6b5a59b897007998ac437914341e2528.ppt
- Количество слайдов: 39
High Throughput Oligonucleotide Analysis Using the Combi. Sep Oligo PRO™ System www. combisep. com
Introduction • Massive quantities of ss. DNA and ss. RNA oligonucleotides are produced daily to support the growth of genomic-related applications • These DNA products need to be characterized to ensure proper sizing and acceptable batch-to-batch purity • Traditional slab gel-based methods for characterizing DNA products are time-consuming, labor intensive, not readily automated, and provide relatively poor resolution • While useful for assessing compound identity, Mass Spectrometry cannot quantitatively assess oligonucleotide purity due to size dependent variations in ionization efficiencies • As a result, a bottleneck exists for characterizing the purity of ss. DNA oligonucleotides
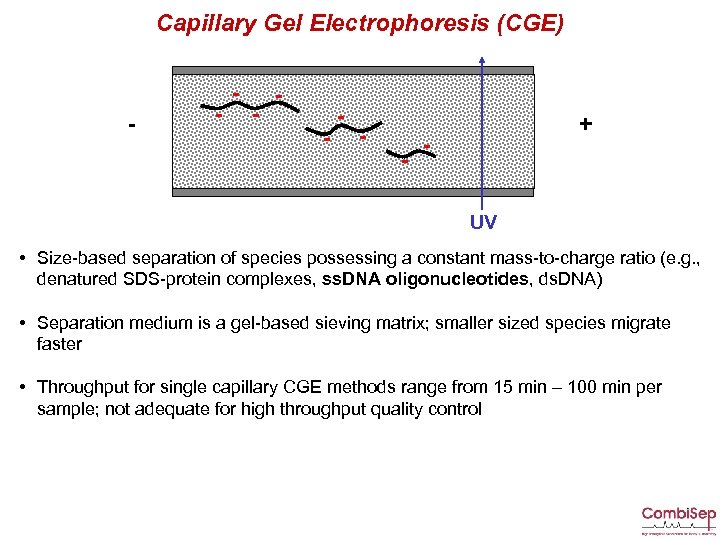
Capillary Gel Electrophoresis (CGE) - - - - + UV • Size-based separation of species possessing a constant mass-to-charge ratio (e. g. , denatured SDS-protein complexes, ss. DNA oligonucleotides, ds. DNA) • Separation medium is a gel-based sieving matrix; smaller sized species migrate faster • Throughput for single capillary CGE methods range from 15 min – 100 min per sample; not adequate for high throughput quality control
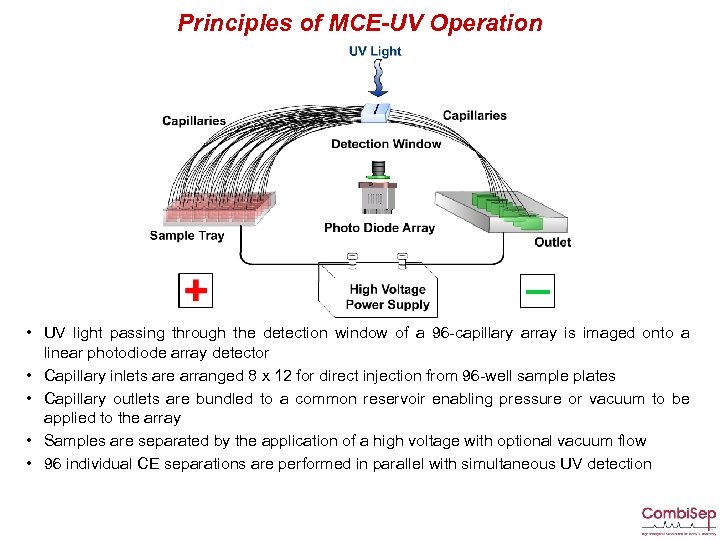
Principles of MCE-UV Operation • UV light passing through the detection window of a 96 -capillary array is imaged onto a linear photodiode array detector • Capillary inlets are arranged 8 x 12 for direct injection from 96 -well sample plates • Capillary outlets are bundled to a common reservoir enabling pressure or vacuum to be applied to the array • Samples are separated by the application of a high voltage with optional vacuum flow • 96 individual CE separations are performed in parallel with simultaneous UV detection
ss. DNA Oligonucleotide Analysis by Multiplexed CGE-UV • CGE-UV is the method of choice for assessing oligonucleotide purity, providing higher resolution and superior quantification vs. slab gels. • Label-free UV detection provides a low cost, toxic free alternative and improved quantification compared to fluorescent label approaches for ss. DNA which may possess size and sequence dependent fluorescent labeling efficiencies • CGE-UV advantages for characterizing oligonucleotide purity include low sample consumption, high resolving power, direct on-line UV detection and automated operation. • A proprietary gel sieving matrix (Oligel ) has been developed for multiplexed operation. When analyzed at low m. M concentrations, single nucleotide resolution can be obtained from 10 mer to 80 mer oligonucleotide lengths, allowing identification of low level n-1, n-2, etc. impurities. • Multiplexed CGE-UV provides a nearly 50 -fold improvement in sample throughput over single capillary CE methods with a minimal loss in separation performance. A throughput of 96 samples/h is achieved for 80 mer length oligonucleotides.
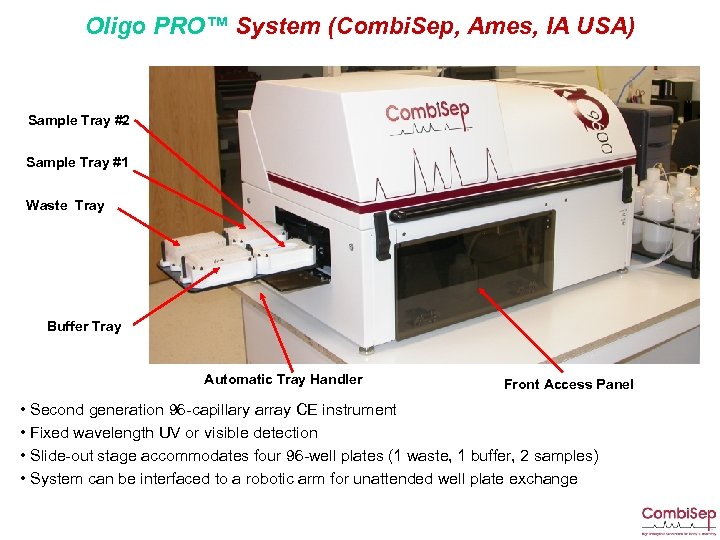
Oligo PRO™ System (Combi. Sep, Ames, IA USA) Sample Tray #2 Sample Tray #1 Waste Tray Buffer Tray Automatic Tray Handler Front Access Panel • Second generation 96 -capillary array CE instrument • Fixed wavelength UV or visible detection • Slide-out stage accommodates four 96 -well plates (1 waste, 1 buffer, 2 samples) • System can be interfaced to a robotic arm for unattended well plate exchange

Inside View of the Oligo PRO™ Instrument Capillary Array Cartridge Lamp Housing Capillary Array Detection Window HV Power Supply Optical Platform Housing Syringe Pump
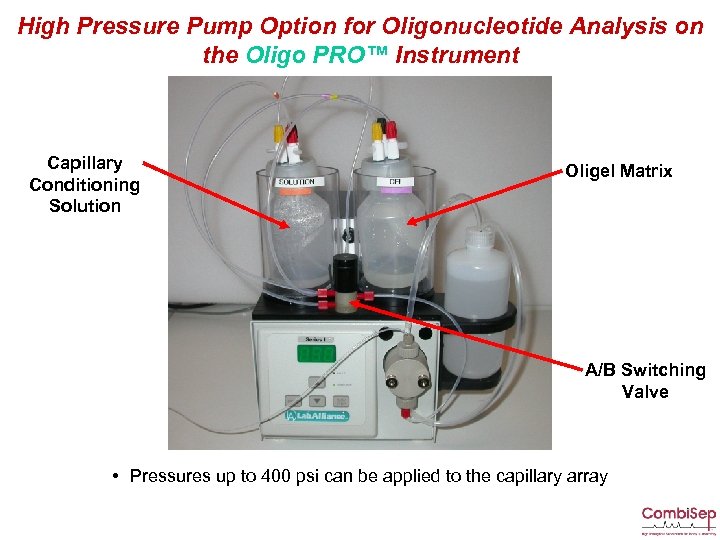
High Pressure Pump Option for Oligonucleotide Analysis on the Oligo PRO™ Instrument Capillary Conditioning Solution Oligel Matrix A/B Switching Valve • Pressures up to 400 psi can be applied to the capillary array
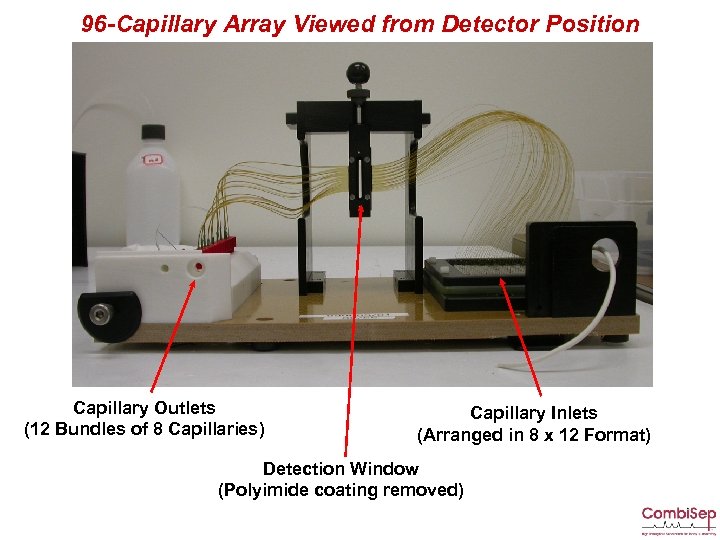
96 -Capillary Array Viewed from Detector Position Capillary Outlets (12 Bundles of 8 Capillaries) Capillary Inlets (Arranged in 8 x 12 Format) Detection Window (Polyimide coating removed)
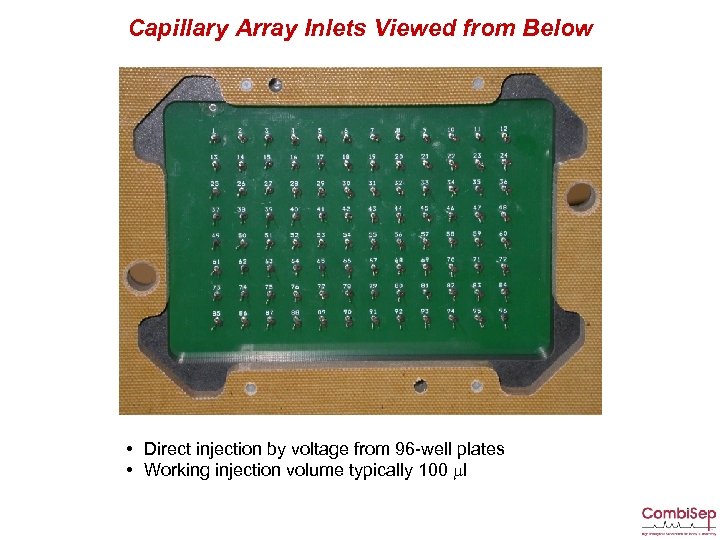
Capillary Array Inlets Viewed from Below • Direct injection by voltage from 96 -well plates • Working injection volume typically 100 ml
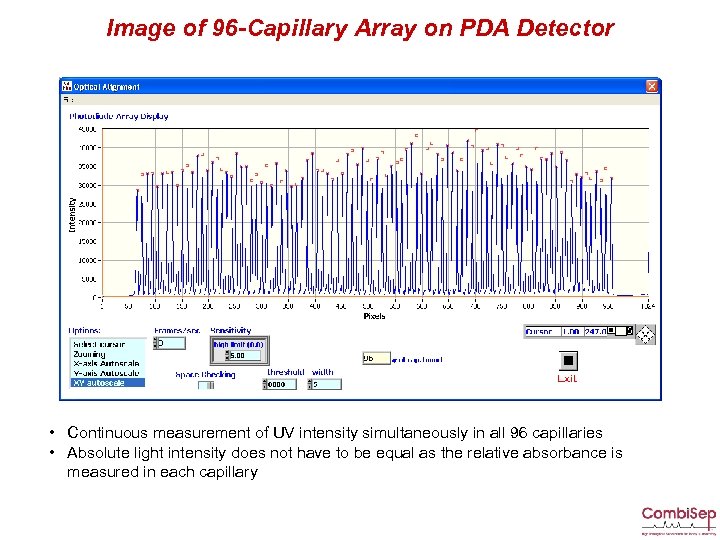
Image of 96 -Capillary Array on PDA Detector • Continuous measurement of UV intensity simultaneously in all 96 capillaries • Absolute light intensity does not have to be equal as the relative absorbance is measured in each capillary
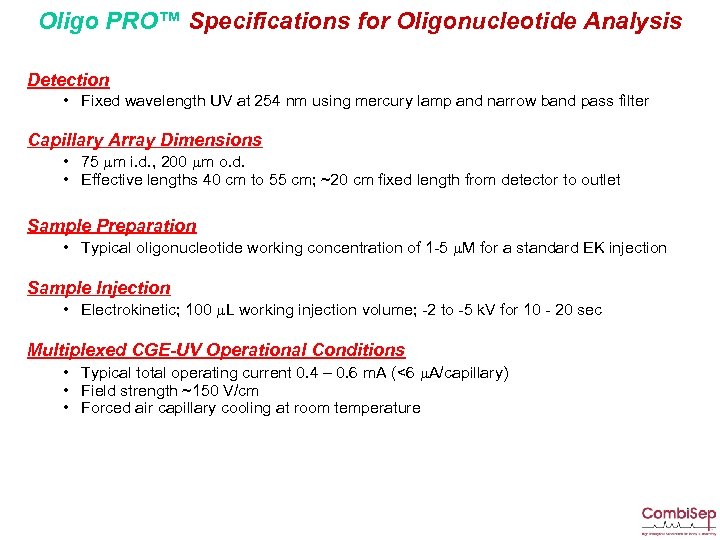
Oligo PRO™ Specifications for Oligonucleotide Analysis Detection • Fixed wavelength UV at 254 nm using mercury lamp and narrow band pass filter Capillary Array Dimensions • 75 mm i. d. , 200 mm o. d. • Effective lengths 40 cm to 55 cm; ~20 cm fixed length from detector to outlet Sample Preparation • Typical oligonucleotide working concentration of 1 -5 m. M for a standard EK injection Sample Injection • Electrokinetic; 100 m. L working injection volume; -2 to -5 k. V for 10 - 20 sec Multiplexed CGE-UV Operational Conditions • Typical total operating current 0. 4 – 0. 6 m. A (<6 m. A/capillary) • Field strength ~150 V/cm • Forced air capillary cooling at room temperature
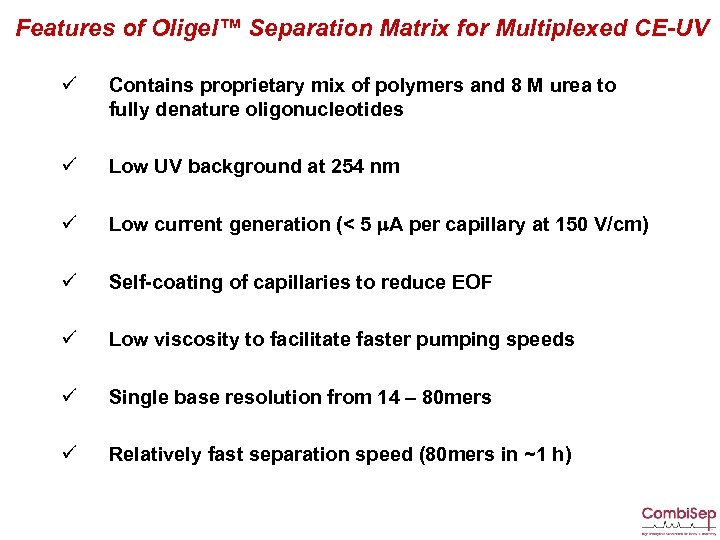
Features of Oligel™ Separation Matrix for Multiplexed CE-UV ü Contains proprietary mix of polymers and 8 M urea to fully denature oligonucleotides ü Low UV background at 254 nm ü Low current generation (< 5 m. A per capillary at 150 V/cm) ü Self-coating of capillaries to reduce EOF ü Low viscosity to facilitate faster pumping speeds ü Single base resolution from 14 – 80 mers ü Relatively fast separation speed (80 mers in ~1 h)
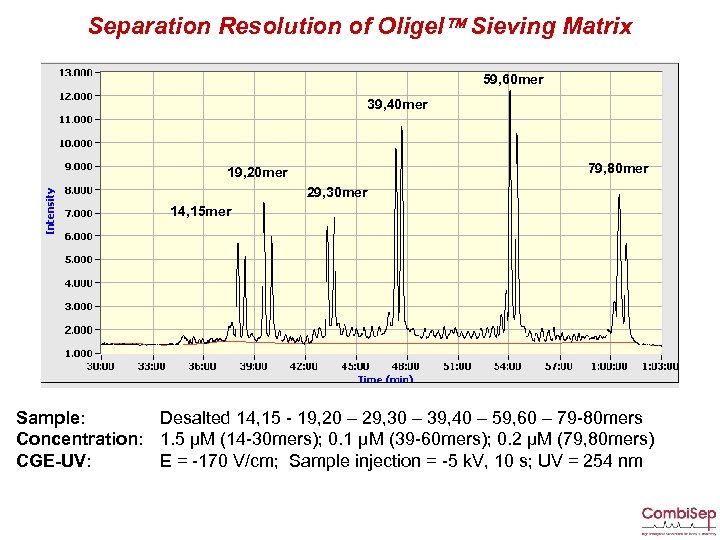
Separation Resolution of Oligel Sieving Matrix 59, 60 mer 39, 40 mer 79, 80 mer 19, 20 mer 29, 30 mer 14, 15 mer Sample: Desalted 14, 15 - 19, 20 – 29, 30 – 39, 40 – 59, 60 – 79 -80 mers Concentration: 1. 5 µM (14 -30 mers); 0. 1 µM (39 -60 mers); 0. 2 µM (79, 80 mers) CGE-UV: E = -170 V/cm; Sample injection = -5 k. V, 10 s; UV = 254 nm
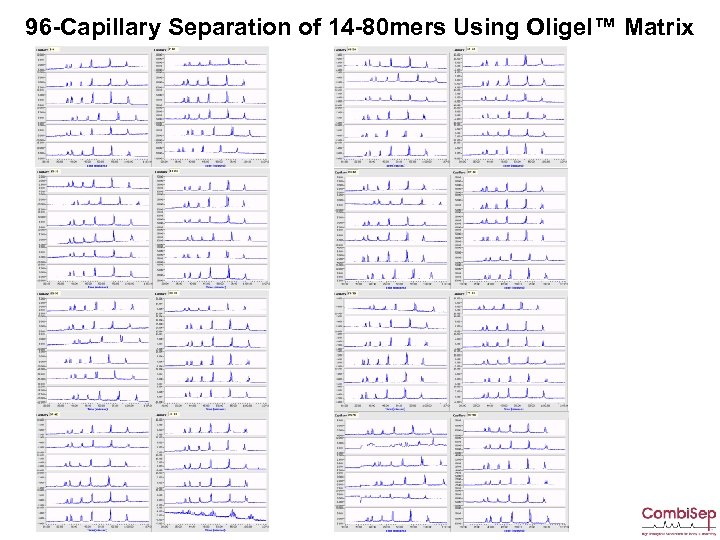
96 -Capillary Separation of 14 -80 mers Using Oligel™ Matrix
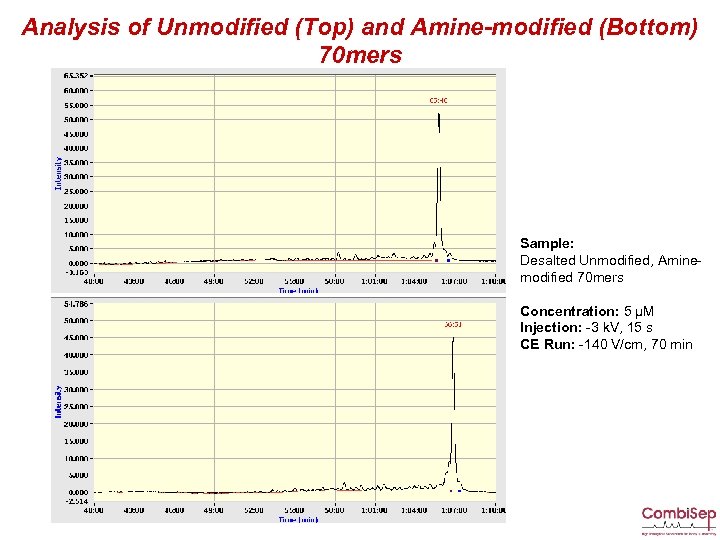
Analysis of Unmodified (Top) and Amine-modified (Bottom) 70 mers Sample: Desalted Unmodified, Aminemodified 70 mers Concentration: 5 µM Injection: -3 k. V, 15 s CE Run: -140 V/cm, 70 min
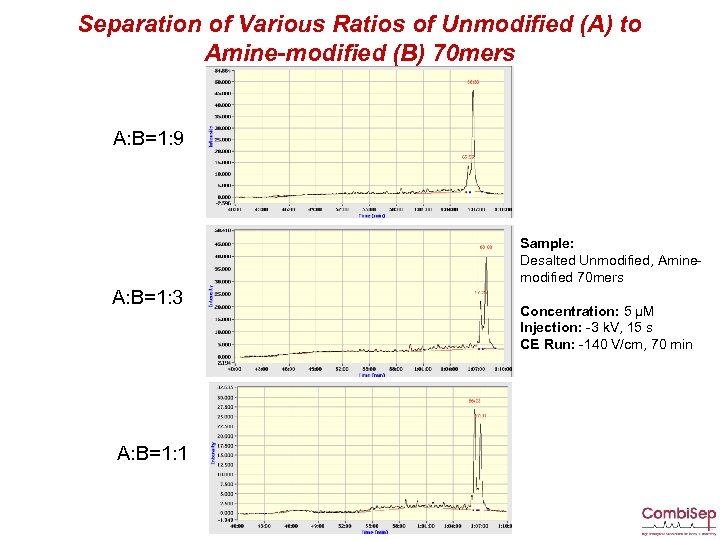
Separation of Various Ratios of Unmodified (A) to Amine-modified (B) 70 mers A: B=1: 9 Sample: Desalted Unmodified, Aminemodified 70 mers A: B=1: 3 A: B=1: 1 Concentration: 5 µM Injection: -3 k. V, 15 s CE Run: -140 V/cm, 70 min
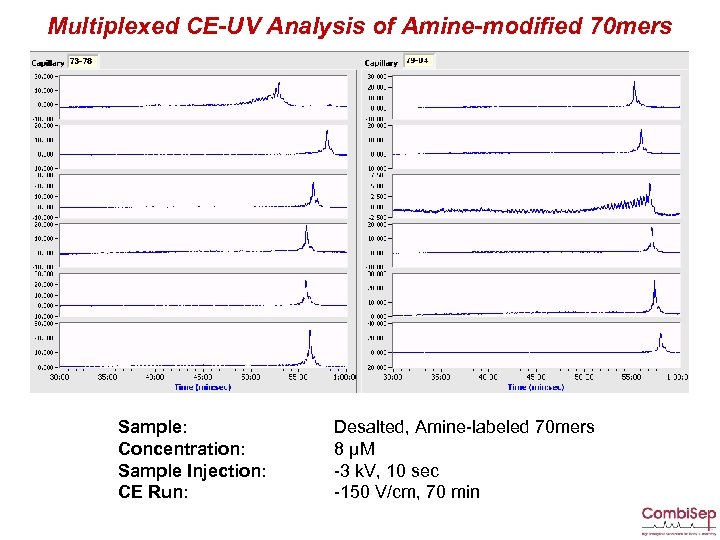
Multiplexed CE-UV Analysis of Amine-modified 70 mers Sample: Concentration: Sample Injection: CE Run: Desalted, Amine-labeled 70 mers 8 µM -3 k. V, 10 sec -150 V/cm, 70 min

Characteristics of Gel Matrices Used in CGE • Chemical Gels – Cross-linked, chemically linked to the capillary wall: not replaceable – Well-defined pore structure but pore size cannot be varied after polymerization – Heat sensitive – Particulates can damage gel matrix • Physical Gels – Not cross-linked, not attached to the capillary wall: replaceable – Entangled polymer networks of linear or branched hydrophilic polymers – Dynamic pore structure – Heat insensitive
Permanent coating vs. Dynamic coating Why: for non-cross-linked gel, EOF reduces separation efficiency How to reduce the EOF? Permanent coating: – Chemical bonding to the capillary wall – Non-replaceable – Limited life time Dynamic coating: – Physical bonding (hydrophobicity and charge) – Replaceable
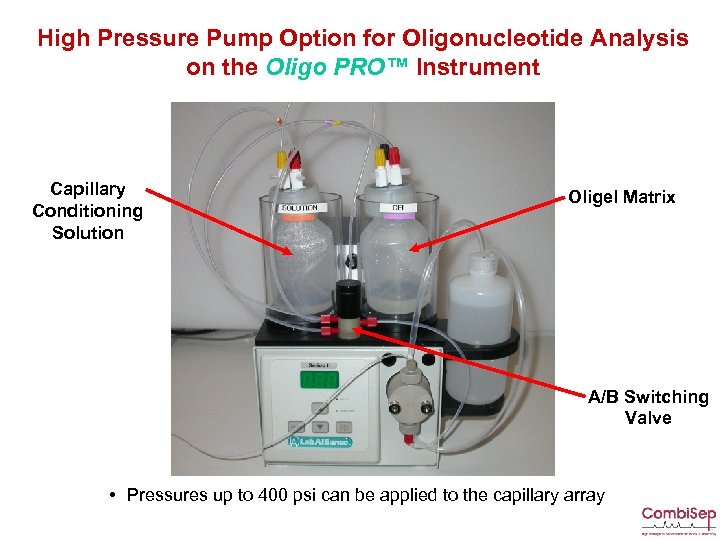
High Pressure Pump Option for Oligonucleotide Analysis on the Oligo PRO™ Instrument Capillary Conditioning Solution Oligel Matrix A/B Switching Valve • Pressures up to 400 psi can be applied to the capillary array
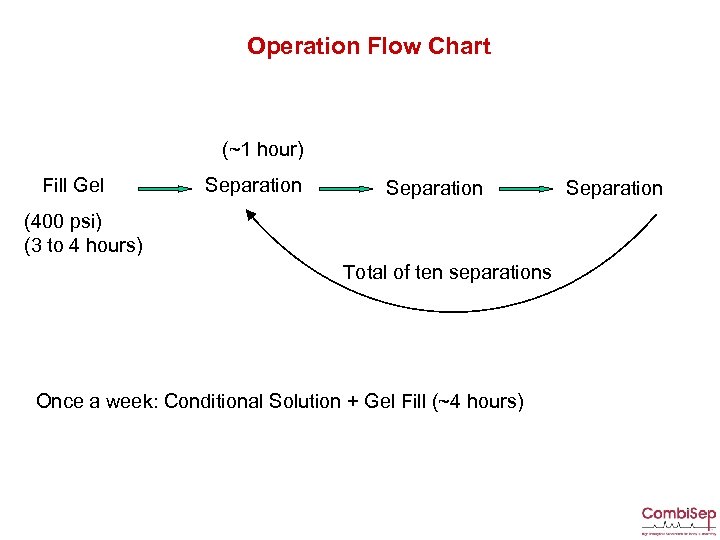
Operation Flow Chart (~1 hour) Fill Gel Separation (400 psi) (3 to 4 hours) Total of ten separations Once a week: Conditional Solution + Gel Fill (~4 hours) Separation
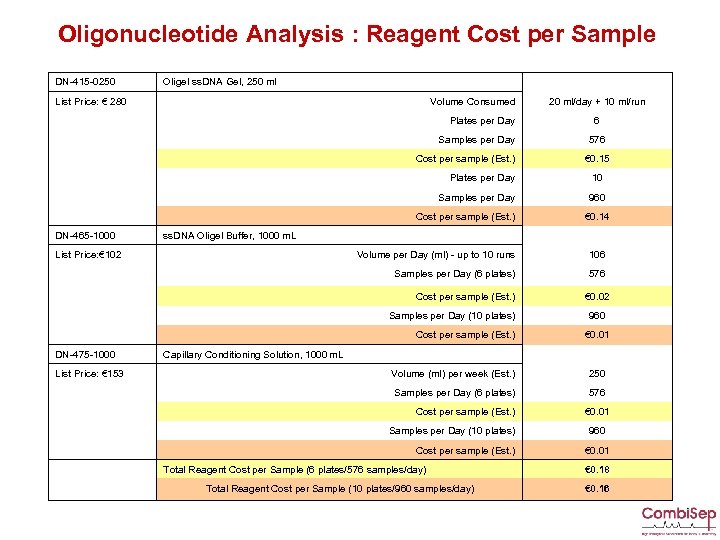
Oligonucleotide Analysis : Reagent Cost per Sample DN-415 -0250 Oligel ss. DNA Gel, 250 ml List Price: € 280 Volume Consumed 20 ml/day + 10 ml/run Plates per Day Samples per Day Cost per sample (Est. ) Plates per Day 10 Samples per Day 960 Cost per sample (Est. ) DN-465 -1000 ss. DNA Oligel Buffer, 1000 m. L List Price: € 102 6 576 € 0. 15 € 0. 14 Volume per Day (ml) - up to 10 runs 106 Samples per Day (6 plates) 576 Cost per sample (Est. ) Samples per Day (10 plates) Cost per sample (Est. ) DN-475 -1000 Capillary Conditioning Solution, 1000 m. L € 0. 02 960 € 0. 01 List Price: € 153 Volume (ml) per week (Est. ) 250 Samples per Day (6 plates) 576 Cost per sample (Est. ) Samples per Day (10 plates) Cost per sample (Est. ) Total Reagent Cost per Sample (6 plates/576 samples/day) Total Reagent Cost per Sample (10 plates/960 samples/day) € 0. 01 960 € 0. 01 € 0. 18 € 0. 16
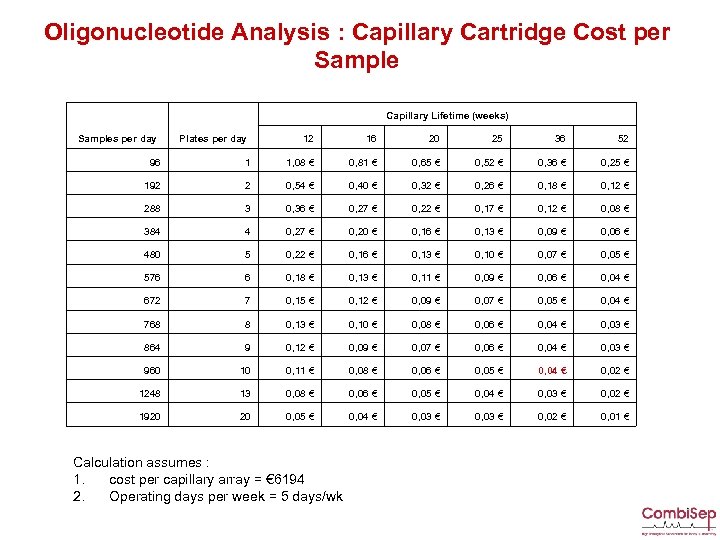
Oligonucleotide Analysis : Capillary Cartridge Cost per Sample Capillary Lifetime (weeks) Samples per day Plates per day 12 16 20 25 36 52 96 1 1, 08 € 0, 81 € 0, 65 € 0, 52 € 0, 36 € 0, 25 € 192 2 0, 54 € 0, 40 € 0, 32 € 0, 26 € 0, 18 € 0, 12 € 288 3 0, 36 € 0, 27 € 0, 22 € 0, 17 € 0, 12 € 0, 08 € 384 4 0, 27 € 0, 20 € 0, 16 € 0, 13 € 0, 09 € 0, 06 € 480 5 0, 22 € 0, 16 € 0, 13 € 0, 10 € 0, 07 € 0, 05 € 576 6 0, 18 € 0, 13 € 0, 11 € 0, 09 € 0, 06 € 0, 04 € 672 7 0, 15 € 0, 12 € 0, 09 € 0, 07 € 0, 05 € 0, 04 € 768 8 0, 13 € 0, 10 € 0, 08 € 0, 06 € 0, 04 € 0, 03 € 864 9 0, 12 € 0, 09 € 0, 07 € 0, 06 € 0, 04 € 0, 03 € 960 10 0, 11 € 0, 08 € 0, 06 € 0, 05 € 0, 04 € 0, 02 € 1248 13 0, 08 € 0, 06 € 0, 05 € 0, 04 € 0, 03 € 0, 02 € 1920 20 0, 05 € 0, 04 € 0, 03 € 0, 02 € 0, 01 € Calculation assumes : 1. cost per capillary array = € 6194 2. Operating days per week = 5 days/wk

Other Technologies • • Slab Gel Single column capillary electrophoresis Mass Spectrometers HPLC
Slab Gel • • Multiple samples analysis Good resolution Very labor intensive Difficult to document the image Semi-quantitation Long separation time Acrylamide

Single Column Capillary Electrophoresis • • Similar resolution as in Oligo PRO™ Better reproducibility Slow (~96 times slower) Sample cost: higher due to the use of coated capillary (limited life time)
Mass Spectrometers – MALDI (matrix assisted laser desorption ionization) • Size determination up to 60’mers • Difficult for quantification – Electrospray ionization • Size determination up to 120’mers • Relatively slow: a couple minutes per sample • Difficult for quantification – HPLC-MS • Size determination & quantification • Slow: half hour to an hour per sample
HPLC (Ion Exchanged Chromatography) • • • Quantitation measurement Separation up to 40’mers Slow: half hour per sample Separation not completely based on size Better reproducibility
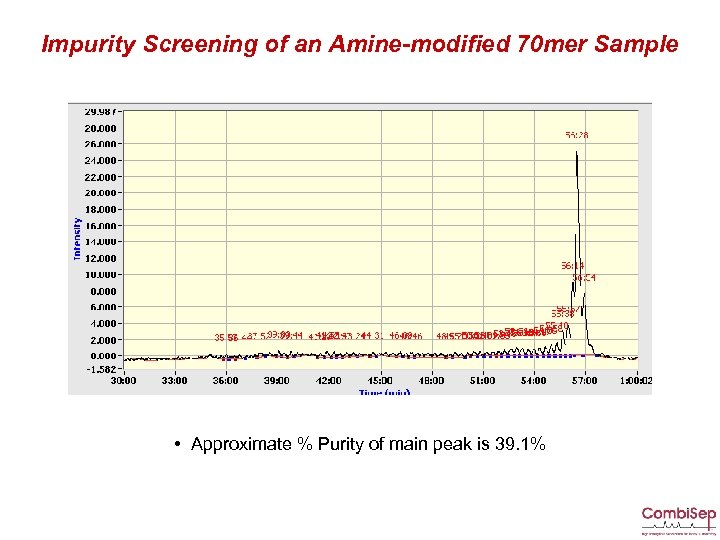
Impurity Screening of an Amine-modified 70 mer Sample • Approximate % Purity of main peak is 39. 1%

Co-Winner Best Poster at Tides 2005 Combi. Sep and IDT Poster Co-Winners of Best Poster at Tides 2005 Conference The poster titled “Comparisons between Multiplexed, Absorbance-Based Capillary Electrophoresis, and Ion Exchange Chromatography for Analysis of n-1 Oligonucleotide Impurities” by Wei, Ho-ming Pang, Dennis Tallman, and Jeremy Kenseth of Combi. Sep, Inc and Lisa Bogh of Integrated DNA Technology was recently selected as the co -winner of the best poster at Tides 2005. The Tides Conference is an industry event for manufacturing and development of oligonucleotide and peptide products. The meeting was held May 1 st – 5 th, 2005 at the Boston Convention & Exhibition Center. The poster award was sponsored by Bio. Process International. The selection criteria was based on novelty, applicability, and clarity of data presented.
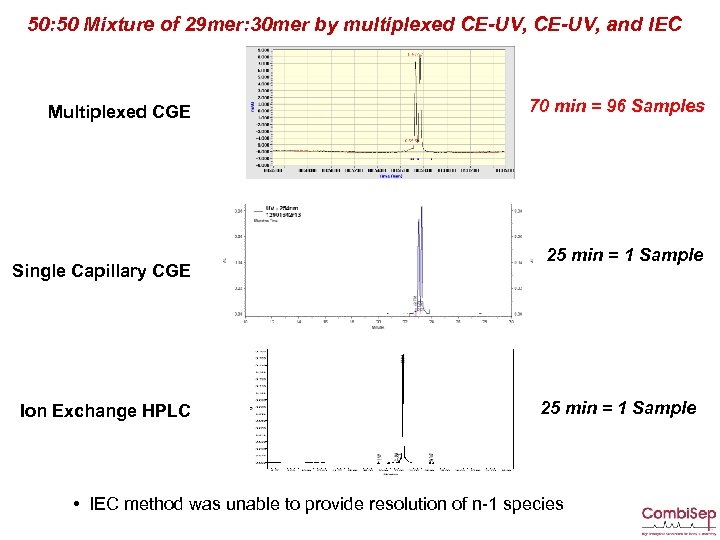
50: 50 Mixture of 29 mer: 30 mer by multiplexed CE-UV, and IEC Multiplexed CGE Single Capillary CGE Ion Exchange HPLC 70 min = 96 Samples 25 min = 1 Sample • IEC method was unable to provide resolution of n-1 species
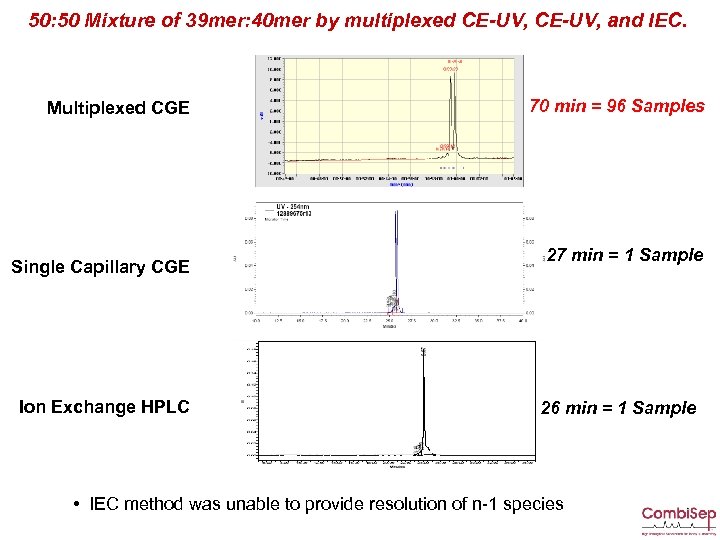
50: 50 Mixture of 39 mer: 40 mer by multiplexed CE-UV, and IEC. Multiplexed CGE Single Capillary CGE Ion Exchange HPLC 70 min = 96 Samples 27 min = 1 Sample 26 min = 1 Sample • IEC method was unable to provide resolution of n-1 species
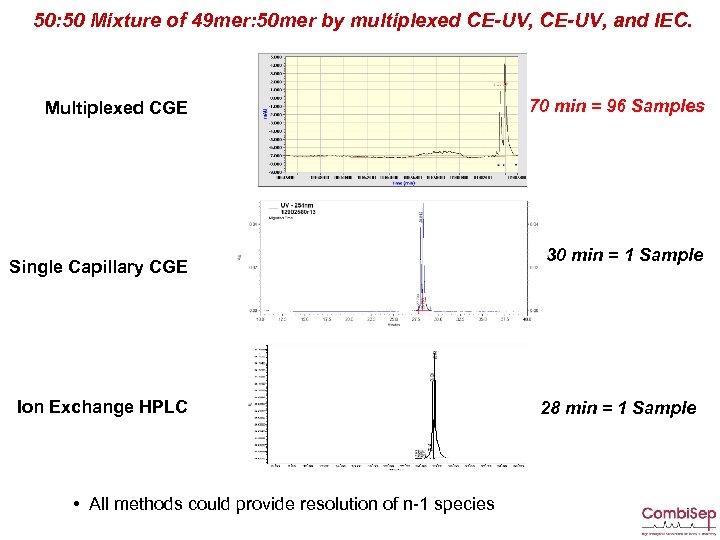
50: 50 Mixture of 49 mer: 50 mer by multiplexed CE-UV, and IEC. Multiplexed CGE Single Capillary CGE Ion Exchange HPLC • All methods could provide resolution of n-1 species 70 min = 96 Samples 30 min = 1 Sample 28 min = 1 Sample
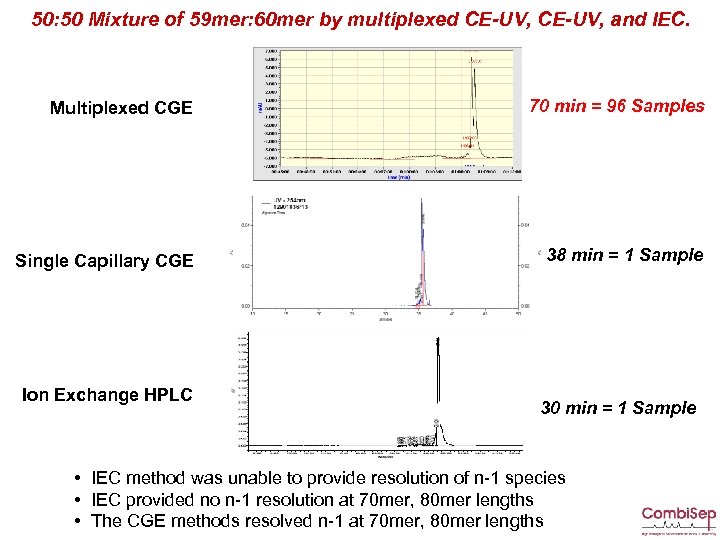
50: 50 Mixture of 59 mer: 60 mer by multiplexed CE-UV, and IEC. Multiplexed CGE 70 min = 96 Samples Single Capillary CGE 38 min = 1 Sample Ion Exchange HPLC 30 min = 1 Sample • IEC method was unable to provide resolution of n-1 species • IEC provided no n-1 resolution at 70 mer, 80 mer lengths • The CGE methods resolved n-1 at 70 mer, 80 mer lengths
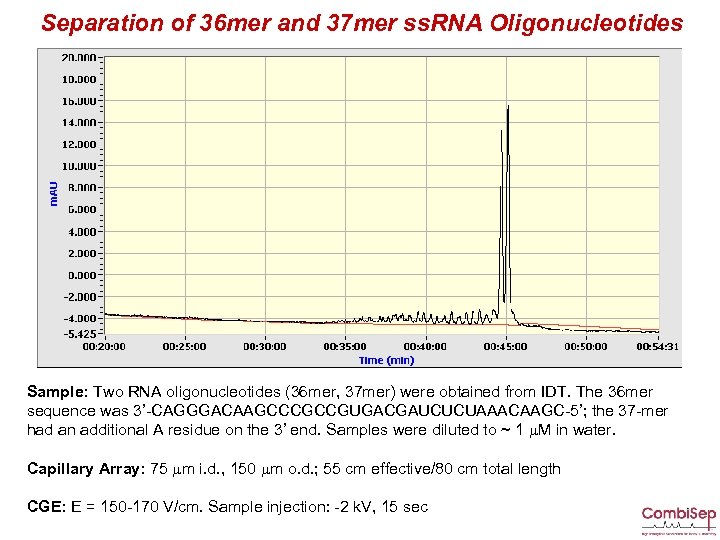
Separation of 36 mer and 37 mer ss. RNA Oligonucleotides Sample: Two RNA oligonucleotides (36 mer, 37 mer) were obtained from IDT. The 36 mer sequence was 3’-CAGGGACAAGCCCGCCGUGACGAUCUCUAAACAAGC-5’; the 37 -mer had an additional A residue on the 3’ end. Samples were diluted to ~ 1 m. M in water. Capillary Array: 75 mm i. d. , 150 mm o. d. ; 55 cm effective/80 cm total length CGE: E = 150 -170 V/cm. Sample injection: -2 k. V, 15 sec

Oligo PROTM Analyzer System • Multiplexed CE-UV system designed and optimized specifically for oligonucleotide analysis and QC • Improved ease-of-use and automation • Enhanced data analysis and report generation capabilities designed with feedback from scientists directly engaged in oligo production • Launched in May 2006
Summary • Multiplexed CGE-UV provides a powerful method for oligonucleotide purity analysis, providing superior resolution, throughput and automation compared to slab gel methods • Only minimal sample preparation is required for analysis • The developed Oligel™ matrix is capable of achieving single base resolution from 14 -80 mers in about 1 h • Multiplexed CGE-UV provides much higher sample throughput and superior resolution to HPLC methods. The standard IEC method could not resolve n-1 species at 30 mer, 40 mer, or above 60 mer lengths • RNA oligonucleotides as well as ds. RNA duplexes can be analyzed for purity in addition to DNA oligonucleotides

Details of Disruptive Technologies Contacts William Amoyal (Sales and Marketing Director) Franck Do Vale (Technical Director) Disruptive Technologies 3 Allée des Camélias 94440 Villecresnes, France Mobile: +33 (0)6 98 64 98 81 Email: wamoyal@disruptechno. com Web: www. disruptechno. com
6b5a59b897007998ac437914341e2528.ppt