lesson2.pptx
- Количество слайдов: 43
 Home reading Guide to Protein Purification, 2 nd Edition / Ed. by R. R. Burgess and M. P. Deutscher. Ch. 18 and 19.
Home reading Guide to Protein Purification, 2 nd Edition / Ed. by R. R. Burgess and M. P. Deutscher. Ch. 18 and 19.
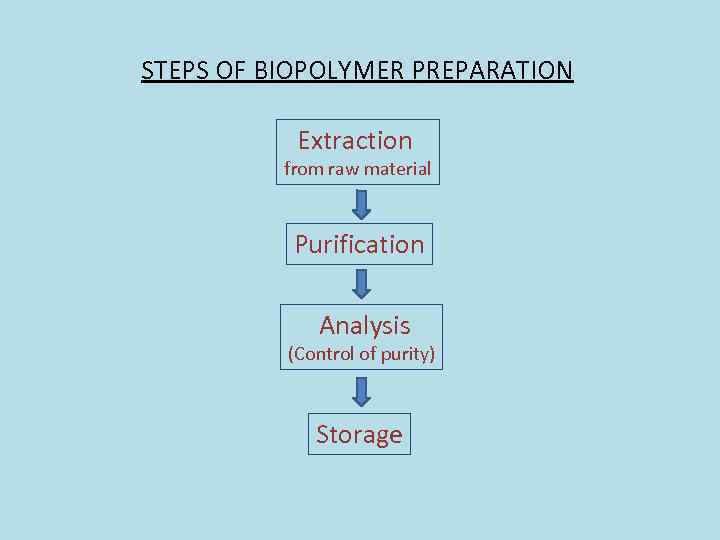 STEPS OF BIOPOLYMER PREPARATION Extraction from raw material Purification Analysis (Control of purity) Storage
STEPS OF BIOPOLYMER PREPARATION Extraction from raw material Purification Analysis (Control of purity) Storage
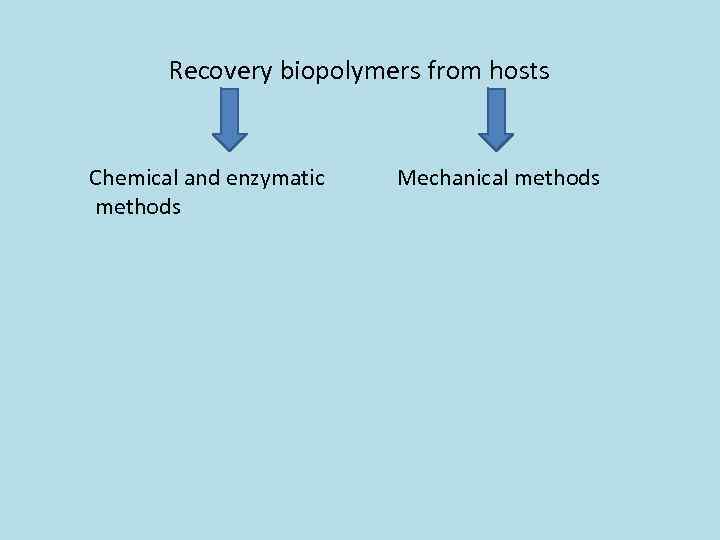 Recovery biopolymers from hosts Chemical and enzymatic methods Mechanical methods
Recovery biopolymers from hosts Chemical and enzymatic methods Mechanical methods
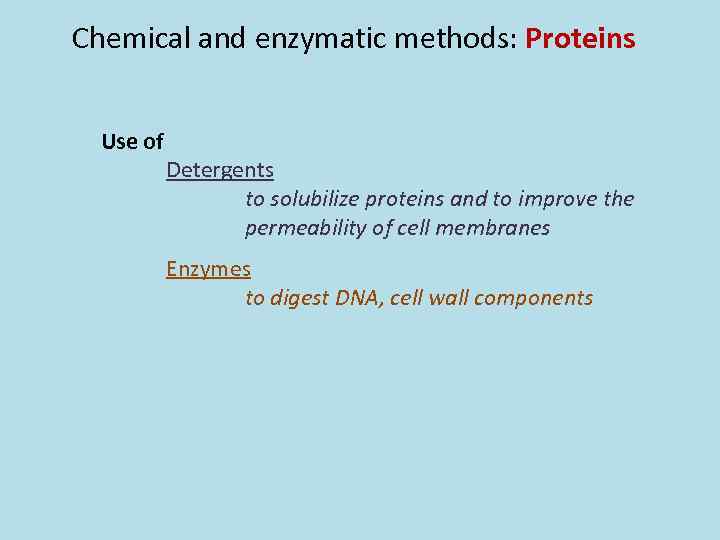 Chemical and enzymatic methods: Proteins Use of Detergents to solubilize proteins and to improve the permeability of cell membranes Enzymes to digest DNA, cell wall components
Chemical and enzymatic methods: Proteins Use of Detergents to solubilize proteins and to improve the permeability of cell membranes Enzymes to digest DNA, cell wall components
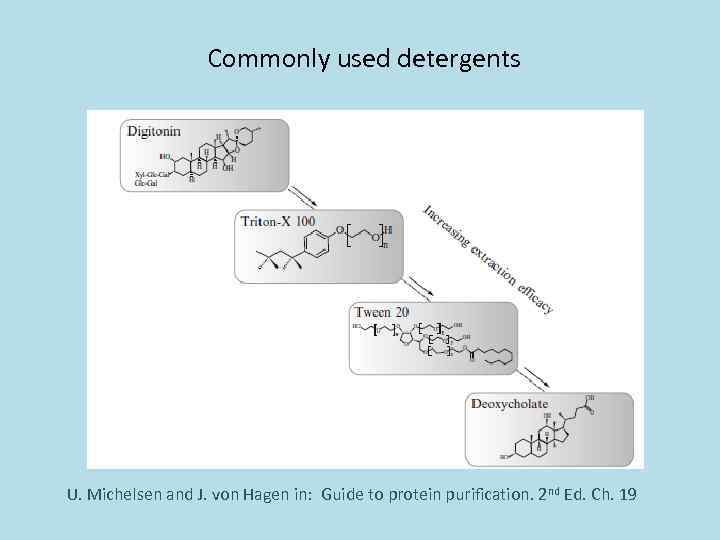 Commonly used detergents U. Michelsen and J. von Hagen in: Guide to protein purification. 2 nd Ed. Ch. 19
Commonly used detergents U. Michelsen and J. von Hagen in: Guide to protein purification. 2 nd Ed. Ch. 19
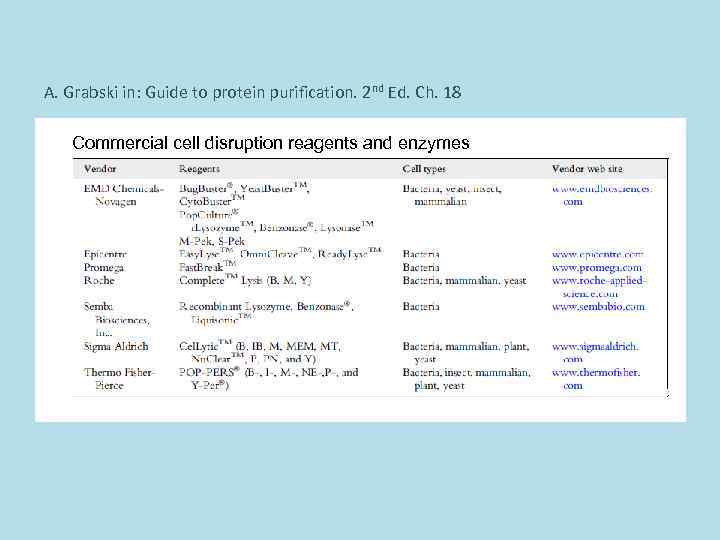 A. Grabski in: Guide to protein purification. 2 nd Ed. Ch. 18 Commercial cell disruption reagents and enzymes
A. Grabski in: Guide to protein purification. 2 nd Ed. Ch. 18 Commercial cell disruption reagents and enzymes
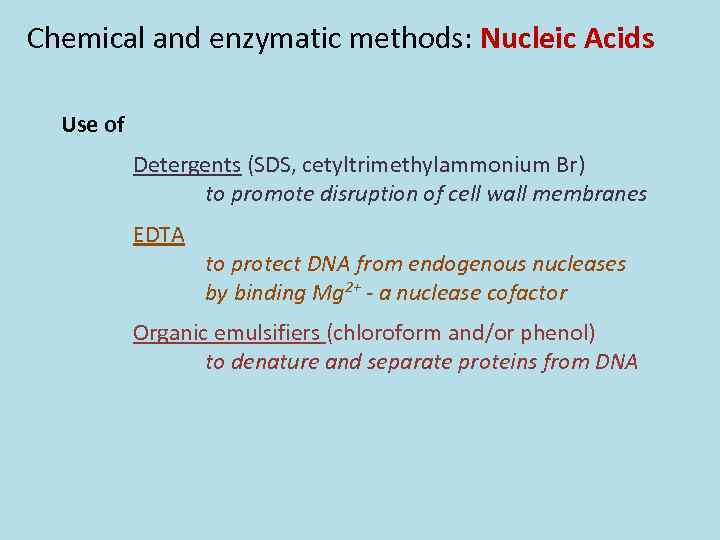 Chemical and enzymatic methods: Nucleic Acids Use of Detergents (SDS, cetyltrimethylammonium Br) to promote disruption of cell wall membranes EDTA to protect DNA from endogenous nucleases by binding Mg 2+ - a nuclease cofactor Organic emulsifiers (chloroform and/or phenol) to denature and separate proteins from DNA
Chemical and enzymatic methods: Nucleic Acids Use of Detergents (SDS, cetyltrimethylammonium Br) to promote disruption of cell wall membranes EDTA to protect DNA from endogenous nucleases by binding Mg 2+ - a nuclease cofactor Organic emulsifiers (chloroform and/or phenol) to denature and separate proteins from DNA
 Recovery biopolymers from hosts Chemical and enzymatic methods Mechanical methods Sonification, 15 -25 MHz High-Pressure Bead Milling Grinder homogenizers
Recovery biopolymers from hosts Chemical and enzymatic methods Mechanical methods Sonification, 15 -25 MHz High-Pressure Bead Milling Grinder homogenizers
 Ultrasonic homogenizers
Ultrasonic homogenizers
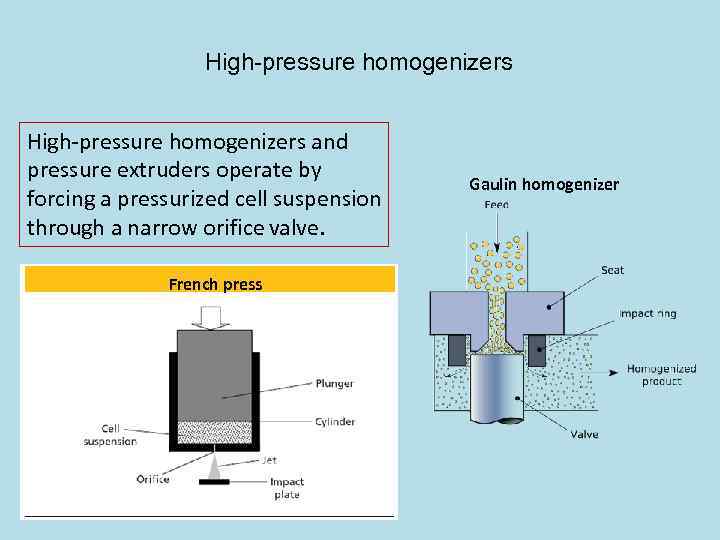 High-pressure homogenizers and pressure extruders operate by forcing a pressurized cell suspension through a narrow orifice valve. French press Gaulin homogenizer
High-pressure homogenizers and pressure extruders operate by forcing a pressurized cell suspension through a narrow orifice valve. French press Gaulin homogenizer
 Bead milling Glass beads
Bead milling Glass beads
 Glass Homogenizers Pestle
Glass Homogenizers Pestle
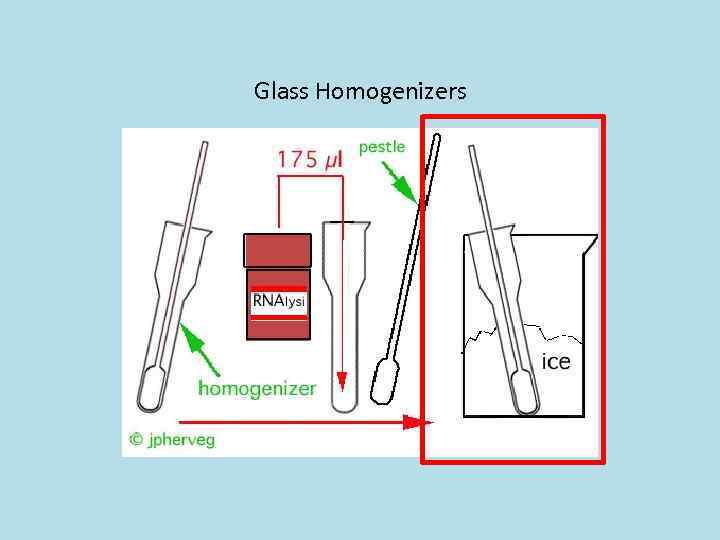 Glass Homogenizers
Glass Homogenizers
 Grinding in liquid nitrogen or with dry ice Pestle Mechanical grinder Mortar
Grinding in liquid nitrogen or with dry ice Pestle Mechanical grinder Mortar
 (c) dnews. com
(c) dnews. com
 After homogenization one has a slurry consisting of the solution of target components, endogenous contaminants, and cell debris Separation liquid from solid matter Filtration Centrifugation
After homogenization one has a slurry consisting of the solution of target components, endogenous contaminants, and cell debris Separation liquid from solid matter Filtration Centrifugation
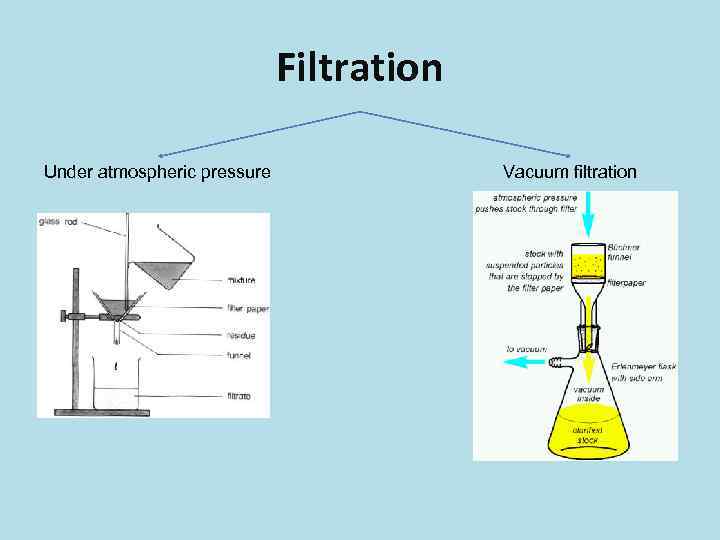 Filtration Under atmospheric pressure Vacuum filtration
Filtration Under atmospheric pressure Vacuum filtration
 Filter paper
Filter paper
 Large scale filters
Large scale filters
 Filtration Removing coarse solids (usually fibrous tissue material) Typical pore size: > 100 mm (cheesecloth) 10 -100 mm (paper filters) Removing fine solids and colloid particles (cell debris, insoluble biocompounds ) Typical pore sizes: 0. 20 mm (membrane filters) 0. 45 mm 1. 00 mm
Filtration Removing coarse solids (usually fibrous tissue material) Typical pore size: > 100 mm (cheesecloth) 10 -100 mm (paper filters) Removing fine solids and colloid particles (cell debris, insoluble biocompounds ) Typical pore sizes: 0. 20 mm (membrane filters) 0. 45 mm 1. 00 mm
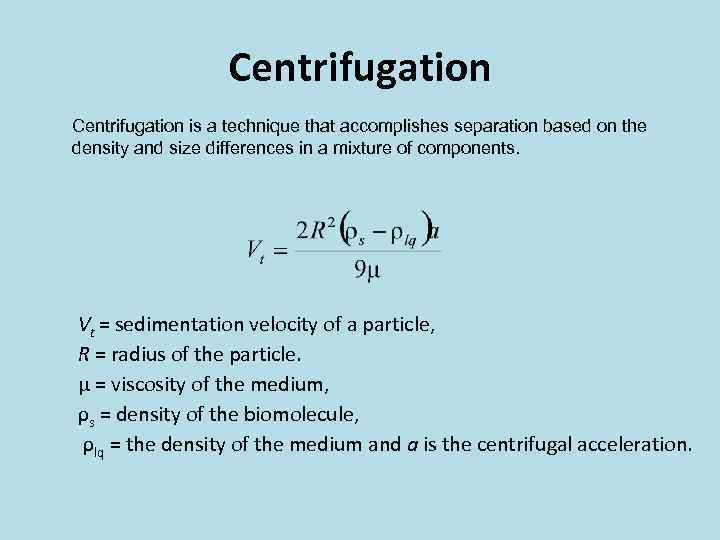 Centrifugation is a technique that accomplishes separation based on the density and size differences in a mixture of components. Vt = sedimentation velocity of a particle, R = radius of the particle. μ = viscosity of the medium, ρs = density of the biomolecule, ρlq = the density of the medium and a is the centrifugal acceleration.
Centrifugation is a technique that accomplishes separation based on the density and size differences in a mixture of components. Vt = sedimentation velocity of a particle, R = radius of the particle. μ = viscosity of the medium, ρs = density of the biomolecule, ρlq = the density of the medium and a is the centrifugal acceleration.
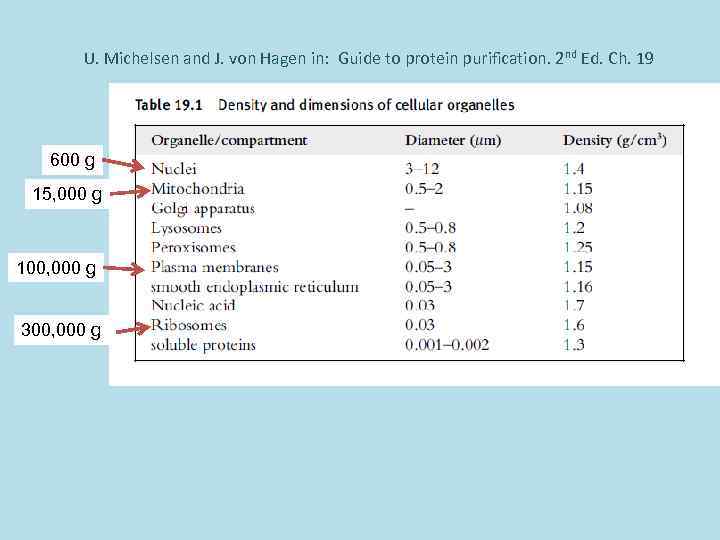 U. Michelsen and J. von Hagen in: Guide to protein purification. 2 nd Ed. Ch. 19 600 g 15, 000 g 100, 000 g 300, 000 g
U. Michelsen and J. von Hagen in: Guide to protein purification. 2 nd Ed. Ch. 19 600 g 15, 000 g 100, 000 g 300, 000 g
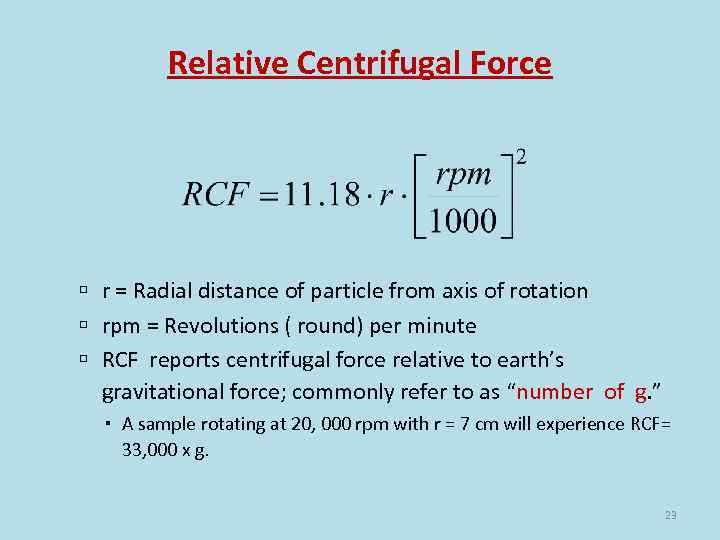 Relative Centrifugal Force r = Radial distance of particle from axis of rotation rpm = Revolutions ( round) per minute RCF reports centrifugal force relative to earth’s gravitational force; commonly refer to as “number of g. ” A sample rotating at 20, 000 rpm with r = 7 cm will experience RCF= 33, 000 x g. 23
Relative Centrifugal Force r = Radial distance of particle from axis of rotation rpm = Revolutions ( round) per minute RCF reports centrifugal force relative to earth’s gravitational force; commonly refer to as “number of g. ” A sample rotating at 20, 000 rpm with r = 7 cm will experience RCF= 33, 000 x g. 23
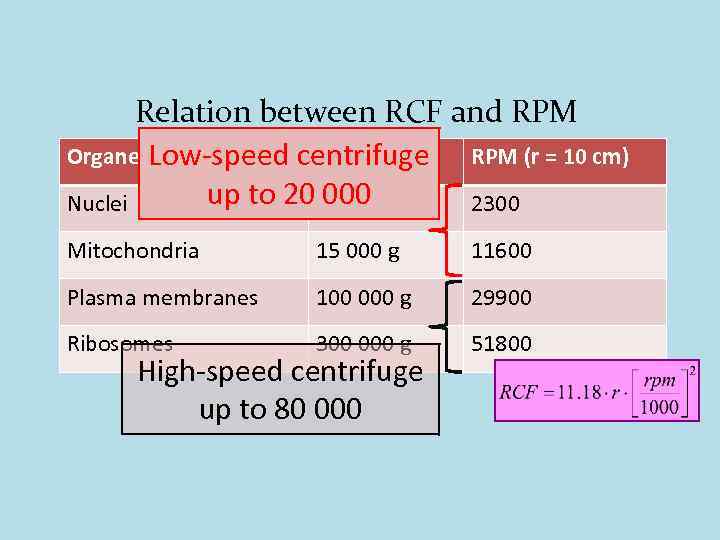 Relation between RCF and RPM Organelle RCF Low-speed centrifuge RPM (r = 10 cm) up to 20 600 g 000 Nuclei 2300 Mitochondria 15 000 g 11600 Plasma membranes 100 000 g 29900 Ribosomes 300 000 g 51800 High-speed centrifuge up to 80 000
Relation between RCF and RPM Organelle RCF Low-speed centrifuge RPM (r = 10 cm) up to 20 600 g 000 Nuclei 2300 Mitochondria 15 000 g 11600 Plasma membranes 100 000 g 29900 Ribosomes 300 000 g 51800 High-speed centrifuge up to 80 000
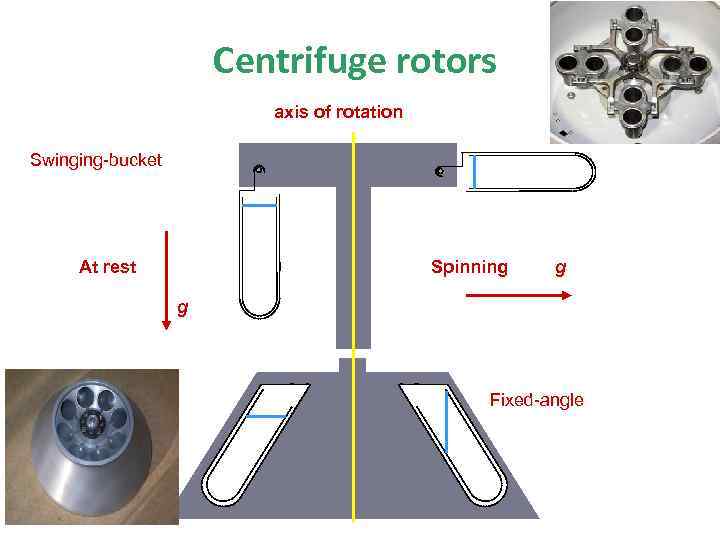 Centrifuge rotors axis of rotation Swinging-bucket Spinning At rest g g Fixed-angle
Centrifuge rotors axis of rotation Swinging-bucket Spinning At rest g g Fixed-angle
 Comparison of rotors
Comparison of rotors
 The sample before and after centrifuge (c) Abdoli et al. Pasteur Institute of Iran
The sample before and after centrifuge (c) Abdoli et al. Pasteur Institute of Iran
 Low-speed centrifuges High-speed centrifuges 28
Low-speed centrifuges High-speed centrifuges 28
 A typical protocol for recovery of DNA from plant tissues by Lodhi et al. Plant Mol. Biol. Reporter, 12 (1994) 6 -13 1. Collect unexpanded young leaves in liquid nitrogen or on ice and store at or below -70 o. C until used. Purpose: Sampling 2. Grind 0. 5 g of leaves using mortar and pestle in the presence of liquid nitrogen. Purpose: Disintegration of tissues 3. Add 5 m. L of extraction buffer to the ground leaves and mix in the mortar. Purpose: Extraction of DNA
A typical protocol for recovery of DNA from plant tissues by Lodhi et al. Plant Mol. Biol. Reporter, 12 (1994) 6 -13 1. Collect unexpanded young leaves in liquid nitrogen or on ice and store at or below -70 o. C until used. Purpose: Sampling 2. Grind 0. 5 g of leaves using mortar and pestle in the presence of liquid nitrogen. Purpose: Disintegration of tissues 3. Add 5 m. L of extraction buffer to the ground leaves and mix in the mortar. Purpose: Extraction of DNA
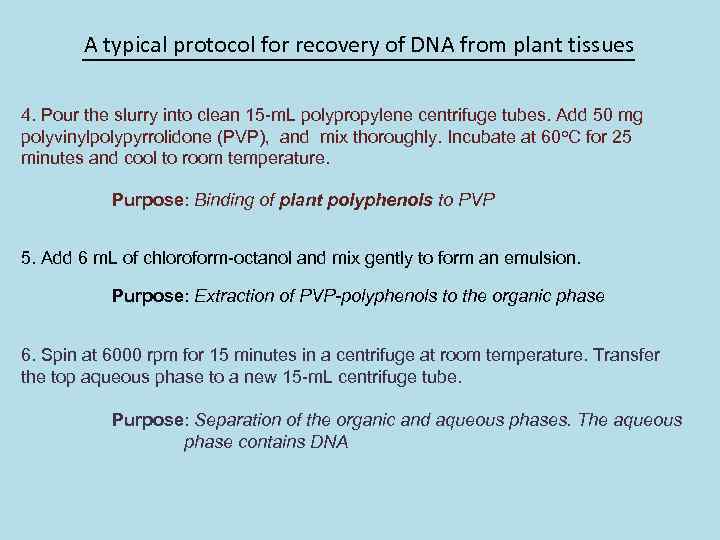 A typical protocol for recovery of DNA from plant tissues 4. Pour the slurry into clean 15 -m. L polypropylene centrifuge tubes. Add 50 mg polyvinylpolypyrrolidone (PVP), and mix thoroughly. Incubate at 60 o. C for 25 minutes and cool to room temperature. Purpose: Binding of plant polyphenols to PVP 5. Add 6 m. L of chloroform-octanol and mix gently to form an emulsion. Purpose: Extraction of PVP-polyphenols to the organic phase 6. Spin at 6000 rpm for 15 minutes in a centrifuge at room temperature. Transfer the top aqueous phase to a new 15 -m. L centrifuge tube. Purpose: Separation of the organic and aqueous phases. The aqueous phase contains DNA
A typical protocol for recovery of DNA from plant tissues 4. Pour the slurry into clean 15 -m. L polypropylene centrifuge tubes. Add 50 mg polyvinylpolypyrrolidone (PVP), and mix thoroughly. Incubate at 60 o. C for 25 minutes and cool to room temperature. Purpose: Binding of plant polyphenols to PVP 5. Add 6 m. L of chloroform-octanol and mix gently to form an emulsion. Purpose: Extraction of PVP-polyphenols to the organic phase 6. Spin at 6000 rpm for 15 minutes in a centrifuge at room temperature. Transfer the top aqueous phase to a new 15 -m. L centrifuge tube. Purpose: Separation of the organic and aqueous phases. The aqueous phase contains DNA
 A typical protocol for recovery of DNA from plant tissues 7. Add 0. 5 volume of 5 M Na. Cl to the aqueous solution recovered from the previous step and mix well. Add two volumes of cold (-20 o. C 95% ethanol) and refrigerate (4 to 6 o. C) until DNA strands begin to appear. Purpose: Pressing out polysaccharides to the ethanol solution as DNA precipitates. 8. Spin at 3000 rpm for three minutes and then increase speed to 5000 rpm for an additional three minutes at room temperature. Purpose: Separation of DNA precipitate from the polysaccharide containing supernatant. The two step centrifugation is necessary to keep DNA at the bottom of centrifuge tube.
A typical protocol for recovery of DNA from plant tissues 7. Add 0. 5 volume of 5 M Na. Cl to the aqueous solution recovered from the previous step and mix well. Add two volumes of cold (-20 o. C 95% ethanol) and refrigerate (4 to 6 o. C) until DNA strands begin to appear. Purpose: Pressing out polysaccharides to the ethanol solution as DNA precipitates. 8. Spin at 3000 rpm for three minutes and then increase speed to 5000 rpm for an additional three minutes at room temperature. Purpose: Separation of DNA precipitate from the polysaccharide containing supernatant. The two step centrifugation is necessary to keep DNA at the bottom of centrifuge tube.
 A typical protocol for recovery of DNA from plant tissues 9. Pour off supernatant and wash pellet with cold (0 to 4 o. C) 76% ethanol. Completely remove ethanol without drying the DNA pellet by leaving the tubes uncovered at 37 o. C for 20 to 30 minutes. Purpose: Removal of supernatant and traces of the solvent. 10. Dissolve in 200 to 300 m. L Tris-EDTA buffer (p. H 8). Treat with 1 m. L RNase A per 100 m. L DNA solution and incubate at 37 o. C for 15 minutes. Purpose: 1. Dissolution of DNA of the pellet. EDTA inactivates endonucleases. p. H 8 serves with purpose too as nucleases are less active at this p. H. 2. Rnase A = Ribonuclease A degrades RNA
A typical protocol for recovery of DNA from plant tissues 9. Pour off supernatant and wash pellet with cold (0 to 4 o. C) 76% ethanol. Completely remove ethanol without drying the DNA pellet by leaving the tubes uncovered at 37 o. C for 20 to 30 minutes. Purpose: Removal of supernatant and traces of the solvent. 10. Dissolve in 200 to 300 m. L Tris-EDTA buffer (p. H 8). Treat with 1 m. L RNase A per 100 m. L DNA solution and incubate at 37 o. C for 15 minutes. Purpose: 1. Dissolution of DNA of the pellet. EDTA inactivates endonucleases. p. H 8 serves with purpose too as nucleases are less active at this p. H. 2. Rnase A = Ribonuclease A degrades RNA
 A typical protocol for recovery of DNA from plant tissues 11. Quantify DNA in a spectrophotometer at l = 260 nm. Purpose: Control yield. 12. Keep DNA at -70 o. C for long-term or -20 o. C for short-term storage.
A typical protocol for recovery of DNA from plant tissues 11. Quantify DNA in a spectrophotometer at l = 260 nm. Purpose: Control yield. 12. Keep DNA at -70 o. C for long-term or -20 o. C for short-term storage.
 Isolation of subcellular organelles Each cell organelle contains its own set of proteins and nucleic acids performing specific physiological functions. Understanding functions of organelles requires the isolation of biopolymers from particular those. For this purpose, the researcher has to be able to isolate cell subunits of interest from a cell lysate. Cell lysates for subcellular fractionation must be obtained by mild lysis methods that would disrupt the cell walls but leave cell compartments untouched.
Isolation of subcellular organelles Each cell organelle contains its own set of proteins and nucleic acids performing specific physiological functions. Understanding functions of organelles requires the isolation of biopolymers from particular those. For this purpose, the researcher has to be able to isolate cell subunits of interest from a cell lysate. Cell lysates for subcellular fractionation must be obtained by mild lysis methods that would disrupt the cell walls but leave cell compartments untouched.
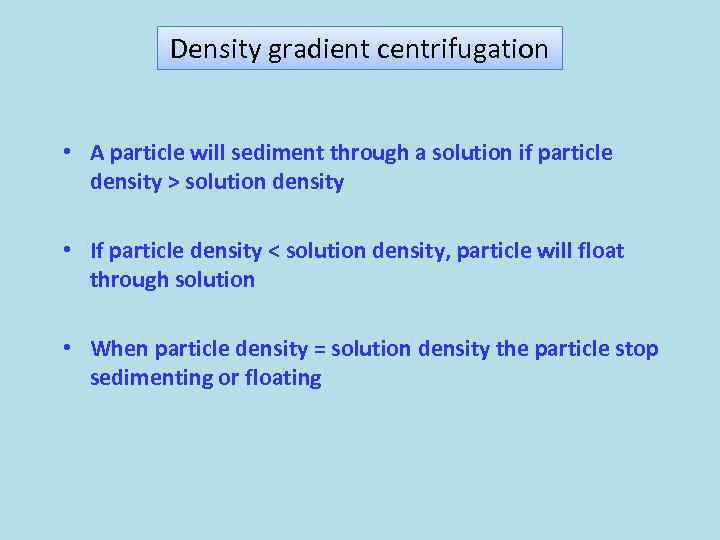 Density gradient centrifugation • A particle will sediment through a solution if particle density > solution density • If particle density < solution density, particle will float through solution • When particle density = solution density the particle stop sedimenting or floating
Density gradient centrifugation • A particle will sediment through a solution if particle density > solution density • If particle density < solution density, particle will float through solution • When particle density = solution density the particle stop sedimenting or floating
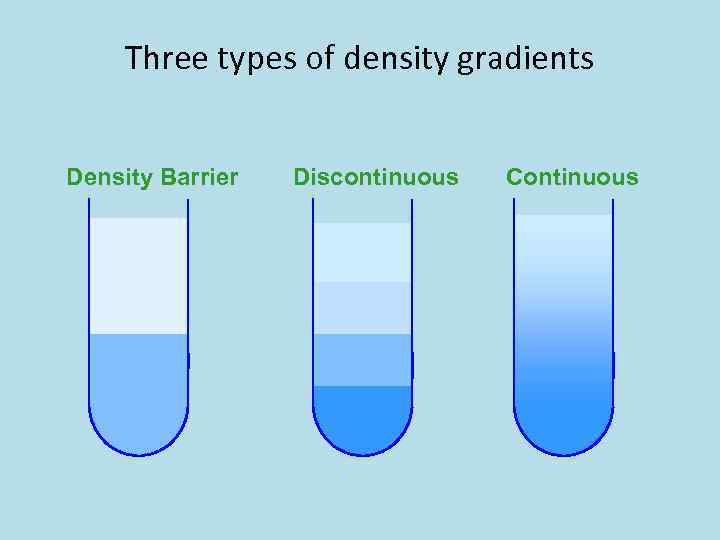 Three types of density gradients Density Barrier Discontinuous Continuous
Three types of density gradients Density Barrier Discontinuous Continuous
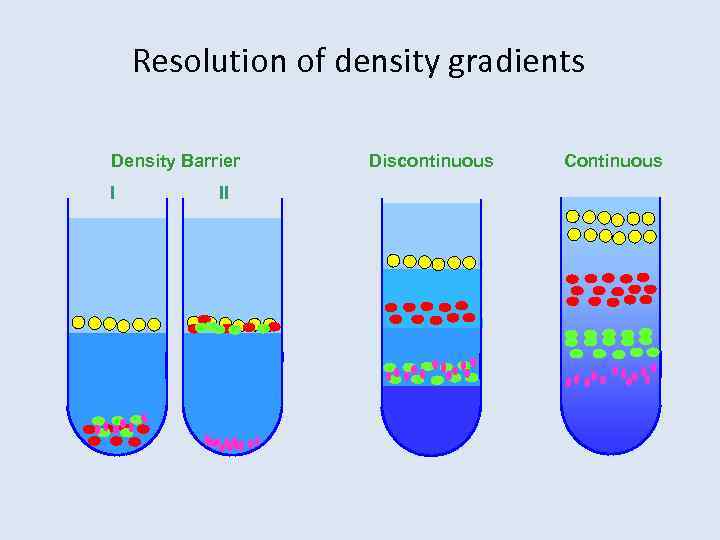 Resolution of density gradients Density Barrier I II Discontinuous Continuous
Resolution of density gradients Density Barrier I II Discontinuous Continuous
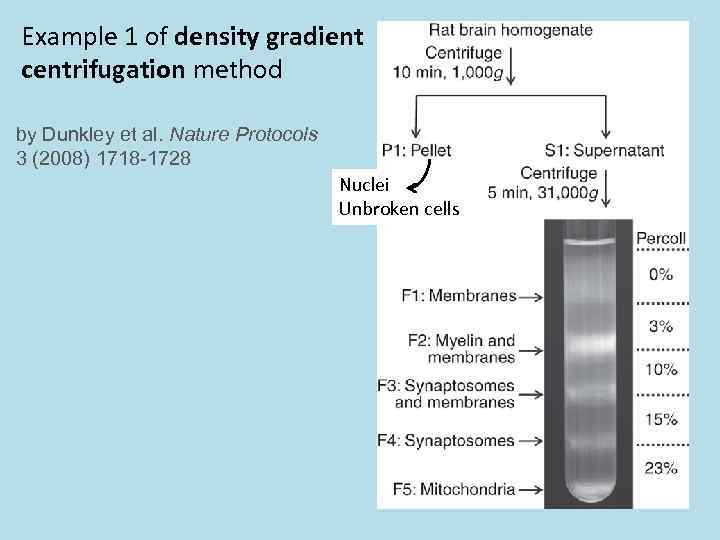 Example 1 of density gradient centrifugation method by Dunkley et al. Nature Protocols 3 (2008) 1718 -1728 Nuclei Unbroken cells
Example 1 of density gradient centrifugation method by Dunkley et al. Nature Protocols 3 (2008) 1718 -1728 Nuclei Unbroken cells
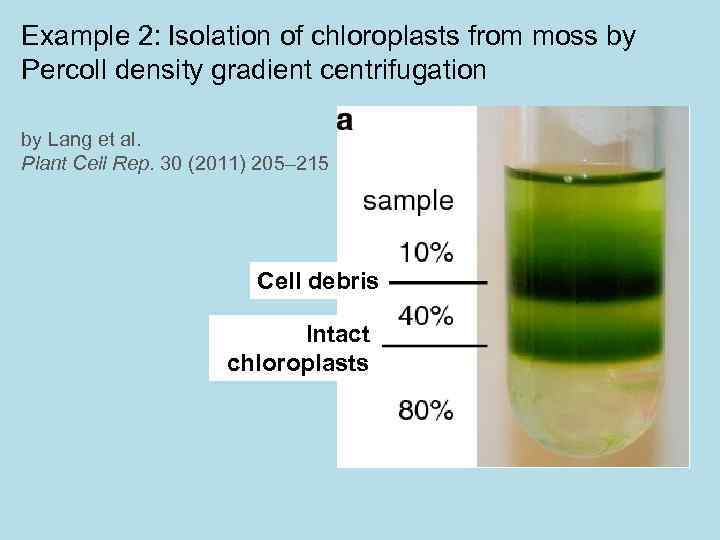 Example 2: Isolation of chloroplasts from moss by Percoll density gradient centrifugation by Lang et al. Plant Cell Rep. 30 (2011) 205– 215 Cell debris Intact chloroplasts
Example 2: Isolation of chloroplasts from moss by Percoll density gradient centrifugation by Lang et al. Plant Cell Rep. 30 (2011) 205– 215 Cell debris Intact chloroplasts
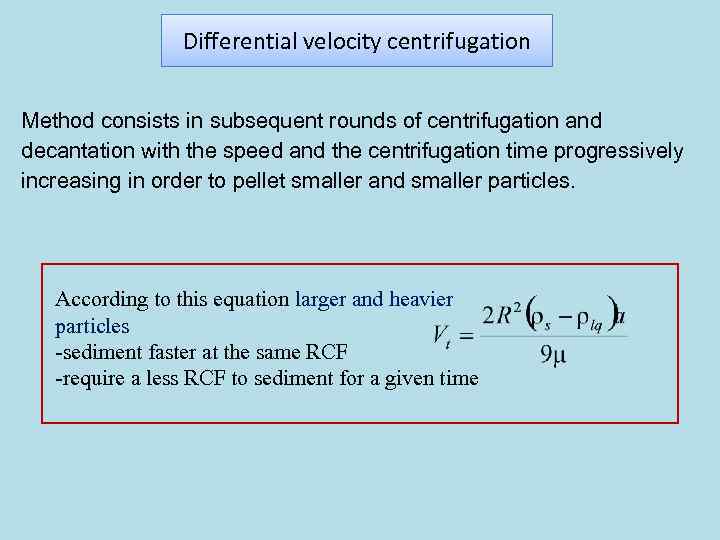 Differential velocity centrifugation Method consists in subsequent rounds of centrifugation and decantation with the speed and the centrifugation time progressively increasing in order to pellet smaller and smaller particles. According to this equation larger and heavier particles -sediment faster at the same RCF -require a less RCF to sediment for a given time
Differential velocity centrifugation Method consists in subsequent rounds of centrifugation and decantation with the speed and the centrifugation time progressively increasing in order to pellet smaller and smaller particles. According to this equation larger and heavier particles -sediment faster at the same RCF -require a less RCF to sediment for a given time
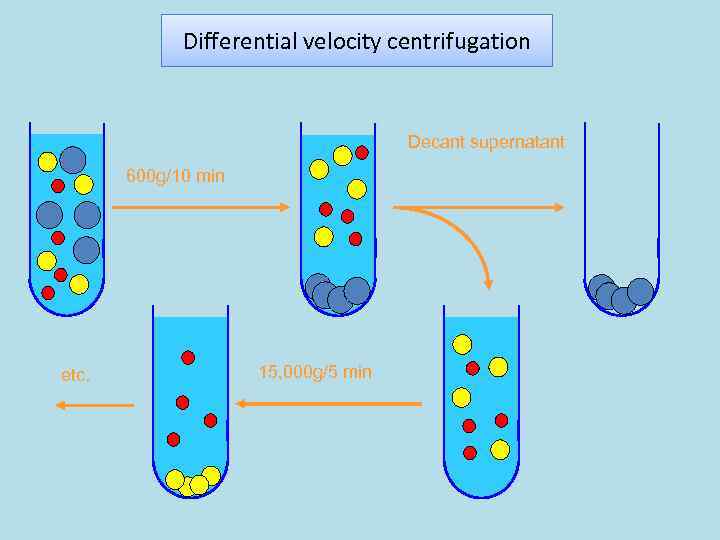 Differential velocity centrifugation Decant supernatant 600 g/10 min etc. 15, 000 g/5 min
Differential velocity centrifugation Decant supernatant 600 g/10 min etc. 15, 000 g/5 min
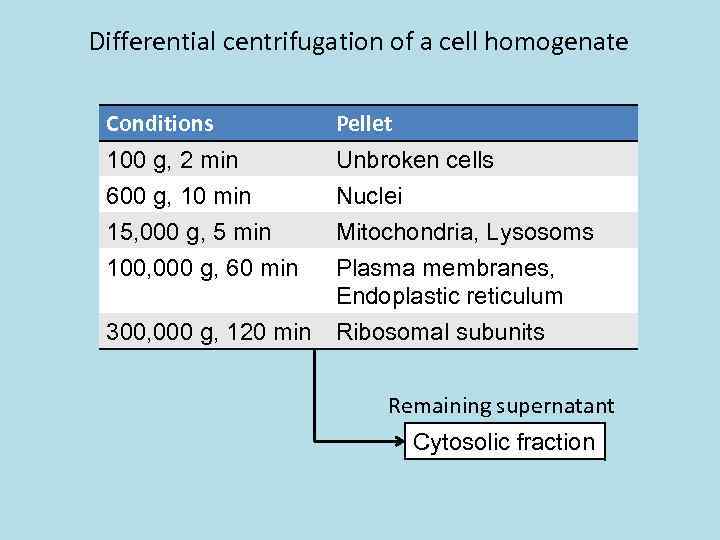 Differential centrifugation of a cell homogenate Conditions Pellet 100 g, 2 min 600 g, 10 min 15, 000 g, 5 min 100, 000 g, 60 min Unbroken cells Nuclei Mitochondria, Lysosoms Plasma membranes, Endoplastic reticulum 300, 000 g, 120 min Ribosomal subunits Remaining supernatant Cytosolic fraction
Differential centrifugation of a cell homogenate Conditions Pellet 100 g, 2 min 600 g, 10 min 15, 000 g, 5 min 100, 000 g, 60 min Unbroken cells Nuclei Mitochondria, Lysosoms Plasma membranes, Endoplastic reticulum 300, 000 g, 120 min Ribosomal subunits Remaining supernatant Cytosolic fraction
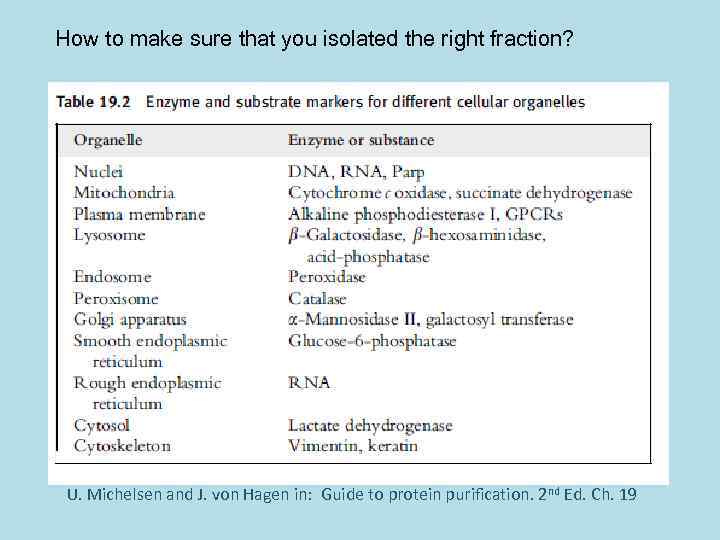 How to make sure that you isolated the right fraction? U. Michelsen and J. von Hagen in: Guide to protein purification. 2 nd Ed. Ch. 19
How to make sure that you isolated the right fraction? U. Michelsen and J. von Hagen in: Guide to protein purification. 2 nd Ed. Ch. 19


