2ae08d899124c4d9cd1dd689d4105da6.ppt
- Количество слайдов: 27

High Throughput Oligonucleotide Analysis Using the Combi. Sep ce. PRO 9600 System™ www. combisep. com
Introduction • Massive quantities of ss. DNA and ss. RNA oligonucleotides are produced daily to support the growth of genomic-related applications • These DNA products need to be characterized to ensure proper sizing and acceptable batch-to-batch purity • Traditional slab gel-based methods for characterizing DNA products are time-consuming, labor intensive, not readily automated, and provide relatively poor resolution • While useful for assessing compound identity, Mass Spectrometry cannot quantitatively assess oligonucleotide purity due to size dependent variations in ionization efficiencies • As a result, a bottleneck exists for characterizing the purity of ss. DNA oligonucleotides
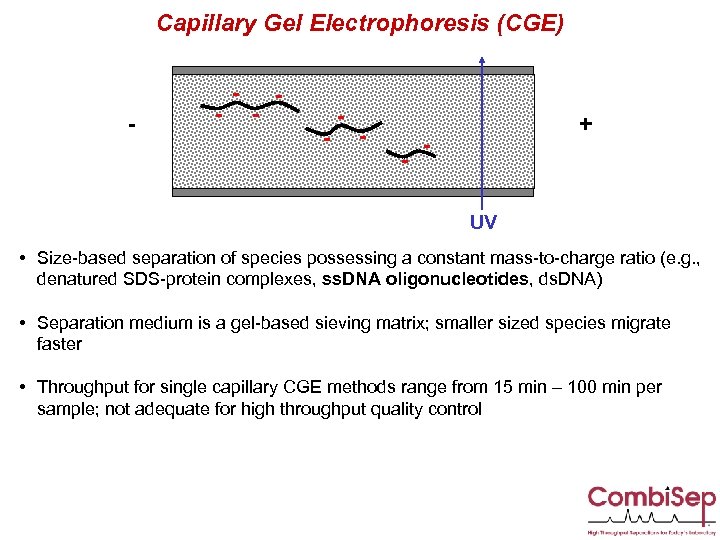
Capillary Gel Electrophoresis (CGE) - - - - + UV • Size-based separation of species possessing a constant mass-to-charge ratio (e. g. , denatured SDS-protein complexes, ss. DNA oligonucleotides, ds. DNA) • Separation medium is a gel-based sieving matrix; smaller sized species migrate faster • Throughput for single capillary CGE methods range from 15 min – 100 min per sample; not adequate for high throughput quality control
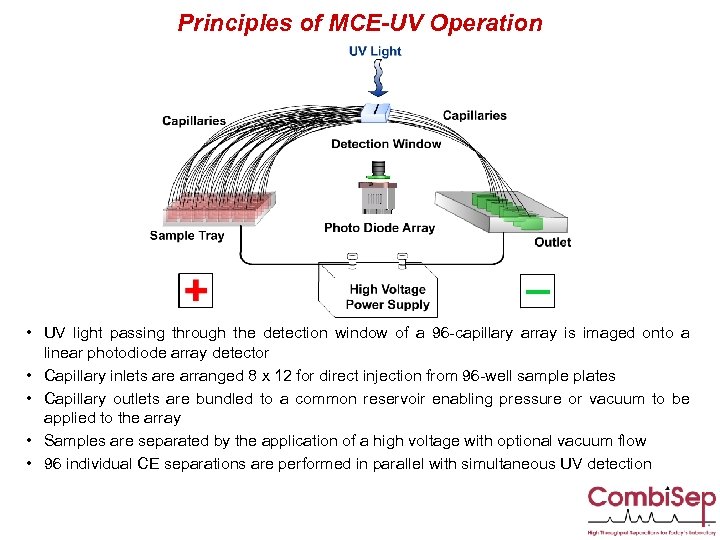
Principles of MCE-UV Operation • UV light passing through the detection window of a 96 -capillary array is imaged onto a linear photodiode array detector • Capillary inlets are arranged 8 x 12 for direct injection from 96 -well sample plates • Capillary outlets are bundled to a common reservoir enabling pressure or vacuum to be applied to the array • Samples are separated by the application of a high voltage with optional vacuum flow • 96 individual CE separations are performed in parallel with simultaneous UV detection
ss. DNA Oligonucleotide Analysis by Multiplexed CGE-UV • CGE-UV is the method of choice for assessing oligonucleotide purity, providing higher resolution and superior quantification vs. slab gels. • Label-free UV detection provides a low cost, toxic free alternative and improved quantification compared to fluorescent label approaches for ss. DNA which may possess size and sequence dependent fluorescent labeling efficiencies • CGE-UV advantages for characterizing oligonucleotide purity include low sample consumption, high resolving power, direct on-line UV detection and automated operation. • A proprietary gel sieving matrix (Oligel ) has been developed for multiplexed operation. When analyzed at low m. M concentrations, single nucleotide resolution can be obtained from 10 mer to 80 mer oligonucleotide lengths, allowing identification of low level n-1, n-2, etc. impurities. • Multiplexed CGE-UV provides a nearly 50 -fold improvement in sample throughput over single capillary CE methods with a minimal loss in separation performance. A throughput of 96 samples/h is achieved for 80 mer length oligonucleotides.
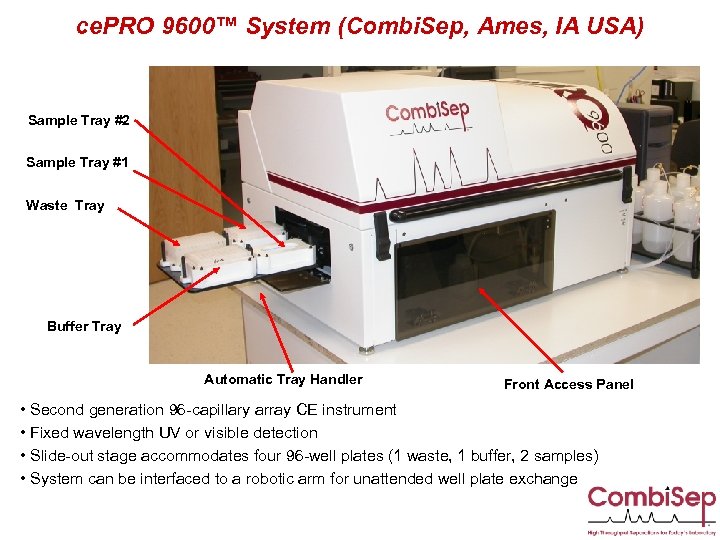
ce. PRO 9600™ System (Combi. Sep, Ames, IA USA) Sample Tray #2 Sample Tray #1 Waste Tray Buffer Tray Automatic Tray Handler Front Access Panel • Second generation 96 -capillary array CE instrument • Fixed wavelength UV or visible detection • Slide-out stage accommodates four 96 -well plates (1 waste, 1 buffer, 2 samples) • System can be interfaced to a robotic arm for unattended well plate exchange

Inside View of the ce. PRO 9600™ Instrument Capillary Array Cartridge Lamp Housing Capillary Array Detection Window HV Power Supply Optical Platform Housing Syringe Pump
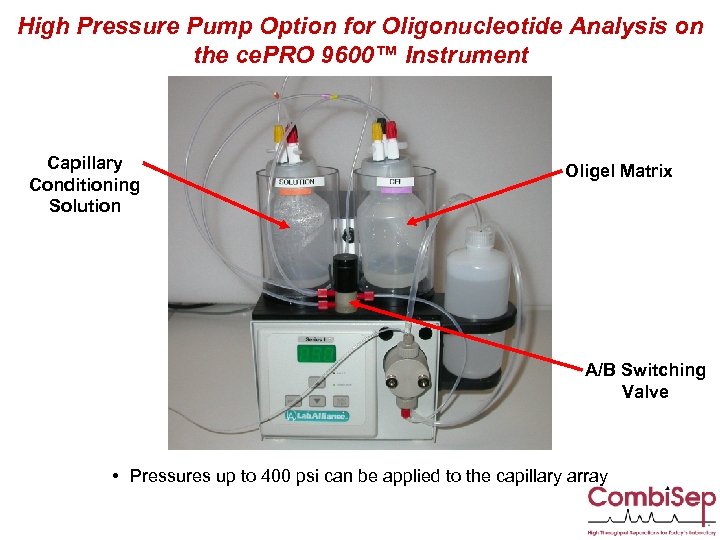
High Pressure Pump Option for Oligonucleotide Analysis on the ce. PRO 9600™ Instrument Capillary Conditioning Solution Oligel Matrix A/B Switching Valve • Pressures up to 400 psi can be applied to the capillary array
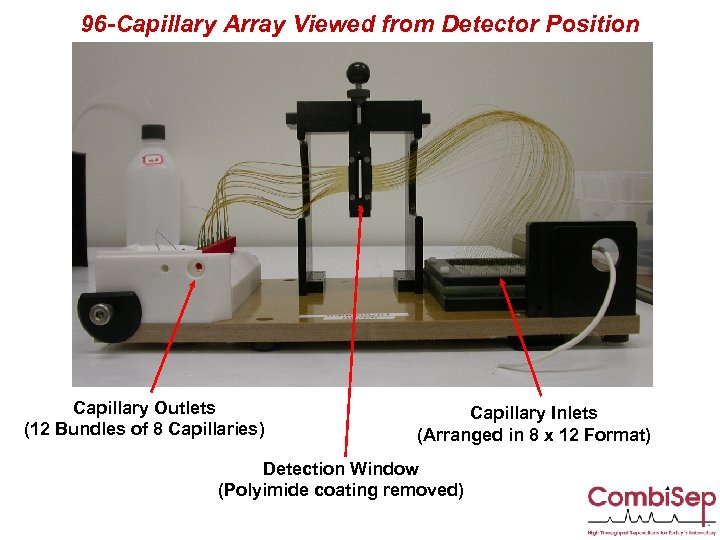
96 -Capillary Array Viewed from Detector Position Capillary Outlets (12 Bundles of 8 Capillaries) Capillary Inlets (Arranged in 8 x 12 Format) Detection Window (Polyimide coating removed)
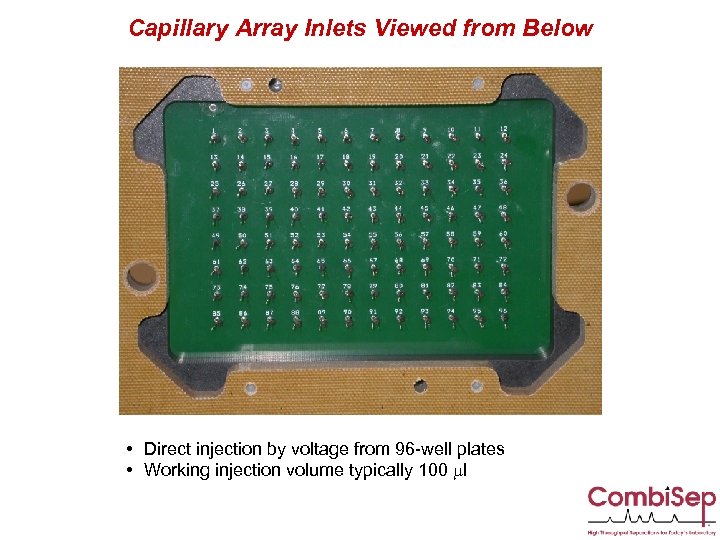
Capillary Array Inlets Viewed from Below • Direct injection by voltage from 96 -well plates • Working injection volume typically 100 ml
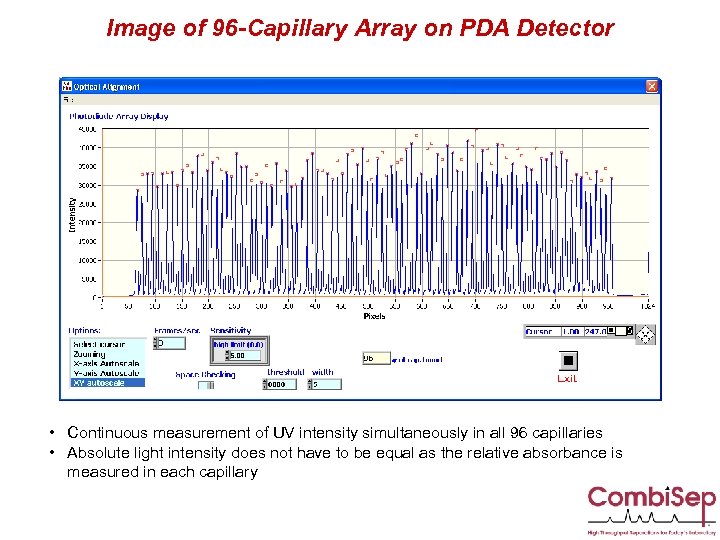
Image of 96 -Capillary Array on PDA Detector • Continuous measurement of UV intensity simultaneously in all 96 capillaries • Absolute light intensity does not have to be equal as the relative absorbance is measured in each capillary
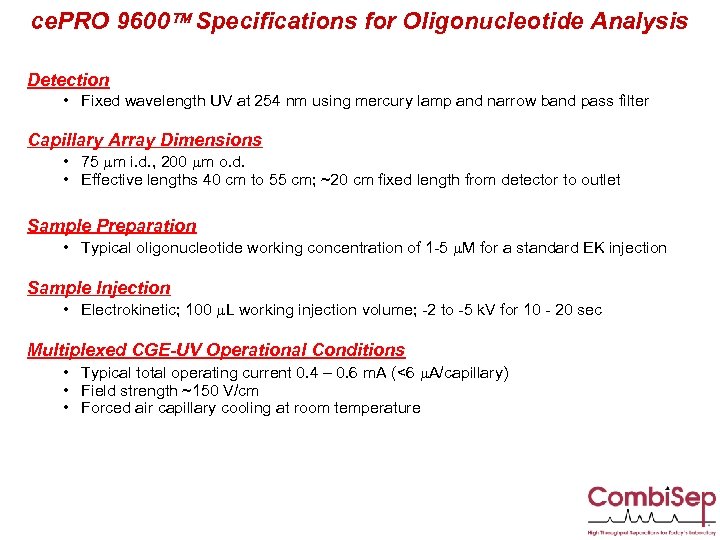
ce. PRO 9600 Specifications for Oligonucleotide Analysis Detection • Fixed wavelength UV at 254 nm using mercury lamp and narrow band pass filter Capillary Array Dimensions • 75 mm i. d. , 200 mm o. d. • Effective lengths 40 cm to 55 cm; ~20 cm fixed length from detector to outlet Sample Preparation • Typical oligonucleotide working concentration of 1 -5 m. M for a standard EK injection Sample Injection • Electrokinetic; 100 m. L working injection volume; -2 to -5 k. V for 10 - 20 sec Multiplexed CGE-UV Operational Conditions • Typical total operating current 0. 4 – 0. 6 m. A (<6 m. A/capillary) • Field strength ~150 V/cm • Forced air capillary cooling at room temperature
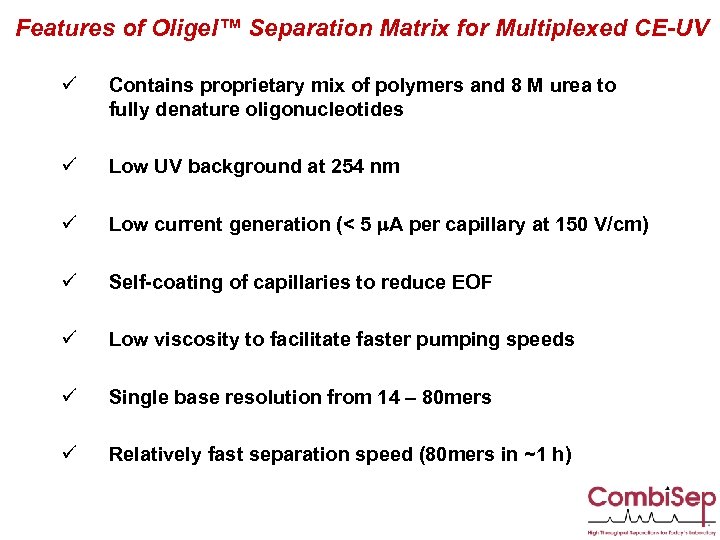
Features of Oligel™ Separation Matrix for Multiplexed CE-UV ü Contains proprietary mix of polymers and 8 M urea to fully denature oligonucleotides ü Low UV background at 254 nm ü Low current generation (< 5 m. A per capillary at 150 V/cm) ü Self-coating of capillaries to reduce EOF ü Low viscosity to facilitate faster pumping speeds ü Single base resolution from 14 – 80 mers ü Relatively fast separation speed (80 mers in ~1 h)
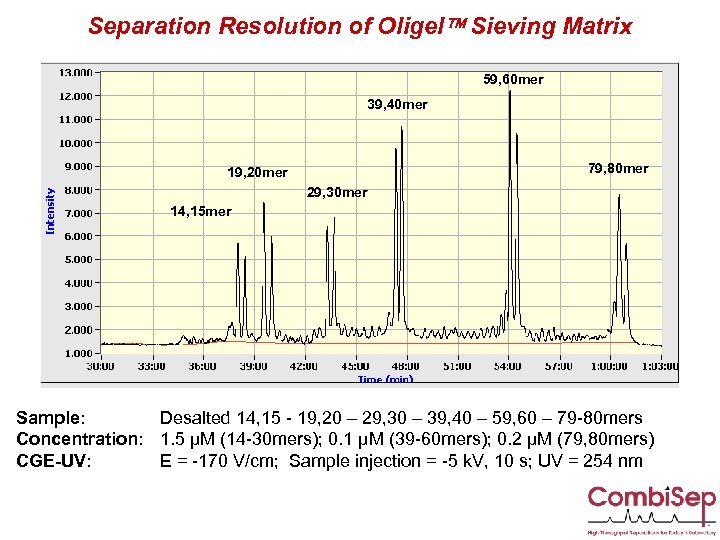
Separation Resolution of Oligel Sieving Matrix 59, 60 mer 39, 40 mer 79, 80 mer 19, 20 mer 29, 30 mer 14, 15 mer Sample: Desalted 14, 15 - 19, 20 – 29, 30 – 39, 40 – 59, 60 – 79 -80 mers Concentration: 1. 5 µM (14 -30 mers); 0. 1 µM (39 -60 mers); 0. 2 µM (79, 80 mers) CGE-UV: E = -170 V/cm; Sample injection = -5 k. V, 10 s; UV = 254 nm
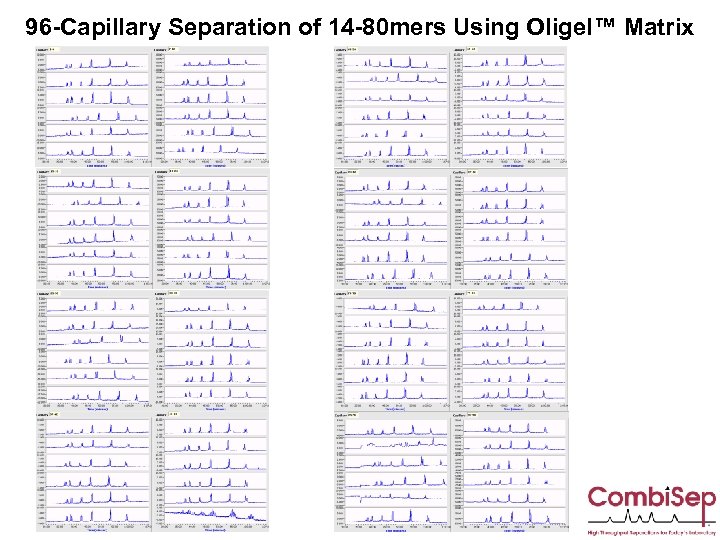
96 -Capillary Separation of 14 -80 mers Using Oligel™ Matrix
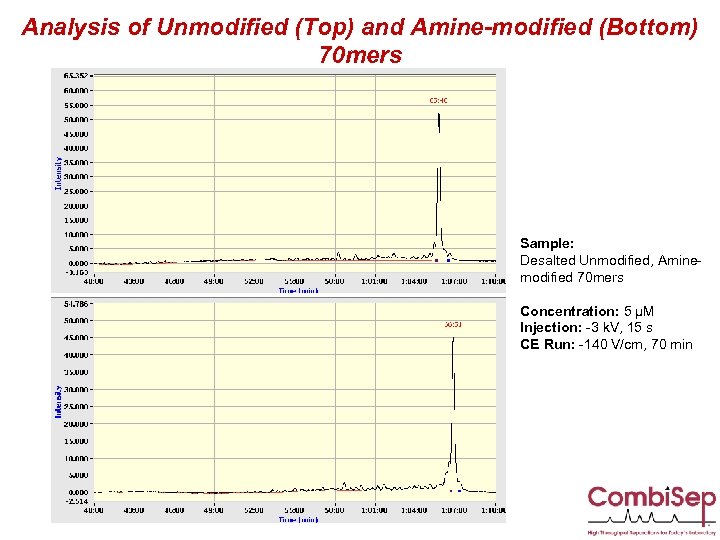
Analysis of Unmodified (Top) and Amine-modified (Bottom) 70 mers Sample: Desalted Unmodified, Aminemodified 70 mers Concentration: 5 µM Injection: -3 k. V, 15 s CE Run: -140 V/cm, 70 min
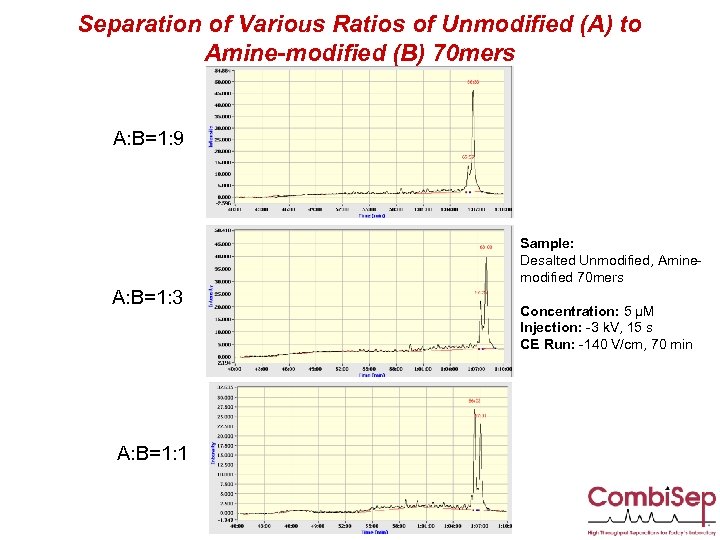
Separation of Various Ratios of Unmodified (A) to Amine-modified (B) 70 mers A: B=1: 9 Sample: Desalted Unmodified, Aminemodified 70 mers A: B=1: 3 A: B=1: 1 Concentration: 5 µM Injection: -3 k. V, 15 s CE Run: -140 V/cm, 70 min
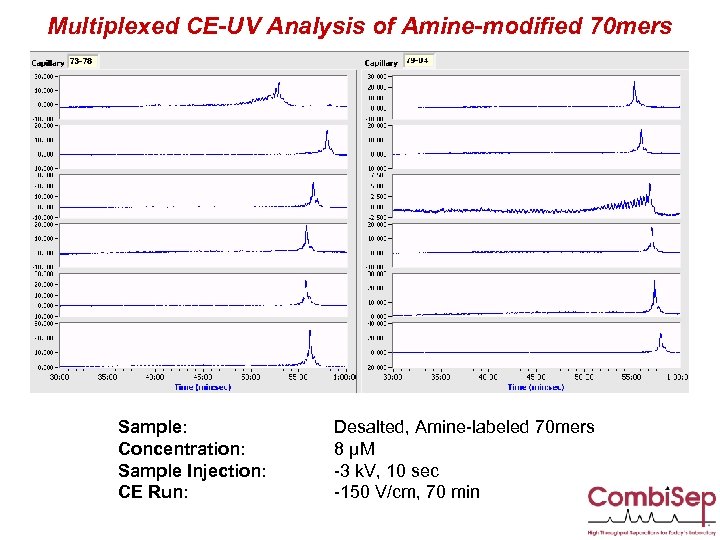
Multiplexed CE-UV Analysis of Amine-modified 70 mers Sample: Concentration: Sample Injection: CE Run: Desalted, Amine-labeled 70 mers 8 µM -3 k. V, 10 sec -150 V/cm, 70 min
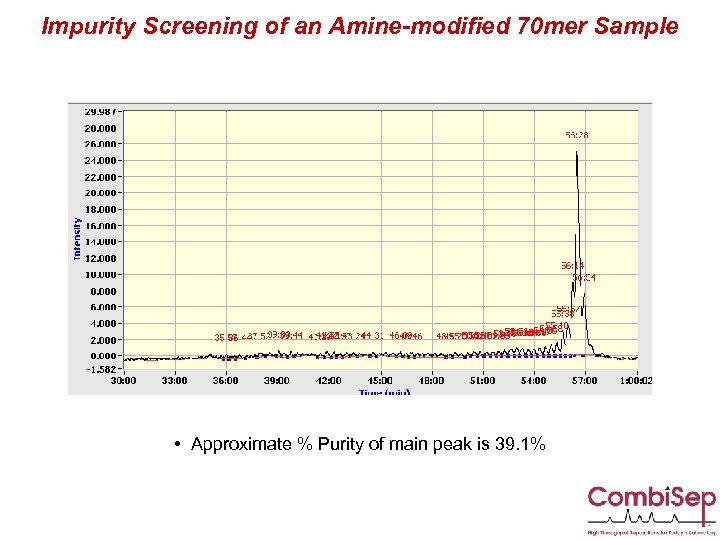
Impurity Screening of an Amine-modified 70 mer Sample • Approximate % Purity of main peak is 39. 1%

Co-Winner Best Poster at Tides 2005 Combi. Sep and IDT Poster Co-Winners of Best Poster at Tides 2005 Conference The poster titled “Comparisons between Multiplexed, Absorbance-Based Capillary Electrophoresis, and Ion Exchange Chromatography for Analysis of n-1 Oligonucleotide Impurities” by Wei, Ho-ming Pang, Dennis Tallman, and Jeremy Kenseth of Combi. Sep, Inc and Lisa Bogh of Integrated DNA Technology was recently selected as the co-winner of the best poster at Tides 2005. The Tides Conference is an industry event for manufacturing and development of oligonucleotide and peptide products. The meeting was held May 1 st – 5 th, 2005 at the Boston Convention & Exhibition Center. The poster award was sponsored by Bio. Process International. The selection criteria was based on novelty, applicability, and clarity of data presented.
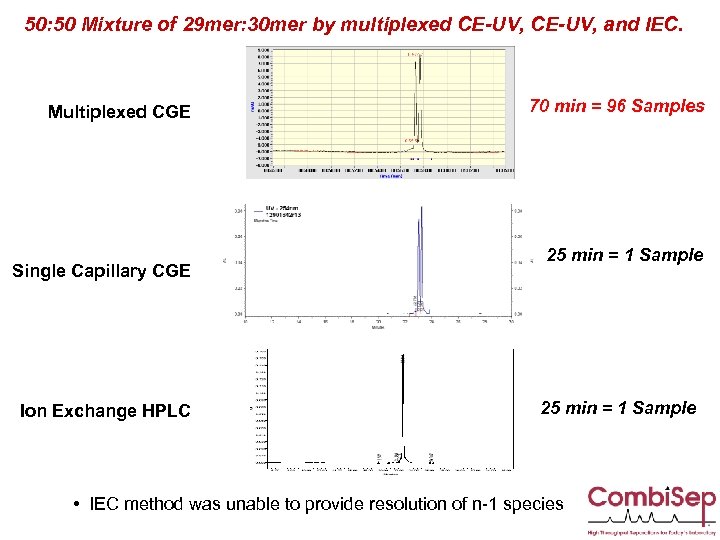
50: 50 Mixture of 29 mer: 30 mer by multiplexed CE-UV, and IEC. Multiplexed CGE Single Capillary CGE Ion Exchange HPLC 70 min = 96 Samples 25 min = 1 Sample • IEC method was unable to provide resolution of n-1 species
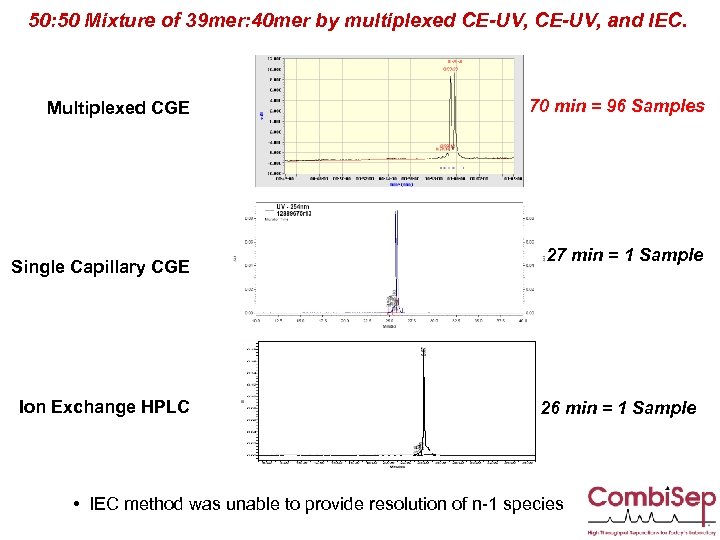
50: 50 Mixture of 39 mer: 40 mer by multiplexed CE-UV, and IEC. Multiplexed CGE Single Capillary CGE Ion Exchange HPLC 70 min = 96 Samples 27 min = 1 Sample 26 min = 1 Sample • IEC method was unable to provide resolution of n-1 species
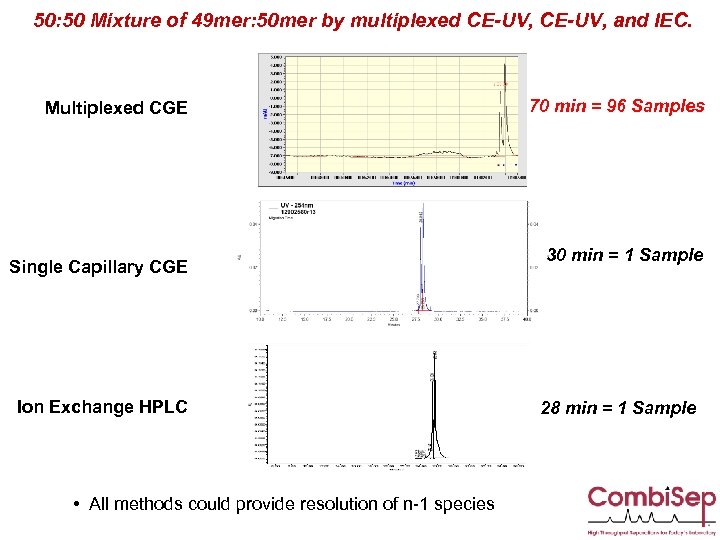
50: 50 Mixture of 49 mer: 50 mer by multiplexed CE-UV, and IEC. Multiplexed CGE Single Capillary CGE Ion Exchange HPLC • All methods could provide resolution of n-1 species 70 min = 96 Samples 30 min = 1 Sample 28 min = 1 Sample
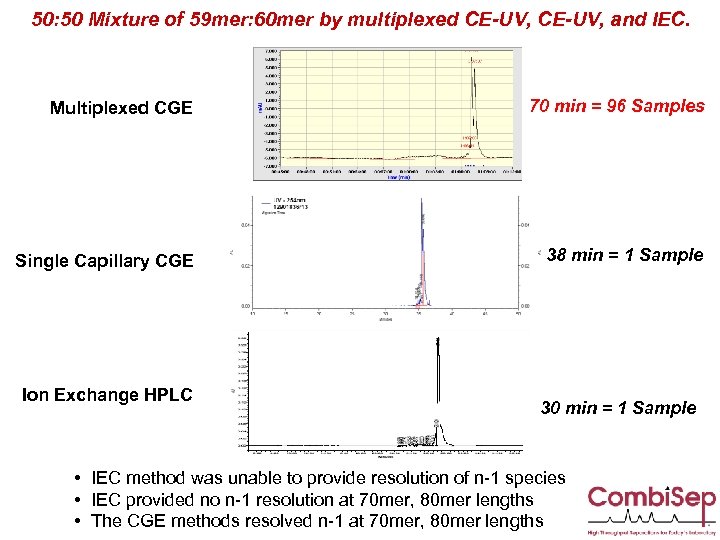
50: 50 Mixture of 59 mer: 60 mer by multiplexed CE-UV, and IEC. Multiplexed CGE 70 min = 96 Samples Single Capillary CGE 38 min = 1 Sample Ion Exchange HPLC 30 min = 1 Sample • IEC method was unable to provide resolution of n-1 species • IEC provided no n-1 resolution at 70 mer, 80 mer lengths • The CGE methods resolved n-1 at 70 mer, 80 mer lengths
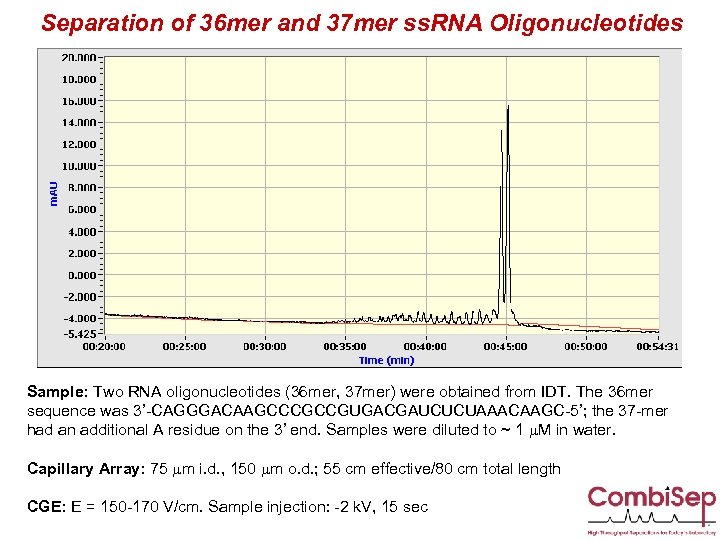
Separation of 36 mer and 37 mer ss. RNA Oligonucleotides Sample: Two RNA oligonucleotides (36 mer, 37 mer) were obtained from IDT. The 36 mer sequence was 3’-CAGGGACAAGCCCGCCGUGACGAUCUCUAAACAAGC-5’; the 37 -mer had an additional A residue on the 3’ end. Samples were diluted to ~ 1 m. M in water. Capillary Array: 75 mm i. d. , 150 mm o. d. ; 55 cm effective/80 cm total length CGE: E = 150 -170 V/cm. Sample injection: -2 k. V, 15 sec

Coming Soon: Oligo Analyzer PRO System • Multiplexed CE-UV system designed and optimized specifically for oligonucleotide analysis • Improved ease-of-use and automation • Enhanced data analysis and report generation capabilities designed with feedback from scientists directly engaged in oligo production • Scheduled for release in Q 2 of 2006
Summary • Multiplexed CGE-UV provides a powerful method for oligonucleotide purity analysis, providing superior resolution, throughput and automation compared to slab gel methods • Only minimal sample preparation is required for analysis • The developed Oligel™ matrix is capable of achieving single base resolution from 14 -80 mers in about 1 h • Multiplexed CGE-UV provides much higher sample throughput and superior resolution to HPLC methods. The standard IEC method could not resolve n-1 species at 30 mer, 40 mer, or above 60 mer lengths • RNA oligonucleotides as well as ds. RNA duplexes can be analyzed for purity in addition to DNA oligonucleotides
2ae08d899124c4d9cd1dd689d4105da6.ppt