97e5e9026168179ed334c96a9ecafb53.ppt
- Количество слайдов: 74

High-throughput sequencing 1

Biological samples Workflows 2

Biological samples Library generation, sequencing, image capture Sequence reads Workflows 3

Sequencing 101 Break up DNA into small fragments 4

Sequencing 101 Cloning 5
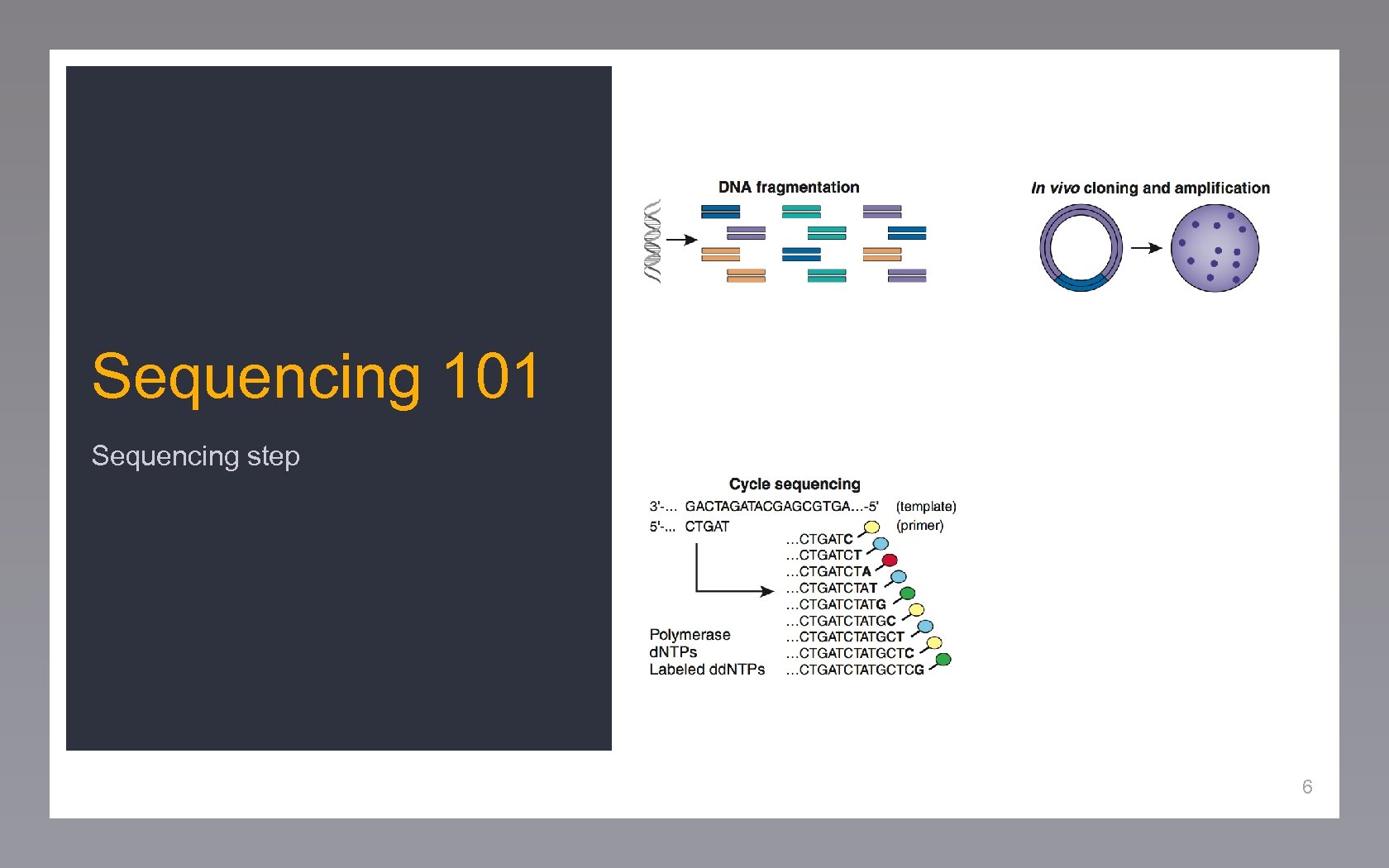
Sequencing 101 Sequencing step 6
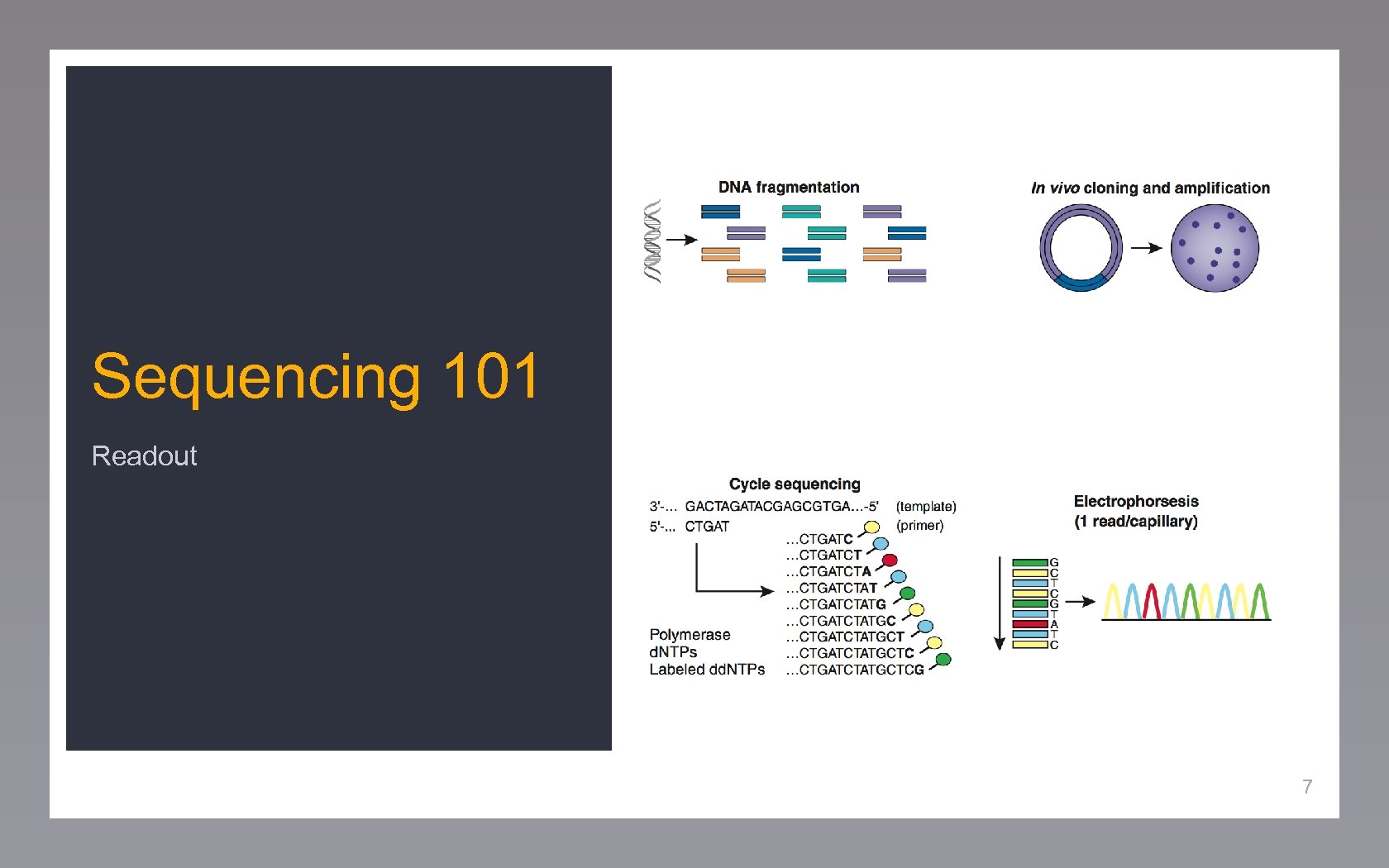
Sequencing 101 Readout 7
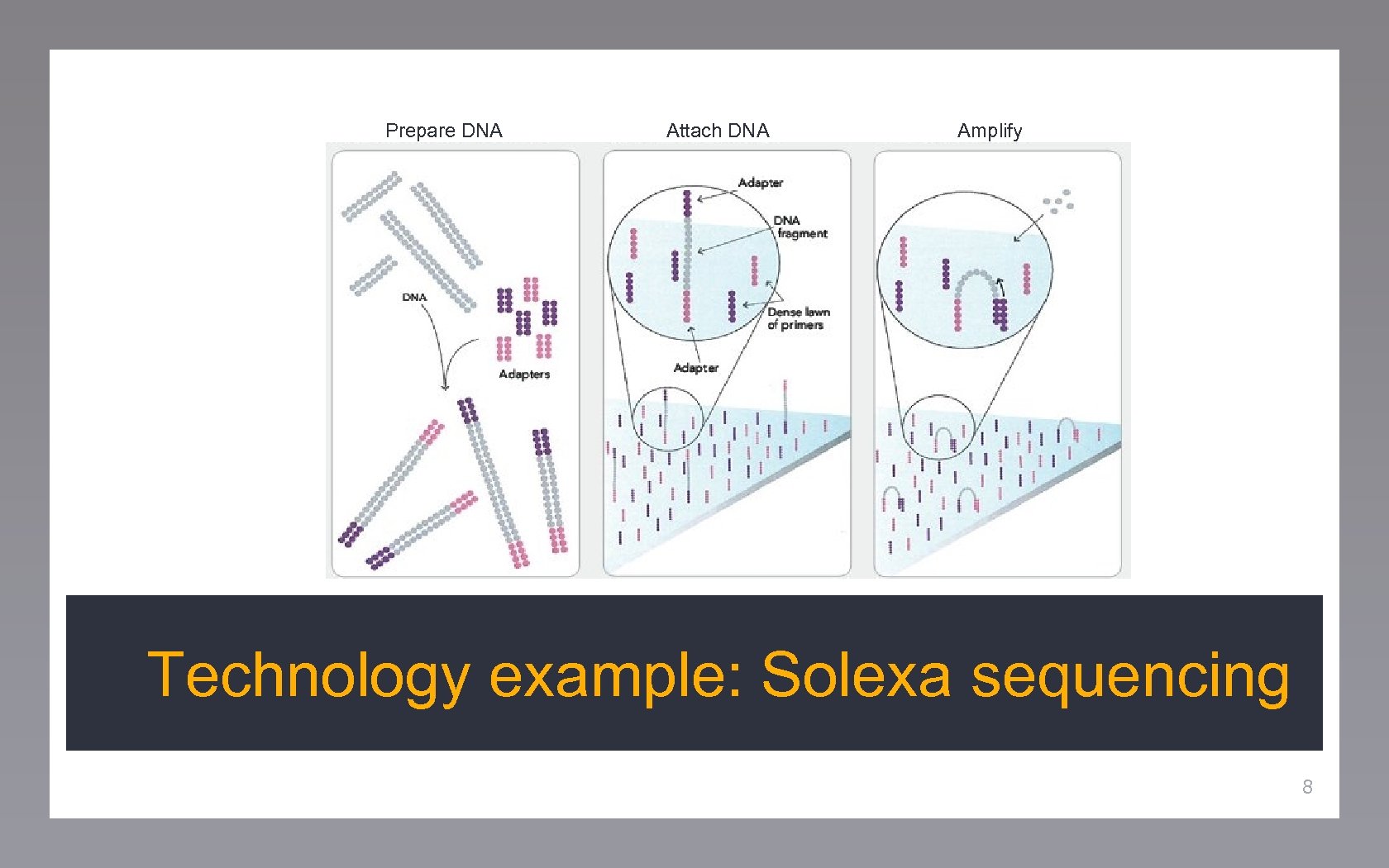
Prepare DNA Attach DNA Amplify Technology example: Solexa sequencing 8
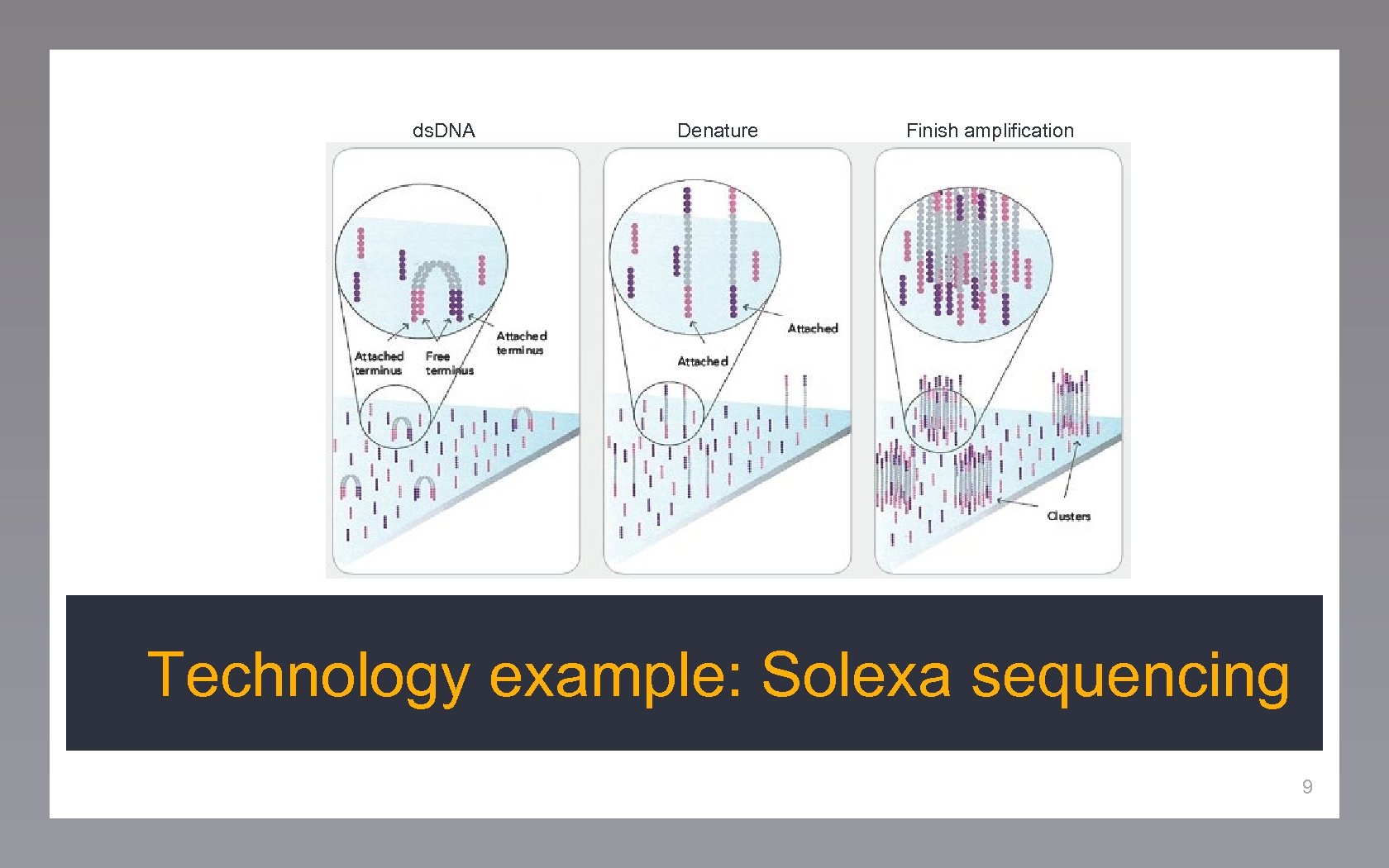
ds. DNA Denature Finish amplification Technology example: Solexa sequencing 9

First nucleotide Readout Second nucleotide Technology example: Solexa sequencing 10

Second readout Repeat Align Technology example: Solexa sequencing 11
Solexa Helicos Other technologies Solexa, Helicos, 454, . . . 12

454/SOLi. D Other technologies Solexa, Helicos, 454, . . . 13
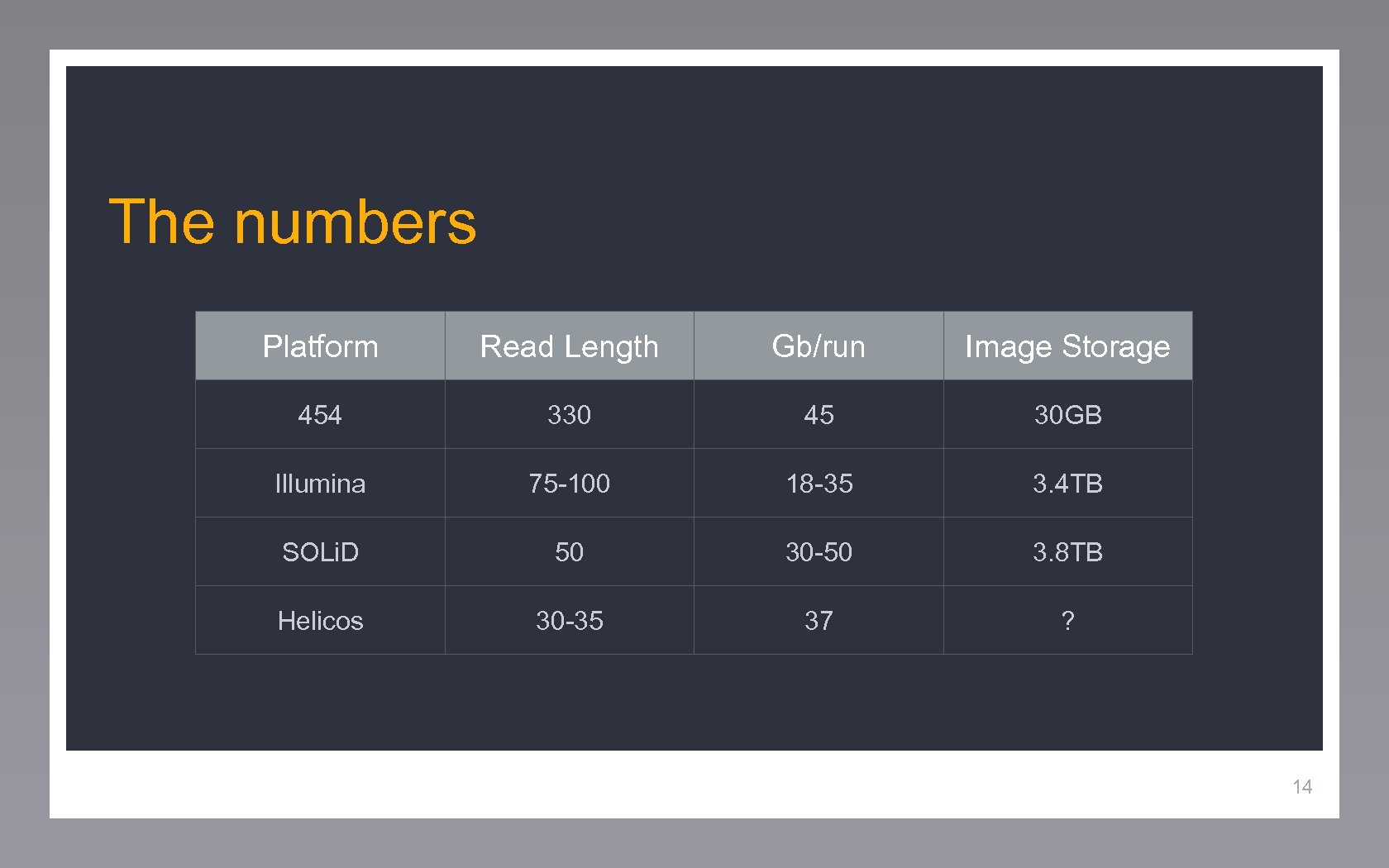
The numbers Platform Read Length Gb/run Image Storage 454 330 45 30 GB Illumina 75 -100 18 -35 3. 4 TB SOLi. D 50 30 -50 3. 8 TB Helicos 30 -35 37 ? 14
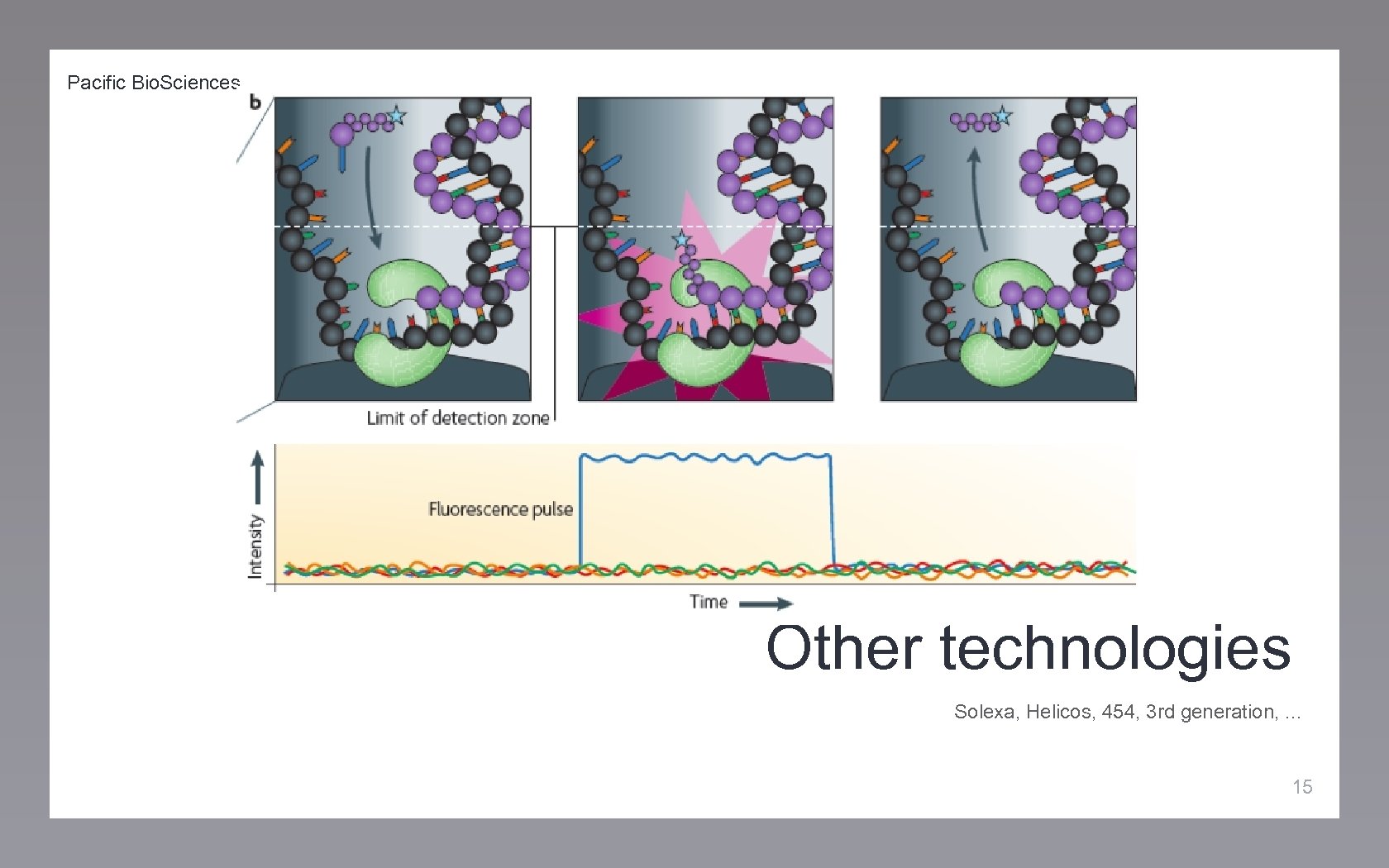
Pacific Bio. Sciences Other technologies Solexa, Helicos, 454, 3 rd generation, . . . 15

Biological samples Library generation, sequencing, image capture Sequence reads Workflows 16

Biological samples Library generation, sequencing, image capture Sequence reads Data storage and initial processing Workflows 17
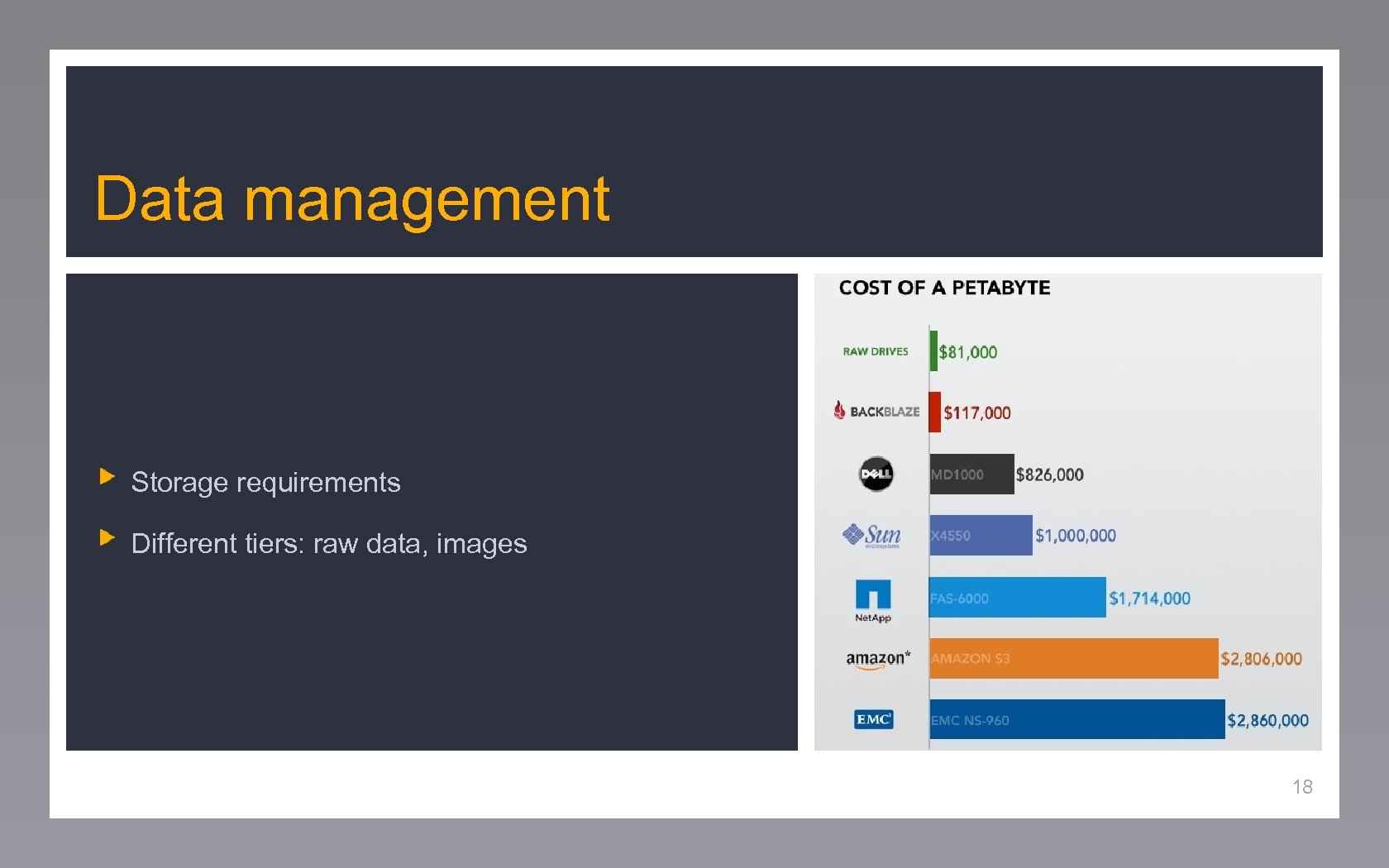
Data management Storage requirements Different tiers: raw data, images 18
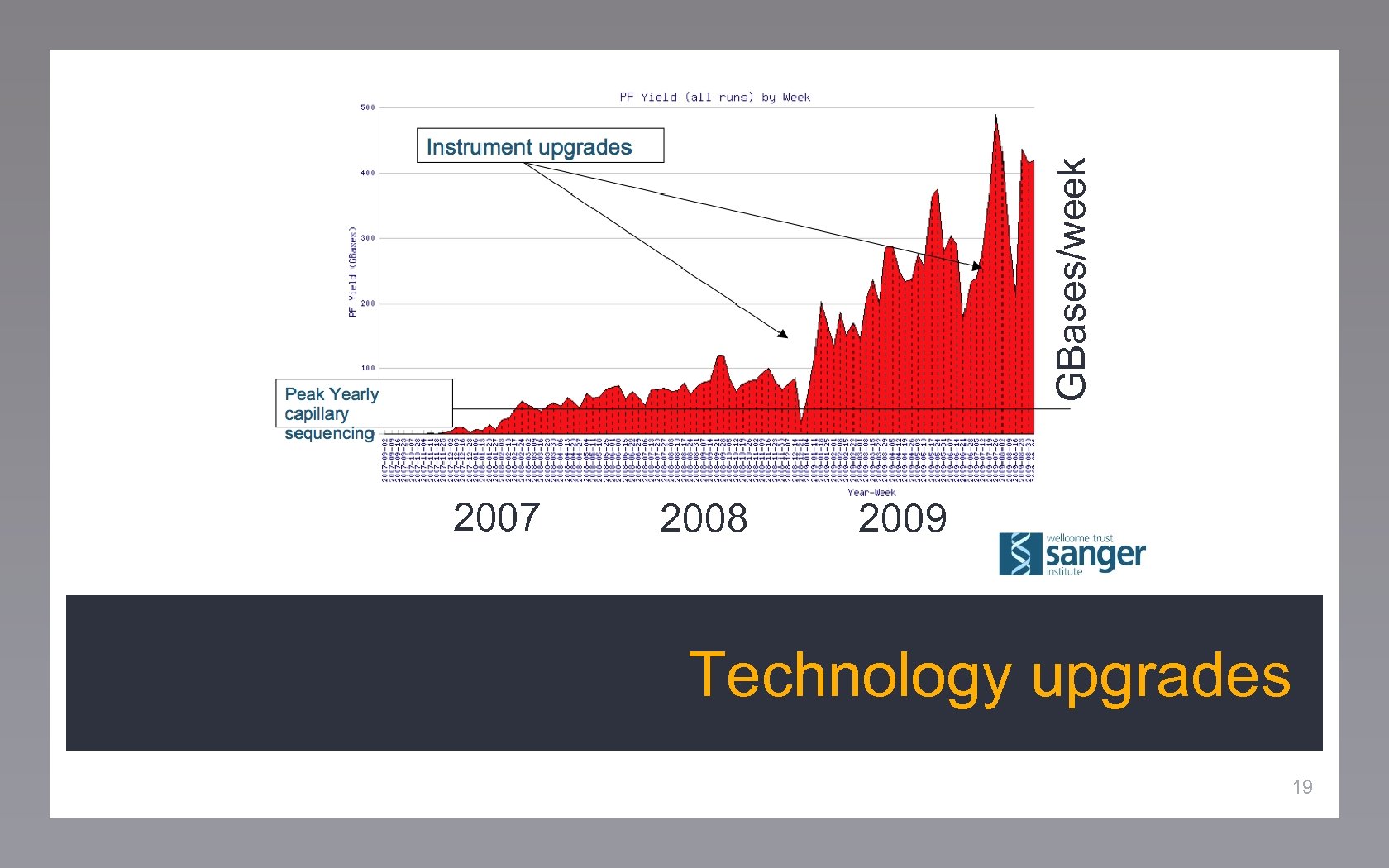
GBases/week 2007 2008 2009 Technology upgrades 19

Technology outpacing hardware 50 MB/s data generation Require real-time data processing CPU demands increase 20
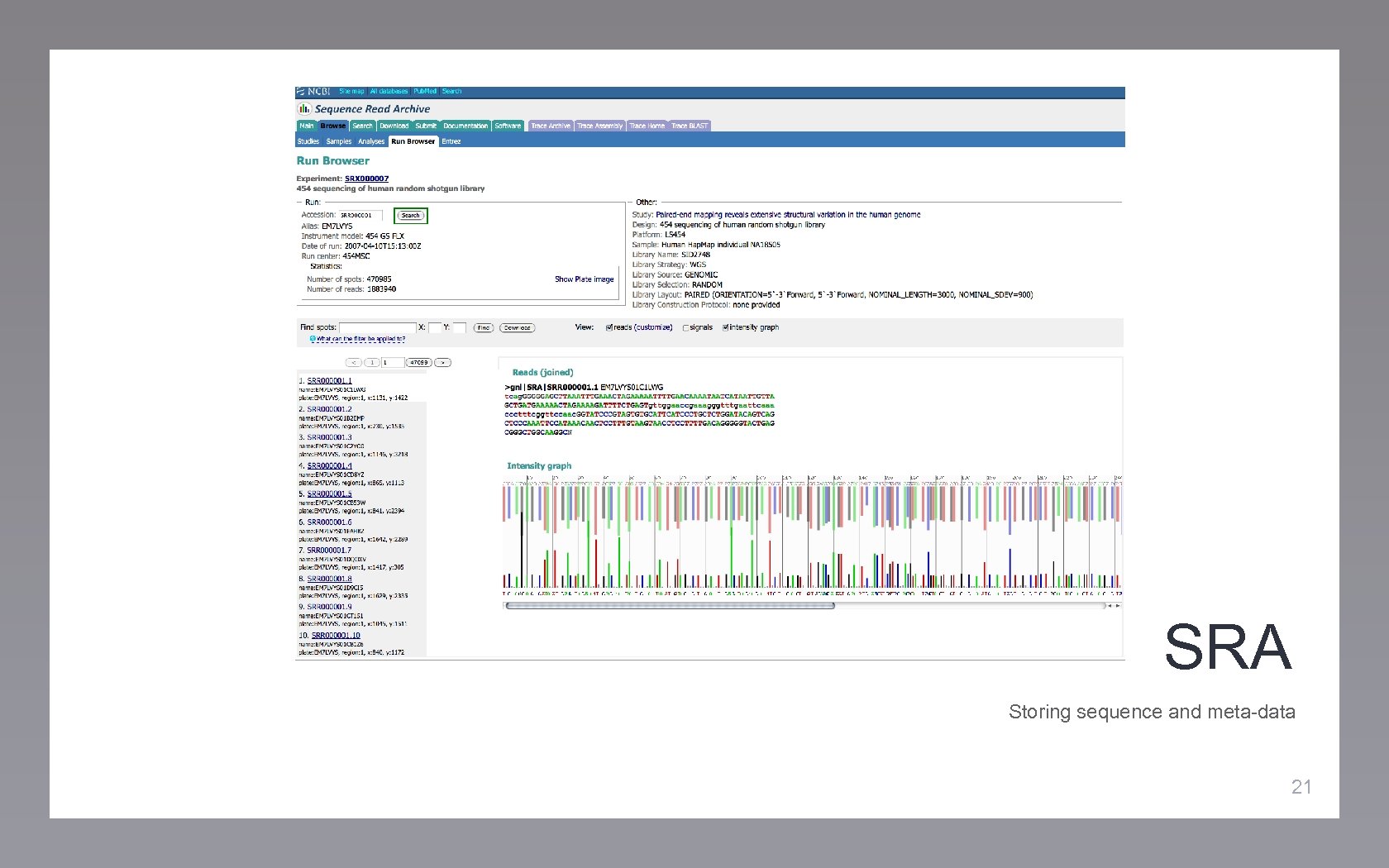
SRA Storing sequence and meta-data 21
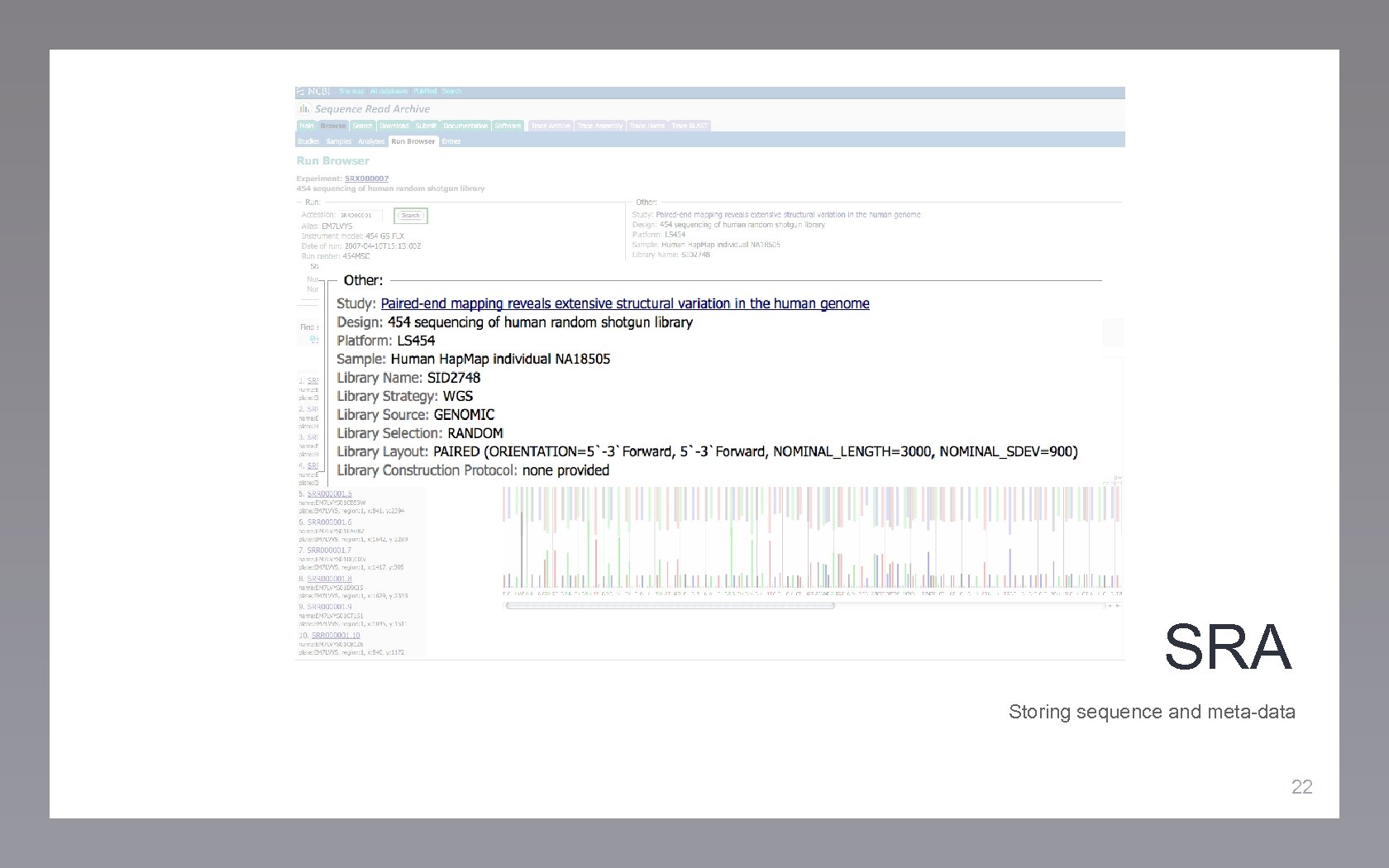
SRA Storing sequence and meta-data 22
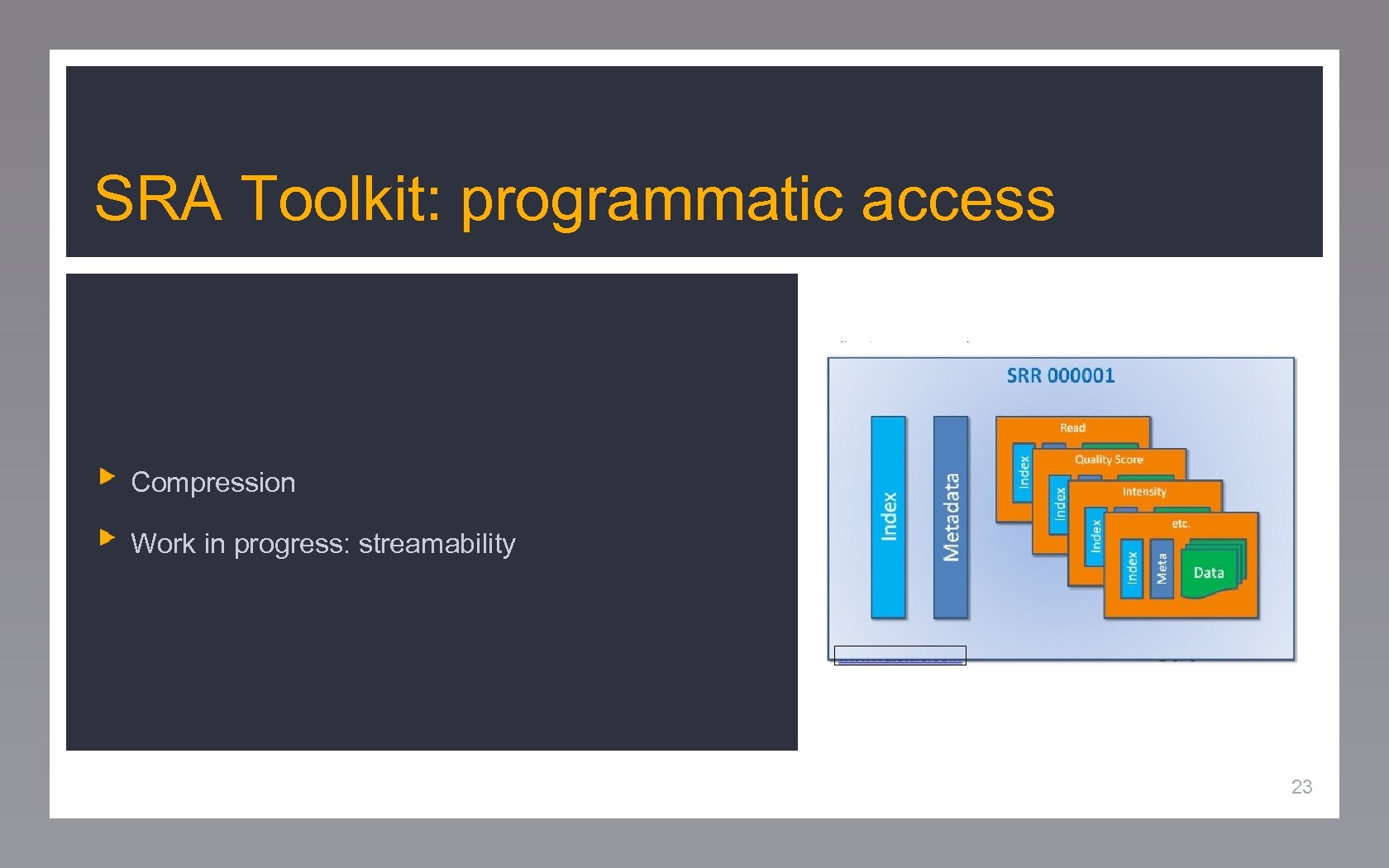
SRA Toolkit: programmatic access Compression Work in progress: streamability 23
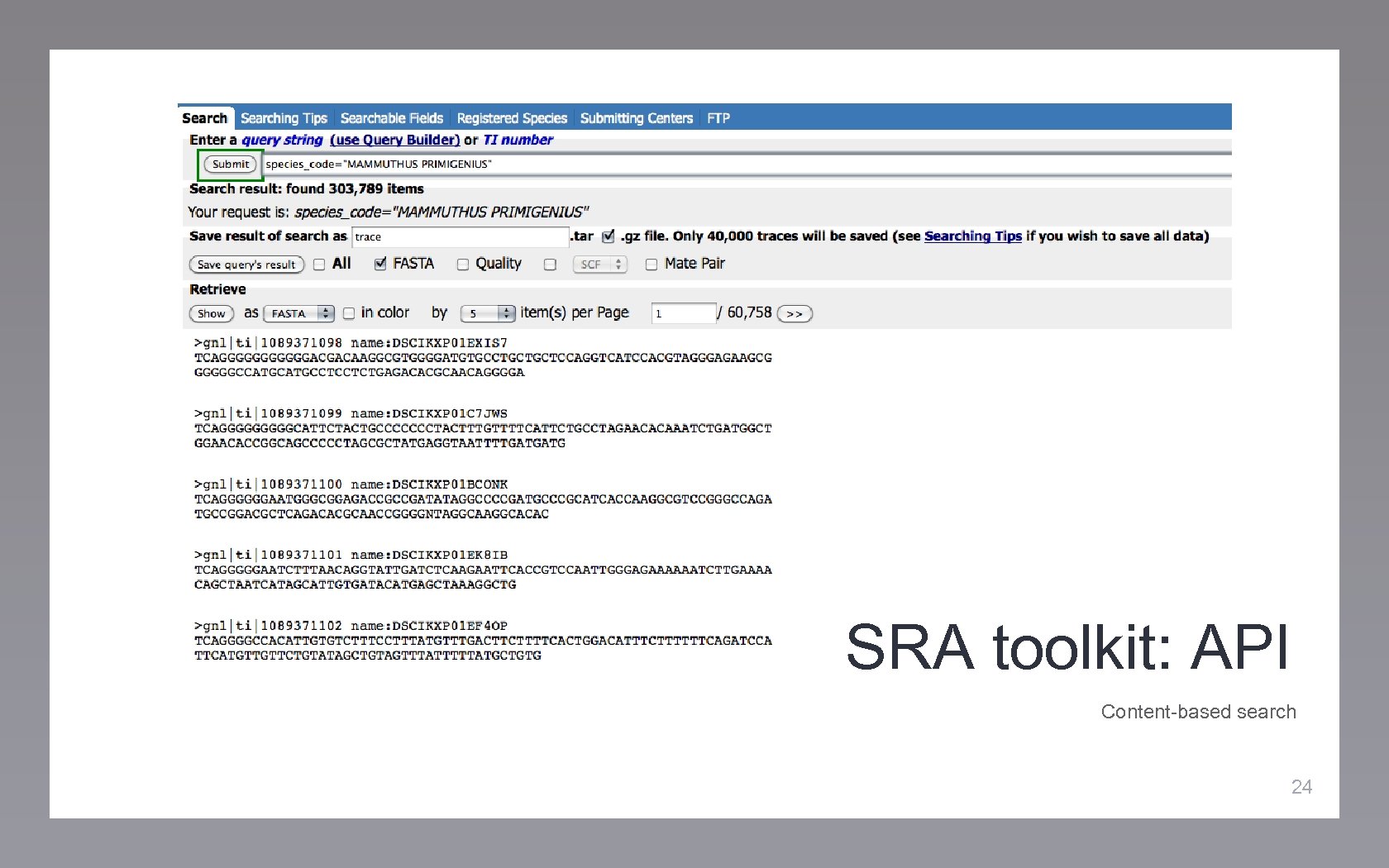
SRA toolkit: API Content-based search 24
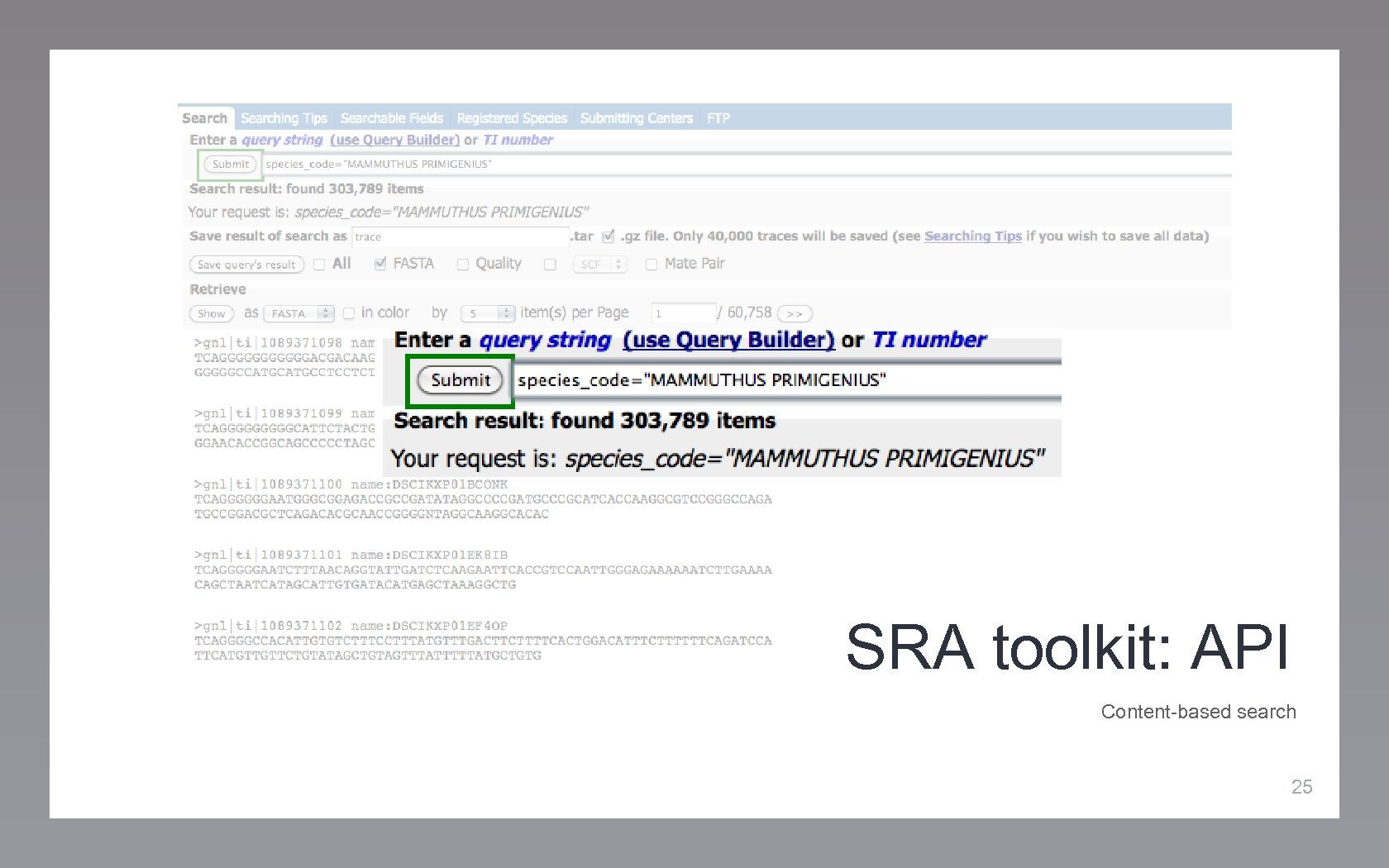
SRA toolkit: API Content-based search 25

Biological samples Library generation, sequencing, image capture Sequence reads Data storage and initial processing Workflows 26
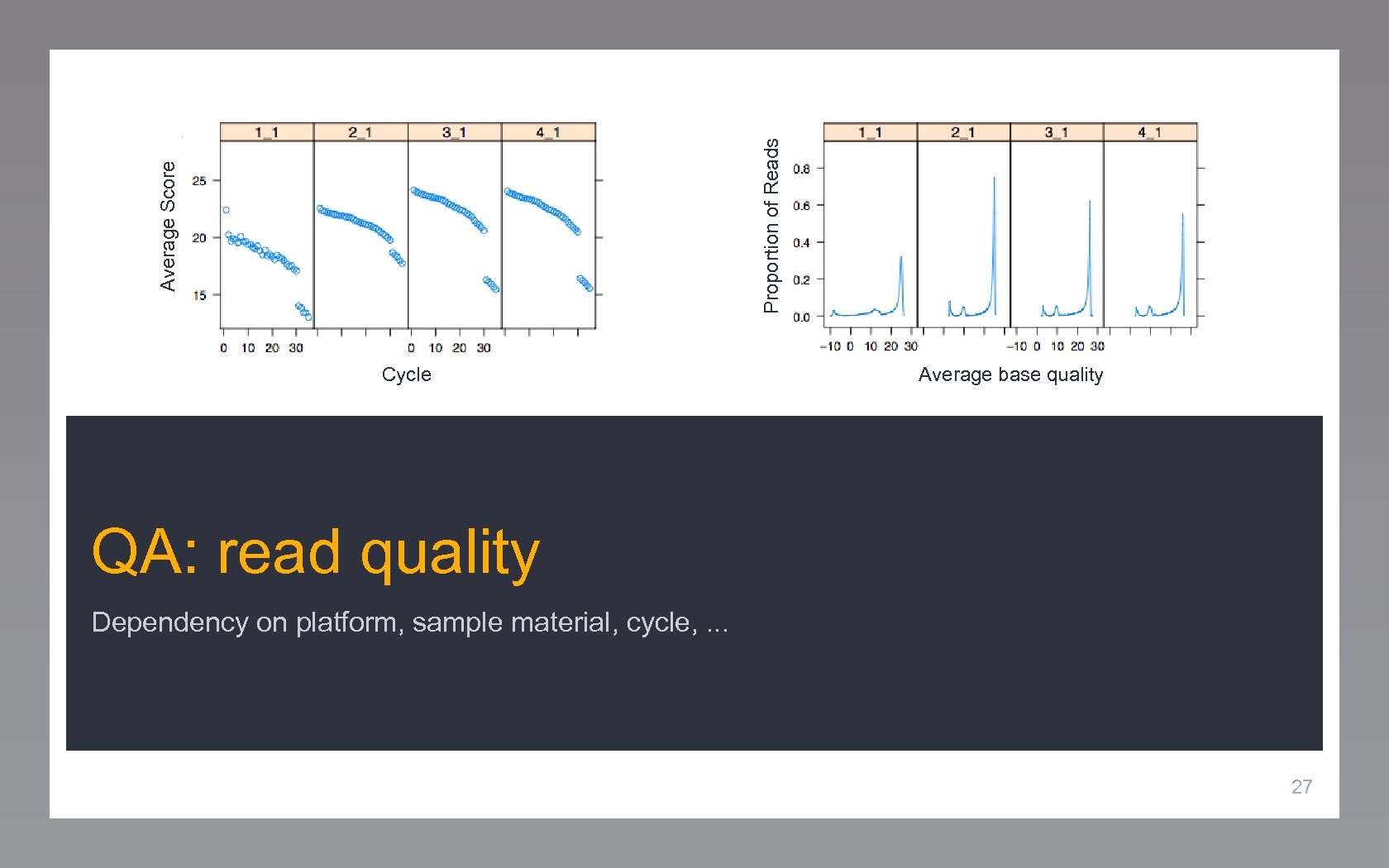
Proportion of Reads Average Score Cycle Average base quality QA: read quality Dependency on platform, sample material, cycle, . . . 27
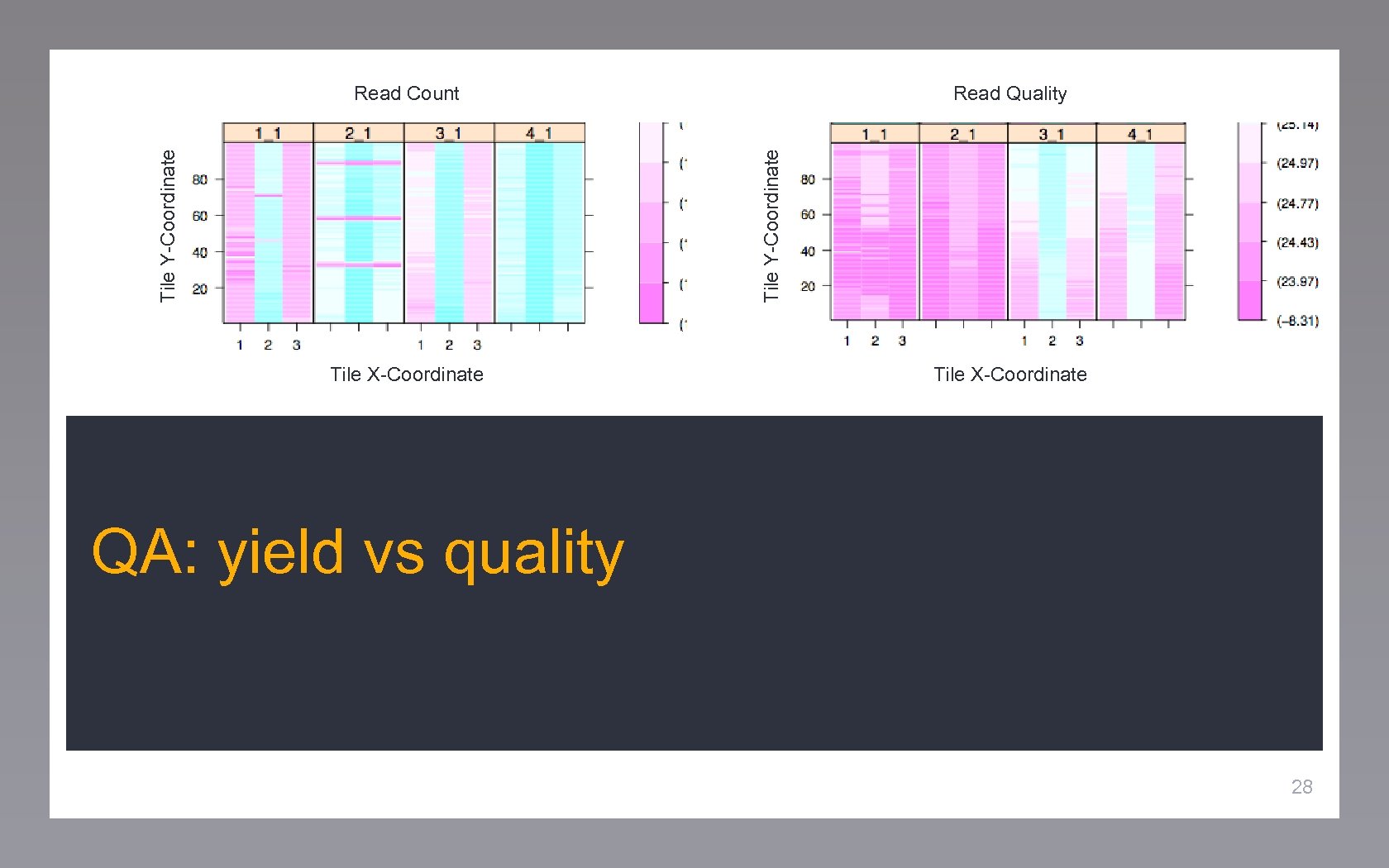
Read Quality Tile Y-Coordinate Read Count Tile X-Coordinate QA: yield vs quality 28

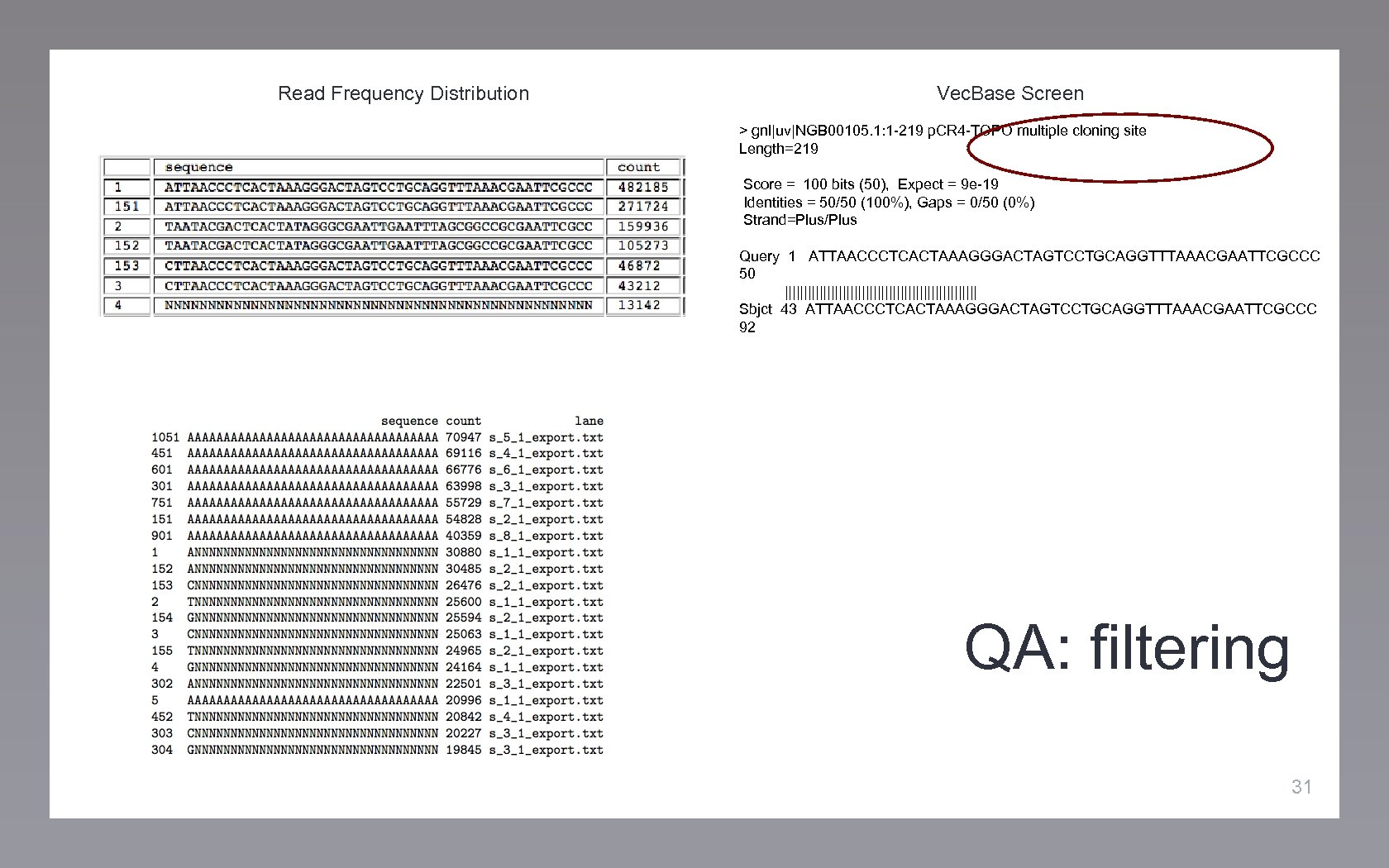
Read Frequency Distribution QA: filtering 29

Read Frequency Distribution Vec. Base Screen > gnl|uv|NGB 00105. 1: 1 -219 p. CR 4 -TOPO multiple cloning site Length=219 Score = 100 bits (50), Expect = 9 e-19 Identities = 50/50 (100%), Gaps = 0/50 (0%) Strand=Plus/Plus Query 1 ATTAACCCTCACTAAAGGGACTAGTCCTGCAGGTTTAAACGAATTCGCCC 50 ||||||||||||||||||||||||| Sbjct 43 ATTAACCCTCACTAAAGGGACTAGTCCTGCAGGTTTAAACGAATTCGCCC 92 QA: filtering 30
Read Frequency Distribution Vec. Base Screen > gnl|uv|NGB 00105. 1: 1 -219 p. CR 4 -TOPO multiple cloning site Length=219 Score = 100 bits (50), Expect = 9 e-19 Identities = 50/50 (100%), Gaps = 0/50 (0%) Strand=Plus/Plus Query 1 ATTAACCCTCACTAAAGGGACTAGTCCTGCAGGTTTAAACGAATTCGCCC 50 ||||||||||||||||||||||||| Sbjct 43 ATTAACCCTCACTAAAGGGACTAGTCCTGCAGGTTTAAACGAATTCGCCC 92 QA: filtering 31
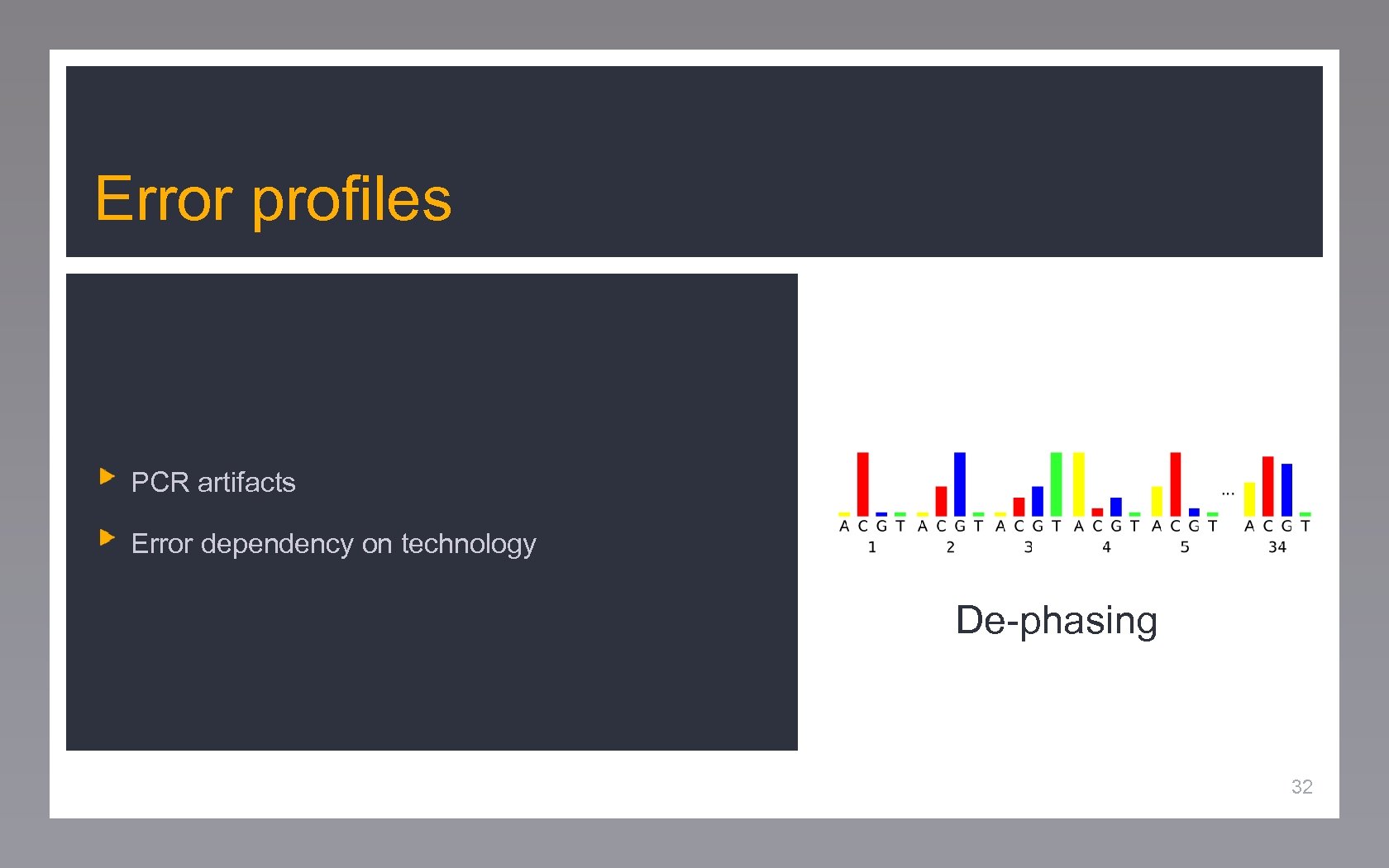
Error profiles PCR artifacts Error dependency on technology De-phasing 32
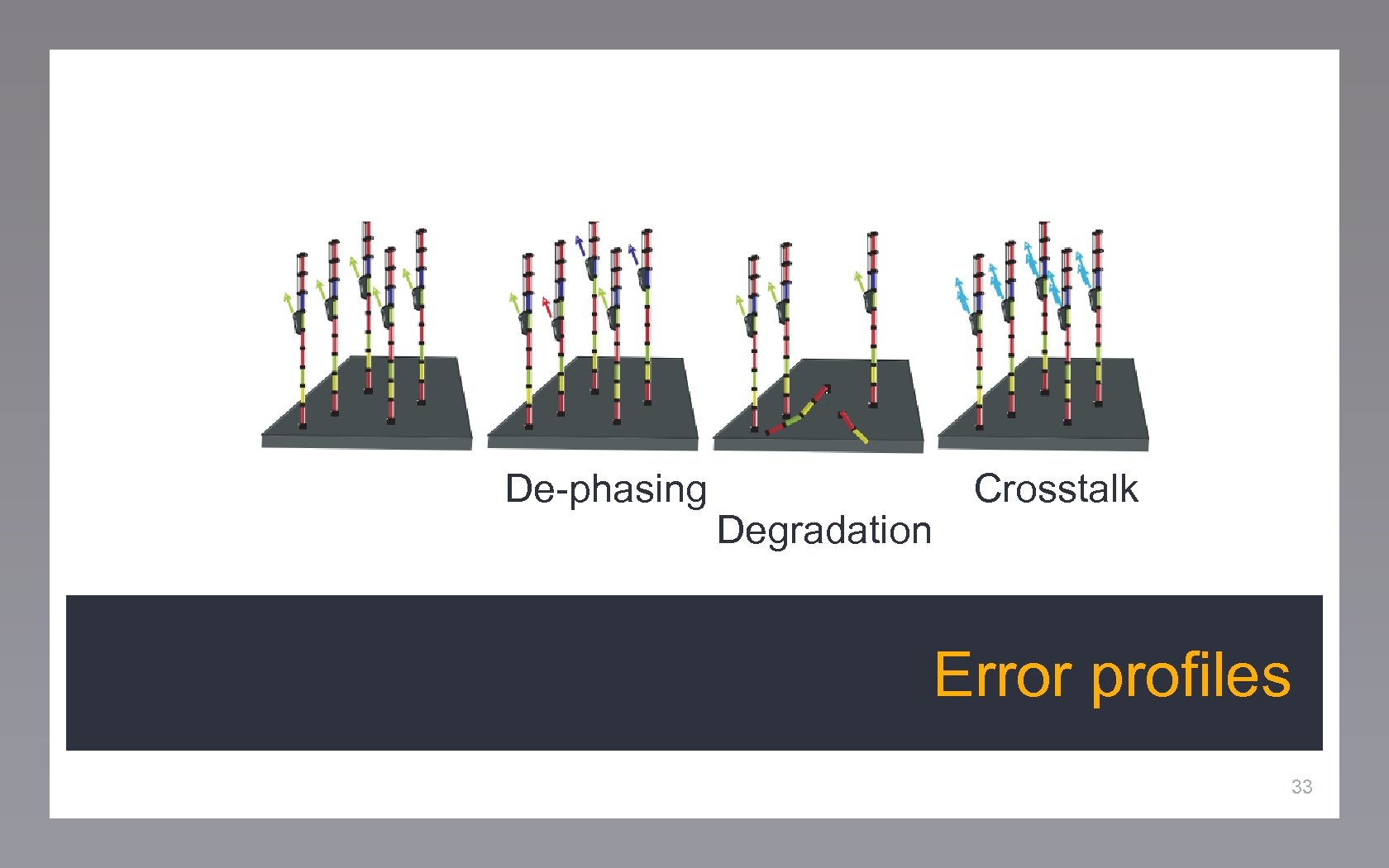
De-phasing Degradation Crosstalk De-phasing Error profiles 33

Biological samples Library generation, sequencing, image capture Sequence reads Data storage and initial processing Quality control Workflows 34
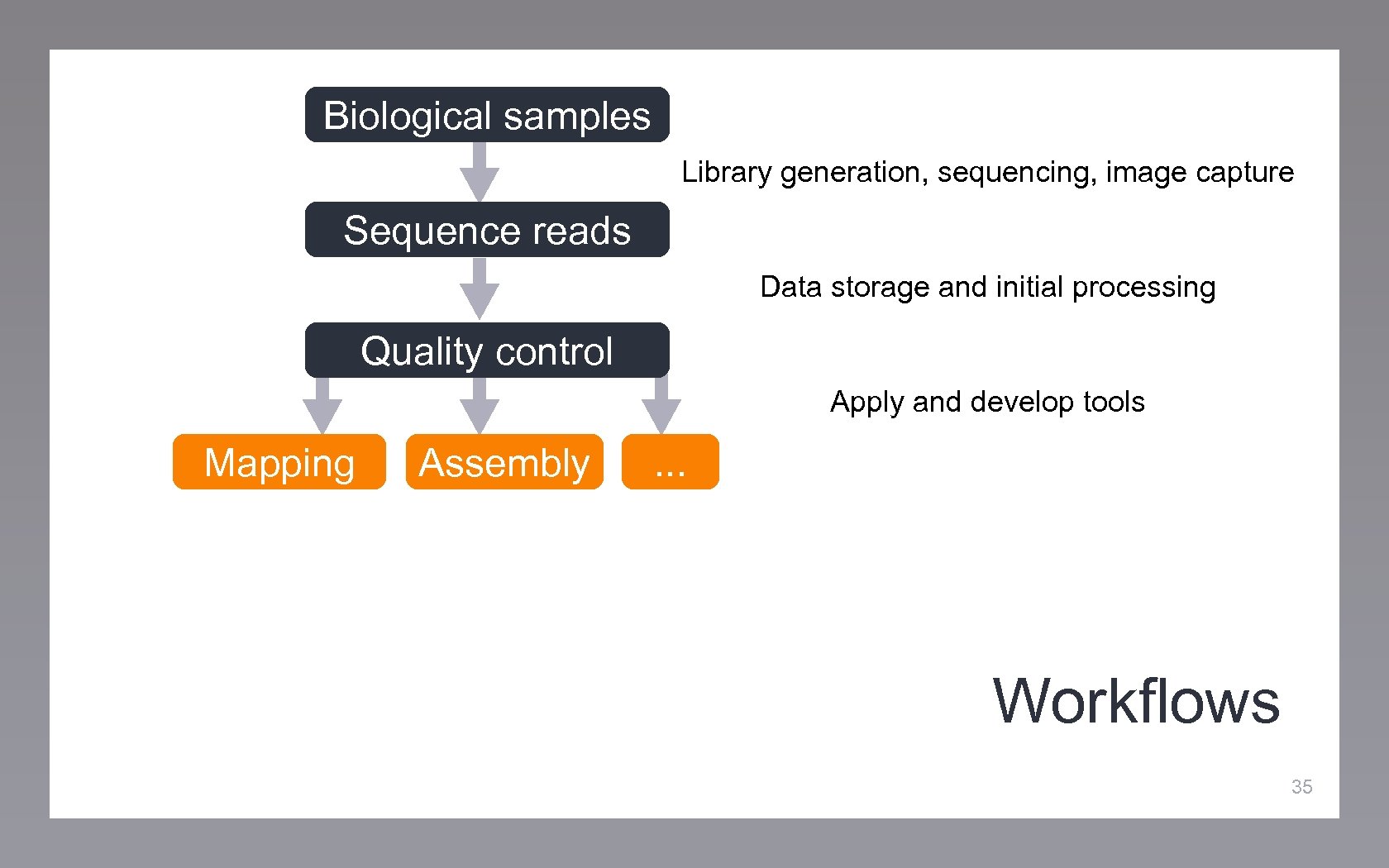
Biological samples Library generation, sequencing, image capture Sequence reads Data storage and initial processing Quality control Apply and develop tools Mapping Assembly . . . Workflows 35

Tool evolution: mapping approaches Variation in algorithm Alignment speed Memory requirements Error tolerance. . . 36

Genome Sequence tag Tool evolution: mapping reads to a genome HSPH Bioinformatics Core 37
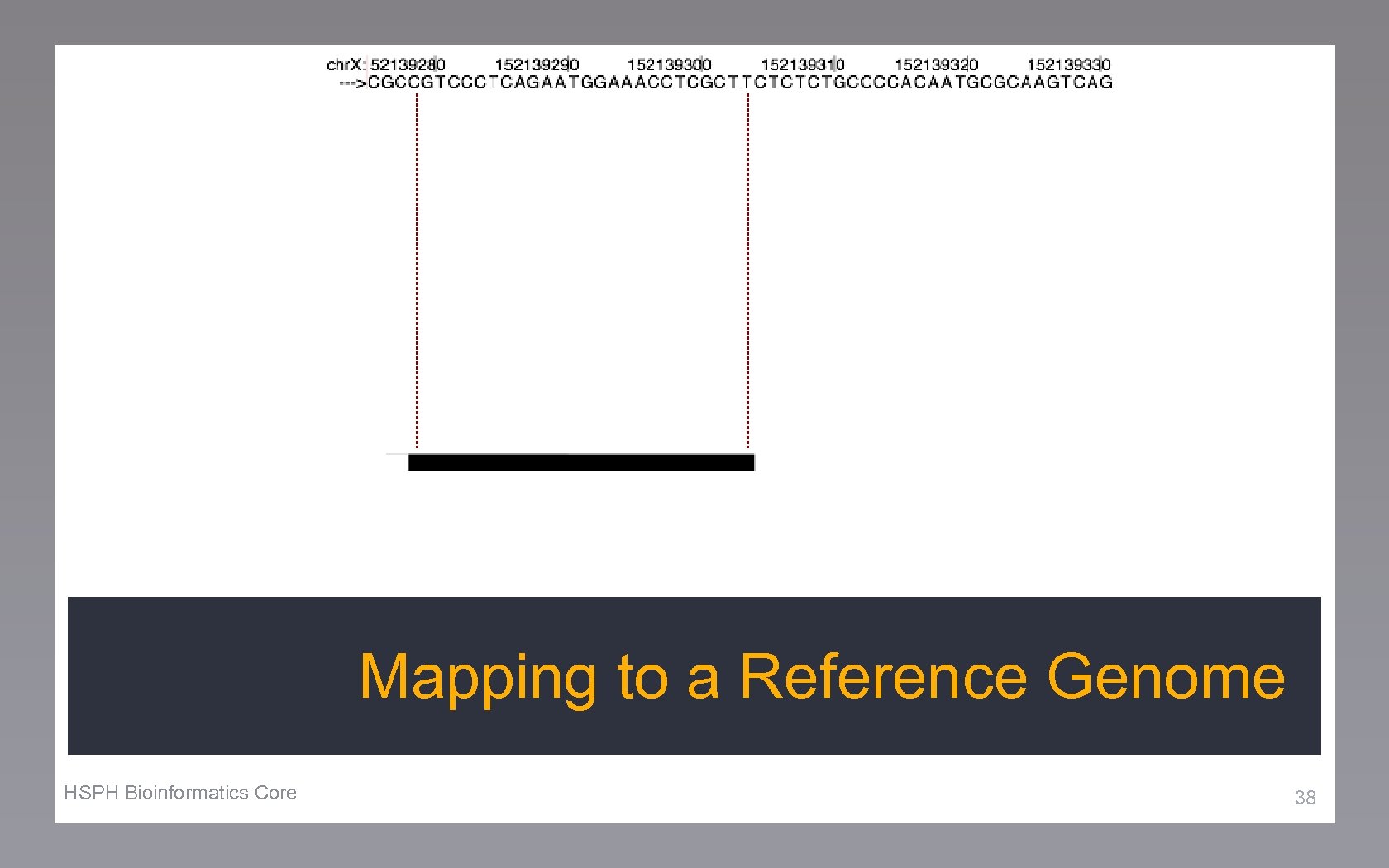
Mapping to a Reference Genome HSPH Bioinformatics Core 38
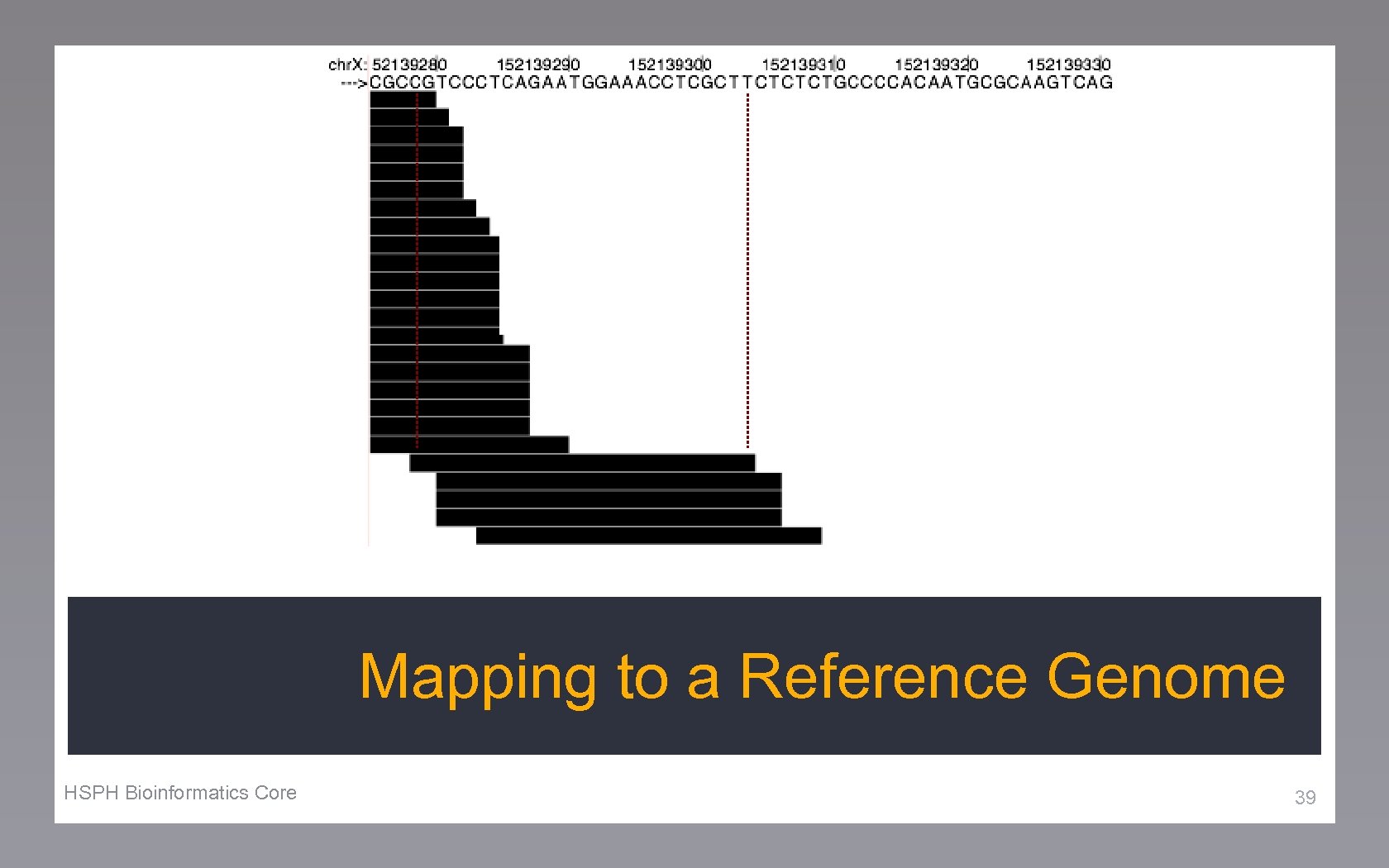
Mapping to a Reference Genome HSPH Bioinformatics Core 39

CGTCCCTCAGATTGGAAACCTCGCTT Mapping to a Reference Genome HSPH Bioinformatics Core 40
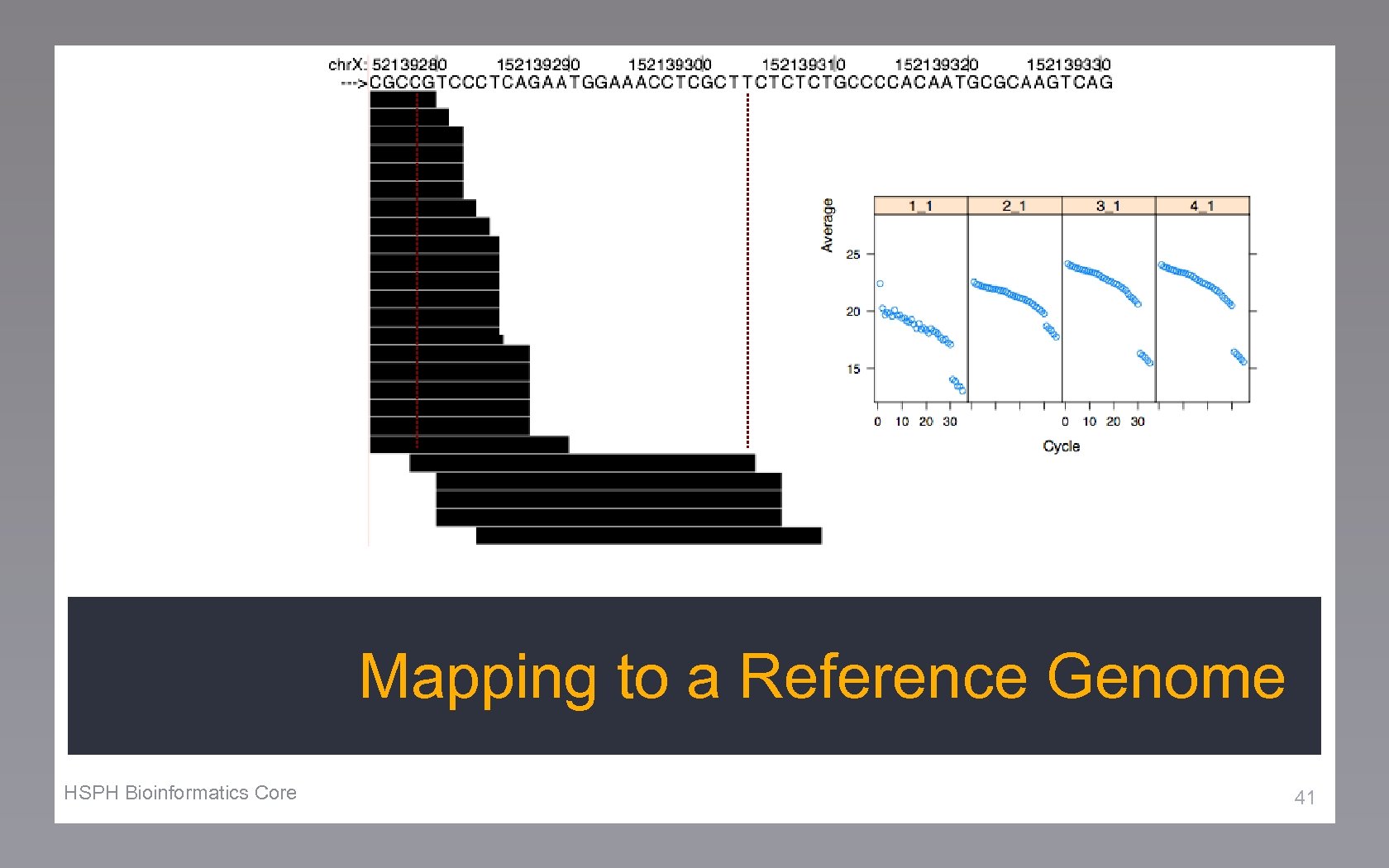
Mapping to a Reference Genome HSPH Bioinformatics Core 41

Reference Genome 42

Reference Genome 43
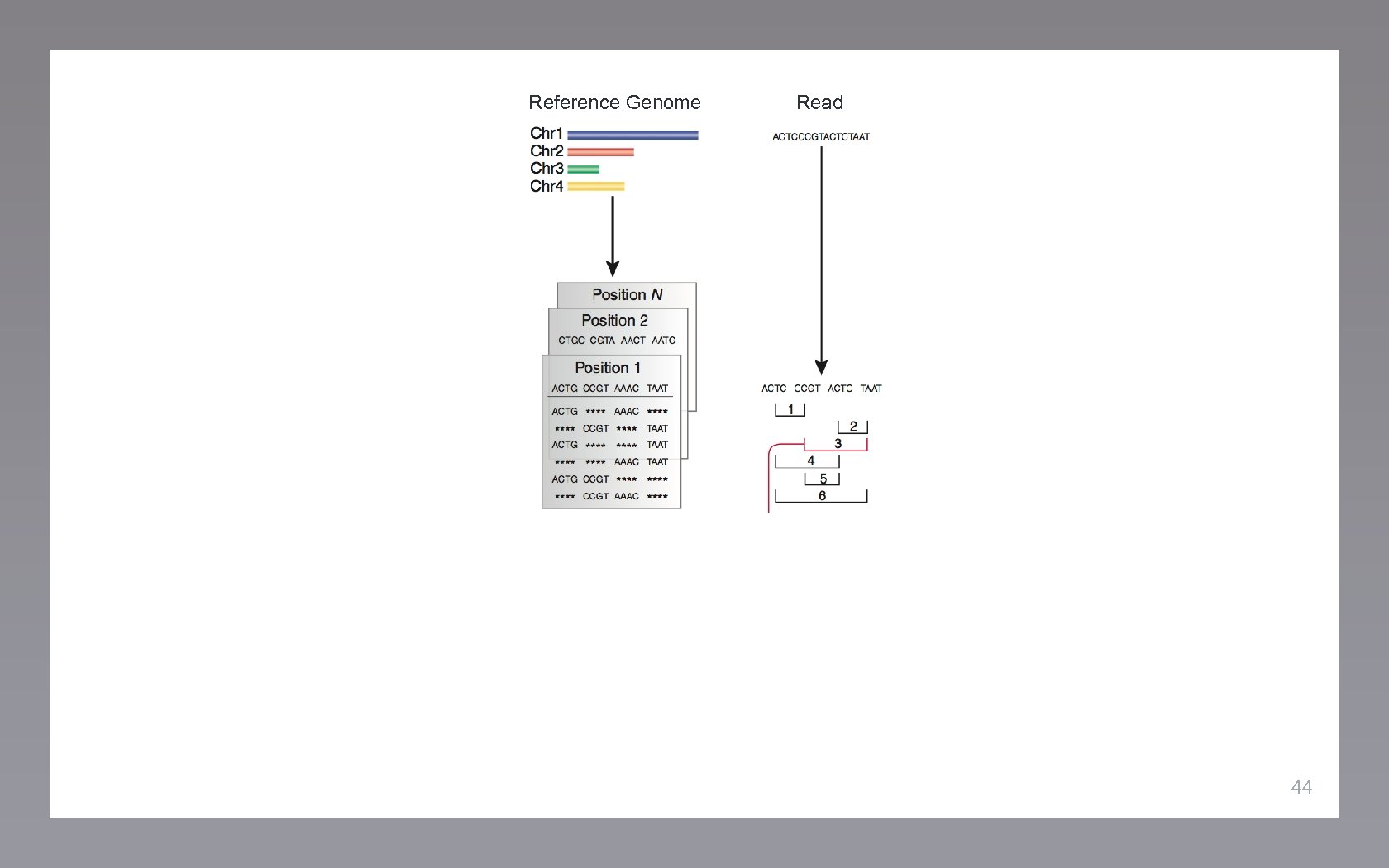
Reference Genome Read 44
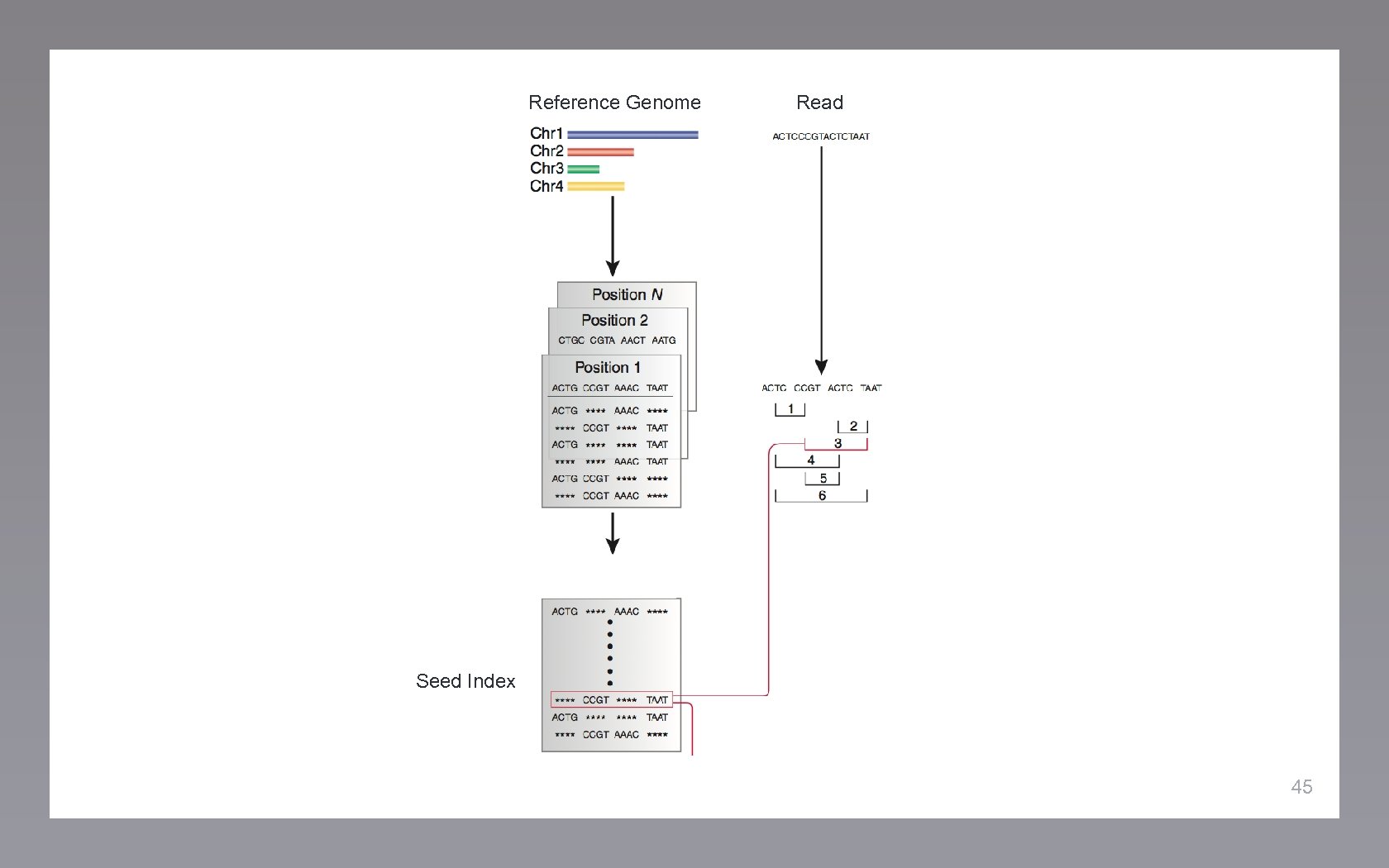
Reference Genome Read Seed Index 45
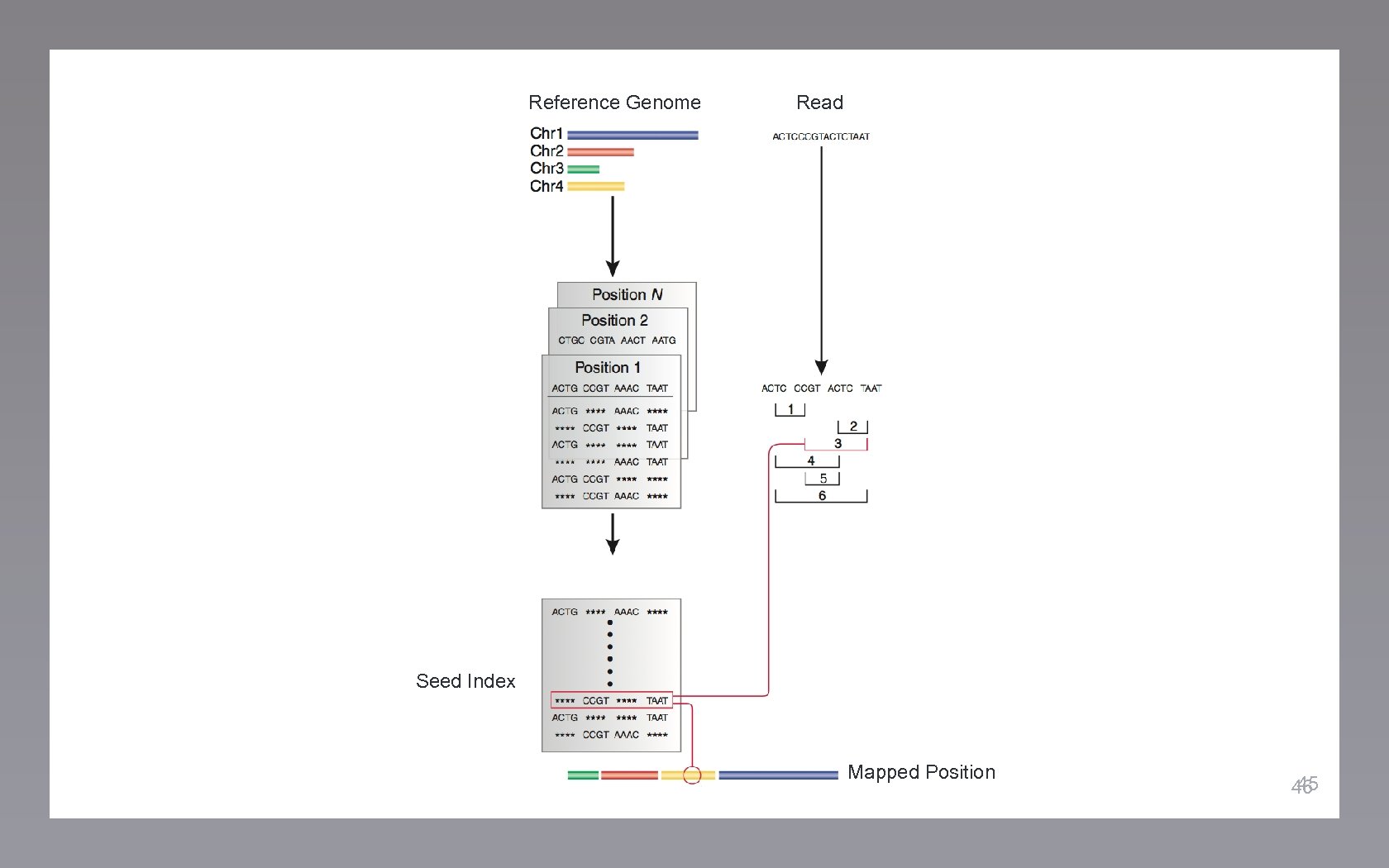
Reference Genome Read Seed Index Mapped Position 45 46
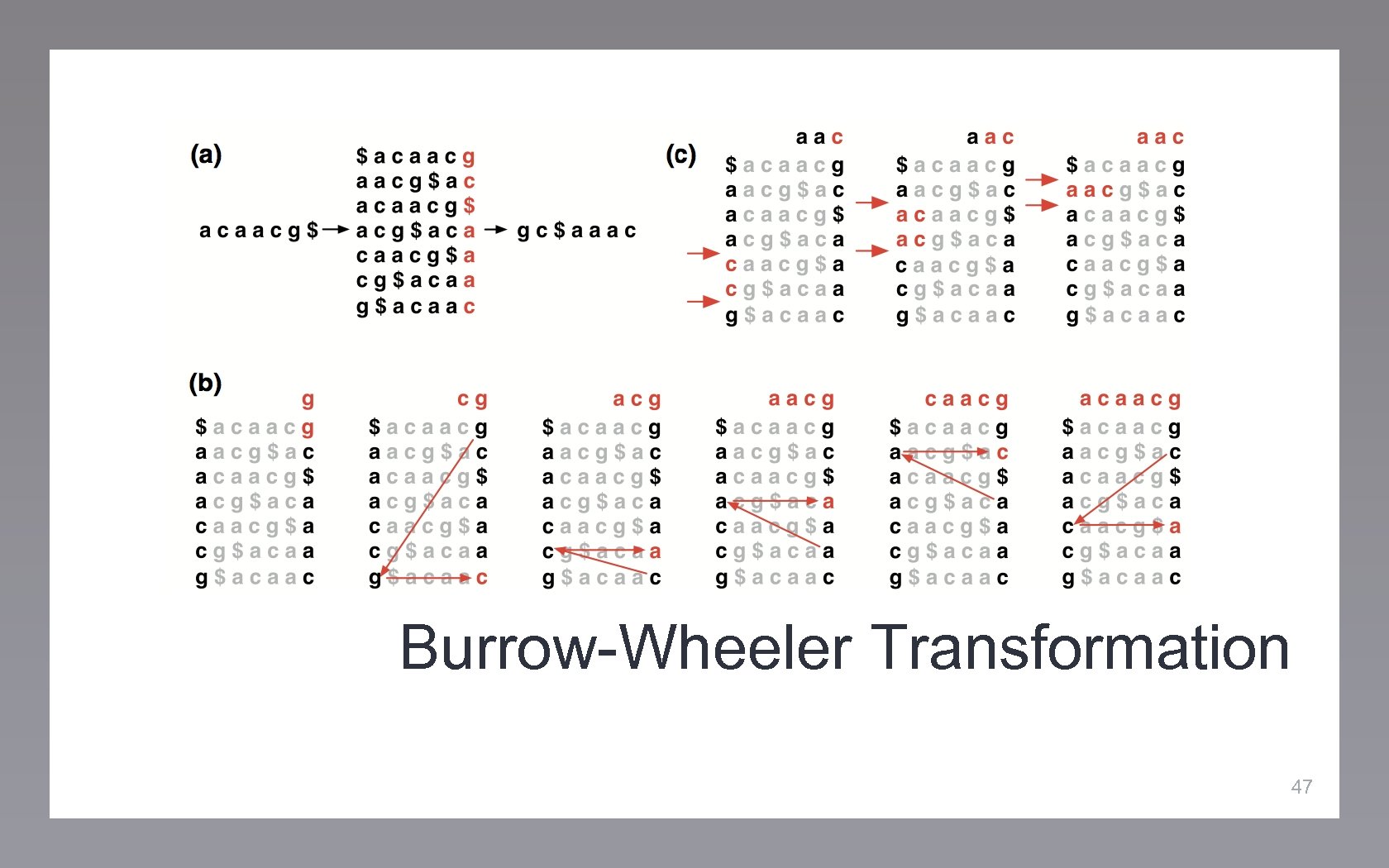
Burrow-Wheeler Transformation 47
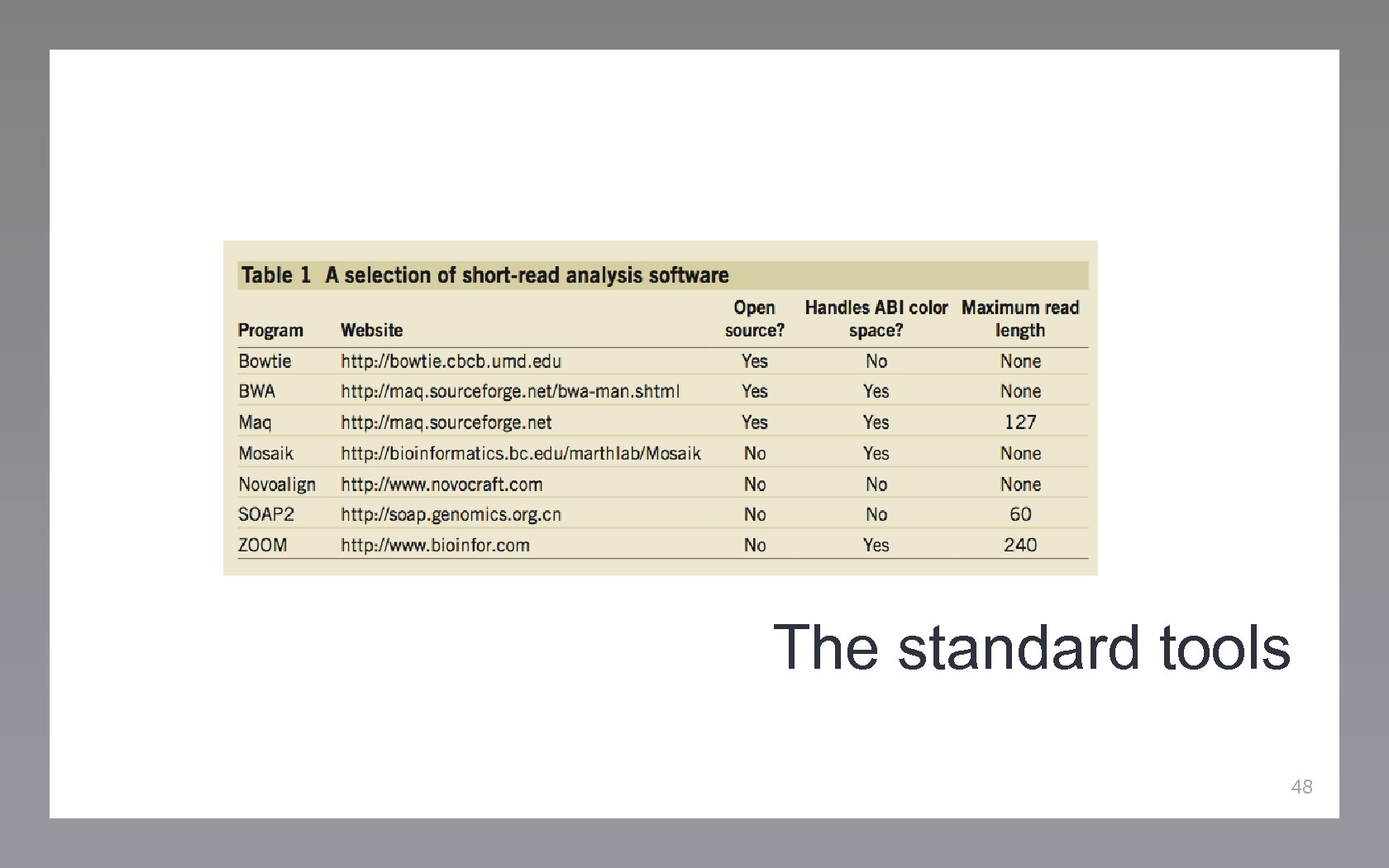
The standard tools 48

http: //seqanswers. com/wiki/SEQanswers The extended selection 220 applications and counting 49
Biological samples Library generation, sequencing, image capture Sequence reads Data storage and initial processing Quality control Apply and develop tools Mapping Assembly . . . Workflows 50
Biological samples Library generation, sequencing, image capture Sequence reads Data storage and initial processing Quality control Apply and develop tools Mapping Peak calling Assembly . . . Variant Detection Contact maps . . . Visualization and processing Workflows 51
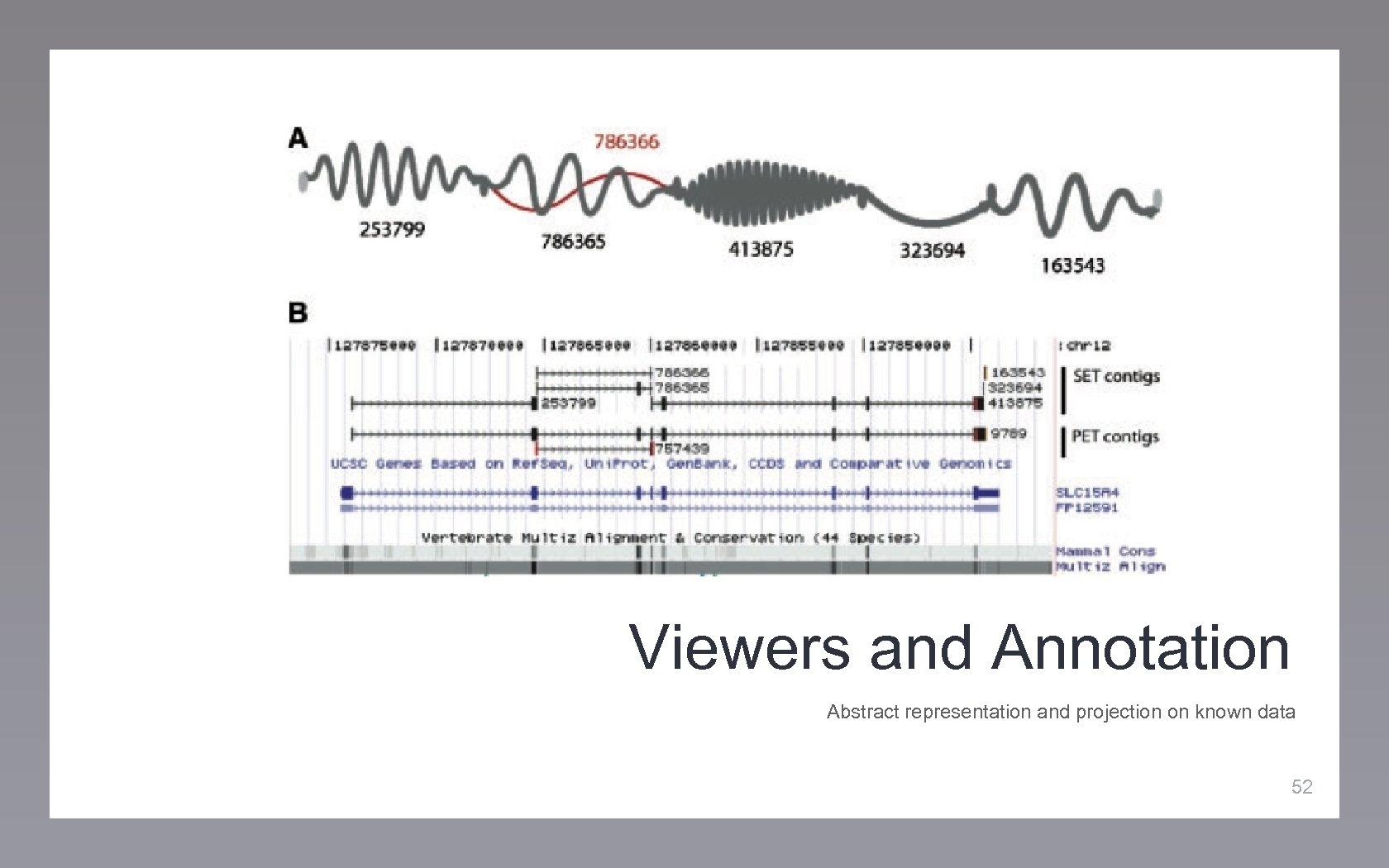
Viewers and Annotation Abstract representation and projection on known data 52
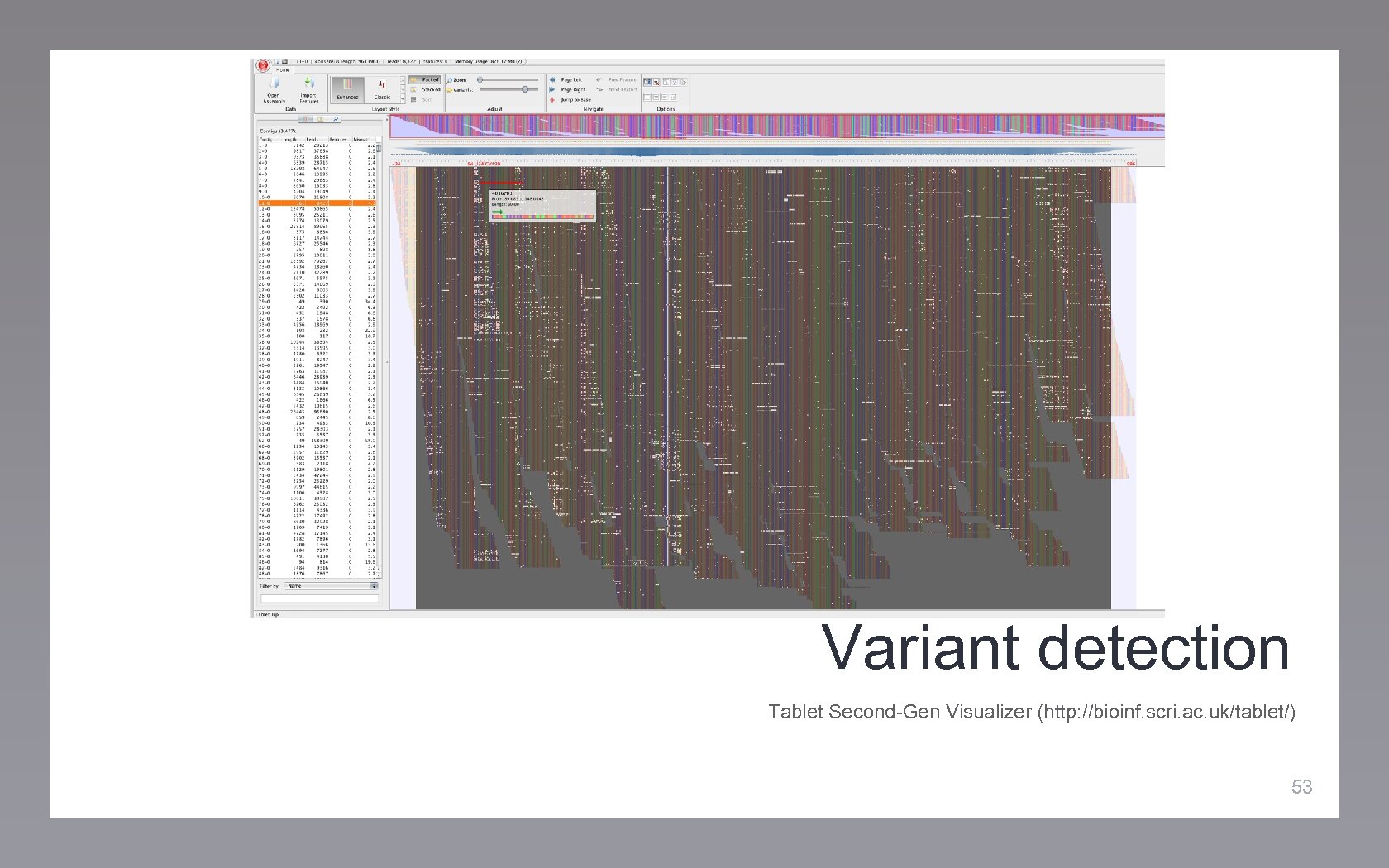
Variant detection Tablet Second-Gen Visualizer (http: //bioinf. scri. ac. uk/tablet/) 53

Variant detection Tablet Second-Gen Visualizer (http: //bioinf. scri. ac. uk/tablet/) 54
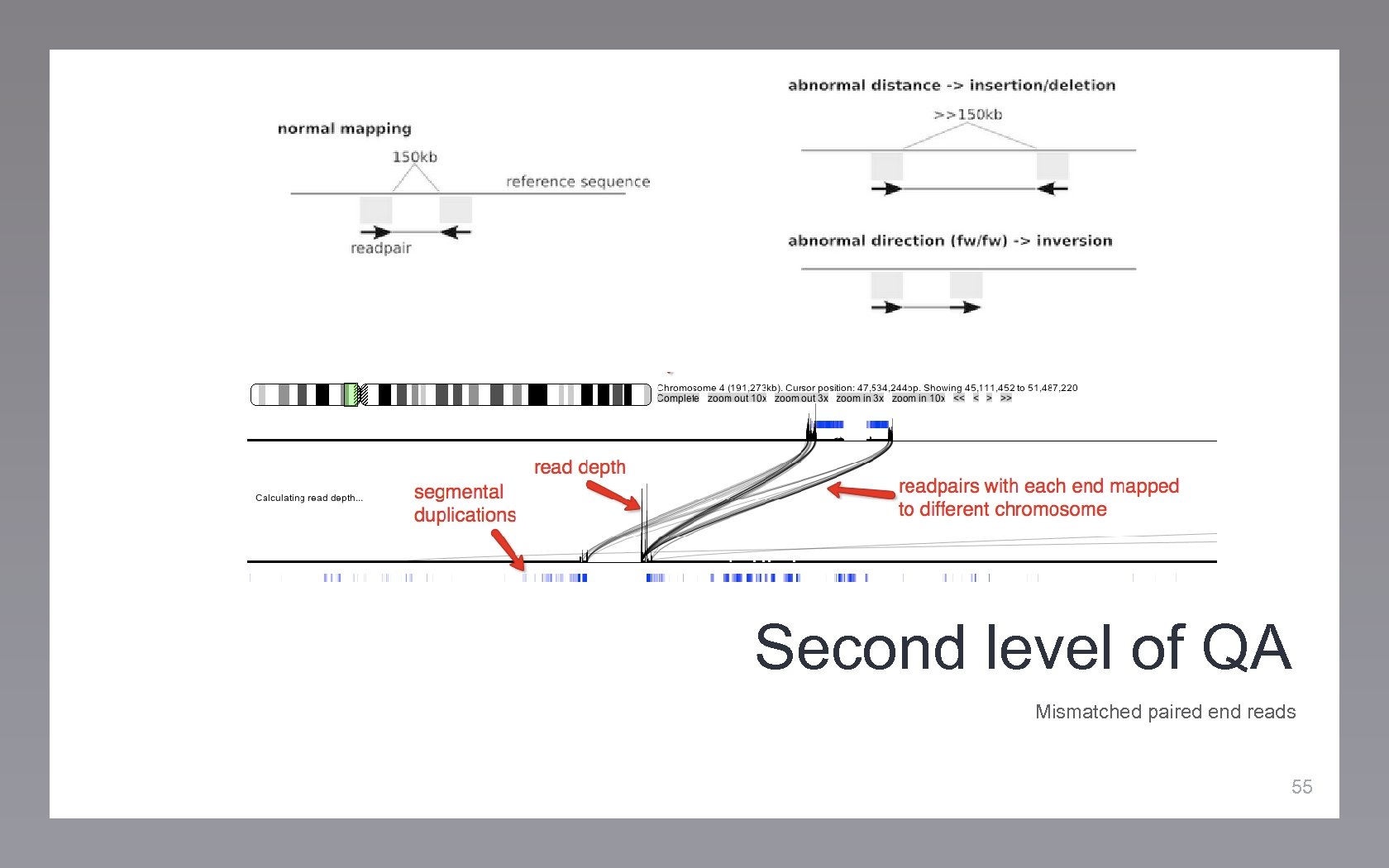
Second level of QA Mismatched paired end reads 55
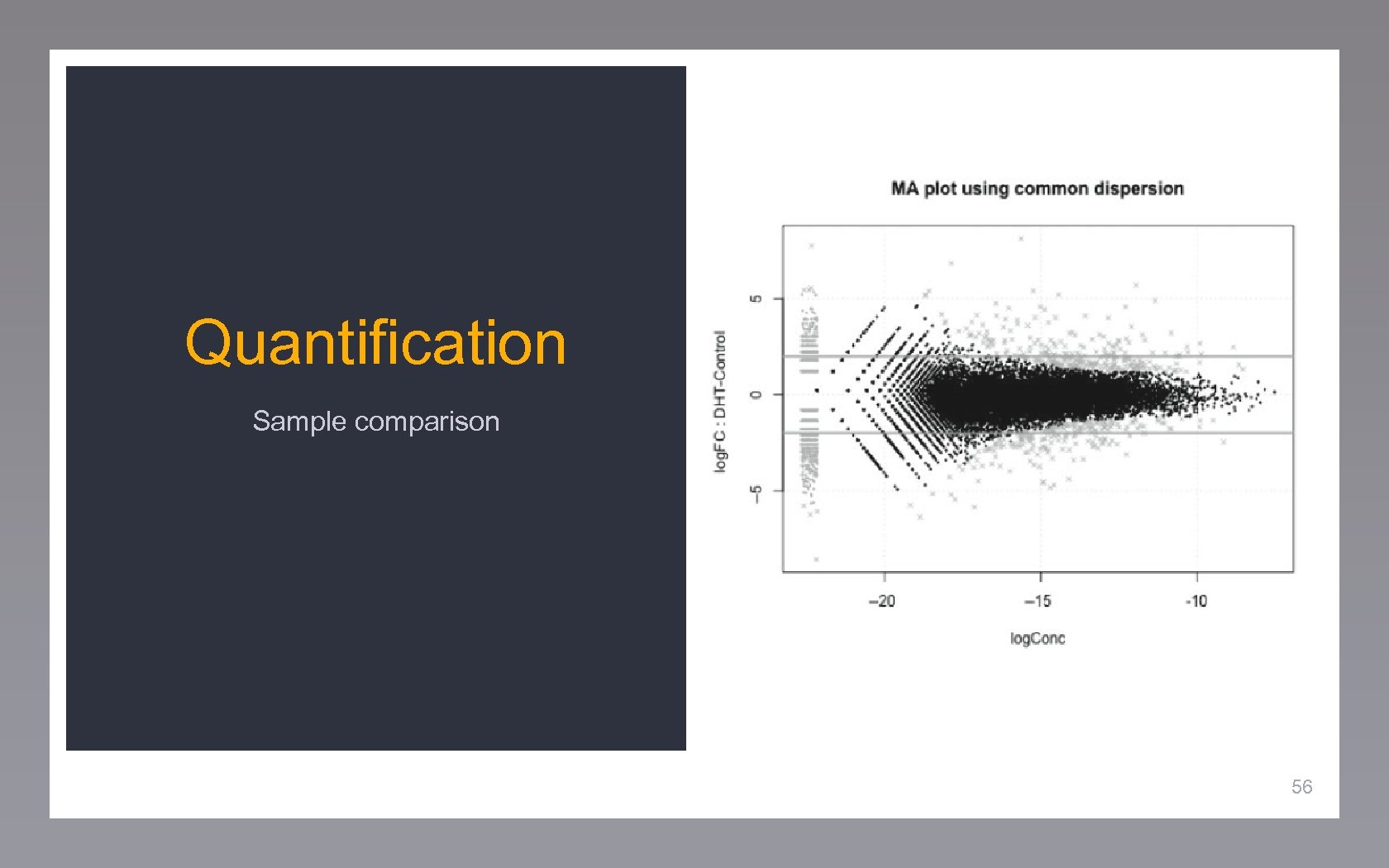
Quantification Sample comparison 56

Transcript discovery 57
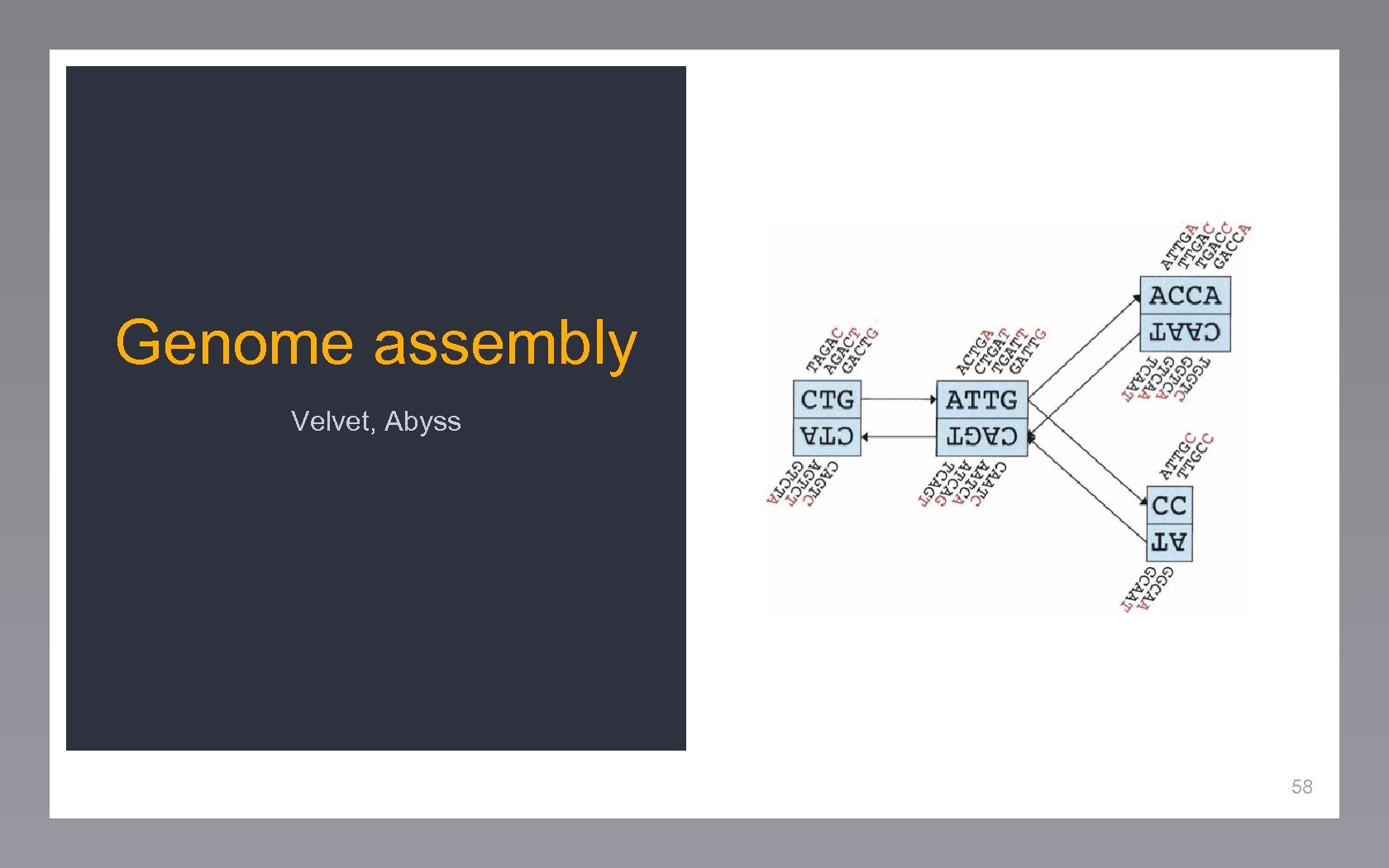
Genome assembly Velvet, Abyss 58
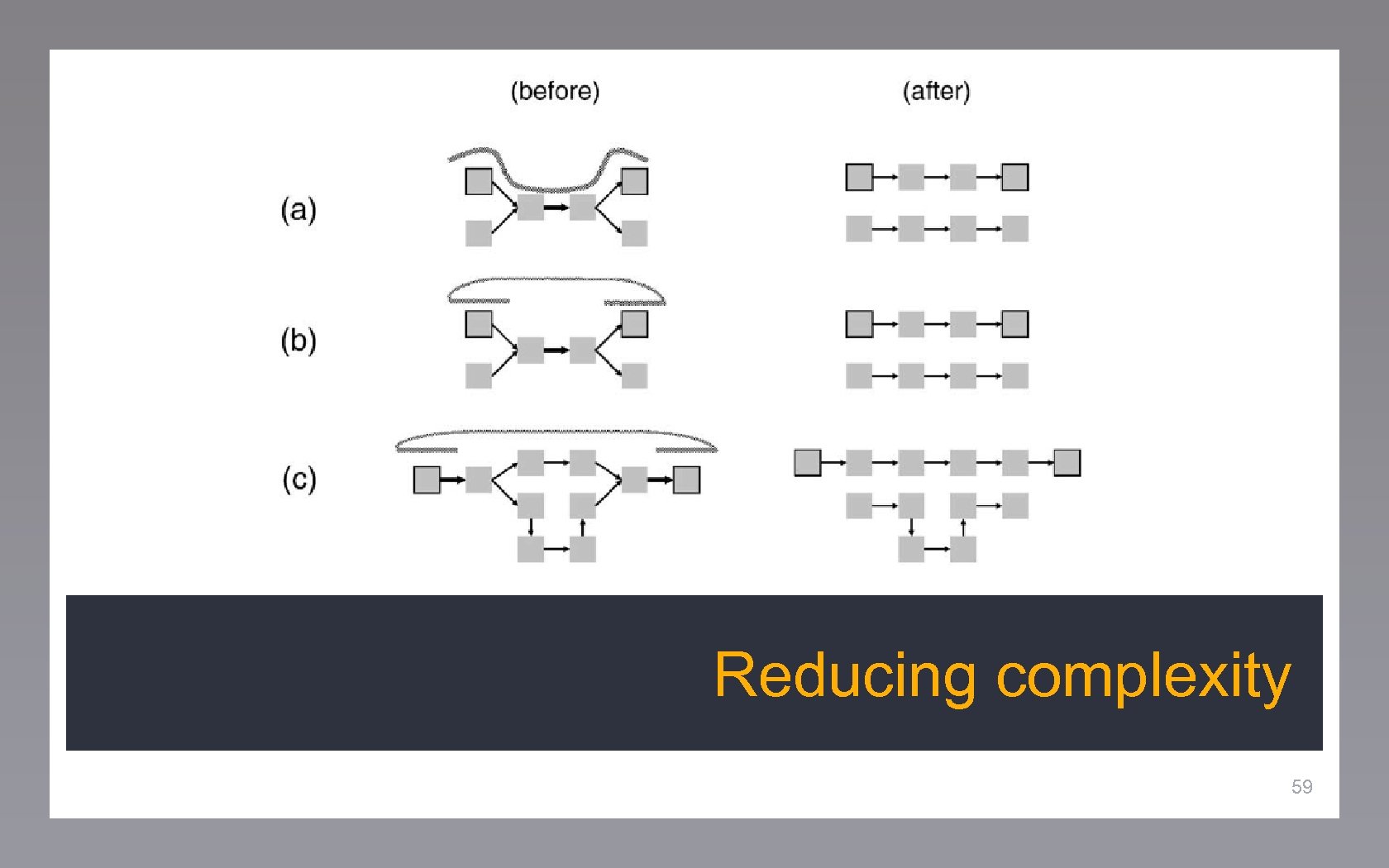
Reducing complexity 59
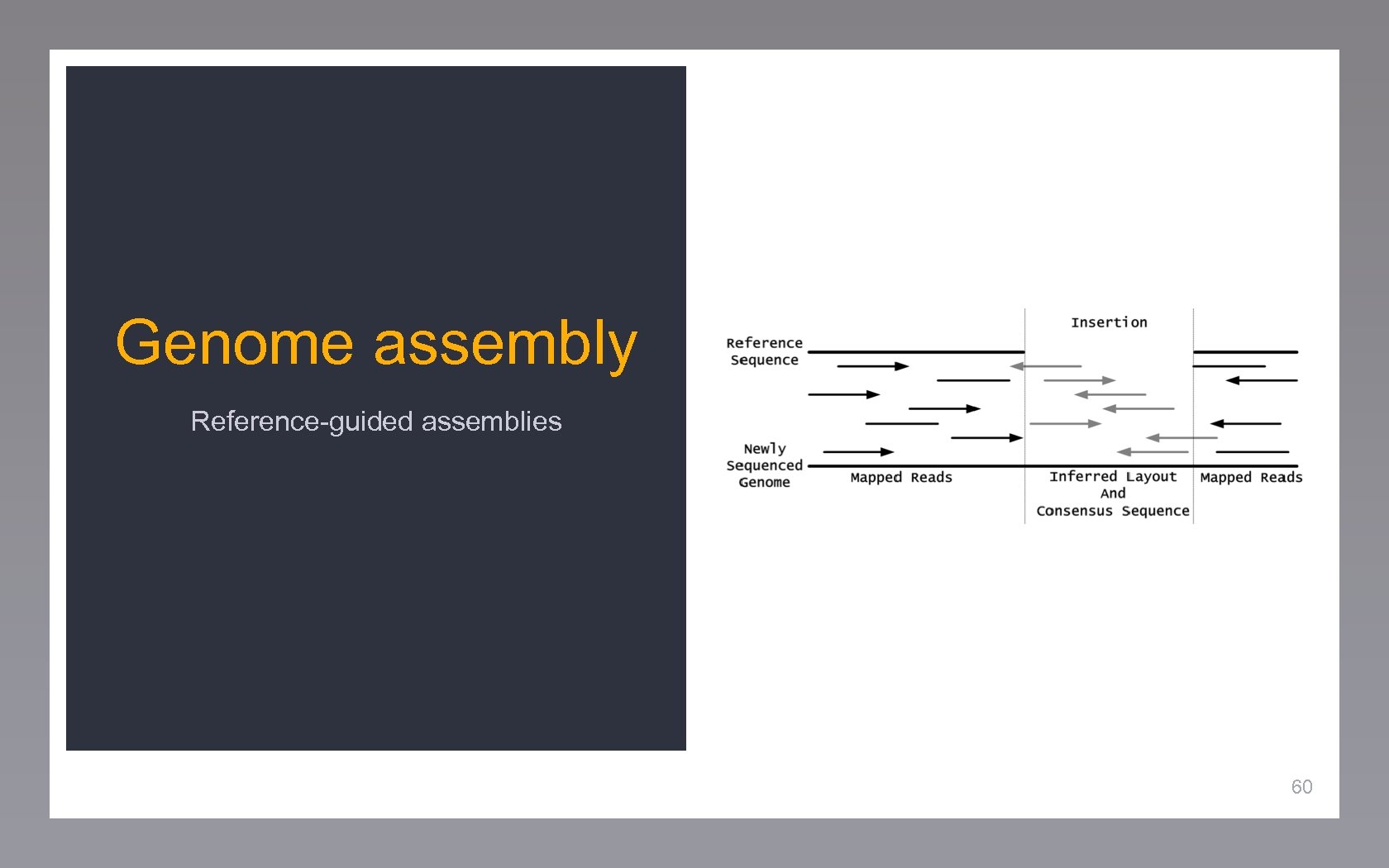
Genome assembly Reference-guided assemblies 60
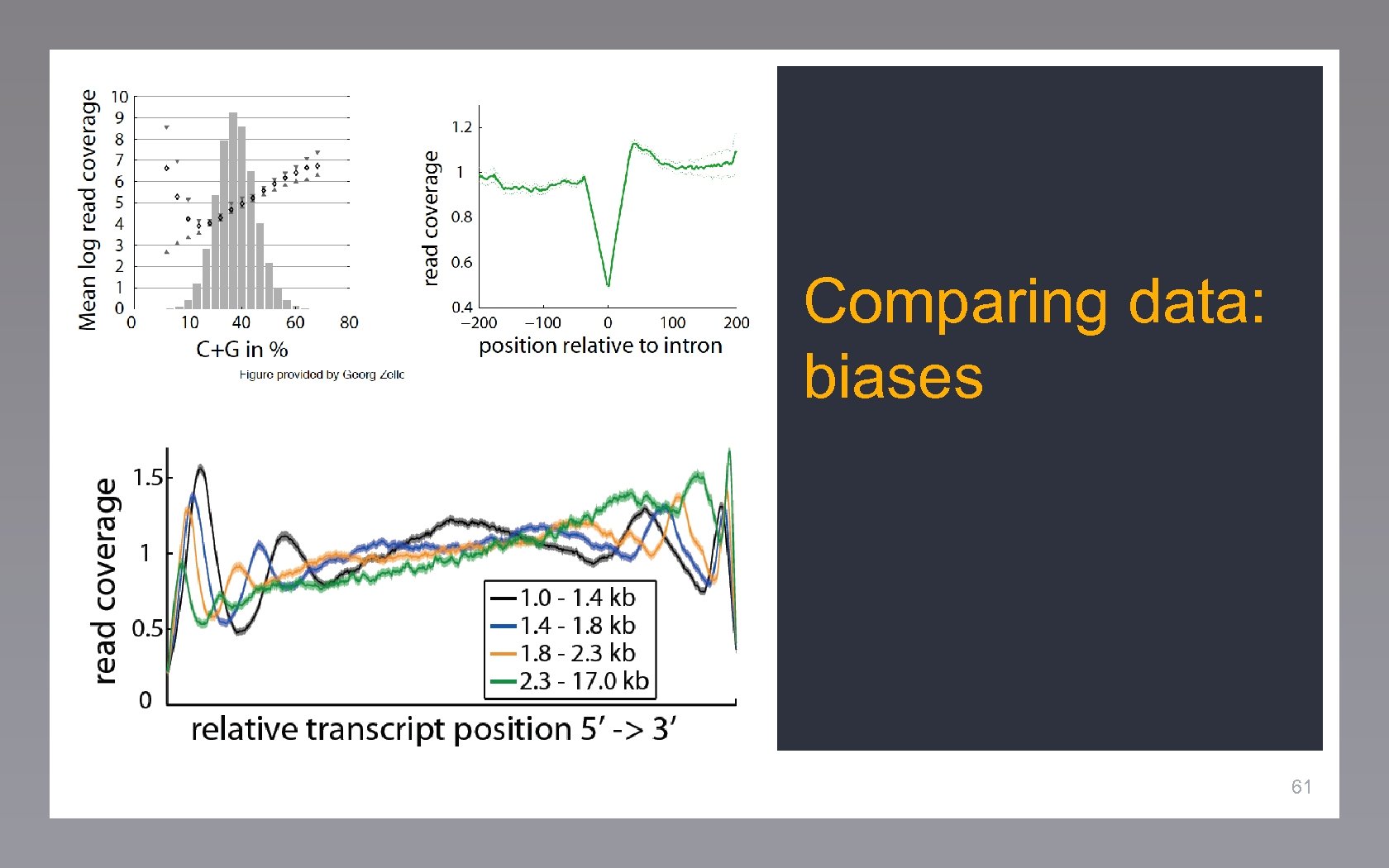
Comparing data: biases 61
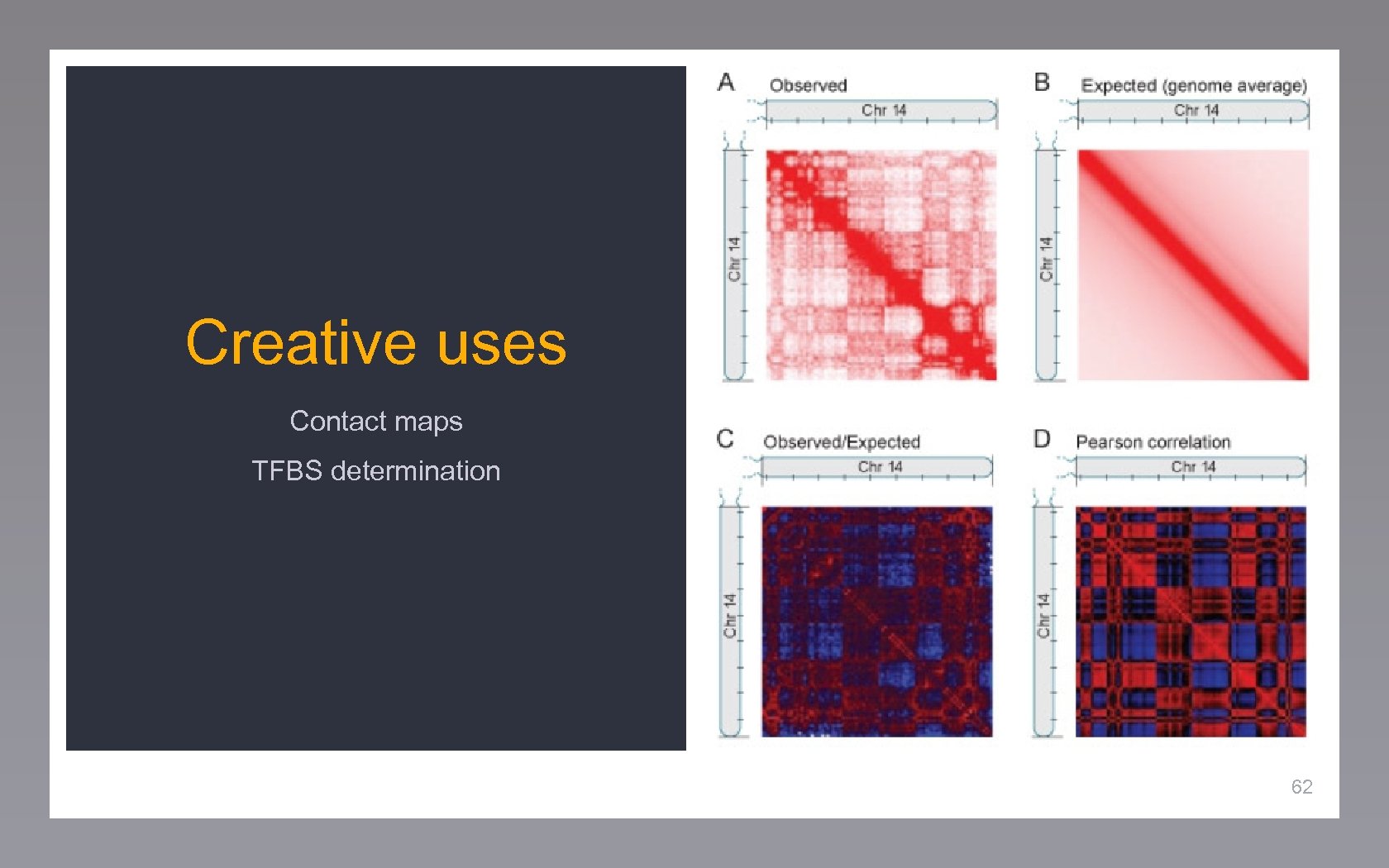
Creative uses Contact maps TFBS determination 62

Need for standards Plug and play: modular approach to tools Vital factor in application acceptance 63
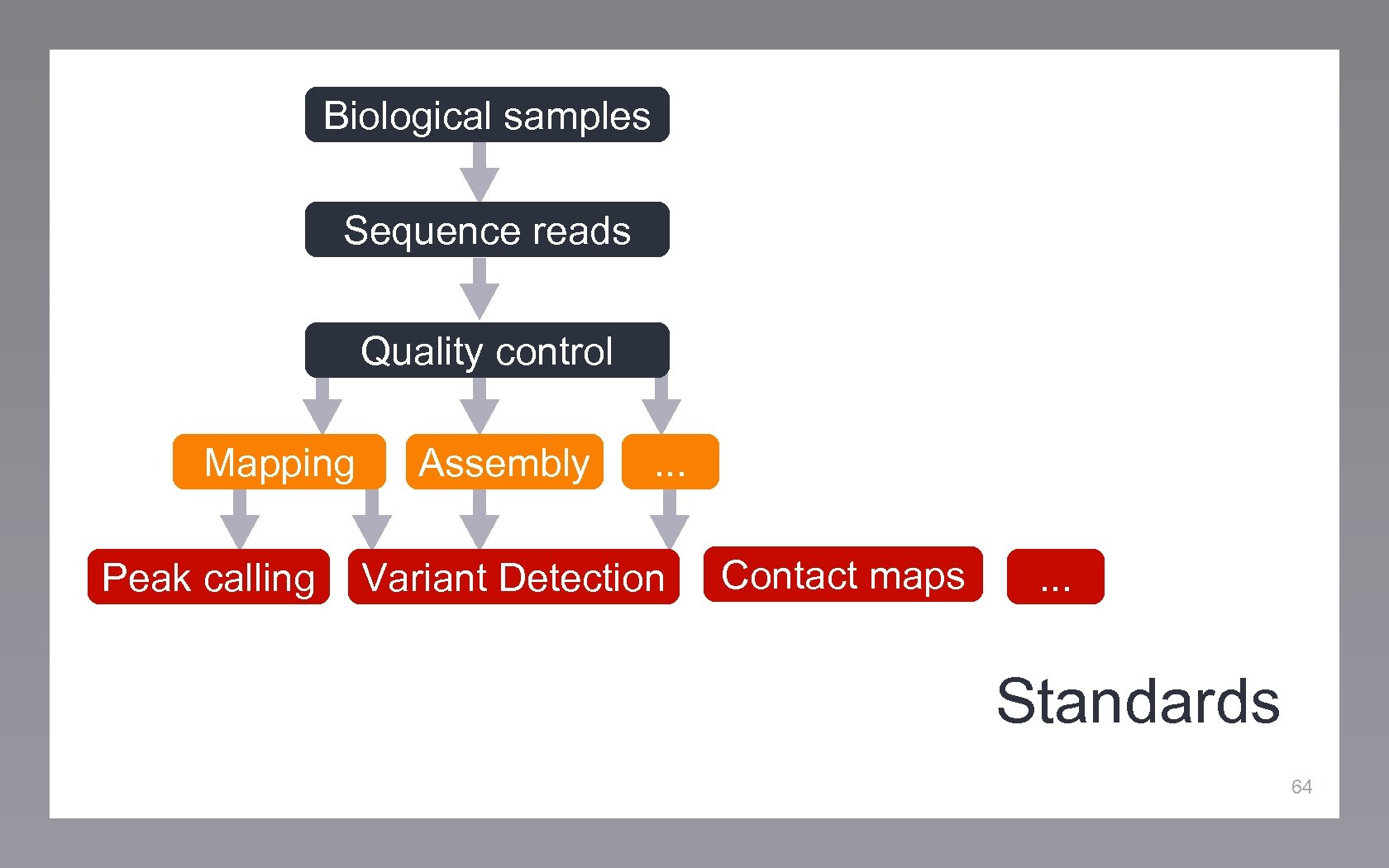
Biological samples Sequence reads Quality control Mapping Peak calling Assembly . . . Variant Detection Contact maps . . . Standards 64
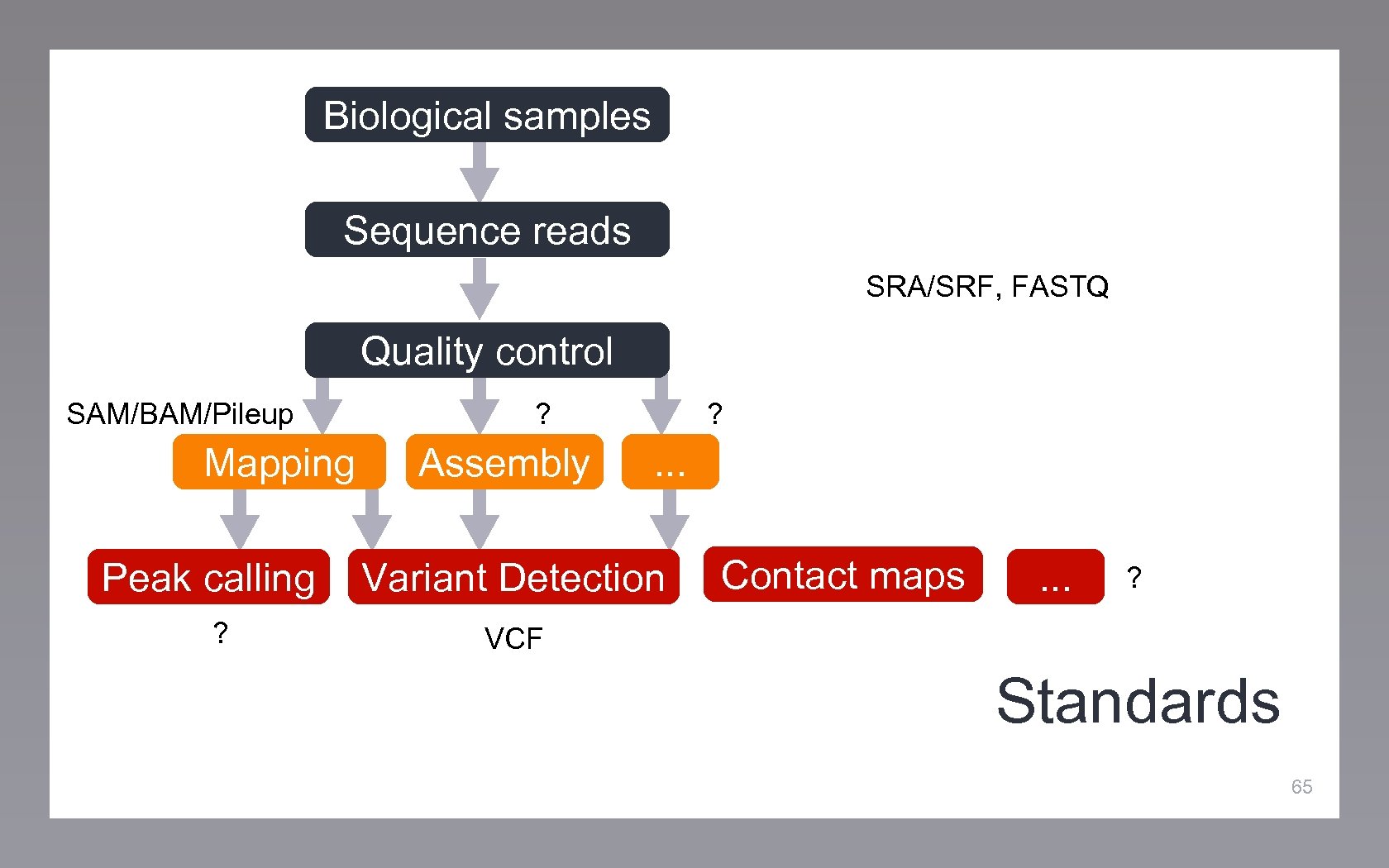
Biological samples Sequence reads SRA/SRF, FASTQ Quality control SAM/BAM/Pileup Mapping Peak calling ? ? Assembly ? . . . Variant Detection Contact maps . . . ? VCF Standards 65

FASTQ: a “standard” Sanger FASTQ, Solexa FASTQ, ABI Colour Space FASTQ, . . . Text @SRR 014849. 1 EIXKN 4201 CFU 84 length=93 GGGGGGGGCTTTTTTTGGAACCGAAAGG GTTTTGAATTTCAAACCCTTTTCGGTTTCCAACCTTCCAA AGCAATGCCAATA +SRR 014849. 1 EIXKN 4201 CFU 84 length=93 3+&$#""""""7 F@71, ’"; C? , B; ? 6 B; : EA 1 EA 5’ 9 B: ? : #9 EA 0 D@2 EA 5’: >5? : %A; A 8 A; ? 9 B; D@ /=<? 7=9<2 A 8== 66
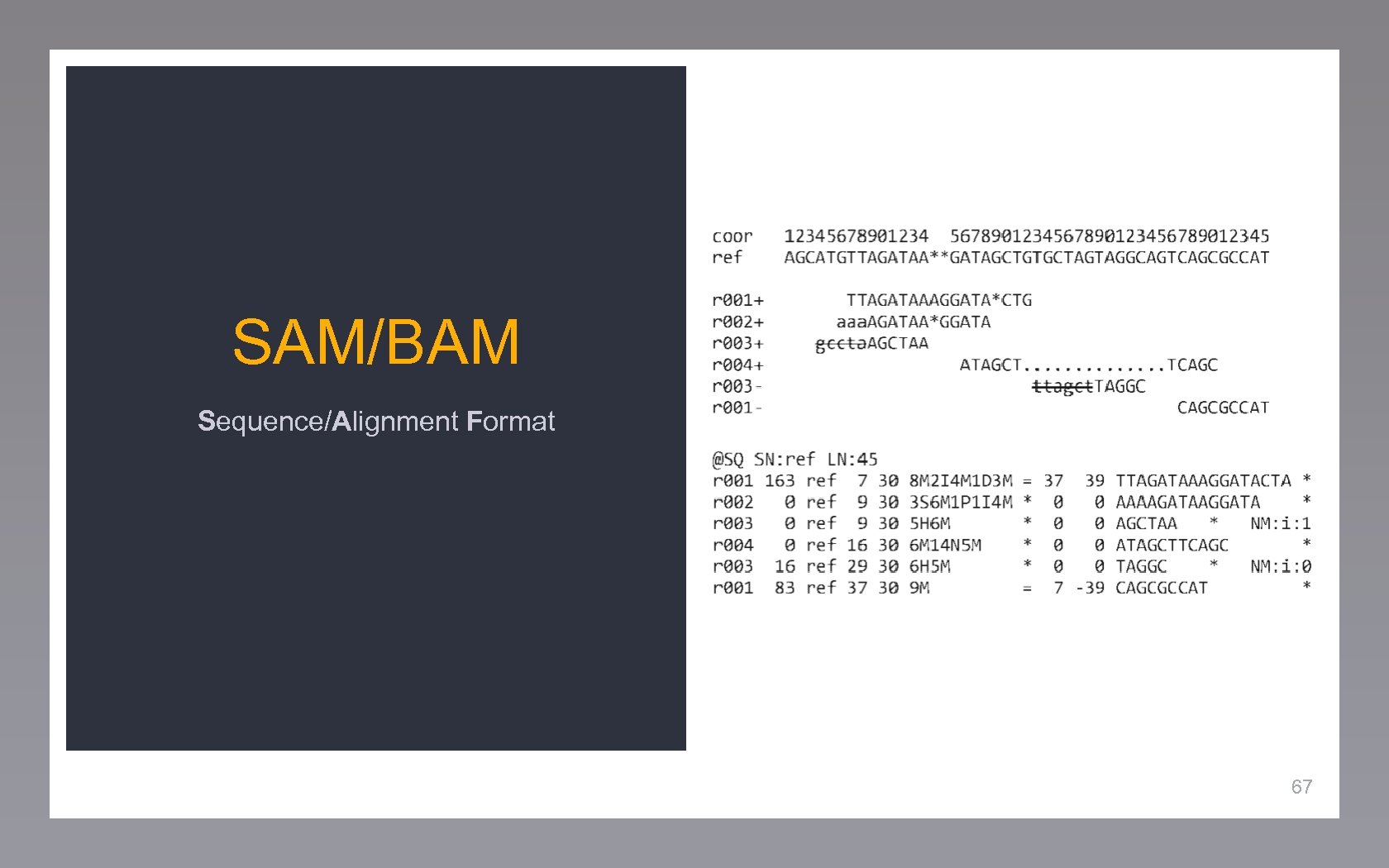
SAM/BAM Sequence/Alignment Format 67
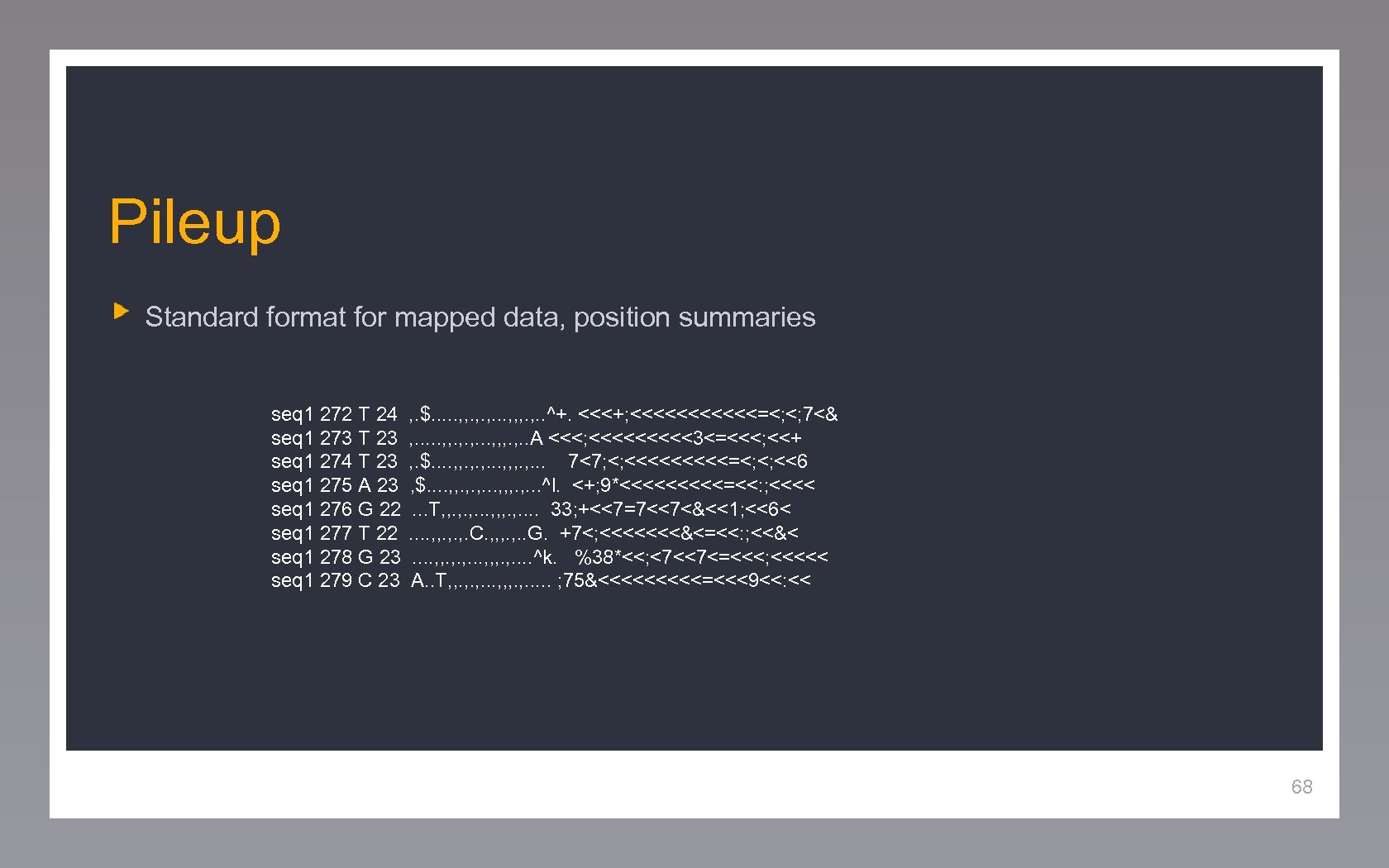
Pileup Standard format for mapped data, position summaries seq 1 272 T 24 seq 1 273 T 23 seq 1 274 T 23 seq 1 275 A 23 seq 1 276 G 22 seq 1 277 T 22 seq 1 278 G 23 seq 1 279 C 23 , . $. . . , , . , . . . , , , . . ^+. <<<+; <<<<<<=<; <; 7<& , . . . , , , . . A <<<; <<<<<3<=<<<; <<+ , . $. . , , . , . . . , , , . . . 7<7; <; <<<<<=<; <; <<6 , $. . , , . , . . . , , , . . . ^l. <+; 9*<<<<<=<<: ; <<<<. . . T, , . , . . . , , , . . . . 33; +<<7=7<<7<&<<1; <<6<. . , , . , . C. , , , . . G. +7<; <<<<<<<&<=<<: ; <<&<. . , , . , . . . , , , . . . . ^k. %38*<<; <7<<7<=<<<; <<<<< A. . T, , . , . . . , , , . . . ; 75&<<<<<=<<<9<<: << Text 68

Variant Call Format ##format=PCFv 1 ##file. Date=20090805 ##source=my. Imputation. Program. V 3. 1 ##reference=1000 Genomes. Pilot-NCBI 36 ##phasing=partial #CHROM POS ID REF ALT QUAL FILTER INFO FORMAT NA 00001 NA 00002 20 14370 rs 6054257 G A 29 0 NS=58; DP=258; AF=0. 786; DB; H 2 GT: GQ: DP: HQ 0|0: 48: 1: 51, 51 1|0: 48: 8: 51, 51 20 13330. T A 3 q 10 NS=55; DP=202; AF=0. 024 GT: GQ: DP: HQ 0|0: 49: 3: 58, 50 0|1: 3: 5: 65, 3 20 1110696 rs 6040355 A G, T 67 0 NS=55; DP=276; AF=0. 421, 0. 579; AA=T; DB GT: GQ: DP: HQ 1|2: 21: 6: 23, 27 2|1: 2: 0: 18, 2 20 10237. T. 47 0 NS=57; DP=257; AA=T GT: GQ: DP: HQ 0|0: 54: 7: 56, 60 0|0: 48: 4: 51, 51 20 123456 microsat 1 G D 4, IGA 50 0 NS=55; DP=250; AA=G GT: GQ: DP 0/1: 35: 4 0/2: 17: 2 69

Evolution of standards Helicos SRF: Helicos “implementation” Goby: tool framework (SOLID, 454, Helicos, Solexa) 70
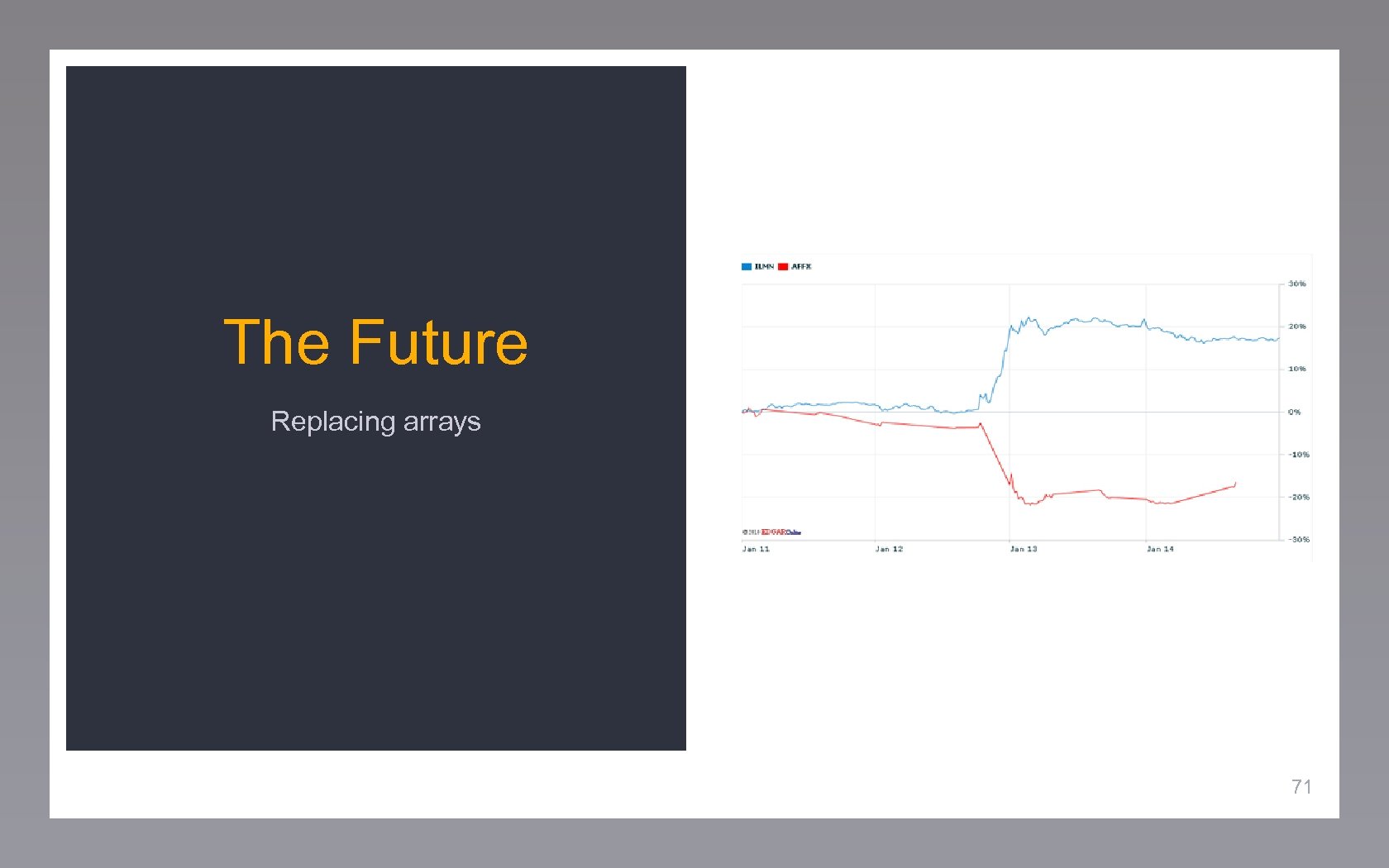
The Future Replacing arrays 71
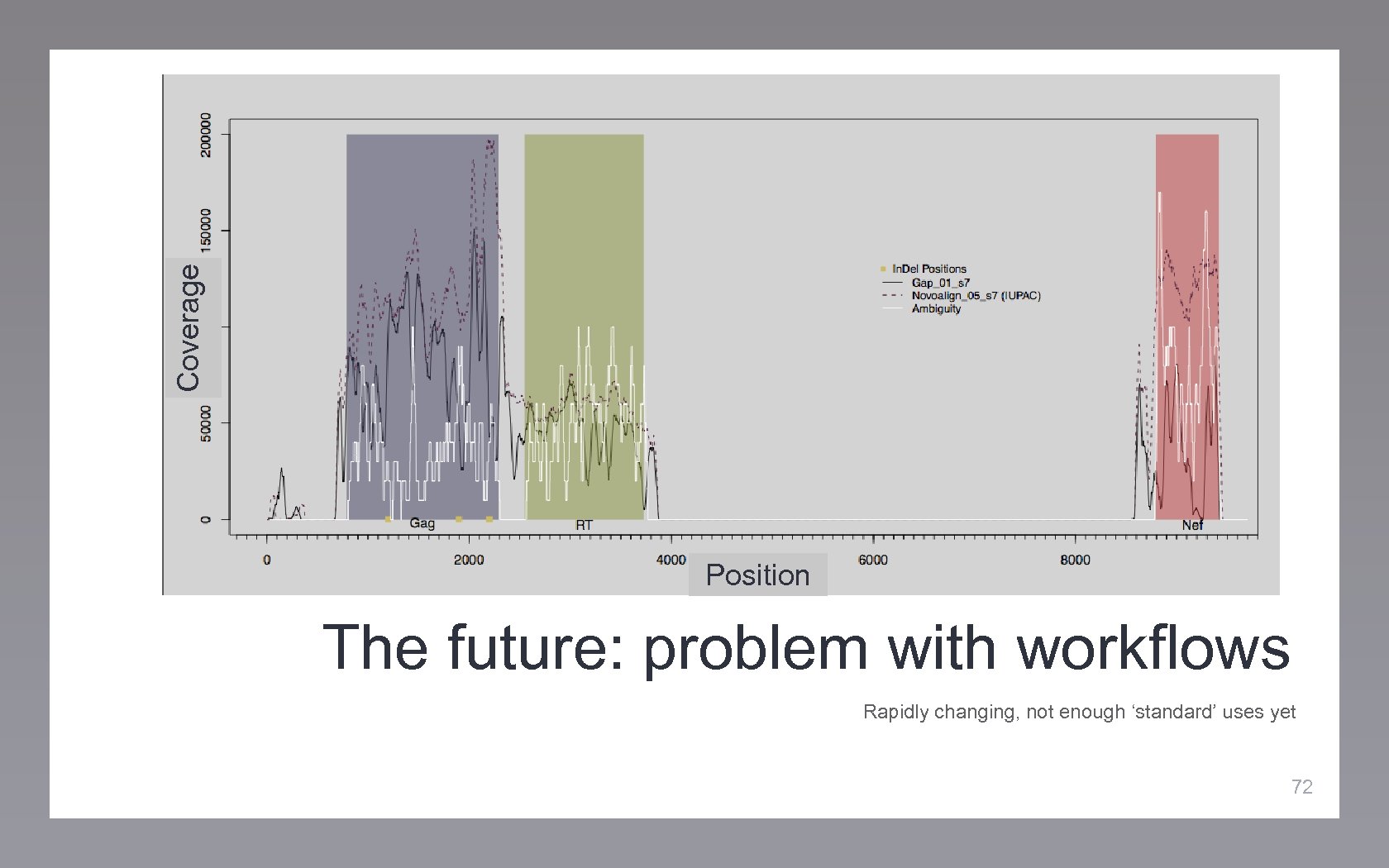
Coverage Position The future: problem with workflows Rapidly changing, not enough ‘standard’ uses yet 72
The future GWAS-by-sequencing Increased read length from 3 rd-generation technologies 73
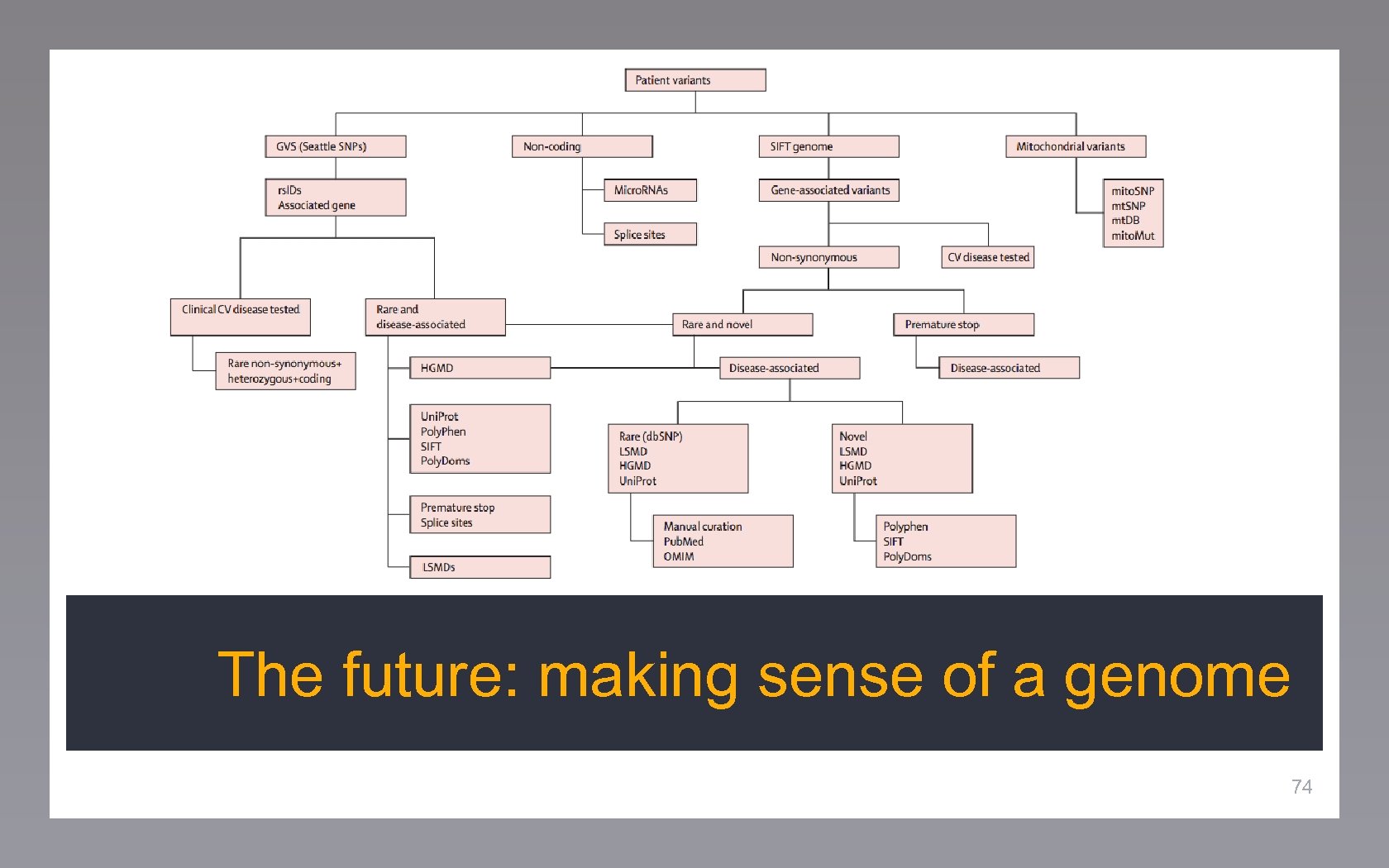
The future: making sense of a genome 74
97e5e9026168179ed334c96a9ecafb53.ppt