4fa64c11c8b12403caf1ef2cffc9d8d4.ppt
- Количество слайдов: 29
 How will we efficiently understand the interactions of ~20, 000 genes, with ~200 million potential pairwise interactions? Minimally, we need to use the information that exists
How will we efficiently understand the interactions of ~20, 000 genes, with ~200 million potential pairwise interactions? Minimally, we need to use the information that exists
 June 1979: 2 relevant papers S. Brenner (Genetics 1974) The genetics of Caenorhabditis elegans J. Sulston & R. Horvitz (Developmental Biology 1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans Jan 2008: >200, 000 relevant papers
June 1979: 2 relevant papers S. Brenner (Genetics 1974) The genetics of Caenorhabditis elegans J. Sulston & R. Horvitz (Developmental Biology 1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans Jan 2008: >200, 000 relevant papers
 Prioritizing high resolution genetic interaction tests by knowledge mining 1 Full text information retrieval Hans-Michael Muller, Arun Rangarajan, Tracy Teal, Kimberly Van Auken, Juancarlos Chan 2 Predicting Gene Interactions from information available in public databases Weiwei Zhong
Prioritizing high resolution genetic interaction tests by knowledge mining 1 Full text information retrieval Hans-Michael Muller, Arun Rangarajan, Tracy Teal, Kimberly Van Auken, Juancarlos Chan 2 Predicting Gene Interactions from information available in public databases Weiwei Zhong
 Textpresso Literature Search Engine www. textpresso. org Scientists spend more time skimming for information than reading papers. Much information are details hidden in the full text, and are neither in the abstract nor captured in Me. SH terms. We designed Textpresso to do automated skimming for researchers and database curators. The output can be used for more sophisticated Language Processing. Natural
Textpresso Literature Search Engine www. textpresso. org Scientists spend more time skimming for information than reading papers. Much information are details hidden in the full text, and are neither in the abstract nor captured in Me. SH terms. We designed Textpresso to do automated skimming for researchers and database curators. The output can be used for more sophisticated Language Processing. Natural
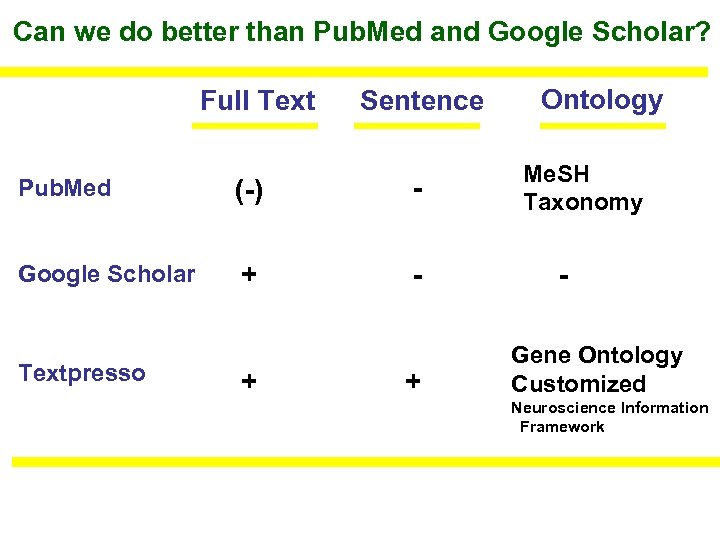 Can we do better than Pub. Med and Google Scholar? Full Text Sentence Pub. Med (-) - Google Scholar + - Textpresso + + Ontology Me. SH Taxonomy Gene Ontology Customized Neuroscience Information Framework
Can we do better than Pub. Med and Google Scholar? Full Text Sentence Pub. Med (-) - Google Scholar + - Textpresso + + Ontology Me. SH Taxonomy Gene Ontology Customized Neuroscience Information Framework
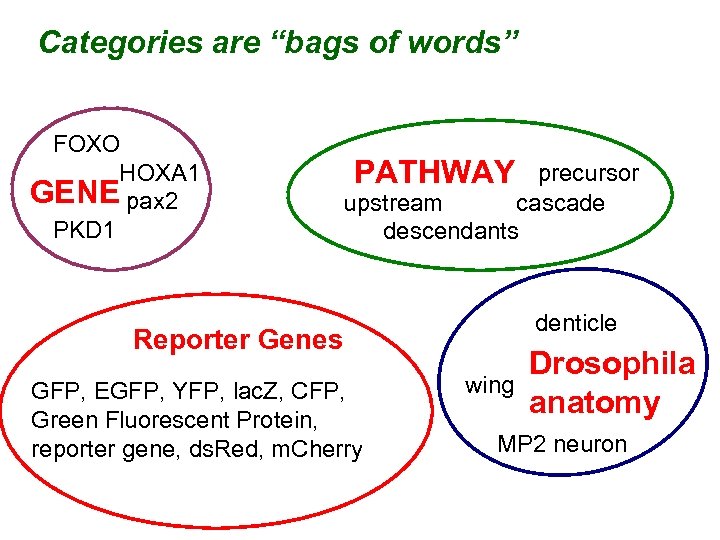 Categories are “bags of words” FOXO HOXA 1 GENE pax 2 PKD 1 PATHWAY precursor upstream cascade descendants denticle Reporter Genes GFP, EGFP, YFP, lac. Z, CFP, Green Fluorescent Protein, reporter gene, ds. Red, m. Cherry wing Drosophila anatomy MP 2 neuron
Categories are “bags of words” FOXO HOXA 1 GENE pax 2 PKD 1 PATHWAY precursor upstream cascade descendants denticle Reporter Genes GFP, EGFP, YFP, lac. Z, CFP, Green Fluorescent Protein, reporter gene, ds. Red, m. Cherry wing Drosophila anatomy MP 2 neuron
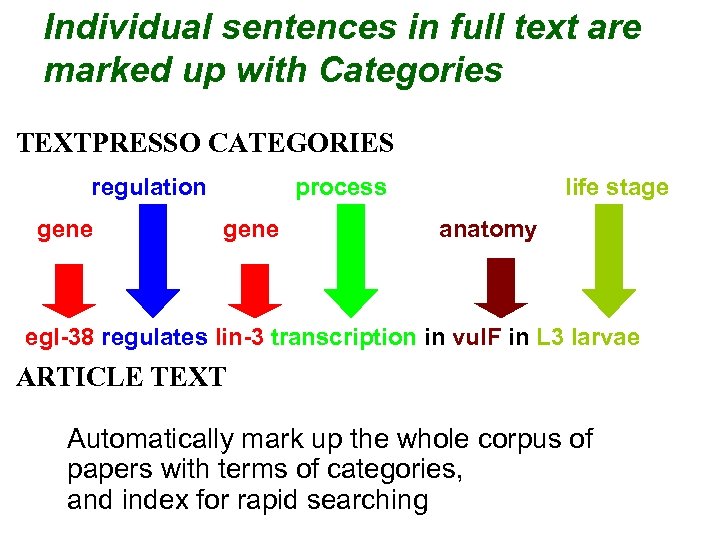 Individual sentences in full text are marked up with Categories TEXTPRESSO CATEGORIES regulation gene process gene life stage anatomy egl-38 regulates lin-3 transcription in vul. F in L 3 larvae ARTICLE TEXT Automatically mark up the whole corpus of papers with terms of categories, and index for rapid searching
Individual sentences in full text are marked up with Categories TEXTPRESSO CATEGORIES regulation gene process gene life stage anatomy egl-38 regulates lin-3 transcription in vul. F in L 3 larvae ARTICLE TEXT Automatically mark up the whole corpus of papers with terms of categories, and index for rapid searching
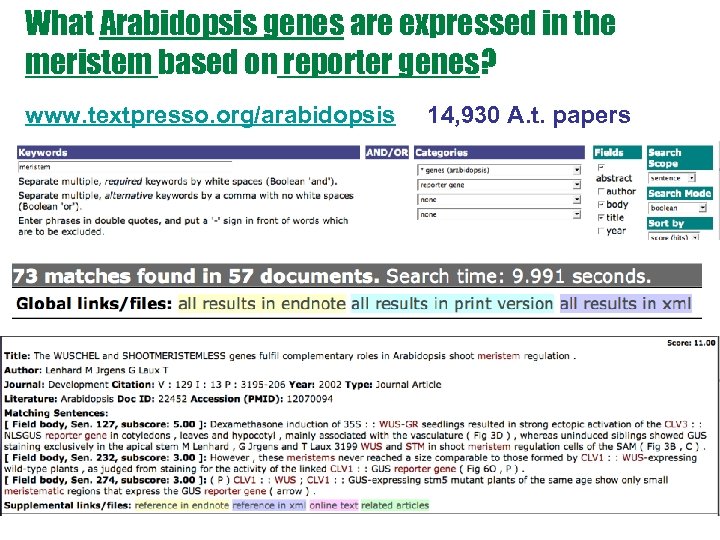 What Arabidopsis genes are expressed in the meristem based on reporter genes? www. textpresso. org/arabidopsis 14, 930 A. t. papers
What Arabidopsis genes are expressed in the meristem based on reporter genes? www. textpresso. org/arabidopsis 14, 930 A. t. papers
 Is a nicotinic receptor associated with Drugs of Abuse other than nicotine? www. textpresso. org/neuroscience 15, 786 papers
Is a nicotinic receptor associated with Drugs of Abuse other than nicotine? www. textpresso. org/neuroscience 15, 786 papers
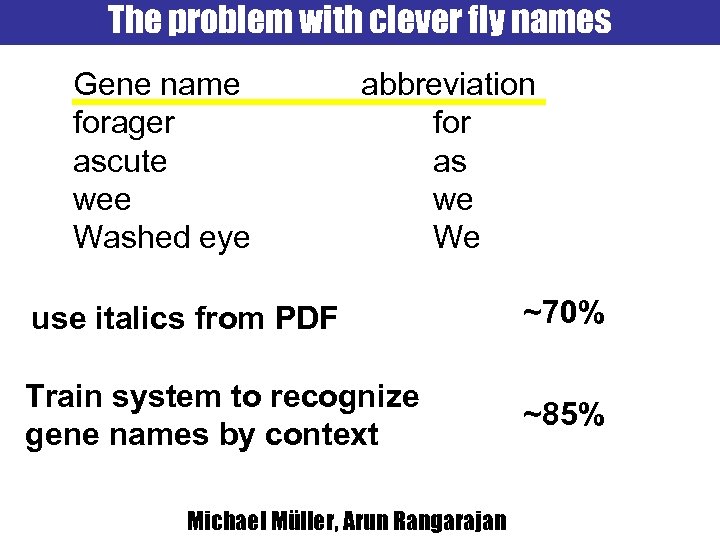 The problem with clever fly names Gene name forager ascute wee Washed eye abbreviation for as we We use italics from PDF ~70% Train system to recognize gene names by context ~85% Michael Müller, Arun Rangarajan
The problem with clever fly names Gene name forager ascute wee Washed eye abbreviation for as we We use italics from PDF ~70% Train system to recognize gene names by context ~85% Michael Müller, Arun Rangarajan
 What reporter genes have been used with Drosophila genes to study human disease? www. textpresso. org/fly 20, 099 full-text fly papers
What reporter genes have been used with Drosophila genes to study human disease? www. textpresso. org/fly 20, 099 full-text fly papers
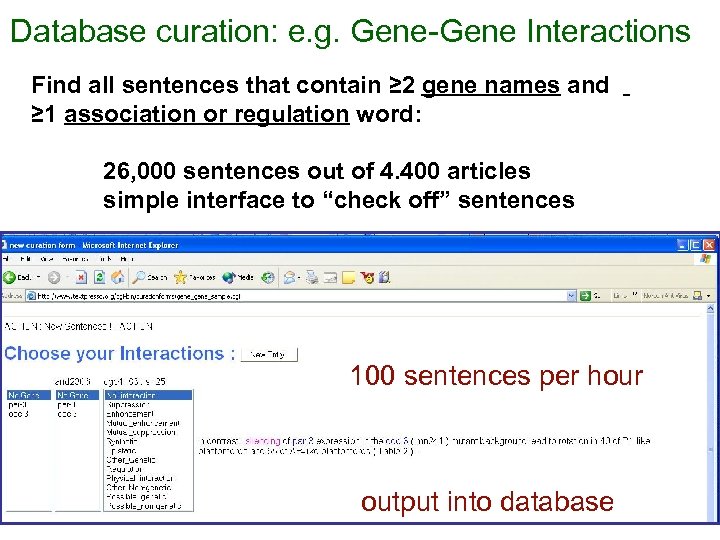 Database curation: e. g. Gene-Gene Interactions Find all sentences that contain ≥ 2 gene names and ≥ 1 association or regulation word: 26, 000 sentences out of 4. 400 articles simple interface to “check off” sentences 100 sentences per hour output into database
Database curation: e. g. Gene-Gene Interactions Find all sentences that contain ≥ 2 gene names and ≥ 1 association or regulation word: 26, 000 sentences out of 4. 400 articles simple interface to “check off” sentences 100 sentences per hour output into database
 Prioritizing high resolution genetic interaction tests by knowledge mining 1 Full text information retrieval Hans-Michael Muller, Arun Rangarajan, Tracy Teal, Kimberly Van Auken, Juancarlos Chan 2 Predicting Gene Interactions from information available in public databases Weiwei Zhong
Prioritizing high resolution genetic interaction tests by knowledge mining 1 Full text information retrieval Hans-Michael Muller, Arun Rangarajan, Tracy Teal, Kimberly Van Auken, Juancarlos Chan 2 Predicting Gene Interactions from information available in public databases Weiwei Zhong
 Training Set Training set § 4775 Positive Interactions § Genetic, Literature curation (1909) § Yeast two-hybrid screen (2933) § 3296 Negative Genetic Interactions § cis doubles in genetic mapping Benchmark § 5515 Positives: KEGG database § 5000 Negatives: Randomly selected
Training Set Training set § 4775 Positive Interactions § Genetic, Literature curation (1909) § Yeast two-hybrid screen (2933) § 3296 Negative Genetic Interactions § cis doubles in genetic mapping Benchmark § 5515 Positives: KEGG database § 5000 Negatives: Randomly selected
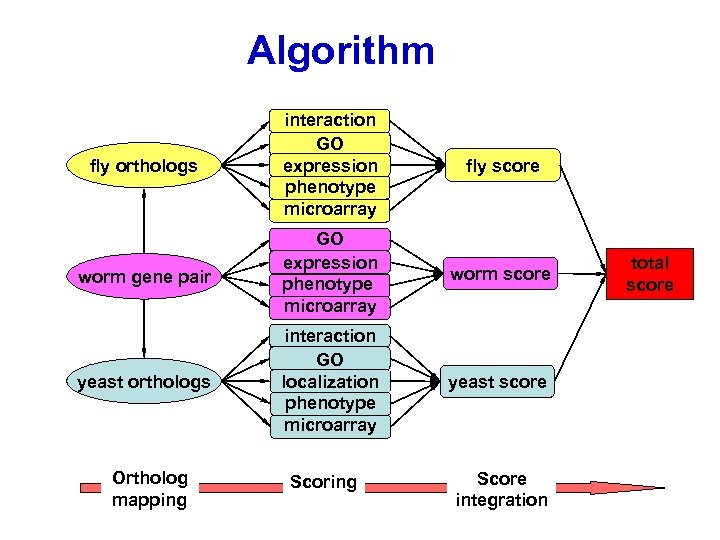 Algorithm fly orthologs interaction GO expression phenotype microarray fly score worm gene pair GO expression phenotype microarray worm score yeast orthologs interaction GO localization phenotype microarray yeast score Ortholog mapping Score integration total score
Algorithm fly orthologs interaction GO expression phenotype microarray fly score worm gene pair GO expression phenotype microarray worm score yeast orthologs interaction GO localization phenotype microarray yeast score Ortholog mapping Score integration total score
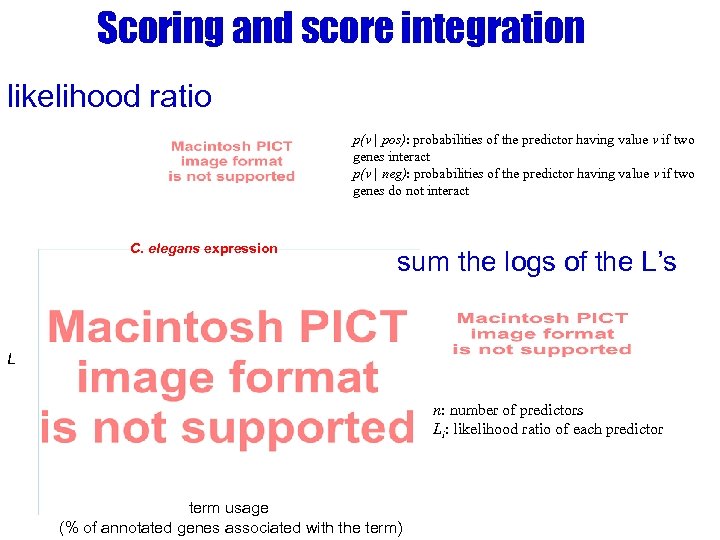 Scoring and score integration likelihood ratio p(v | pos): probabilities of the predictor having value v if two genes interact p(v | neg): probabilities of the predictor having value v if two genes do not interact C. elegans expression sum the logs of the L’s L n: number of predictors Li: likelihood ratio of each predictor term usage (% of annotated genes associated with the term)
Scoring and score integration likelihood ratio p(v | pos): probabilities of the predictor having value v if two genes interact p(v | neg): probabilities of the predictor having value v if two genes do not interact C. elegans expression sum the logs of the L’s L n: number of predictors Li: likelihood ratio of each predictor term usage (% of annotated genes associated with the term)



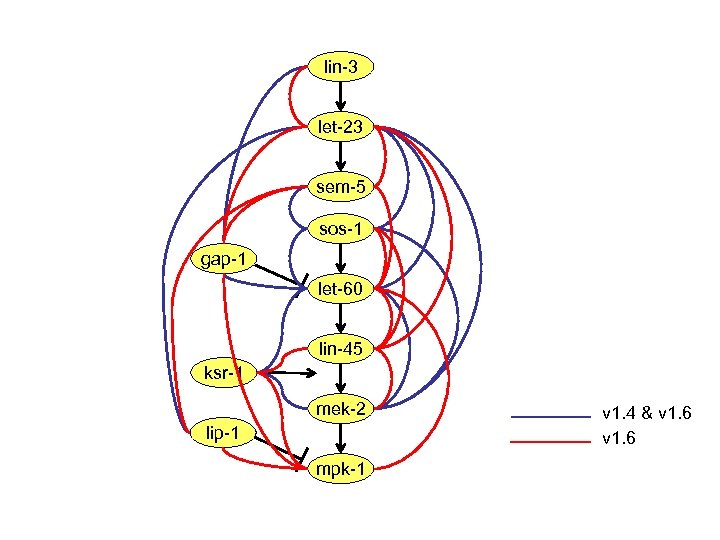 lin-3 let-23 sem-5 sos-1 gap-1 let-60 lin-45 ksr-1 mek-2 lip-1 mpk-1 v 1. 4 & v 1. 6
lin-3 let-23 sem-5 sos-1 gap-1 let-60 lin-45 ksr-1 mek-2 lip-1 mpk-1 v 1. 4 & v 1. 6
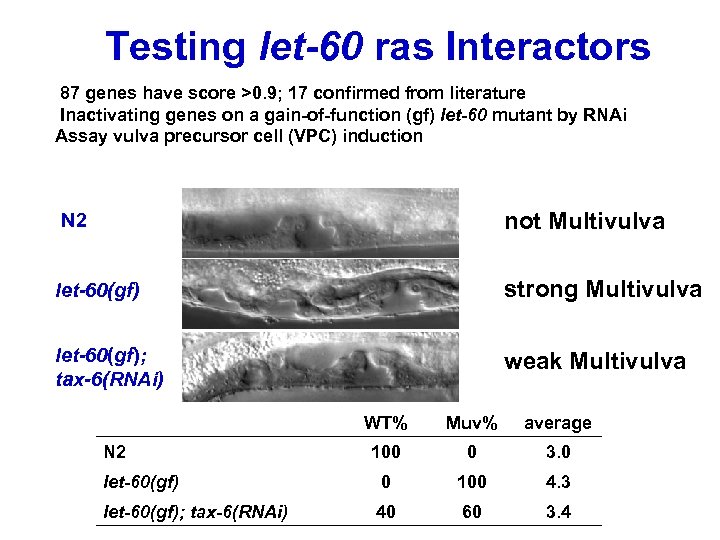 Testing let-60 ras Interactors 87 genes have score >0. 9; 17 confirmed from literature Inactivating genes on a gain-of-function (gf) let-60 mutant by RNAi Assay vulva precursor cell (VPC) induction N 2 not Multivulva let-60(gf) strong Multivulva let-60(gf); tax-6(RNAi) weak Multivulva WT% Muv% average 100 0 3. 0 let-60(gf) 0 100 4. 3 let-60(gf); tax-6(RNAi) 40 60 3. 4 N 2
Testing let-60 ras Interactors 87 genes have score >0. 9; 17 confirmed from literature Inactivating genes on a gain-of-function (gf) let-60 mutant by RNAi Assay vulva precursor cell (VPC) induction N 2 not Multivulva let-60(gf) strong Multivulva let-60(gf); tax-6(RNAi) weak Multivulva WT% Muv% average 100 0 3. 0 let-60(gf) 0 100 4. 3 let-60(gf); tax-6(RNAi) 40 60 3. 4 N 2
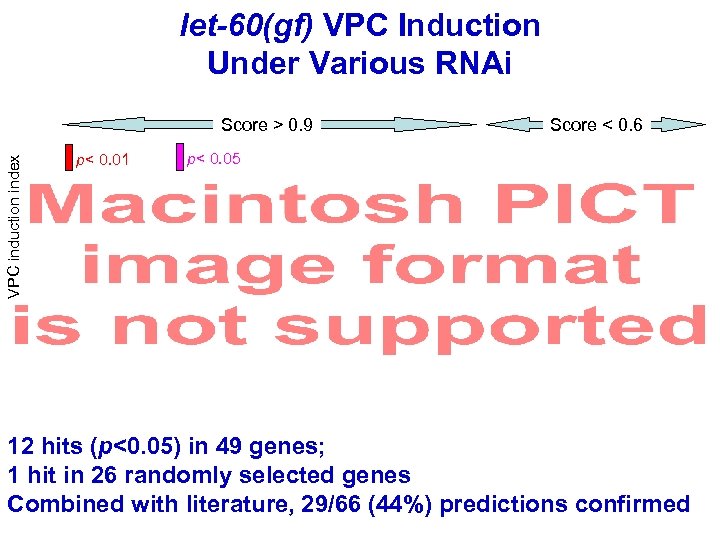 let-60(gf) VPC Induction Under Various RNAi VPC induction index Score > 0. 9 p< 0. 01 Score < 0. 6 p< 0. 05 12 hits (p<0. 05) in 49 genes; 1 hit in 26 randomly selected genes Combined with literature, 29/66 (44%) predictions confirmed
let-60(gf) VPC Induction Under Various RNAi VPC induction index Score > 0. 9 p< 0. 01 Score < 0. 6 p< 0. 05 12 hits (p<0. 05) in 49 genes; 1 hit in 26 randomly selected genes Combined with literature, 29/66 (44%) predictions confirmed
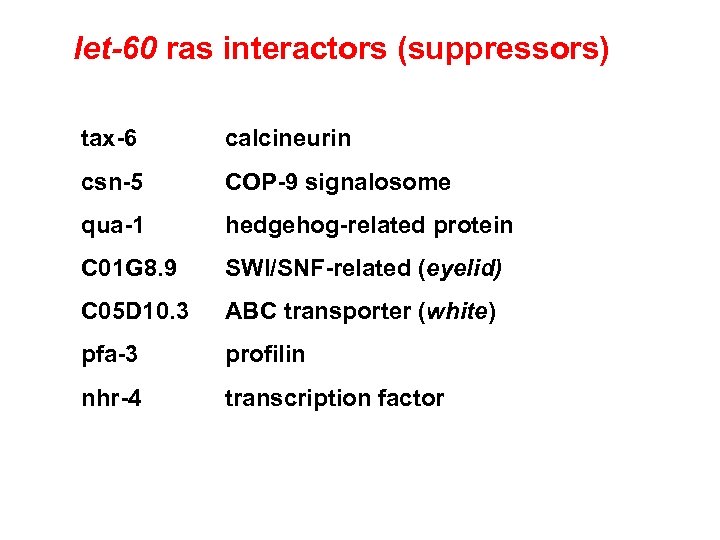 let-60 ras interactors (suppressors) tax-6 calcineurin csn-5 COP-9 signalosome qua-1 hedgehog-related protein C 01 G 8. 9 SWI/SNF-related (eyelid) C 05 D 10. 3 ABC transporter (white) pfa-3 profilin nhr-4 transcription factor
let-60 ras interactors (suppressors) tax-6 calcineurin csn-5 COP-9 signalosome qua-1 hedgehog-related protein C 01 G 8. 9 SWI/SNF-related (eyelid) C 05 D 10. 3 ABC transporter (white) pfa-3 profilin nhr-4 transcription factor
 C. elegans Interactions Input 4, 726 known interactions among 2, 713 genes Predict additional 18, 863 for total of 23, 589 interactions among 4, 408 genes
C. elegans Interactions Input 4, 726 known interactions among 2, 713 genes Predict additional 18, 863 for total of 23, 589 interactions among 4, 408 genes
 for Drosophila
for Drosophila

 D. melanogaster interactions Input 4, 180 known interactions among 1, 262 genes, Predict 13, 126 for 17, 306 interactions among 6, 044 genes
D. melanogaster interactions Input 4, 180 known interactions among 1, 262 genes, Predict 13, 126 for 17, 306 interactions among 6, 044 genes
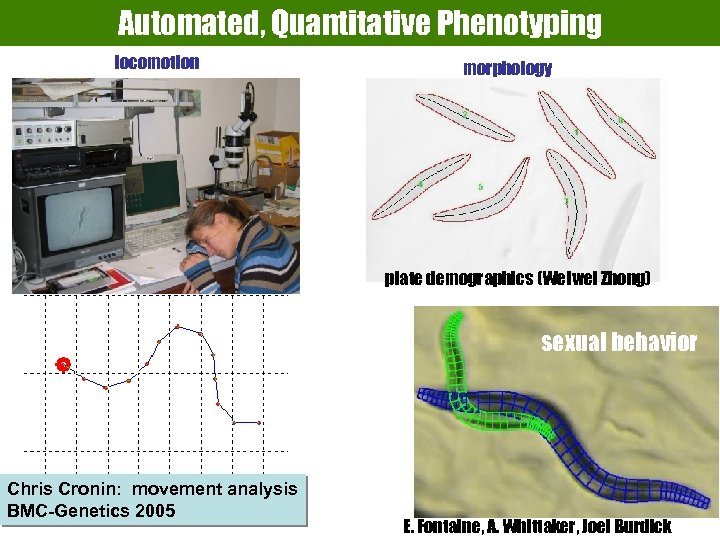 Automated, Quantitative Phenotyping locomotion morphology generative graphics plate demographics (Weiwei Zhong) sexual behavior Chris Cronin: movement analysis BMC-Genetics 2005 E. Fontaine, A. Whittaker, Joel Burdick
Automated, Quantitative Phenotyping locomotion morphology generative graphics plate demographics (Weiwei Zhong) sexual behavior Chris Cronin: movement analysis BMC-Genetics 2005 E. Fontaine, A. Whittaker, Joel Burdick
 Prioritizing high resolution genetic interaction tests by knowledge mining 1 Full text information retrieval Hans-Michael Muller, Arun Rangarajan, Tracy Teal, Kimberly Van Auken, Juancarlos Chan 2 Predicting Gene Interactions from information available in public databases Weiwei Zhong
Prioritizing high resolution genetic interaction tests by knowledge mining 1 Full text information retrieval Hans-Michael Muller, Arun Rangarajan, Tracy Teal, Kimberly Van Auken, Juancarlos Chan 2 Predicting Gene Interactions from information available in public databases Weiwei Zhong


