fc06eed009e3e30aa58785a56261aeec.ppt
- Количество слайдов: 15

HIV-1 Drug Resistance Genotyping Dried Blood Spots Compared with Plasma Paul Sandstrom National HIV and Retrovirology Laboratories Centre for Infectious Disease Prevention and Control, Public Health Agency of Canada

The Problem While plasma and serum specimens are considered the gold standard for genotyping, they are not practical for many resource-limited settings and unsuitable for serving remote populations. logistic difficulties, • lack of a cold chain • lack of secure sample transport/transport of dangerous goods , • requirement for trained phlebotomist • restricted field collection may introduce sampling bias • Expense • Laboratory required for pre-storage specimen processing • Compliance from research subjects
DBS in HIV Surveillance and Monitoring DBS have been reported in peer reviewed scientific literature to be suitable for: ØHIV serology ØHIV molecular diagnostics ØHIV viral RNA quantification (viral load). ØCD 4 lymphocyte enumeration However because DBS remain non traditional specimens for commercial kit based testing, proficiency testing programs will be required.

Objectives § To assess the feasibility of genotyping HIV-1 from DBS collected in the field and stored under extreme environmental conditions § Concordance between sequences as well as DR genotypes obtained from plasma vs DBS § No attempt to delineate the nature of the genetic material (RNA vs DNA) being preferentially amplified from DBS.

Study Design § Part of a large WHO survey to evaluate prevalence of HIVDR in 10 testing and counselling (T&C) sites in Central Mexico § Prospectively collected paired plasma and DBS from consecutive newly diagnosed, ARV-naïve HIV+ Mexican subjects (n=558). § Whole blood (EDTA anticoagulated) was spotted onto S&S 903 cards, dried at room temperature and stored with desiccant at 37°C and 87% humidity for 3 months in sealed plastic bags.
Laboratory Methods Amplification and Sequencing of DBS § Amplification of the protease and RT regions of the pol gene based on an in house nested RT-PCR method. § Target template was amplified in two overlapping pieces using Qiagen One. Step RT-PCR § Nested PCR was performed using Ampli. Taq Gold (ABI) in 50ųl reactions containing 5ųl RT-PCR product § Viral load determination in the matched plasma specimen for those DBS that could not generate PCR products (Amplicor HIV 1 Monitor)
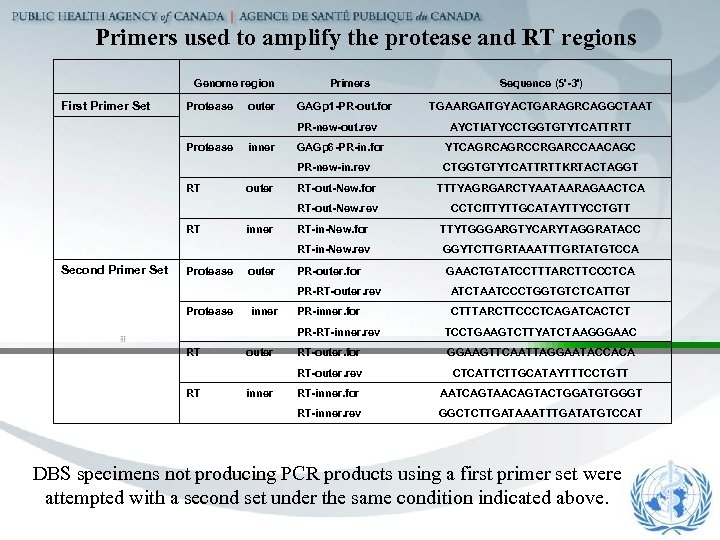
Primers used to amplify the protease and RT regions Genome region Primers Sequence (5'-3') First Primer Set Protease outer GAGp 1 -PR-out. for PR-new-out. rev Protease inner GAGp 6 -PR-in. for YTCAGRCCRGARCCAACAGC PR-new-in. rev CTGGTGTYTCATTRTTKRTACTAGGT RT outer RT-out-New. for TTTYAGRGARCTYAATAARAGAACTCA RT-out-New. rev CCTCITTYTTGCATAYTTYCCTGTT RT inner RT-in-New. for TTYTGGGARGTYCARYTAGGRATACC RT-in-New. rev GGYTCTTGRTAAATTTGRTATGTCCA Second Primer Set Protease outer PR-outer. for PR-RT-outer. rev ATCTAATCCCTGGTGTCTCATTGT Protease inner PR-inner. for CTTTARCTTCCCTCAGATCACTCT PR-RT-inner. rev TCCTGAAGTCTTYATCTAAGGGAAC RT outer RT-outer. for GGAAGTTCAATTAGGAATACCACA RT-outer. rev CTCATTCTTGCATAYTTTCCTGTT RT inner RT-inner. for AATCAGTAACAGTACTGGATGTGGGT RT-inner. rev GGCTCTTGATAAATTTGATATGTCCAT TGAARGAITGYACTGARAGRCAGGCTAAT AYCTIATYCCTGGTGTYTCATTRTT GAACTGTATCCTTTARCTTCCCTCA DBS specimens not producing PCR products using a first primer set were attempted with a second set under the same condition indicated above.
Methods Amplification and Sequencing of DBS § All specimens for which PCR products for both the protease and the RT regions could be generated were sequenced (Big. Dye v 3. 1 (ABI) and run on an ABI Prism 3100 analyzer). § Sequences were aligned to HXB 2 pol reference sequence using Seq. Scape software (ABI) and a contiguous sequence generated. § Sequences were queried against the Stanford University HIV Drug Resistance Database
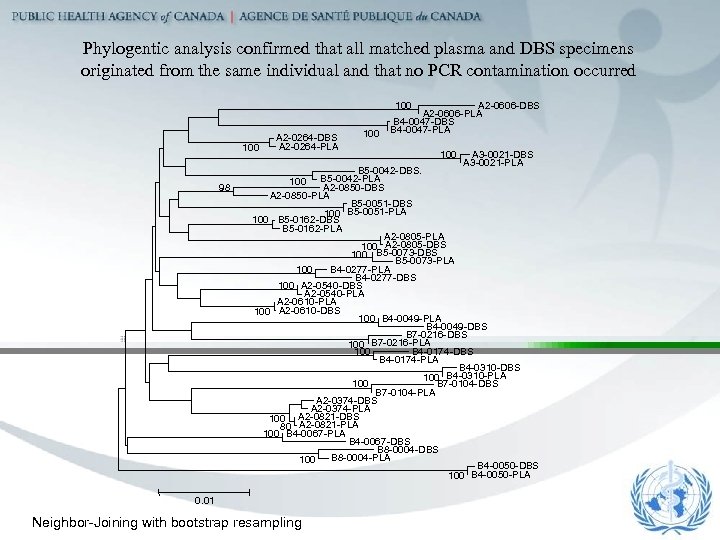
Phylogentic analysis confirmed that all matched plasma and DBS specimens originated from the same individual and that no PCR contamination occurred 100 98 A 2 -0264 -DBS A 2 -0264 -PLA 100 A 2 -0606 -DBS A 2 -0606 -PLA B 4 -0047 -DBS B 4 -0047 -PLA 100 A 3 -0021 -DBS A 3 -0021 -PLA B 5 -0042 -DBS. 100 B 5 -0042 -PLA A 2 -0850 -DBS A 2 -0850 -PLA B 5 -0051 -DBS 100 B 5 -0051 -PLA 100 B 5 -0162 -DBS B 5 -0162 -PLA A 2 -0805 -PLA 100 A 2 -0805 -DBS 100 B 5 -0073 -DBS B 5 -0073 -PLA 100 B 4 -0277 -PLA B 4 -0277 -DBS 100 A 2 -0540 -DBS A 2 -0540 -PLA A 2 -0610 -PLA 100 A 2 -0610 -DBS 100 B 4 -0049 -PLA B 4 -0049 -DBS B 7 -0216 -DBS 100 B 7 -0216 -PLA B 4 -0174 -DBS 100 B 4 -0174 -PLA B 4 -0310 -DBS 100 B 4 -0310 -PLA 100 B 7 -0104 -DBS B 7 -0104 -PLA A 2 -0374 -DBS A 2 -0374 -PLA 100 A 2 -0821 -DBS 80 A 2 -0821 -PLA 100 B 4 -0067 -PLA B 4 -0067 -DBS B 8 -0004 -PLA 100 B 4 -0050 -DBS 100 B 4 -0050 -PLA 0. 01 Neighbor-Joining with bootstrap resampling
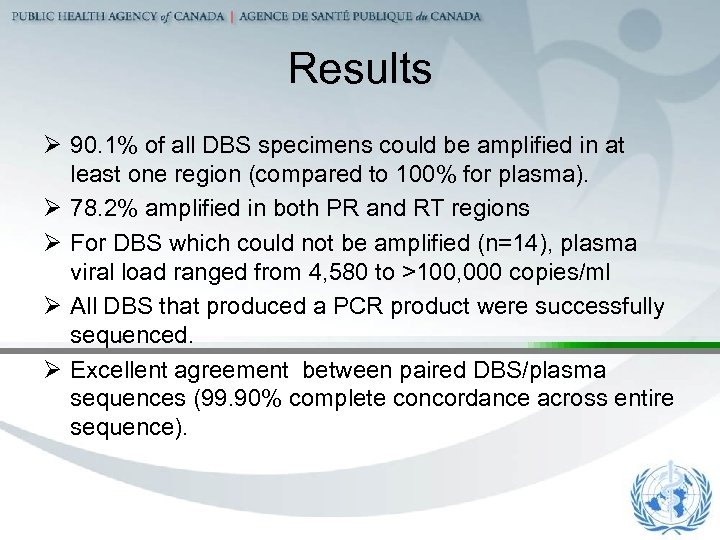
Results Ø 90. 1% of all DBS specimens could be amplified in at least one region (compared to 100% for plasma). Ø 78. 2% amplified in both PR and RT regions Ø For DBS which could not be amplified (n=14), plasma viral load ranged from 4, 580 to >100, 000 copies/ml Ø All DBS that produced a PCR product were successfully sequenced. Ø Excellent agreement between paired DBS/plasma sequences (99. 90% complete concordance across entire sequence).
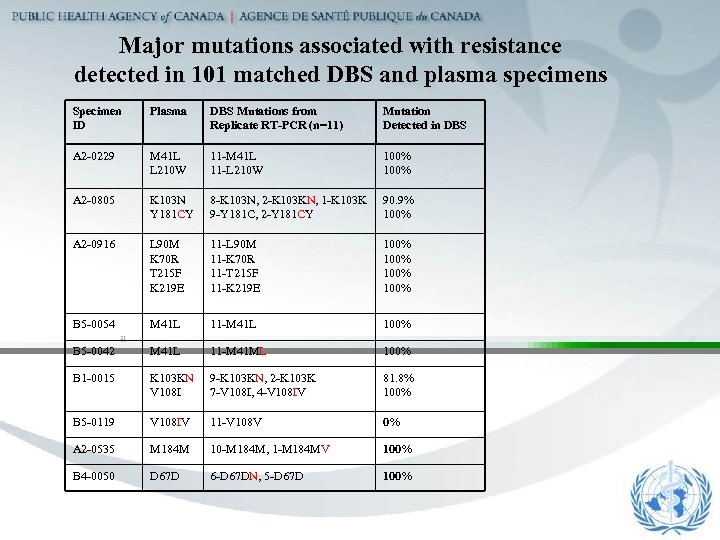
Major mutations associated with resistance detected in 101 matched DBS and plasma specimens Specimen ID Plasma DBS Mutations from Replicate RT-PCR (n=11) Mutation Detected in DBS A 2 -0229 M 41 L L 210 W 11 -M 41 L 11 -L 210 W 100% A 2 -0805 K 103 N Y 181 CY 8 -K 103 N, 2 -K 103 KN, 1 -K 103 K 9 -Y 181 C, 2 -Y 181 CY 90. 9% 100% A 2 -0916 L 90 M K 70 R T 215 F K 219 E 11 -L 90 M 11 -K 70 R 11 -T 215 F 11 -K 219 E 100% B 5 -0054 M 41 L 11 -M 41 L 100% B 5 -0042 M 41 L 11 -M 41 ML 100% B 1 -0015 K 103 KN V 108 I 9 -K 103 KN, 2 -K 103 K 7 -V 108 I, 4 -V 108 IV 81. 8% 100% B 5 -0119 V 108 IV 11 -V 108 V 0% A 2 -0535 M 184 M 10 -M 184 M, 1 -M 184 MV 100% B 4 -0050 D 67 D 6 -D 67 DN, 5 -D 67 D 100%
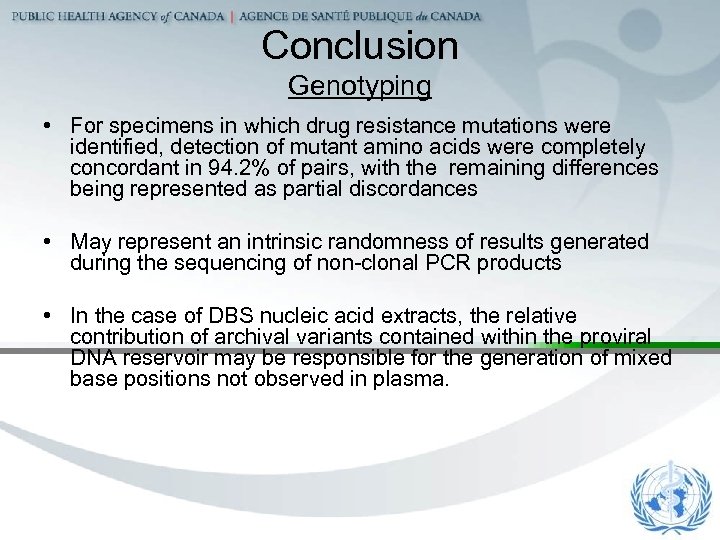
Conclusion Genotyping • For specimens in which drug resistance mutations were identified, detection of mutant amino acids were completely concordant in 94. 2% of pairs, with the remaining differences being represented as partial discordances • May represent an intrinsic randomness of results generated during the sequencing of non-clonal PCR products • In the case of DBS nucleic acid extracts, the relative contribution of archival variants contained within the proviral DNA reservoir may be responsible for the generation of mixed base positions not observed in plasma.
Conclusions Success Rate-PCR The observed differences between DBS and plasma amplification rates may be due to the independent or combined effects of several factors including: Ø Differences in primers and amplification conditions employed at each site Ø Lower sample volume used for the extraction of DBS (100 ul) as compared to plasma (200 -500 ul) Ø The partial degradation of RNA could have occurred under extreme storage conditions.
Conclusions General Ø DBS appear to be a promising public health tool for HIVDR surveillance of treatment-naïve subjects, especially in regions where specimens might be exposed to severe environmental conditions. Ø More study is needed to validate DBS for patient monitoring. Ø Suitability of DBS for commercial genotyping assays remains under investigation

Acknowledgments World Health Organization Silvia Bertagnolio Donald Sutherland Instituto Nacional Ciencias Medicas, México Luis Soto-Ramirez Roberto Rodriguez Monica Viveros Luis Fuentes BC Centre for Excellence Richard Harrigan Theresa Mo Public Health Agency of Canada Richard Pilon Linda Ares
fc06eed009e3e30aa58785a56261aeec.ppt