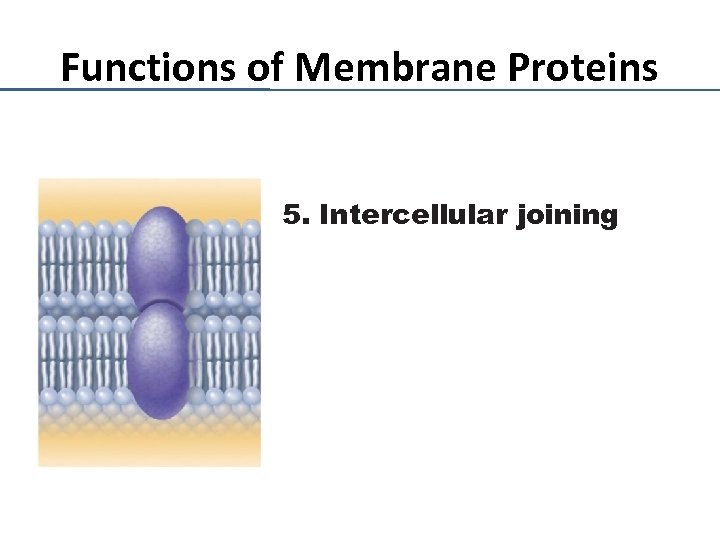 Functions of Membrane Proteins 5. Intercellular joining
Functions of Membrane Proteins 5. Intercellular joining
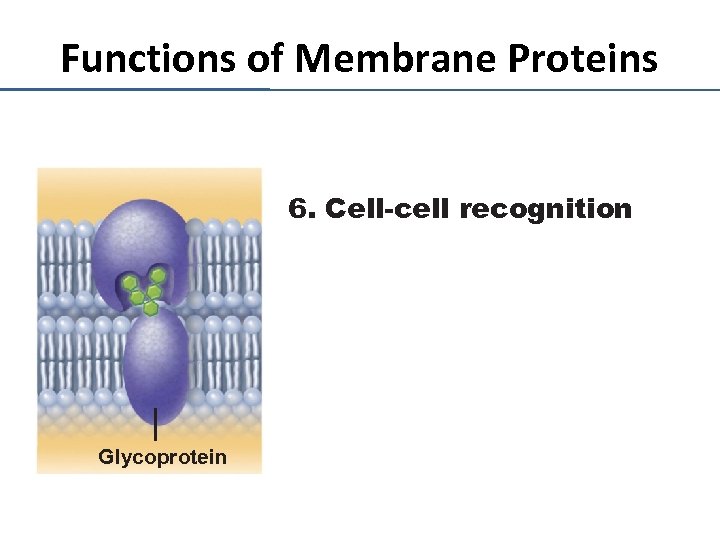 Functions of Membrane Proteins 6. Cell-cell recognition Glycoprotein
Functions of Membrane Proteins 6. Cell-cell recognition Glycoprotein
 Membrane Junctions • Three types – Tight junction – Desmosome – Gap junction
Membrane Junctions • Three types – Tight junction – Desmosome – Gap junction
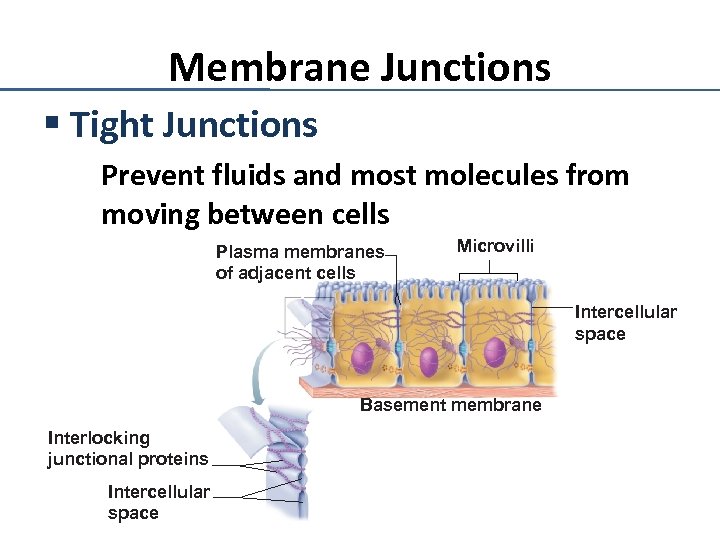 Membrane Junctions § Tight Junctions Prevent fluids and most molecules from moving between cells Plasma membranes of adjacent cells Microvilli Intercellular space Basement membrane Interlocking junctional proteins Intercellular space
Membrane Junctions § Tight Junctions Prevent fluids and most molecules from moving between cells Plasma membranes of adjacent cells Microvilli Intercellular space Basement membrane Interlocking junctional proteins Intercellular space
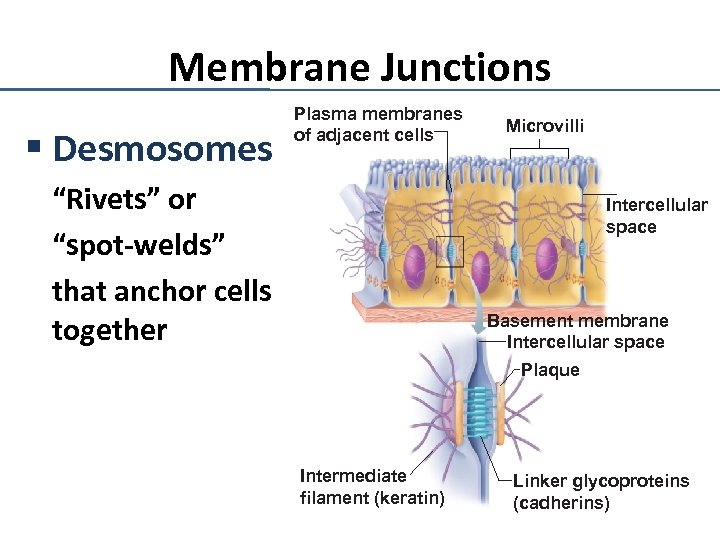 Membrane Junctions § Desmosomes Plasma membranes of adjacent cells “Rivets” or “spot-welds” that anchor cells together Microvilli Intercellular space Basement membrane Intercellular space Plaque Intermediate filament (keratin) Linker glycoproteins (cadherins)
Membrane Junctions § Desmosomes Plasma membranes of adjacent cells “Rivets” or “spot-welds” that anchor cells together Microvilli Intercellular space Basement membrane Intercellular space Plaque Intermediate filament (keratin) Linker glycoproteins (cadherins)
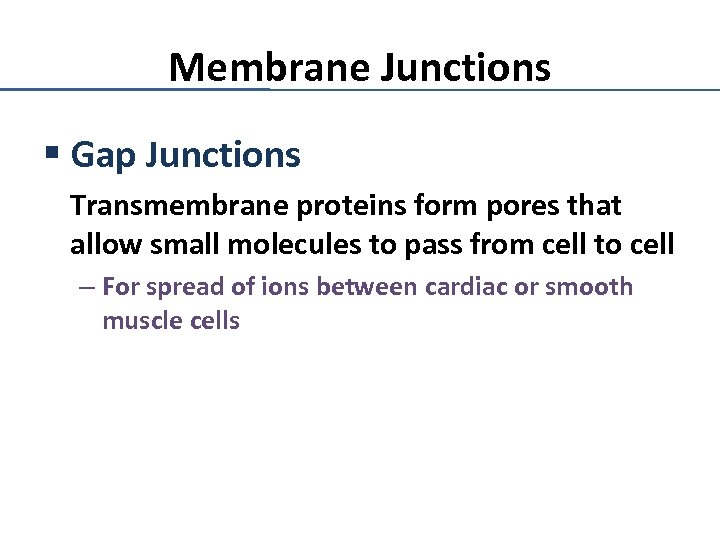 Membrane Junctions § Gap Junctions Transmembrane proteins form pores that allow small molecules to pass from cell to cell – For spread of ions between cardiac or smooth muscle cells
Membrane Junctions § Gap Junctions Transmembrane proteins form pores that allow small molecules to pass from cell to cell – For spread of ions between cardiac or smooth muscle cells
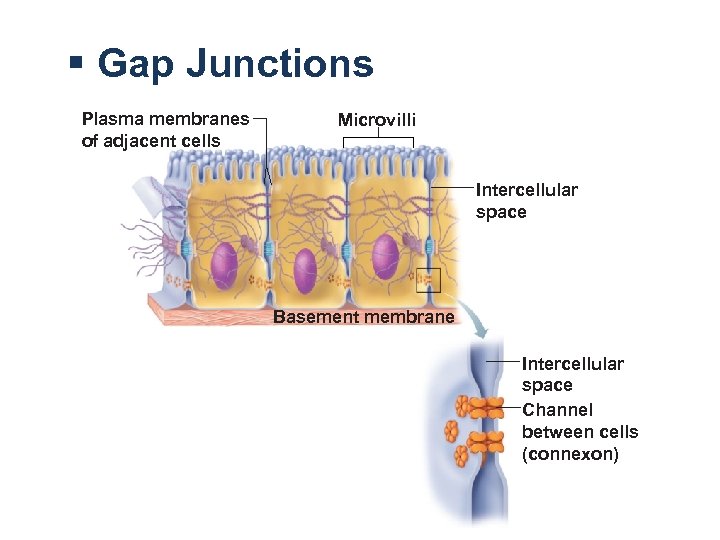 § Gap Junctions Plasma membranes of adjacent cells Microvilli Intercellular space Basement membrane Intercellular space Channel between cells (connexon)
§ Gap Junctions Plasma membranes of adjacent cells Microvilli Intercellular space Basement membrane Intercellular space Channel between cells (connexon)
 Membrane Transport § Plasma membranes are selectively permeable § Some molecules easily pass through the membrane; others do not
Membrane Transport § Plasma membranes are selectively permeable § Some molecules easily pass through the membrane; others do not
 Types of Membrane Transport § Passive processes – No cellular energy (ATP) required – Substance moves down its concentration gradient § Active processes – Energy (ATP) required – Occurs only in living cell membranes
Types of Membrane Transport § Passive processes – No cellular energy (ATP) required – Substance moves down its concentration gradient § Active processes – Energy (ATP) required – Occurs only in living cell membranes
 Passive Transport Four Types 1) Simple diffusion 2) Carrier-mediated facilitated diffusion 3) Channel-mediated facilitated diffusion 4) Osmosis
Passive Transport Four Types 1) Simple diffusion 2) Carrier-mediated facilitated diffusion 3) Channel-mediated facilitated diffusion 4) Osmosis
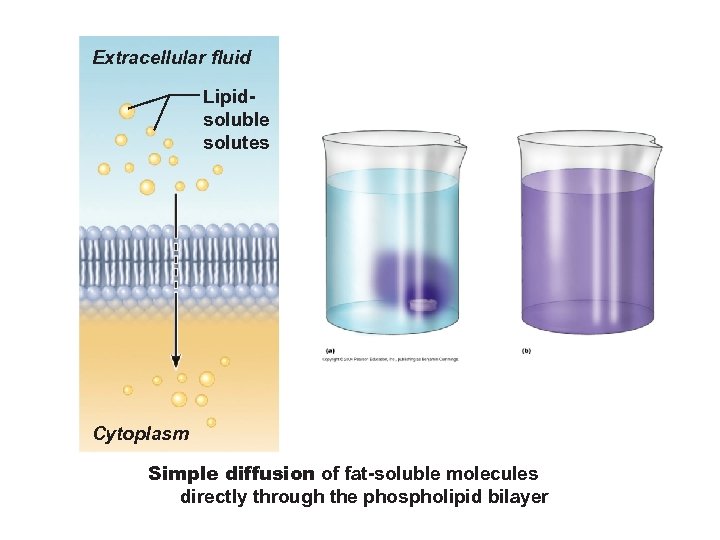 Extracellular fluid Lipidsoluble solutes Cytoplasm Simple diffusion of fat-soluble molecules directly through the phospholipid bilayer
Extracellular fluid Lipidsoluble solutes Cytoplasm Simple diffusion of fat-soluble molecules directly through the phospholipid bilayer
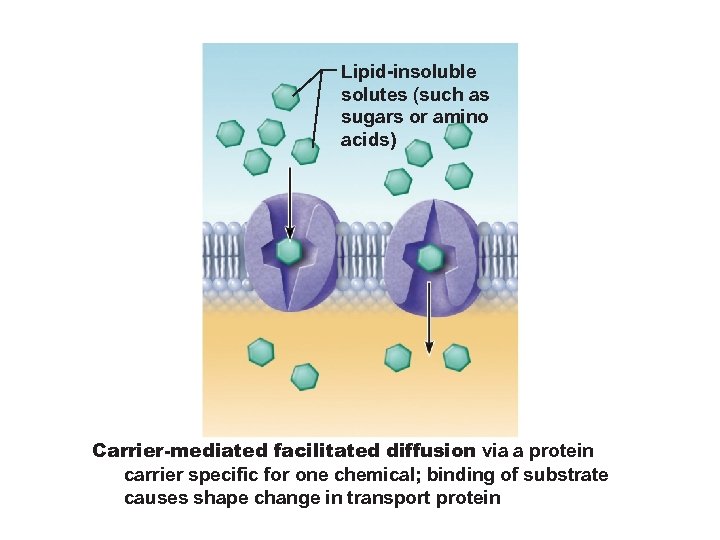 Lipid-insoluble solutes (such as sugars or amino acids) Carrier-mediated facilitated diffusion via a protein carrier specific for one chemical; binding of substrate causes shape change in transport protein
Lipid-insoluble solutes (such as sugars or amino acids) Carrier-mediated facilitated diffusion via a protein carrier specific for one chemical; binding of substrate causes shape change in transport protein
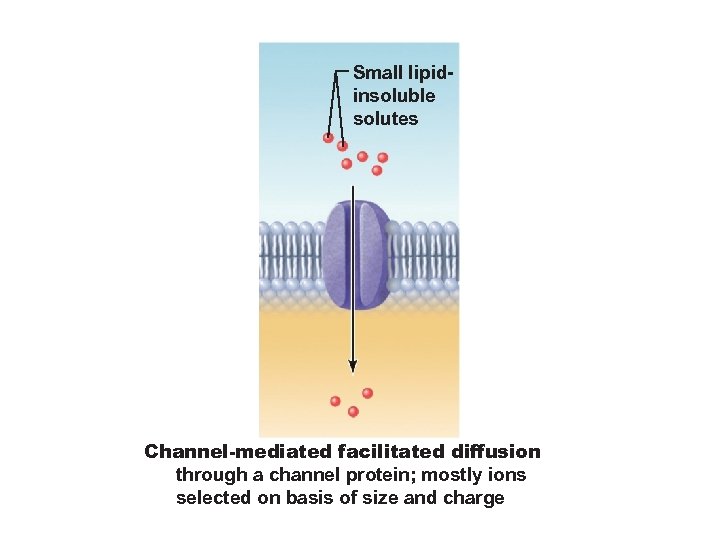 Small lipidinsoluble solutes Channel-mediated facilitated diffusion through a channel protein; mostly ions selected on basis of size and charge
Small lipidinsoluble solutes Channel-mediated facilitated diffusion through a channel protein; mostly ions selected on basis of size and charge
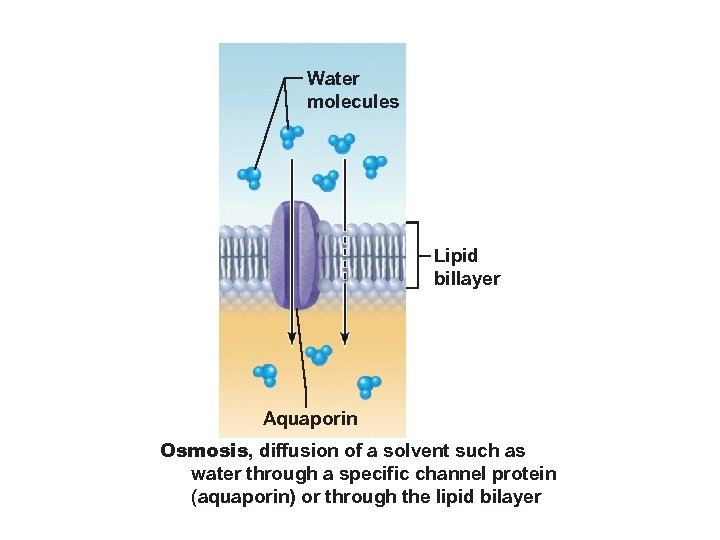 Water molecules Lipid billayer Aquaporin Osmosis, diffusion of a solvent such as water through a specific channel protein (aquaporin) or through the lipid bilayer
Water molecules Lipid billayer Aquaporin Osmosis, diffusion of a solvent such as water through a specific channel protein (aquaporin) or through the lipid bilayer
 Which of the following is a cell junction that allows cell-to-cell communication? A. Tight junction B. Gap junction C. Desmosome
Which of the following is a cell junction that allows cell-to-cell communication? A. Tight junction B. Gap junction C. Desmosome
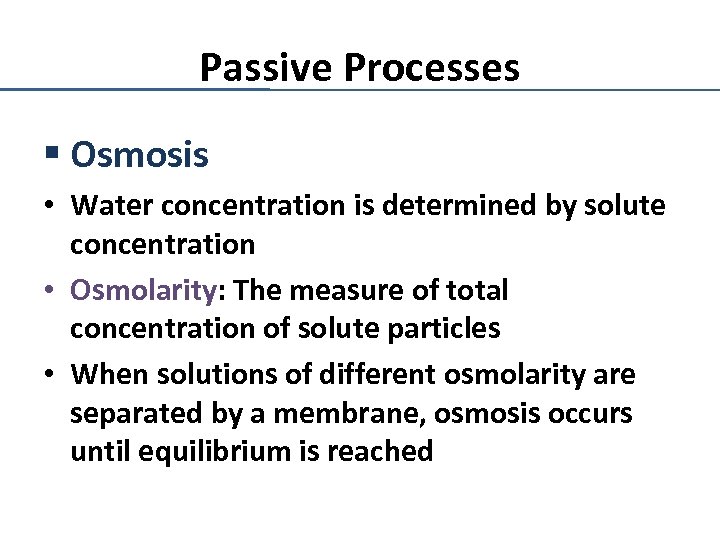 Passive Processes § Osmosis • Water concentration is determined by solute concentration • Osmolarity: The measure of total concentration of solute particles • When solutions of different osmolarity are separated by a membrane, osmosis occurs until equilibrium is reached
Passive Processes § Osmosis • Water concentration is determined by solute concentration • Osmolarity: The measure of total concentration of solute particles • When solutions of different osmolarity are separated by a membrane, osmosis occurs until equilibrium is reached
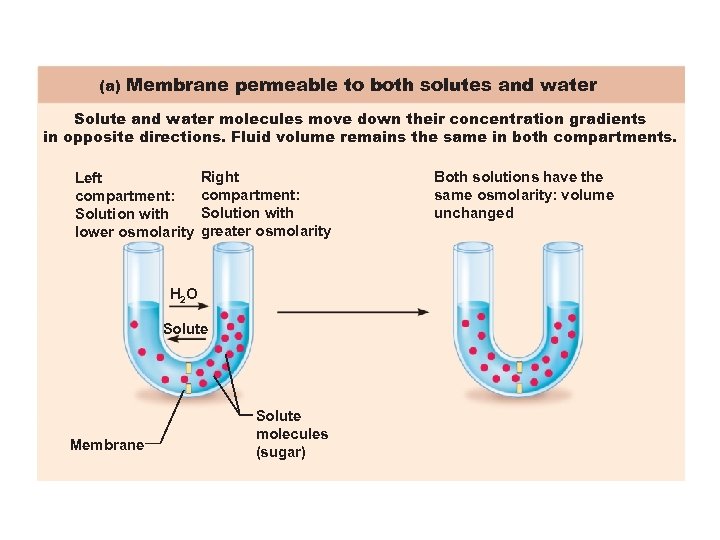 (a) Membrane permeable to both solutes and water Solute and water molecules move down their concentration gradients in opposite directions. Fluid volume remains the same in both compartments. Left compartment: Solution with lower osmolarity Right compartment: Solution with greater osmolarity H 2 O Solute Membrane Solute molecules (sugar) Both solutions have the same osmolarity: volume unchanged
(a) Membrane permeable to both solutes and water Solute and water molecules move down their concentration gradients in opposite directions. Fluid volume remains the same in both compartments. Left compartment: Solution with lower osmolarity Right compartment: Solution with greater osmolarity H 2 O Solute Membrane Solute molecules (sugar) Both solutions have the same osmolarity: volume unchanged
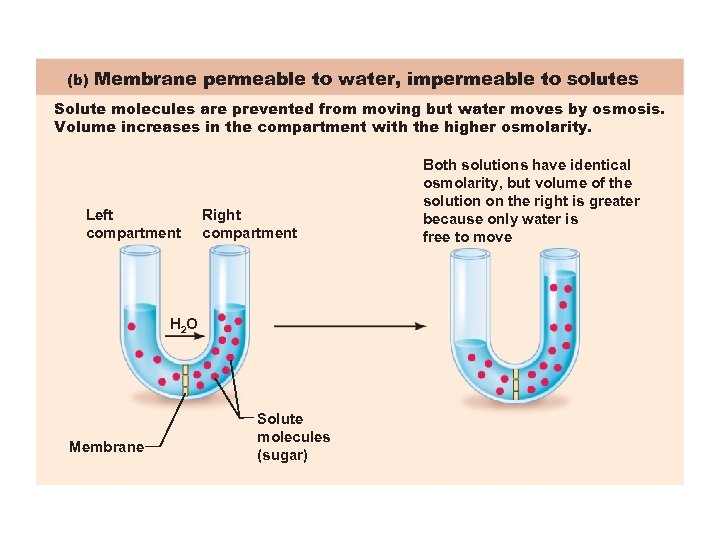 (b) Membrane permeable to water, impermeable to solutes Solute molecules are prevented from moving but water moves by osmosis. Volume increases in the compartment with the higher osmolarity. Left compartment Right compartment H 2 O Membrane Solute molecules (sugar) Both solutions have identical osmolarity, but volume of the solution on the right is greater because only water is free to move
(b) Membrane permeable to water, impermeable to solutes Solute molecules are prevented from moving but water moves by osmosis. Volume increases in the compartment with the higher osmolarity. Left compartment Right compartment H 2 O Membrane Solute molecules (sugar) Both solutions have identical osmolarity, but volume of the solution on the right is greater because only water is free to move
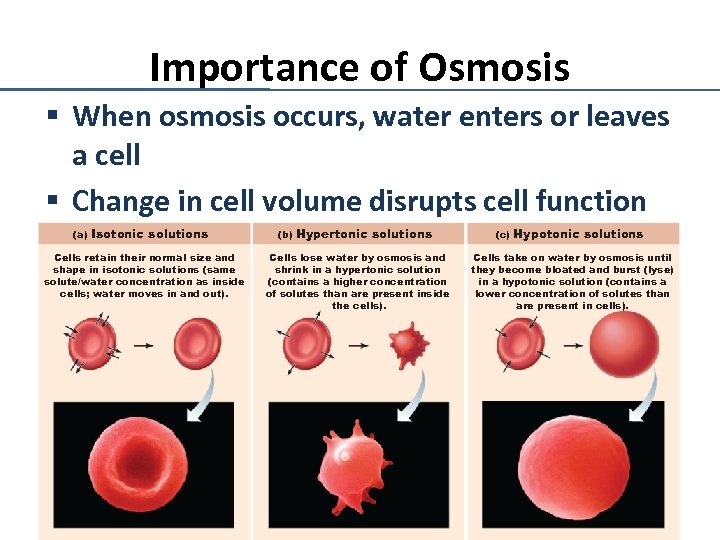 Importance of Osmosis § When osmosis occurs, water enters or leaves a cell § Change in cell volume disrupts cell function (a) Isotonic solutions Cells retain their normal size and shape in isotonic solutions (same solute/water concentration as inside cells; water moves in and out). (b) Hypertonic solutions Cells lose water by osmosis and shrink in a hypertonic solution (contains a higher concentration of solutes than are present inside the cells). (c) Hypotonic solutions Cells take on water by osmosis until they become bloated and burst (lyse) in a hypotonic solution (contains a lower concentration of solutes than are present in cells).
Importance of Osmosis § When osmosis occurs, water enters or leaves a cell § Change in cell volume disrupts cell function (a) Isotonic solutions Cells retain their normal size and shape in isotonic solutions (same solute/water concentration as inside cells; water moves in and out). (b) Hypertonic solutions Cells lose water by osmosis and shrink in a hypertonic solution (contains a higher concentration of solutes than are present inside the cells). (c) Hypotonic solutions Cells take on water by osmosis until they become bloated and burst (lyse) in a hypotonic solution (contains a lower concentration of solutes than are present in cells).
 During Osmosis, _____ move(s) across a semi-permeable membrane to the side with higher ______. A. Solutes, Solutes B. Solutes, Water C. Water, Water D. Water, Solutes
During Osmosis, _____ move(s) across a semi-permeable membrane to the side with higher ______. A. Solutes, Solutes B. Solutes, Water C. Water, Water D. Water, Solutes
 Active Processes • Two types – Active transport – Vesicular transport Both use ATP to move solutes across a living plasma membrane Examples: ions, some sugars and amino acids
Active Processes • Two types – Active transport – Vesicular transport Both use ATP to move solutes across a living plasma membrane Examples: ions, some sugars and amino acids
 Active Transport • Requires carrier proteins (solute pumps) • Moves solutes against a concentration gradient
Active Transport • Requires carrier proteins (solute pumps) • Moves solutes against a concentration gradient
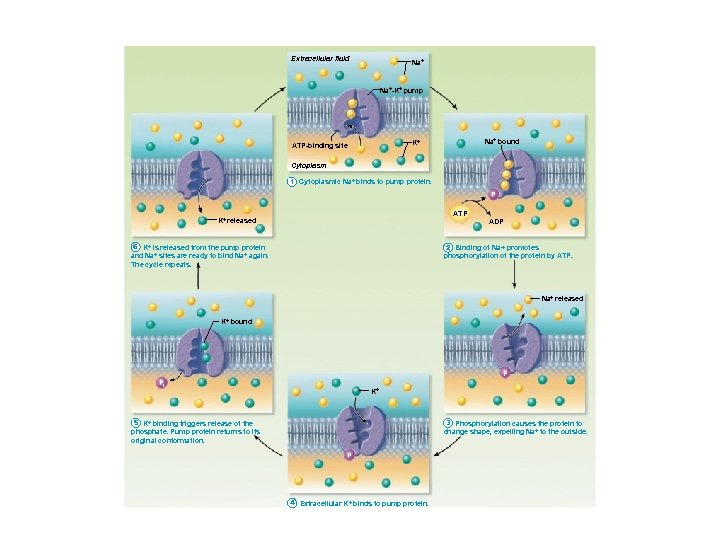 Extracellular fluid Na+-K+ pump Na+ bound K+ ATP-binding site Cytoplasm 1 Cytoplasmic Na+ binds to pump protein. P ATP K+ released ADP 6 K + is released from the pump protein and Na+ sites are ready to bind Na + again. The cycle repeats. 2 Binding of Na+ promotes phosphorylation of the protein by ATP. Na+ released K+ bound P Pi K+ 5 K + binding triggers release of the phosphate. Pump protein returns to its original conformation. 3 Phosphorylation causes the protein to change shape, expelling Na+ to the outside. P 4 Extracellular K+ binds to pump protein.
Extracellular fluid Na+-K+ pump Na+ bound K+ ATP-binding site Cytoplasm 1 Cytoplasmic Na+ binds to pump protein. P ATP K+ released ADP 6 K + is released from the pump protein and Na+ sites are ready to bind Na + again. The cycle repeats. 2 Binding of Na+ promotes phosphorylation of the protein by ATP. Na+ released K+ bound P Pi K+ 5 K + binding triggers release of the phosphate. Pump protein returns to its original conformation. 3 Phosphorylation causes the protein to change shape, expelling Na+ to the outside. P 4 Extracellular K+ binds to pump protein.
 Vesicular Transport • Transport of large particles, macromolecules, and fluids across plasma membranes • Requires cellular energy (ATP)
Vesicular Transport • Transport of large particles, macromolecules, and fluids across plasma membranes • Requires cellular energy (ATP)
 Endocytosis § Phagocytosis—pseudopods engulf solids and bring them into cell’s interior – Macrophages and some white blood cells § Pinocytosis - fluid-phase endocytosis plasma membrane infolds, bringing extracellular fluid and solutes into interior of the cell – Nutrient absorption in the small intestine
Endocytosis § Phagocytosis—pseudopods engulf solids and bring them into cell’s interior – Macrophages and some white blood cells § Pinocytosis - fluid-phase endocytosis plasma membrane infolds, bringing extracellular fluid and solutes into interior of the cell – Nutrient absorption in the small intestine
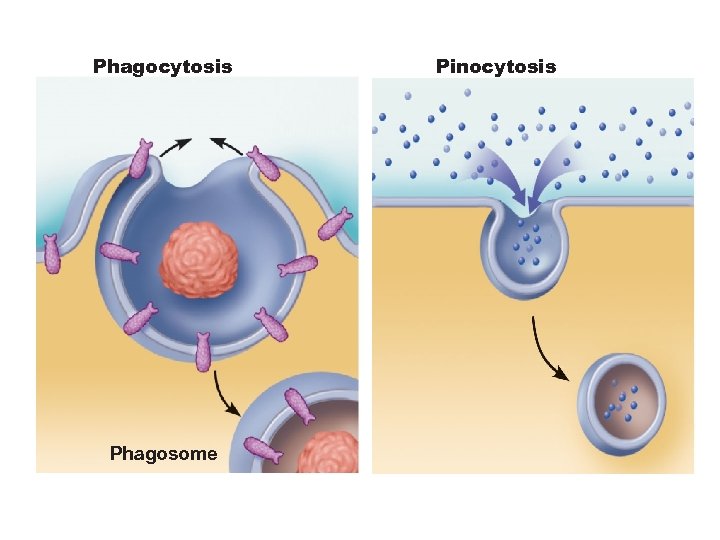 Phagocytosis Phagosome Pinocytosis
Phagocytosis Phagosome Pinocytosis
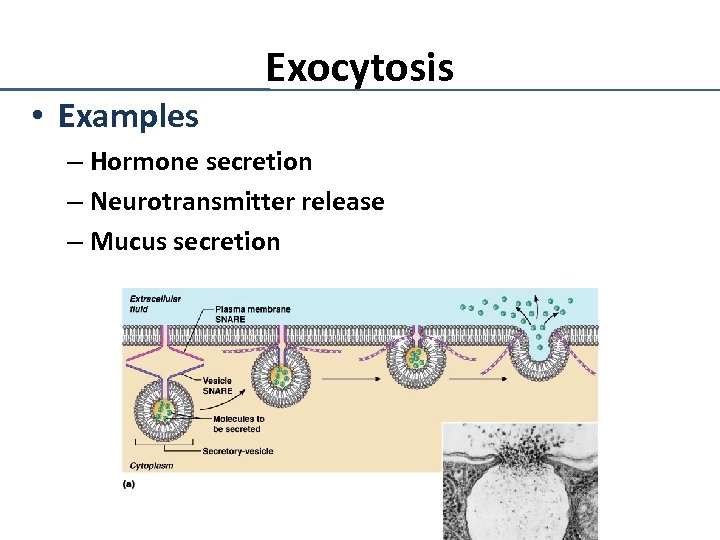 • Examples Exocytosis – Hormone secretion – Neurotransmitter release – Mucus secretion
• Examples Exocytosis – Hormone secretion – Neurotransmitter release – Mucus secretion
 Did You Know? • Humans have an estimated 10 trillion cells • The largest known cell is an unfertilized ostrich egg cell • The word cell comes from Latin cellula, meaning, small room • One single cell contains two meters of DNA http: //wiki. answers. com
Did You Know? • Humans have an estimated 10 trillion cells • The largest known cell is an unfertilized ostrich egg cell • The word cell comes from Latin cellula, meaning, small room • One single cell contains two meters of DNA http: //wiki. answers. com
 Cytoplasm • Located between plasma membrane and nucleus Cytosol –Water with solutes (protein, salts, sugars, etc. ) Cytoplasmic organelles –Metabolic machinery of cell Inclusions – Granules of glycogen or pigments, lipid droplets, vacuoles, and crystals
Cytoplasm • Located between plasma membrane and nucleus Cytosol –Water with solutes (protein, salts, sugars, etc. ) Cytoplasmic organelles –Metabolic machinery of cell Inclusions – Granules of glycogen or pigments, lipid droplets, vacuoles, and crystals
 Cytoplasmic Organelles • Membranous – Mitochondria – Peroxisomes – Lysosomes – Endoplasmic reticulum – Golgi apparatus • Nonmembranous – Cytoskeleton – Centrioles – Ribosomes
Cytoplasmic Organelles • Membranous – Mitochondria – Peroxisomes – Lysosomes – Endoplasmic reticulum – Golgi apparatus • Nonmembranous – Cytoskeleton – Centrioles – Ribosomes
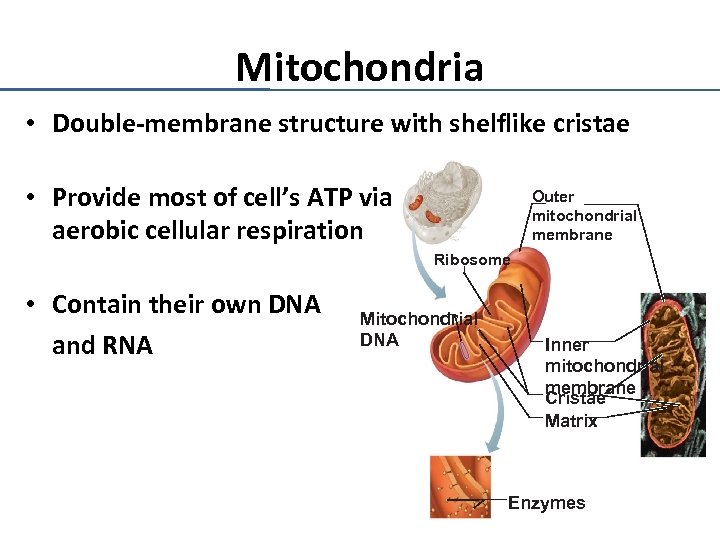 Mitochondria • Double-membrane structure with shelflike cristae • Provide most of cell’s ATP via aerobic cellular respiration Outer mitochondrial membrane Ribosome • Contain their own DNA and RNA Mitochondrial DNA Inner mitochondrial membrane Cristae Matrix Enzymes
Mitochondria • Double-membrane structure with shelflike cristae • Provide most of cell’s ATP via aerobic cellular respiration Outer mitochondrial membrane Ribosome • Contain their own DNA and RNA Mitochondrial DNA Inner mitochondrial membrane Cristae Matrix Enzymes
 Ribosomes • • Granules containing protein and r. RNA Site of protein synthesis Free ribosomes synthesize soluble proteins Membrane-bound ribosomes (on rough ER) synthesize proteins to be incorporated into membranes or exported from the cell
Ribosomes • • Granules containing protein and r. RNA Site of protein synthesis Free ribosomes synthesize soluble proteins Membrane-bound ribosomes (on rough ER) synthesize proteins to be incorporated into membranes or exported from the cell
 Endoplasmic Reticulum (ER) • Interconnected tubes and parallel membranes enclosing cisternae • Continuous with nuclear membrane • Two varieties – Rough ER – Smooth ER
Endoplasmic Reticulum (ER) • Interconnected tubes and parallel membranes enclosing cisternae • Continuous with nuclear membrane • Two varieties – Rough ER – Smooth ER
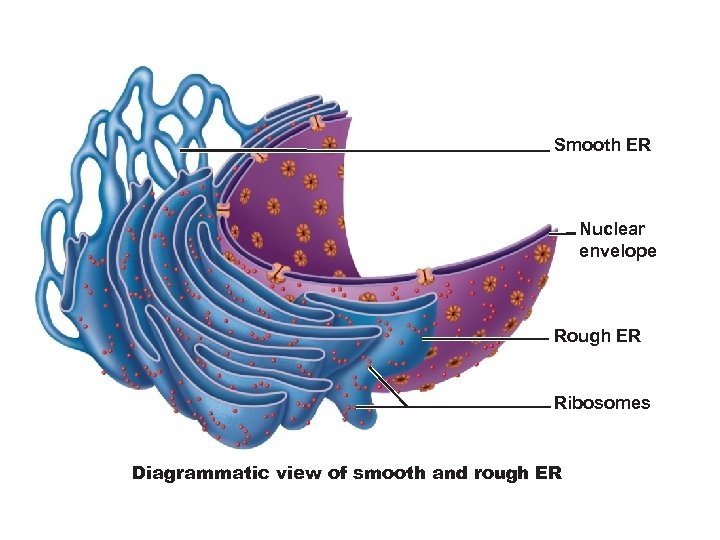 Smooth ER Nuclear envelope Rough ER Ribosomes Diagrammatic view of smooth and rough ER
Smooth ER Nuclear envelope Rough ER Ribosomes Diagrammatic view of smooth and rough ER
 Rough ER • External surface studded with ribosomes • Manufactures all secreted proteins • Synthesizes membrane integral proteins and phospholipids
Rough ER • External surface studded with ribosomes • Manufactures all secreted proteins • Synthesizes membrane integral proteins and phospholipids
 Smooth ER • Tubules arranged in a looping network • Enzyme (integral protein) functions – In the liver—lipid and cholesterol metabolism, breakdown of glycogen, and, along with kidneys, detoxification of drugs, pesticides, and carcinogens – Synthesis of steroid-based hormones – In intestinal cells—absorption, synthesis, and transport of fats – In skeletal and cardiac muscle—storage and release of calcium
Smooth ER • Tubules arranged in a looping network • Enzyme (integral protein) functions – In the liver—lipid and cholesterol metabolism, breakdown of glycogen, and, along with kidneys, detoxification of drugs, pesticides, and carcinogens – Synthesis of steroid-based hormones – In intestinal cells—absorption, synthesis, and transport of fats – In skeletal and cardiac muscle—storage and release of calcium
 Golgi Apparatus • Stacked and flattened membranous sacs • Modifies, concentrates, and packages proteins and lipids
Golgi Apparatus • Stacked and flattened membranous sacs • Modifies, concentrates, and packages proteins and lipids
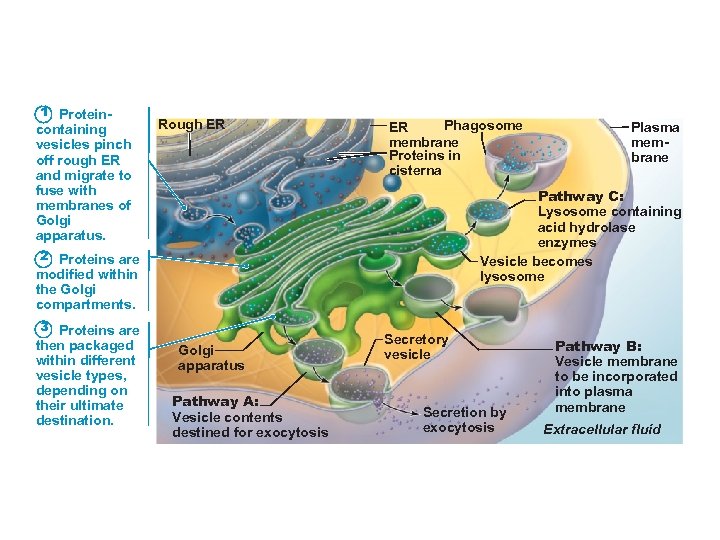 1 Protein- containing vesicles pinch off rough ER and migrate to fuse with membranes of Golgi apparatus. Rough ER Phagosome ER membrane Proteins in cisterna Pathway C: Lysosome containing acid hydrolase enzymes Vesicle becomes lysosome 2 Proteins are modified within the Golgi compartments. 3 Proteins are then packaged within different vesicle types, depending on their ultimate destination. Plasma membrane Golgi apparatus Pathway A: Vesicle contents destined for exocytosis Secretory vesicle Secretion by exocytosis Pathway B: Vesicle membrane to be incorporated into plasma membrane Extracellular fluid
1 Protein- containing vesicles pinch off rough ER and migrate to fuse with membranes of Golgi apparatus. Rough ER Phagosome ER membrane Proteins in cisterna Pathway C: Lysosome containing acid hydrolase enzymes Vesicle becomes lysosome 2 Proteins are modified within the Golgi compartments. 3 Proteins are then packaged within different vesicle types, depending on their ultimate destination. Plasma membrane Golgi apparatus Pathway A: Vesicle contents destined for exocytosis Secretory vesicle Secretion by exocytosis Pathway B: Vesicle membrane to be incorporated into plasma membrane Extracellular fluid
 Lysosomes • Spherical membranous bags containing digestive enzymes (acid hydrolases) • Digest ingested bacteria, viruses, and toxins • Degrade nonfunctional organelles • Break down and release glycogen • Break down bone to release Ca 2+ • Destroy cells in injured or nonuseful tissue (autolysis)
Lysosomes • Spherical membranous bags containing digestive enzymes (acid hydrolases) • Digest ingested bacteria, viruses, and toxins • Degrade nonfunctional organelles • Break down and release glycogen • Break down bone to release Ca 2+ • Destroy cells in injured or nonuseful tissue (autolysis)
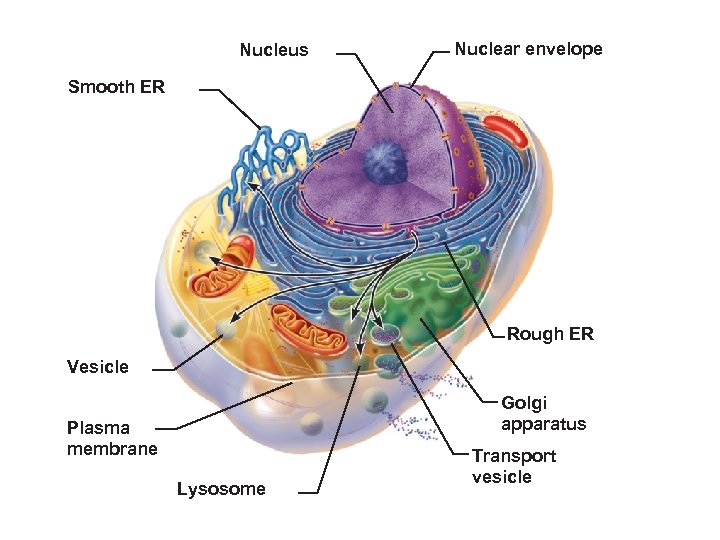 Nucleus Nuclear envelope Smooth ER Rough ER Vesicle Golgi apparatus Plasma membrane Lysosome Transport vesicle
Nucleus Nuclear envelope Smooth ER Rough ER Vesicle Golgi apparatus Plasma membrane Lysosome Transport vesicle
 Peroxisomes • Membranous sacs containing powerful oxidases and catalases • Detoxify harmful or toxic substances • Neutralize dangerous free radicals (highly reactive chemicals with unpaired electrons)
Peroxisomes • Membranous sacs containing powerful oxidases and catalases • Detoxify harmful or toxic substances • Neutralize dangerous free radicals (highly reactive chemicals with unpaired electrons)
 What organelle is important for protein synthesis? A. Ribosomes B. Nucleus C. Mitochondria D. Lysosome
What organelle is important for protein synthesis? A. Ribosomes B. Nucleus C. Mitochondria D. Lysosome
 Cytoskeleton • Elaborate series of rods throughout cytosol – Microtubules – Microfilaments – Intermediate filaments
Cytoskeleton • Elaborate series of rods throughout cytosol – Microtubules – Microfilaments – Intermediate filaments
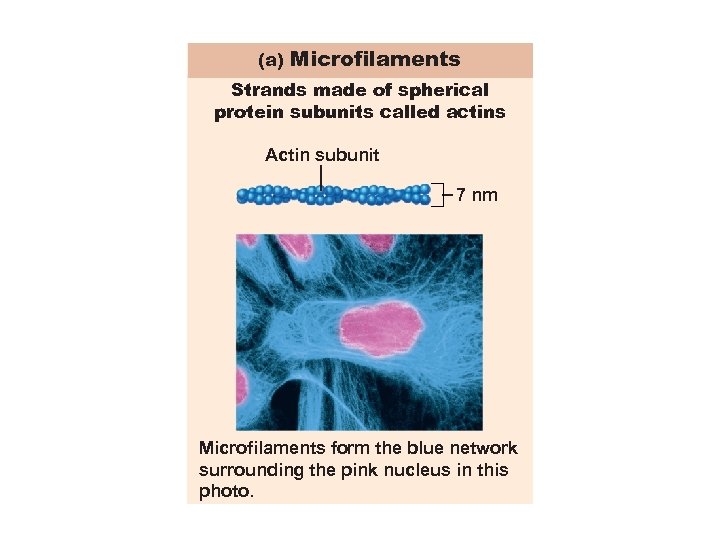 (a) Microfilaments Strands made of spherical protein subunits called actins Actin subunit 7 nm Microfilaments form the blue network surrounding the pink nucleus in this photo.
(a) Microfilaments Strands made of spherical protein subunits called actins Actin subunit 7 nm Microfilaments form the blue network surrounding the pink nucleus in this photo.
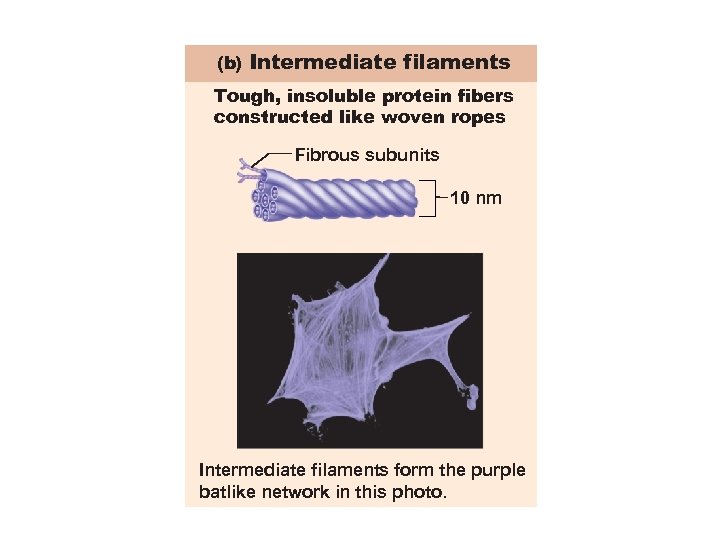 (b) Intermediate filaments Tough, insoluble protein fibers constructed like woven ropes Fibrous subunits 10 nm Intermediate filaments form the purple batlike network in this photo.
(b) Intermediate filaments Tough, insoluble protein fibers constructed like woven ropes Fibrous subunits 10 nm Intermediate filaments form the purple batlike network in this photo.
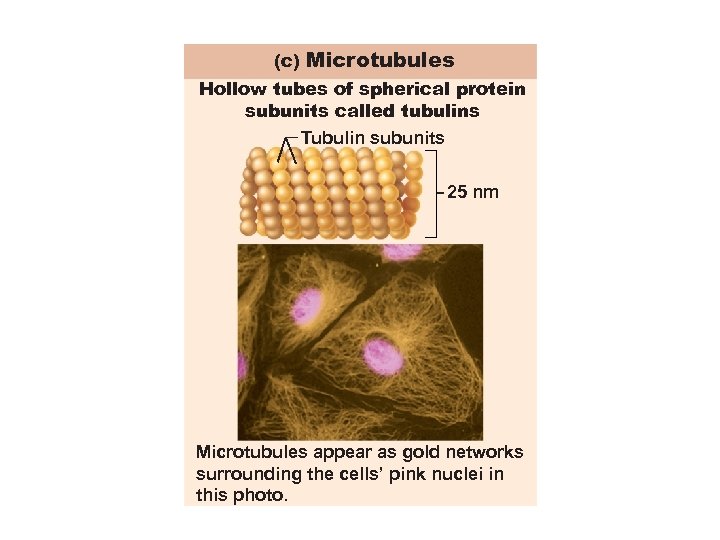 (c) Microtubules Hollow tubes of spherical protein subunits called tubulins Tubulin subunits 25 nm Microtubules appear as gold networks surrounding the cells’ pink nuclei in this photo.
(c) Microtubules Hollow tubes of spherical protein subunits called tubulins Tubulin subunits 25 nm Microtubules appear as gold networks surrounding the cells’ pink nuclei in this photo.
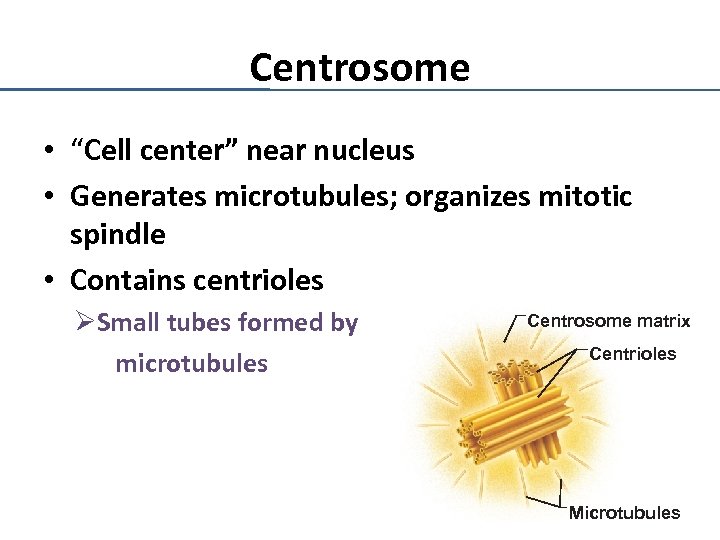 Centrosome • “Cell center” near nucleus • Generates microtubules; organizes mitotic spindle • Contains centrioles ØSmall tubes formed by microtubules Centrosome matrix Centrioles Microtubules
Centrosome • “Cell center” near nucleus • Generates microtubules; organizes mitotic spindle • Contains centrioles ØSmall tubes formed by microtubules Centrosome matrix Centrioles Microtubules
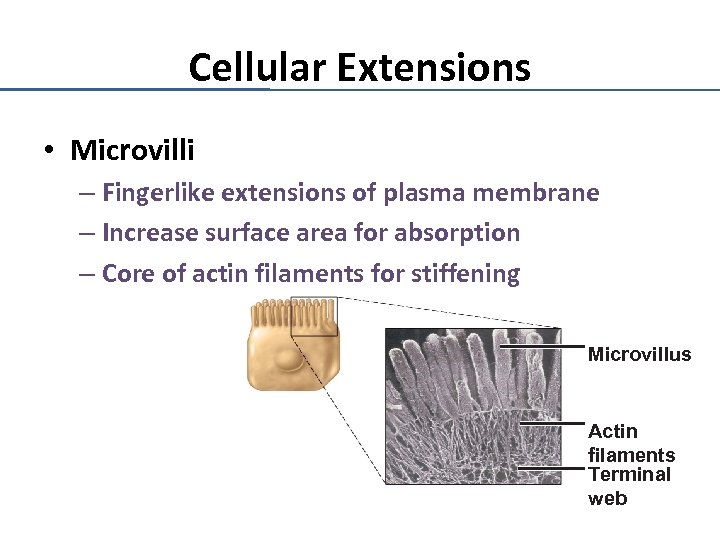 Cellular Extensions • Microvilli – Fingerlike extensions of plasma membrane – Increase surface area for absorption – Core of actin filaments for stiffening Microvillus Actin filaments Terminal web
Cellular Extensions • Microvilli – Fingerlike extensions of plasma membrane – Increase surface area for absorption – Core of actin filaments for stiffening Microvillus Actin filaments Terminal web
 Nucleus • Genetic library with blueprints for nearly all cellular proteins • Responds to signals and dictates kinds and amounts of proteins to be synthesized • Most cells have one nucleus
Nucleus • Genetic library with blueprints for nearly all cellular proteins • Responds to signals and dictates kinds and amounts of proteins to be synthesized • Most cells have one nucleus
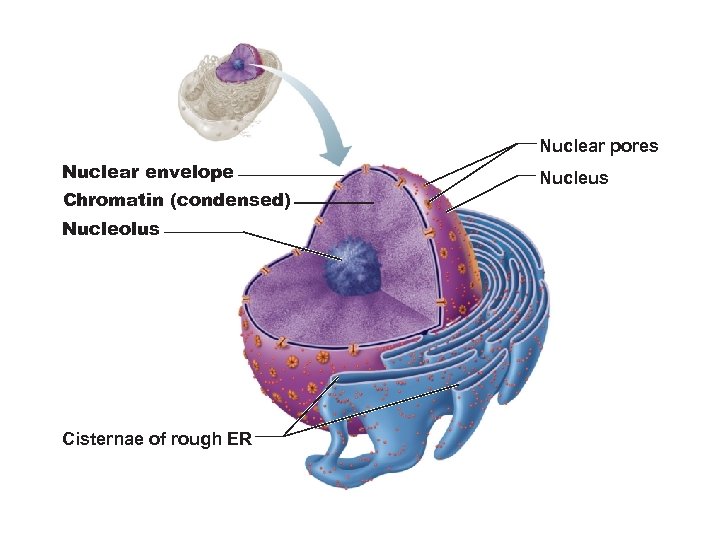 Nuclear pores Nuclear envelope Chromatin (condensed) Nucleolus Cisternae of rough ER Nucleus
Nuclear pores Nuclear envelope Chromatin (condensed) Nucleolus Cisternae of rough ER Nucleus
 Nuclear Envelope • Double-membrane barrier containing pores • Outer layer is continuous with rough ER and bears ribosomes • Inner lining (nuclear lamina) maintains shape of nucleus • Pore complex regulates transport of large molecules into and out of nucleus
Nuclear Envelope • Double-membrane barrier containing pores • Outer layer is continuous with rough ER and bears ribosomes • Inner lining (nuclear lamina) maintains shape of nucleus • Pore complex regulates transport of large molecules into and out of nucleus
 Nucleoli • Dark-staining spherical bodies within nucleus • Involved in r. RNA synthesis and ribosome subunit assembly
Nucleoli • Dark-staining spherical bodies within nucleus • Involved in r. RNA synthesis and ribosome subunit assembly
 Cell Cycle • Two Major Parts – Interphase – Cell division (Mitotic phase)
Cell Cycle • Two Major Parts – Interphase – Cell division (Mitotic phase)
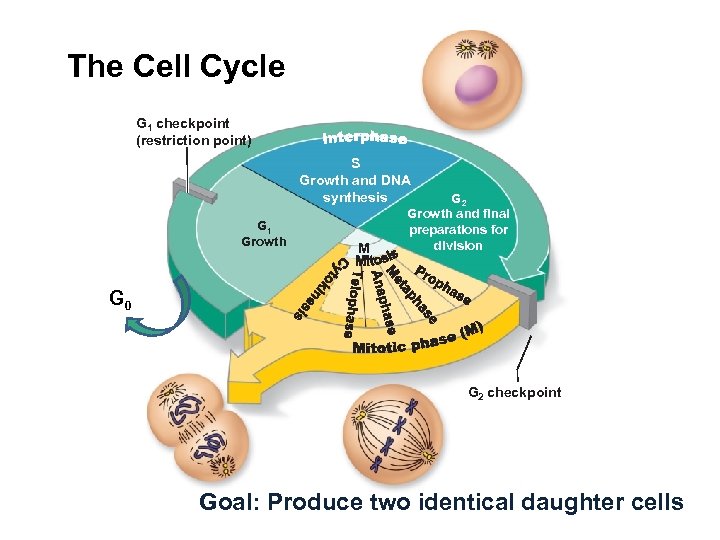 The Cell Cycle G 1 checkpoint (restriction point) S Growth and DNA synthesis G 1 Growth M G 2 Growth and final preparations for division G 0 G 2 checkpoint Goal: Produce two identical daughter cells
The Cell Cycle G 1 checkpoint (restriction point) S Growth and DNA synthesis G 1 Growth M G 2 Growth and final preparations for division G 0 G 2 checkpoint Goal: Produce two identical daughter cells
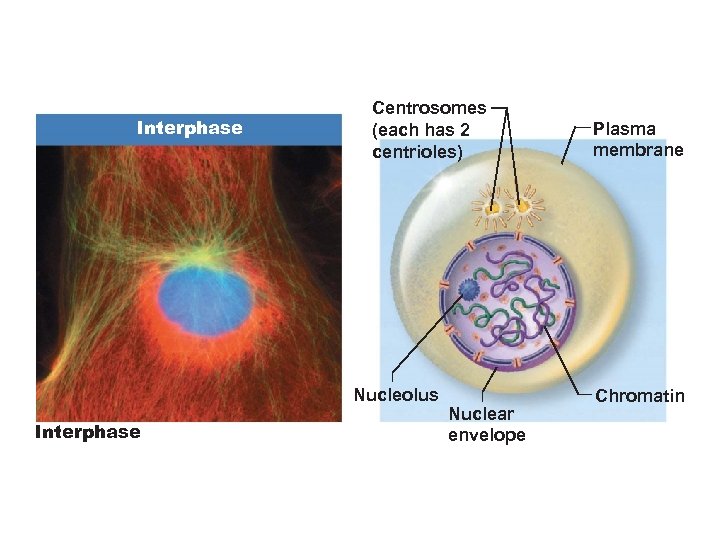 Interphase Centrosomes (each has 2 centrioles) Nucleolus Interphase Nuclear envelope Plasma membrane Chromatin
Interphase Centrosomes (each has 2 centrioles) Nucleolus Interphase Nuclear envelope Plasma membrane Chromatin
 Mitosis Four Parts • • Prophase Metaphase Anaphase Telophase
Mitosis Four Parts • • Prophase Metaphase Anaphase Telophase
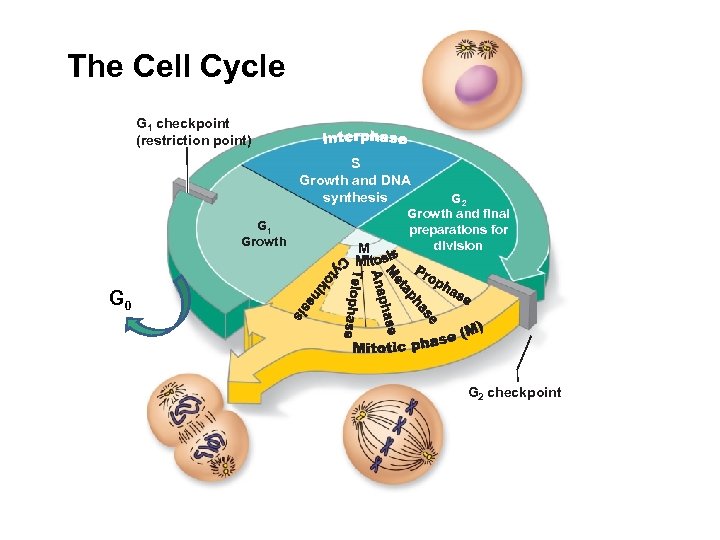 The Cell Cycle G 1 checkpoint (restriction point) S Growth and DNA synthesis G 1 Growth M G 2 Growth and final preparations for division G 0 G 2 checkpoint
The Cell Cycle G 1 checkpoint (restriction point) S Growth and DNA synthesis G 1 Growth M G 2 Growth and final preparations for division G 0 G 2 checkpoint
 Prophase • Chromosomes become visible, each with two chromatids joined at a centromere • Centrosomes separate and migrate toward opposite poles • Mitotic spindles and asters form
Prophase • Chromosomes become visible, each with two chromatids joined at a centromere • Centrosomes separate and migrate toward opposite poles • Mitotic spindles and asters form
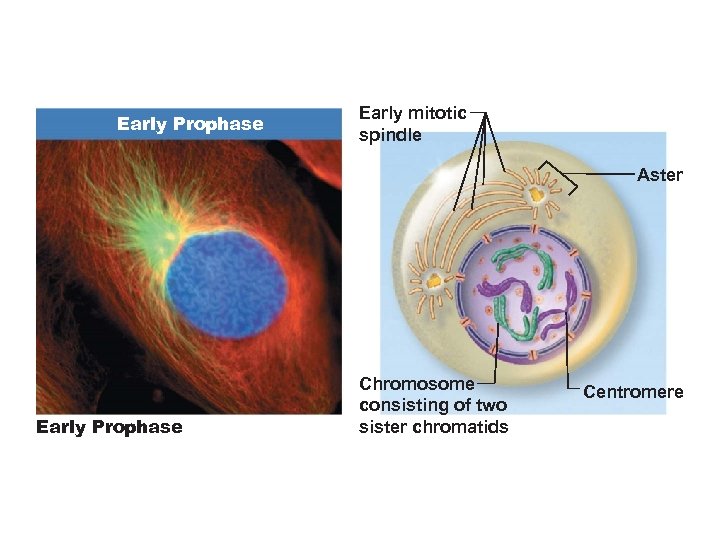 Early Prophase Early mitotic spindle Aster Early Prophase Chromosome consisting of two sister chromatids Centromere
Early Prophase Early mitotic spindle Aster Early Prophase Chromosome consisting of two sister chromatids Centromere
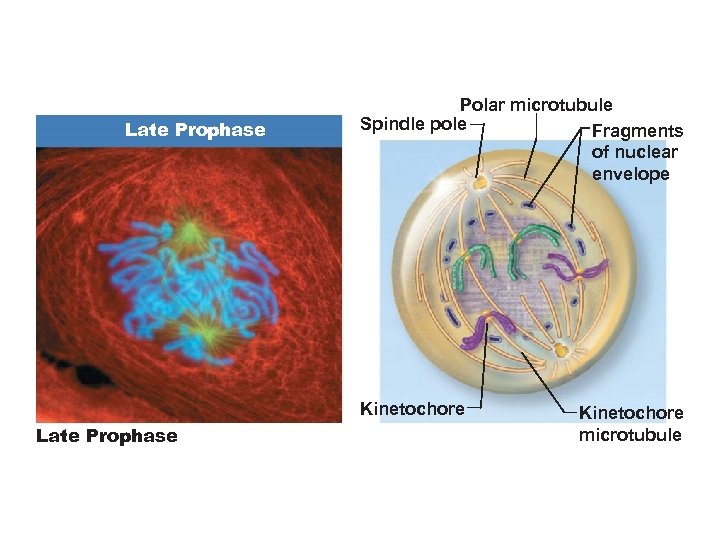 Late Prophase Polar microtubule Spindle pole Fragments of nuclear envelope Kinetochore Late Prophase Kinetochore microtubule
Late Prophase Polar microtubule Spindle pole Fragments of nuclear envelope Kinetochore Late Prophase Kinetochore microtubule
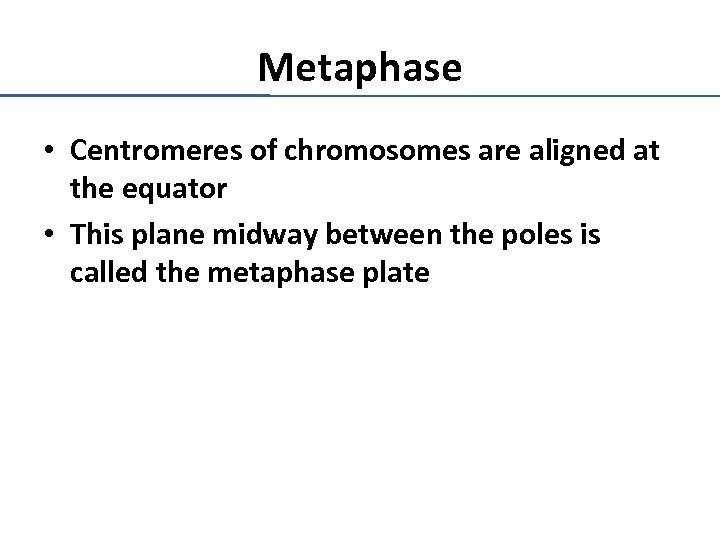 Metaphase • Centromeres of chromosomes are aligned at the equator • This plane midway between the poles is called the metaphase plate
Metaphase • Centromeres of chromosomes are aligned at the equator • This plane midway between the poles is called the metaphase plate
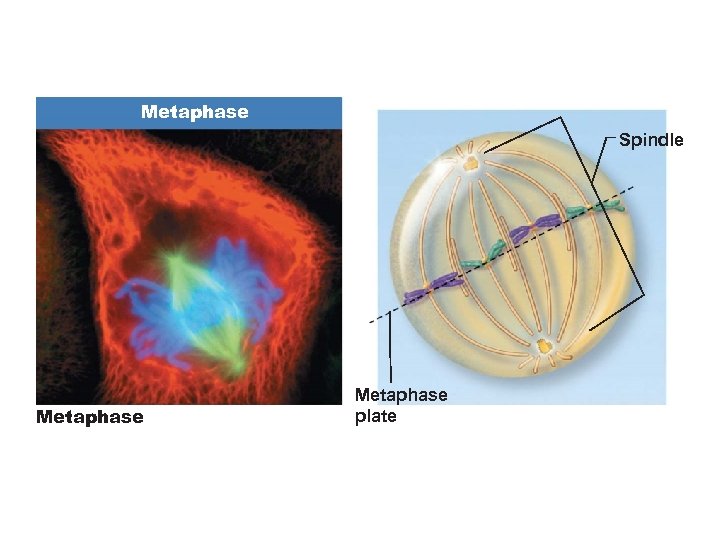 Metaphase Spindle Metaphase plate
Metaphase Spindle Metaphase plate
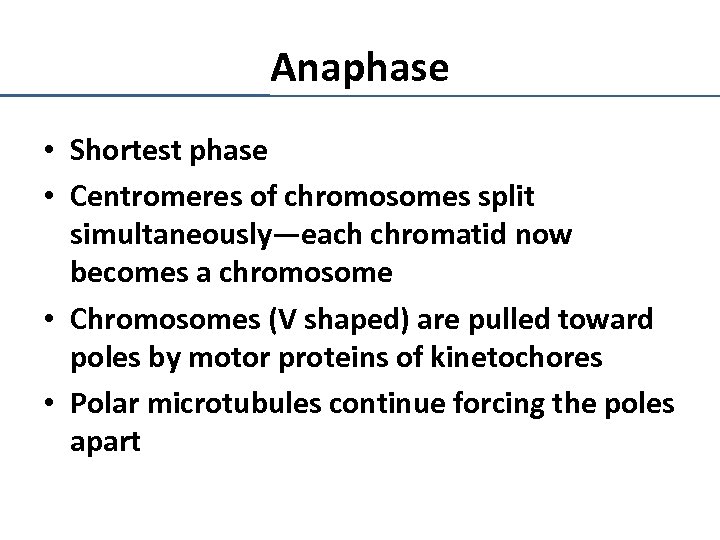 Anaphase • Shortest phase • Centromeres of chromosomes split simultaneously—each chromatid now becomes a chromosome • Chromosomes (V shaped) are pulled toward poles by motor proteins of kinetochores • Polar microtubules continue forcing the poles apart
Anaphase • Shortest phase • Centromeres of chromosomes split simultaneously—each chromatid now becomes a chromosome • Chromosomes (V shaped) are pulled toward poles by motor proteins of kinetochores • Polar microtubules continue forcing the poles apart
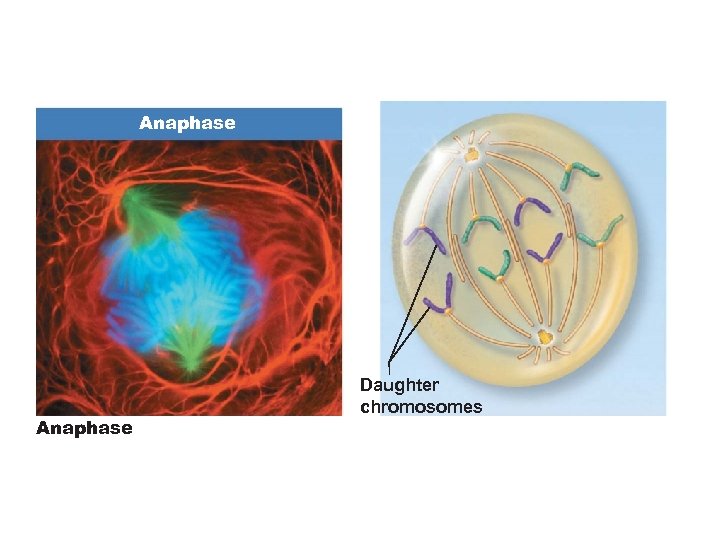 Anaphase Daughter chromosomes
Anaphase Daughter chromosomes
 Telophase • Begins when chromosome movement stops • The two sets of chromosomes uncoil to form chromatin • New nuclear membrane forms around each chromatin mass • Nucleoli reappear • Spindles disappear
Telophase • Begins when chromosome movement stops • The two sets of chromosomes uncoil to form chromatin • New nuclear membrane forms around each chromatin mass • Nucleoli reappear • Spindles disappear
 Cytokinesis • Begins during late anaphase • Ring of actin microfilaments contracts to form a cleavage furrow • Two daughter cells are pinched apart, each containing a nucleus identical to the original
Cytokinesis • Begins during late anaphase • Ring of actin microfilaments contracts to form a cleavage furrow • Two daughter cells are pinched apart, each containing a nucleus identical to the original
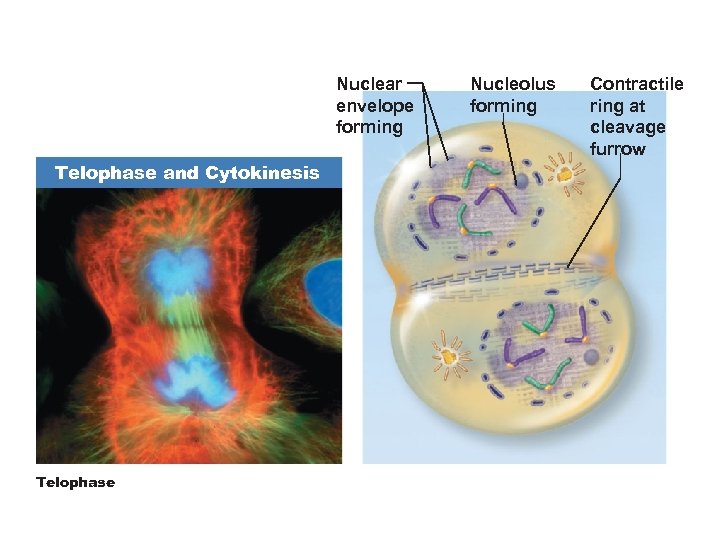 Nuclear envelope forming Telophase and Cytokinesis Telophase Nucleolus forming Contractile ring at cleavage furrow
Nuclear envelope forming Telophase and Cytokinesis Telophase Nucleolus forming Contractile ring at cleavage furrow
 During what stage of the cell cycle is DNA replicated? A. Metaphase B. Interphase C. Prophase D. Telophase
During what stage of the cell cycle is DNA replicated? A. Metaphase B. Interphase C. Prophase D. Telophase
 Cellular Respiration - How the body produces energy (ATP) from glucose • Aerobic Respiration – respiration in the presence of oxygen • Glucose + Oxygen Water + Carbon Dioxide + ATP • Anaerobic Respiration – respiration in the absence of oxygen • Glucose Lactic Acid + ATP
Cellular Respiration - How the body produces energy (ATP) from glucose • Aerobic Respiration – respiration in the presence of oxygen • Glucose + Oxygen Water + Carbon Dioxide + ATP • Anaerobic Respiration – respiration in the absence of oxygen • Glucose Lactic Acid + ATP
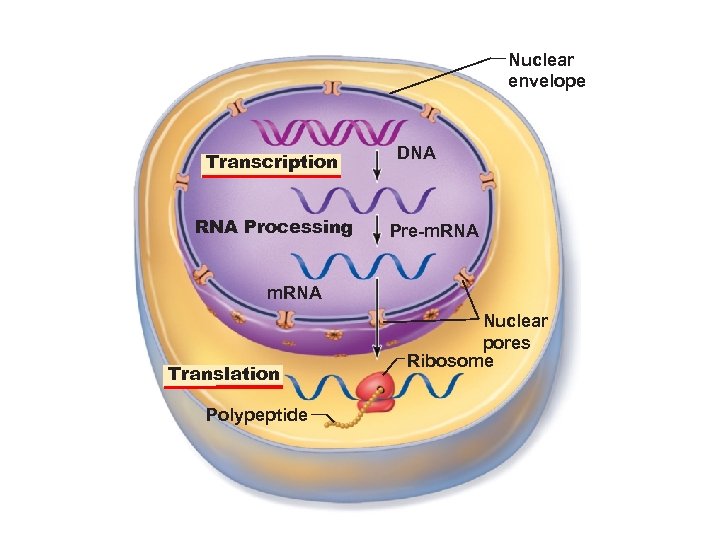 Nuclear envelope Transcription RNA Processing DNA Pre-m. RNA Translation Polypeptide Nuclear pores Ribosome
Nuclear envelope Transcription RNA Processing DNA Pre-m. RNA Translation Polypeptide Nuclear pores Ribosome
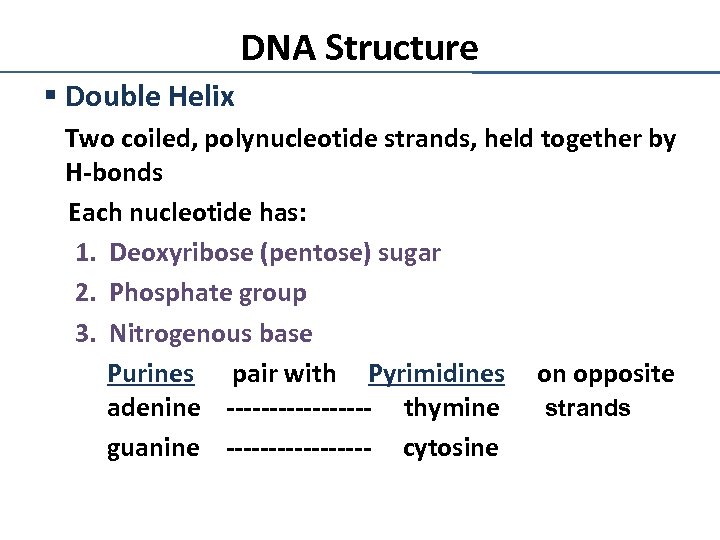 DNA Structure § Double Helix Two coiled, polynucleotide strands, held together by H-bonds Each nucleotide has: 1. Deoxyribose (pentose) sugar 2. Phosphate group 3. Nitrogenous base Purines pair with Pyrimidines on opposite adenine --------- thymine strands guanine --------- cytosine
DNA Structure § Double Helix Two coiled, polynucleotide strands, held together by H-bonds Each nucleotide has: 1. Deoxyribose (pentose) sugar 2. Phosphate group 3. Nitrogenous base Purines pair with Pyrimidines on opposite adenine --------- thymine strands guanine --------- cytosine
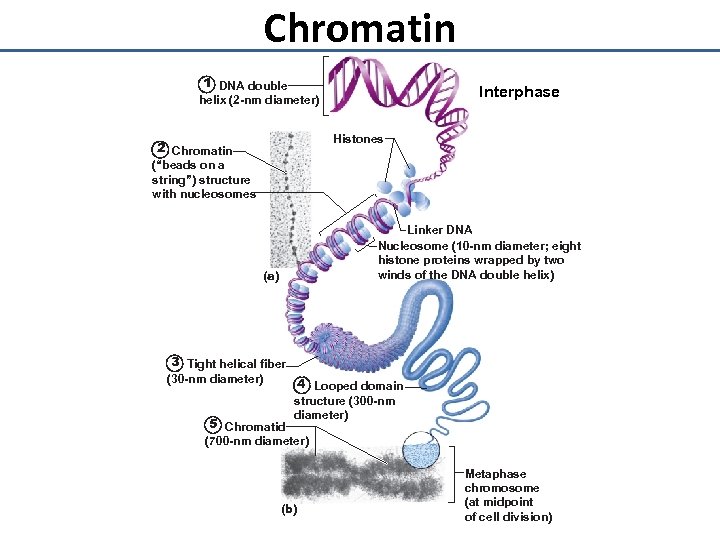 Chromatin 1 DNA double Interphase helix (2 -nm diameter) Histones 2 Chromatin (“beads on a string”) structure with nucleosomes Linker DNA Nucleosome (10 -nm diameter; eight histone proteins wrapped by two winds of the DNA double helix) (a) 3 Tight helical fiber (30 -nm diameter) 4 Looped domain 5 Chromatid structure (300 -nm diameter) (700 -nm diameter) (b) Metaphase chromosome (at midpoint of cell division)
Chromatin 1 DNA double Interphase helix (2 -nm diameter) Histones 2 Chromatin (“beads on a string”) structure with nucleosomes Linker DNA Nucleosome (10 -nm diameter; eight histone proteins wrapped by two winds of the DNA double helix) (a) 3 Tight helical fiber (30 -nm diameter) 4 Looped domain 5 Chromatid structure (300 -nm diameter) (700 -nm diameter) (b) Metaphase chromosome (at midpoint of cell division)
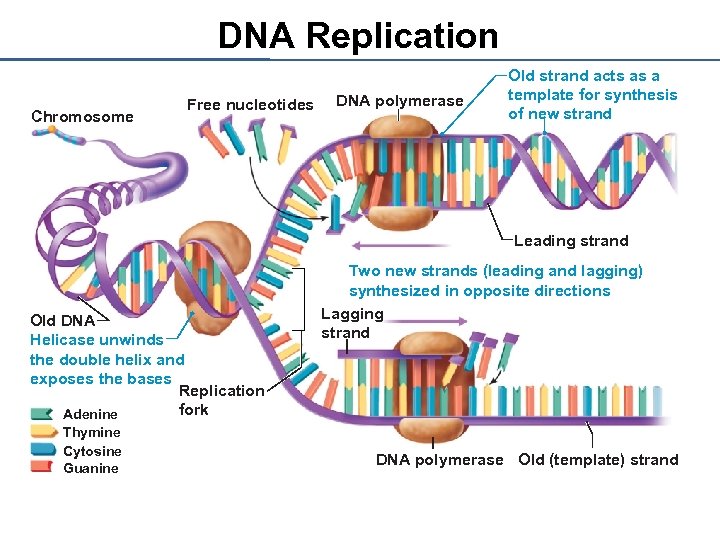 DNA Replication Chromosome Free nucleotides DNA polymerase Old strand acts as a template for synthesis of new strand Leading strand Old DNA Helicase unwinds the double helix and exposes the bases Replication fork Adenine Thymine Cytosine Guanine Two new strands (leading and lagging) synthesized in opposite directions Lagging strand DNA polymerase Old (template) strand
DNA Replication Chromosome Free nucleotides DNA polymerase Old strand acts as a template for synthesis of new strand Leading strand Old DNA Helicase unwinds the double helix and exposes the bases Replication fork Adenine Thymine Cytosine Guanine Two new strands (leading and lagging) synthesized in opposite directions Lagging strand DNA polymerase Old (template) strand
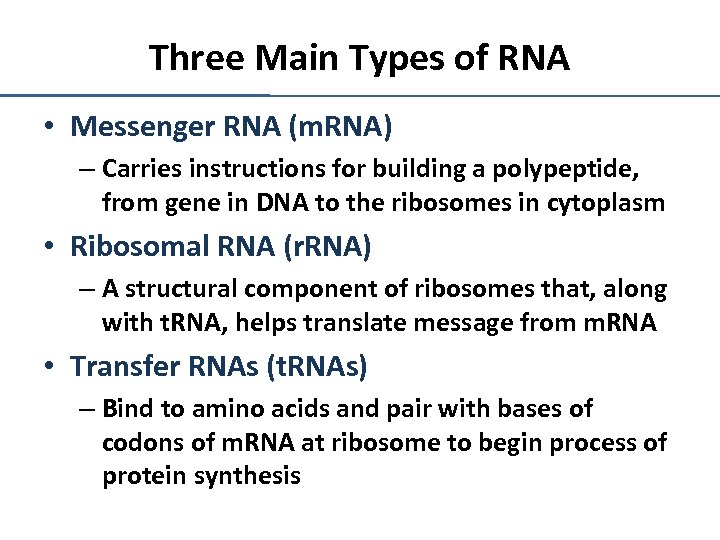 Three Main Types of RNA • Messenger RNA (m. RNA) – Carries instructions for building a polypeptide, from gene in DNA to the ribosomes in cytoplasm • Ribosomal RNA (r. RNA) – A structural component of ribosomes that, along with t. RNA, helps translate message from m. RNA • Transfer RNAs (t. RNAs) – Bind to amino acids and pair with bases of codons of m. RNA at ribosome to begin process of protein synthesis
Three Main Types of RNA • Messenger RNA (m. RNA) – Carries instructions for building a polypeptide, from gene in DNA to the ribosomes in cytoplasm • Ribosomal RNA (r. RNA) – A structural component of ribosomes that, along with t. RNA, helps translate message from m. RNA • Transfer RNAs (t. RNAs) – Bind to amino acids and pair with bases of codons of m. RNA at ribosome to begin process of protein synthesis
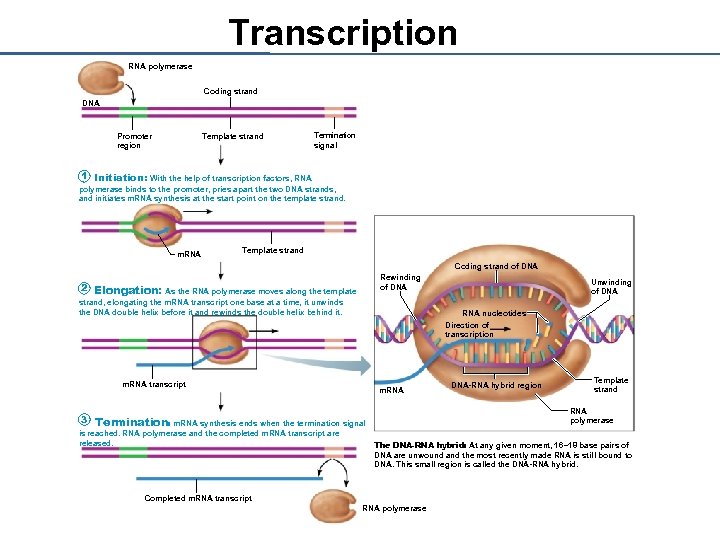 Transcription RNA polymerase Coding strand DNA Promoter region Template strand Termination signal 1 Initiation: With the help of transcription factors, RNA polymerase binds to the promoter, pries apart the two DNA strands, and initiates m. RNA synthesis at the start point on the template strand. m. RNA Template strand Coding strand of DNA 2 Rewinding of DNA Elongation: As the RNA polymerase moves along the template strand, elongating the m. RNA transcript one base at a time, it unwinds the DNA double helix before it and rewinds the double helix behind it. RNA nucleotides Direction of transcription m. RNA transcript 3 m. RNA DNA-RNA hybrid region Template strand RNA polymerase Termination: m. RNA synthesis ends when the termination signal is reached. RNA polymerase and the completed m. RNA transcript are released. Unwinding of DNA The DNA-RNA hybrid: At any given moment, 16– 18 base pairs of DNA are unwound and the most recently made RNA is still bound to DNA. This small region is called the DNA-RNA hybrid. Completed m. RNA transcript RNA polymerase
Transcription RNA polymerase Coding strand DNA Promoter region Template strand Termination signal 1 Initiation: With the help of transcription factors, RNA polymerase binds to the promoter, pries apart the two DNA strands, and initiates m. RNA synthesis at the start point on the template strand. m. RNA Template strand Coding strand of DNA 2 Rewinding of DNA Elongation: As the RNA polymerase moves along the template strand, elongating the m. RNA transcript one base at a time, it unwinds the DNA double helix before it and rewinds the double helix behind it. RNA nucleotides Direction of transcription m. RNA transcript 3 m. RNA DNA-RNA hybrid region Template strand RNA polymerase Termination: m. RNA synthesis ends when the termination signal is reached. RNA polymerase and the completed m. RNA transcript are released. Unwinding of DNA The DNA-RNA hybrid: At any given moment, 16– 18 base pairs of DNA are unwound and the most recently made RNA is still bound to DNA. This small region is called the DNA-RNA hybrid. Completed m. RNA transcript RNA polymerase
 Translation • Converts base sequence of nucleic acids into the amino acid sequence of proteins • Involves m. RNAs, t. RNAs, and r. RNAs • Takes place in the cytoplasm
Translation • Converts base sequence of nucleic acids into the amino acid sequence of proteins • Involves m. RNAs, t. RNAs, and r. RNAs • Takes place in the cytoplasm
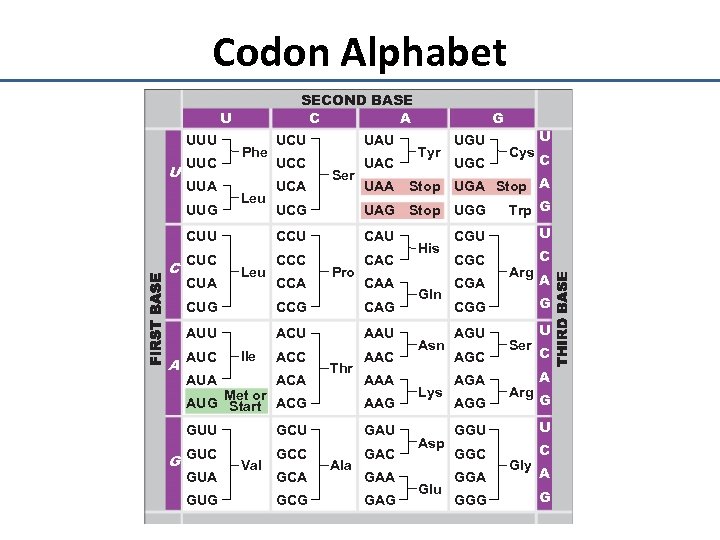 Codon Alphabet SECOND BASE C A U UUU U UUC Phe UCU UAU UCC UAC Ser G Tyr UGU UGC Cys U C UCG Stop UGA Stop A UAG Stop UGG Trp G CUU CCU CAU CUC CCC CAC UUA UUG C CUA Leu UCA CCA Pro UAA CUG CAG AUU A CCG ACU AAU ACC AAC AUC Ile AUA ACA Thr Met or AUG Start ACG AAA AAG GUU G GCU GAU GUC GCC GAC GUA GUG Val GCA GCG Ala GAA GAG His Gln Asn Lys Asp Glu CGU U CGC C CGA Arg A CGG G AGU U AGC AGA AGG Ser Arg C A G GGU U GGC C GGA GGG Gly A G
Codon Alphabet SECOND BASE C A U UUU U UUC Phe UCU UAU UCC UAC Ser G Tyr UGU UGC Cys U C UCG Stop UGA Stop A UAG Stop UGG Trp G CUU CCU CAU CUC CCC CAC UUA UUG C CUA Leu UCA CCA Pro UAA CUG CAG AUU A CCG ACU AAU ACC AAC AUC Ile AUA ACA Thr Met or AUG Start ACG AAA AAG GUU G GCU GAU GUC GCC GAC GUA GUG Val GCA GCG Ala GAA GAG His Gln Asn Lys Asp Glu CGU U CGC C CGA Arg A CGG G AGU U AGC AGA AGG Ser Arg C A G GGU U GGC C GGA GGG Gly A G
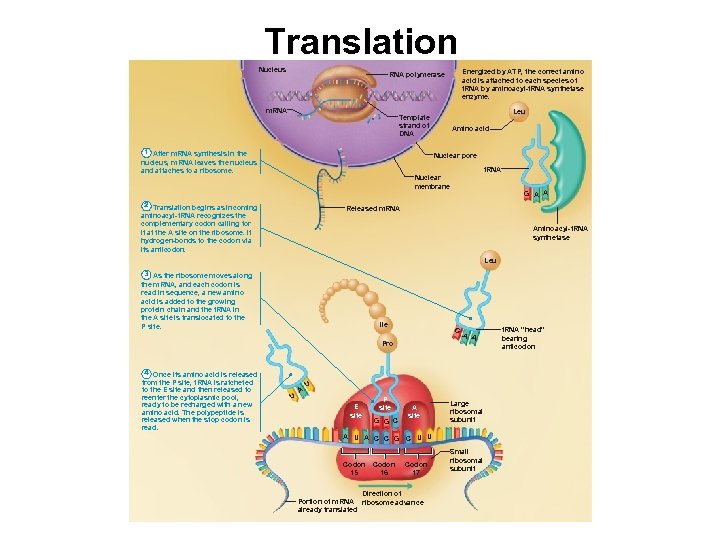 Translation Nucleus Energized by ATP, the correct amino acid is attached to each species of t. RNA by aminoacyl-t. RNA synthetase enzyme. RNA polymerase m. RNA Leu Template strand of DNA 1 After m. RNA synthesis in the nucleus, m. RNA leaves the nucleus and attaches to a ribosome. Amino acid Nuclear pore t. RNA Nuclear membrane 2 Translation begins as incoming aminoacyl-t. RNA recognizes the complementary codon calling for it at the A site on the ribosome. It hydrogen-bonds to the codon via its anticodon. G A A Released m. RNA Aminoacyl-t. RNA synthetase Leu 3 As the ribosome moves along the m. RNA, and each codon is read in sequence, a new amino acid is added to the growing protein chain and the t. RNA in the A site is translocated to the P site. Ile G Pro 4 Once its amino acid is released from the P site, t. RNA is ratcheted to the E site and then released to reenter the cytoplasmic pool, ready to be recharged with a new amino acid. The polypeptide is released when the stop codon is read. U A A A U E site P site G G C A site A U A C C G C U U Codon 15 Codon 17 Large ribosomal subunit Codon 16 Direction of Portion of m. RNA ribosome advance already translated Small ribosomal subunit t. RNA “head” bearing anticodon
Translation Nucleus Energized by ATP, the correct amino acid is attached to each species of t. RNA by aminoacyl-t. RNA synthetase enzyme. RNA polymerase m. RNA Leu Template strand of DNA 1 After m. RNA synthesis in the nucleus, m. RNA leaves the nucleus and attaches to a ribosome. Amino acid Nuclear pore t. RNA Nuclear membrane 2 Translation begins as incoming aminoacyl-t. RNA recognizes the complementary codon calling for it at the A site on the ribosome. It hydrogen-bonds to the codon via its anticodon. G A A Released m. RNA Aminoacyl-t. RNA synthetase Leu 3 As the ribosome moves along the m. RNA, and each codon is read in sequence, a new amino acid is added to the growing protein chain and the t. RNA in the A site is translocated to the P site. Ile G Pro 4 Once its amino acid is released from the P site, t. RNA is ratcheted to the E site and then released to reenter the cytoplasmic pool, ready to be recharged with a new amino acid. The polypeptide is released when the stop codon is read. U A A A U E site P site G G C A site A U A C C G C U U Codon 15 Codon 17 Large ribosomal subunit Codon 16 Direction of Portion of m. RNA ribosome advance already translated Small ribosomal subunit t. RNA “head” bearing anticodon
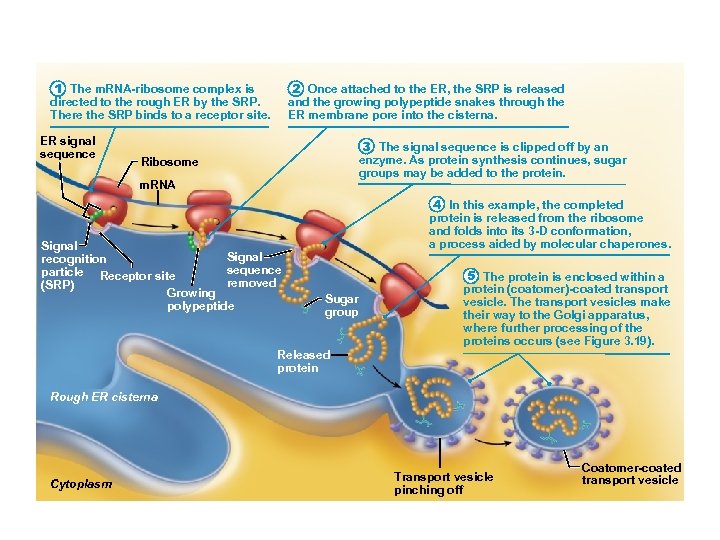 1 The m. RNA-ribosome complex is directed to the rough ER by the SRP. There the SRP binds to a receptor site. ER signal sequence 2 Once attached to the ER, the SRP is released and the growing polypeptide snakes through the ER membrane pore into the cisterna. 3 The signal sequence is clipped off by an enzyme. As protein synthesis continues, sugar groups may be added to the protein. Ribosome m. RNA Signal recognition sequence particle Receptor site removed (SRP) Growing polypeptide 4 In this example, the completed protein is released from the ribosome and folds into its 3 -D conformation, a process aided by molecular chaperones. Sugar group 5 The protein is enclosed within a protein (coatomer)-coated transport vesicle. The transport vesicles make their way to the Golgi apparatus, where further processing of the proteins occurs (see Figure 3. 19). Released protein Rough ER cisterna Cytoplasm Transport vesicle pinching off Coatomer-coated transport vesicle
1 The m. RNA-ribosome complex is directed to the rough ER by the SRP. There the SRP binds to a receptor site. ER signal sequence 2 Once attached to the ER, the SRP is released and the growing polypeptide snakes through the ER membrane pore into the cisterna. 3 The signal sequence is clipped off by an enzyme. As protein synthesis continues, sugar groups may be added to the protein. Ribosome m. RNA Signal recognition sequence particle Receptor site removed (SRP) Growing polypeptide 4 In this example, the completed protein is released from the ribosome and folds into its 3 -D conformation, a process aided by molecular chaperones. Sugar group 5 The protein is enclosed within a protein (coatomer)-coated transport vesicle. The transport vesicles make their way to the Golgi apparatus, where further processing of the proteins occurs (see Figure 3. 19). Released protein Rough ER cisterna Cytoplasm Transport vesicle pinching off Coatomer-coated transport vesicle


