5975572b71460c51076af2dd3201069c.ppt
- Количество слайдов: 66
 HGP (Human Genome Project) HPP (Human Proteome Proyect) D: SPLASH. EXE
HGP (Human Genome Project) HPP (Human Proteome Proyect) D: SPLASH. EXE
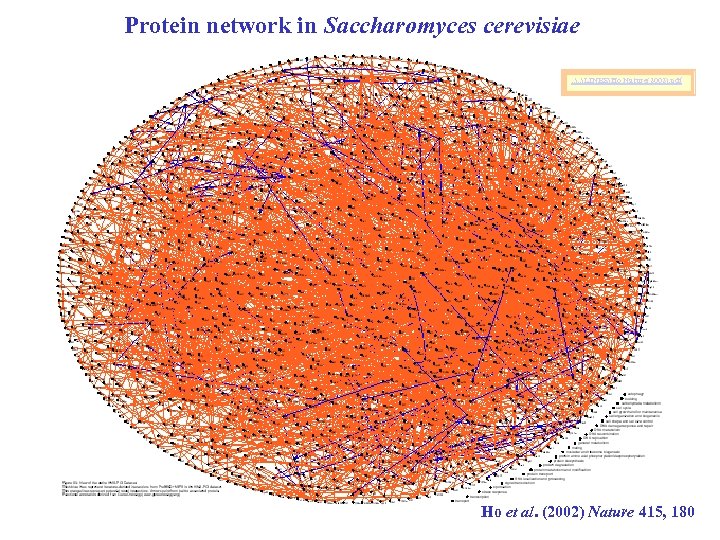 Protein network in Saccharomyces cerevisiae. . LINKSHo Nature(2002). pdf Ho et al. (2002) Nature 415, 180
Protein network in Saccharomyces cerevisiae. . LINKSHo Nature(2002). pdf Ho et al. (2002) Nature 415, 180
 Determination of Protein Structures The two most common methods used to investigate molecular structures are: 1. X-ray crystallography (also called X-ray diffraction) 2. Nuclear magnetic resonance (NMR) spectroscopy National Institutes of Health, USA
Determination of Protein Structures The two most common methods used to investigate molecular structures are: 1. X-ray crystallography (also called X-ray diffraction) 2. Nuclear magnetic resonance (NMR) spectroscopy National Institutes of Health, USA
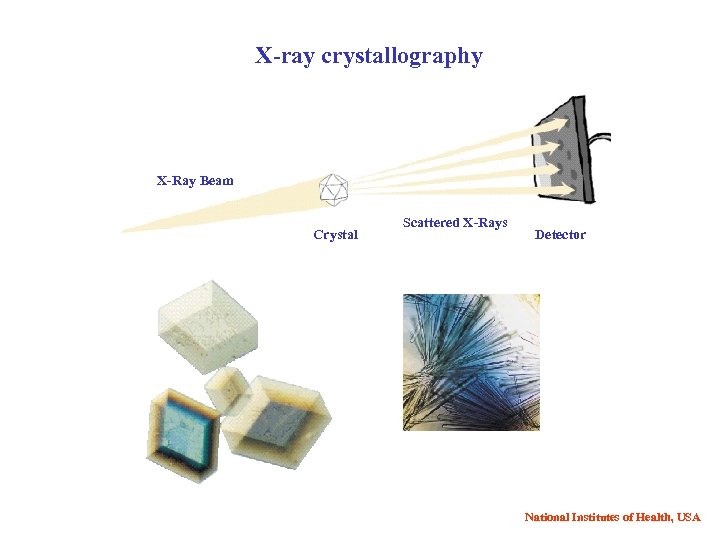 X-ray crystallography X-Ray Beam Crystal Scattered X-Rays Detector National Institutes of Health, USA
X-ray crystallography X-Ray Beam Crystal Scattered X-Rays Detector National Institutes of Health, USA
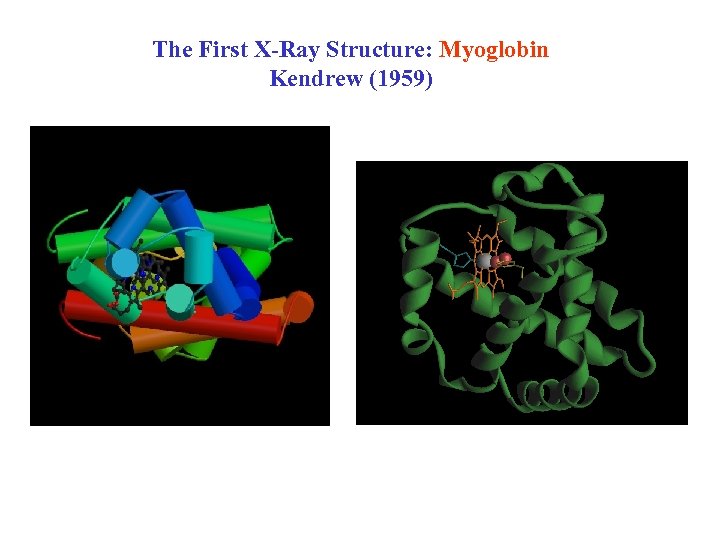 The First X-Ray Structure: Myoglobin Kendrew (1959)
The First X-Ray Structure: Myoglobin Kendrew (1959)
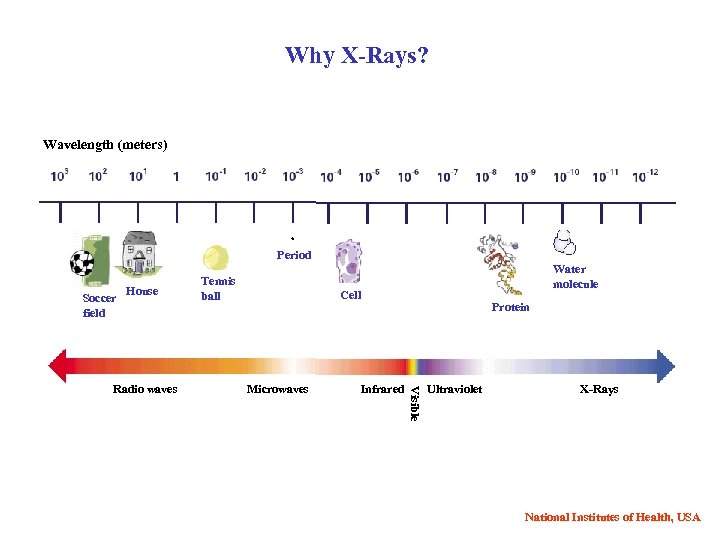 Why X-Rays? Wavelength (meters) Period Soccer field House Water molecule Cell Microwaves Infrared Protein Visible Radio waves Tennis ball Ultraviolet X-Rays National Institutes of Health, USA
Why X-Rays? Wavelength (meters) Period Soccer field House Water molecule Cell Microwaves Infrared Protein Visible Radio waves Tennis ball Ultraviolet X-Rays National Institutes of Health, USA
 ESRF - The European Synchrotron Radiation Facility Grenoble, Francia National Institutes of Health, USA
ESRF - The European Synchrotron Radiation Facility Grenoble, Francia National Institutes of Health, USA
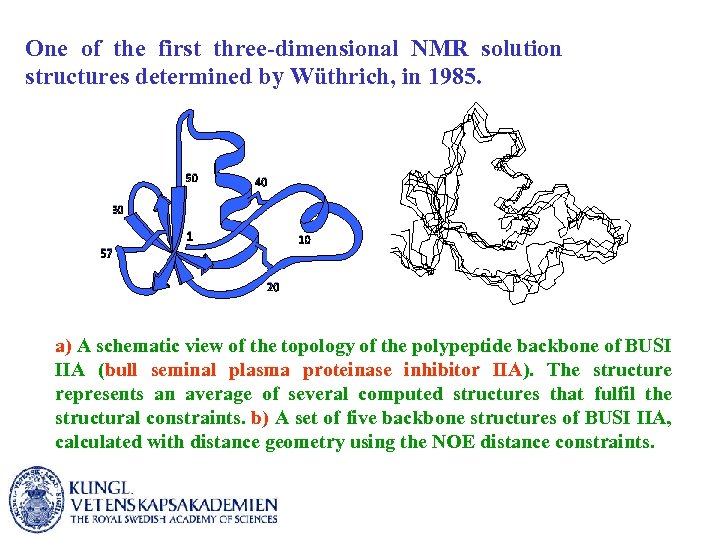 One of the first three-dimensional NMR solution structures determined by Wüthrich, in 1985. a) A schematic view of the topology of the polypeptide backbone of BUSI IIA (bull seminal plasma proteinase inhibitor IIA). The structure represents an average of several computed structures that fulfil the structural constraints. b) A set of five backbone structures of BUSI IIA, calculated with distance geometry using the NOE distance constraints.
One of the first three-dimensional NMR solution structures determined by Wüthrich, in 1985. a) A schematic view of the topology of the polypeptide backbone of BUSI IIA (bull seminal plasma proteinase inhibitor IIA). The structure represents an average of several computed structures that fulfil the structural constraints. b) A set of five backbone structures of BUSI IIA, calculated with distance geometry using the NOE distance constraints.
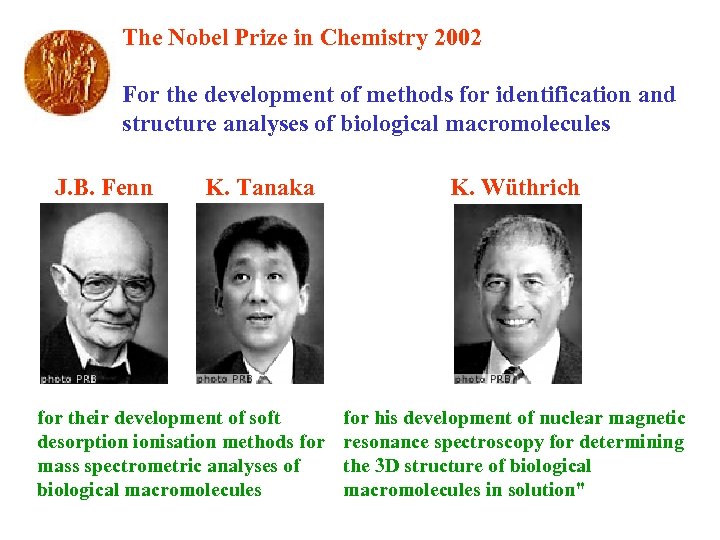 The Nobel Prize in Chemistry 2002 For the development of methods for identification and structure analyses of biological macromolecules J. B. Fenn K. Tanaka for their development of soft desorption ionisation methods for mass spectrometric analyses of biological macromolecules K. Wüthrich for his development of nuclear magnetic resonance spectroscopy for determining the 3 D structure of biological macromolecules in solution"
The Nobel Prize in Chemistry 2002 For the development of methods for identification and structure analyses of biological macromolecules J. B. Fenn K. Tanaka for their development of soft desorption ionisation methods for mass spectrometric analyses of biological macromolecules K. Wüthrich for his development of nuclear magnetic resonance spectroscopy for determining the 3 D structure of biological macromolecules in solution"
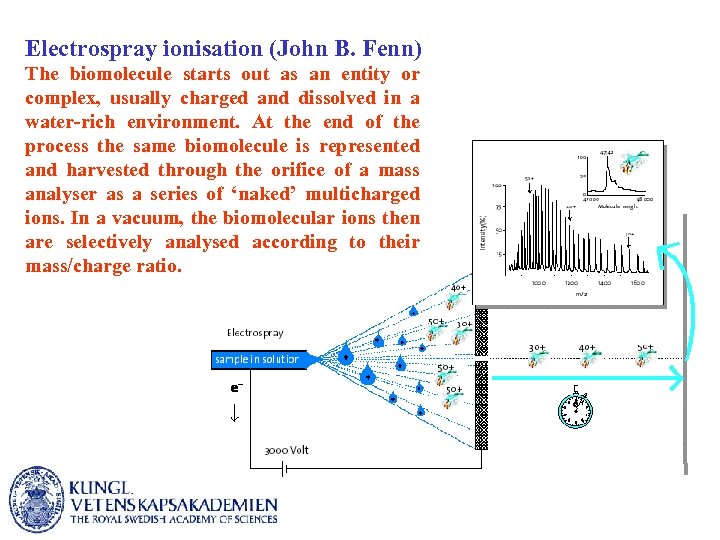 Electrospray ionisation (John B. Fenn) The biomolecule starts out as an entity or complex, usually charged and dissolved in a water-rich environment. At the end of the process the same biomolecule is represented and harvested through the orifice of a mass analyser as a series of ‘naked’ multicharged ions. In a vacuum, the biomolecular ions then are selectively analysed according to their mass/charge ratio.
Electrospray ionisation (John B. Fenn) The biomolecule starts out as an entity or complex, usually charged and dissolved in a water-rich environment. At the end of the process the same biomolecule is represented and harvested through the orifice of a mass analyser as a series of ‘naked’ multicharged ions. In a vacuum, the biomolecular ions then are selectively analysed according to their mass/charge ratio.
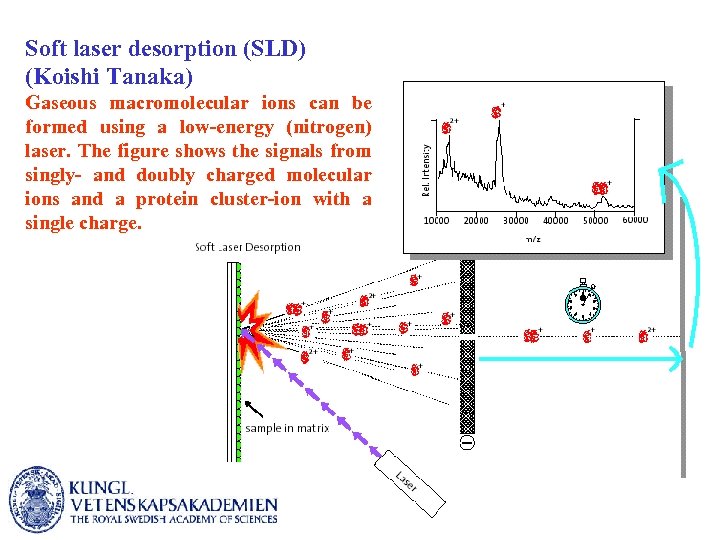 Soft laser desorption (SLD) (Koishi Tanaka) Gaseous macromolecular ions can be formed using a low-energy (nitrogen) laser. The figure shows the signals from singly- and doubly charged molecular ions and a protein cluster-ion with a single charge.
Soft laser desorption (SLD) (Koishi Tanaka) Gaseous macromolecular ions can be formed using a low-energy (nitrogen) laser. The figure shows the signals from singly- and doubly charged molecular ions and a protein cluster-ion with a single charge.
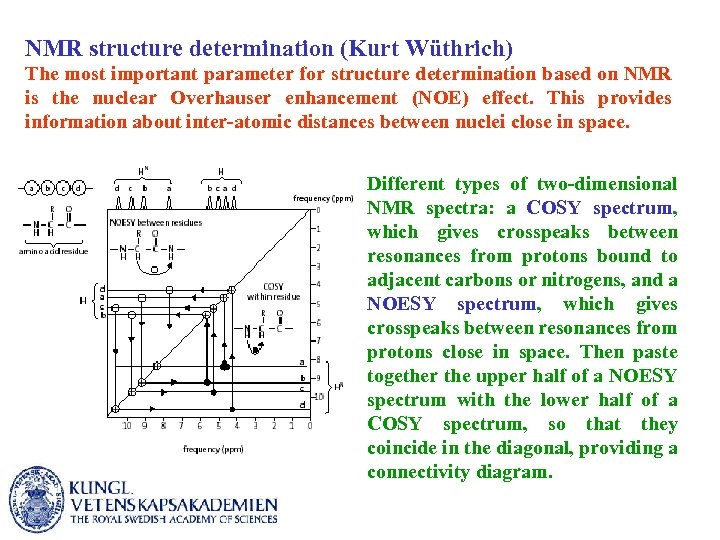 NMR structure determination (Kurt Wüthrich) The most important parameter for structure determination based on NMR is the nuclear Overhauser enhancement (NOE) effect. This provides information about inter-atomic distances between nuclei close in space. Different types of two-dimensional NMR spectra: a COSY spectrum, which gives crosspeaks between resonances from protons bound to adjacent carbons or nitrogens, and a NOESY spectrum, which gives crosspeaks between resonances from protons close in space. Then paste together the upper half of a NOESY spectrum with the lower half of a COSY spectrum, so that they coincide in the diagonal, providing a connectivity diagram.
NMR structure determination (Kurt Wüthrich) The most important parameter for structure determination based on NMR is the nuclear Overhauser enhancement (NOE) effect. This provides information about inter-atomic distances between nuclei close in space. Different types of two-dimensional NMR spectra: a COSY spectrum, which gives crosspeaks between resonances from protons bound to adjacent carbons or nitrogens, and a NOESY spectrum, which gives crosspeaks between resonances from protons close in space. Then paste together the upper half of a NOESY spectrum with the lower half of a COSY spectrum, so that they coincide in the diagonal, providing a connectivity diagram.
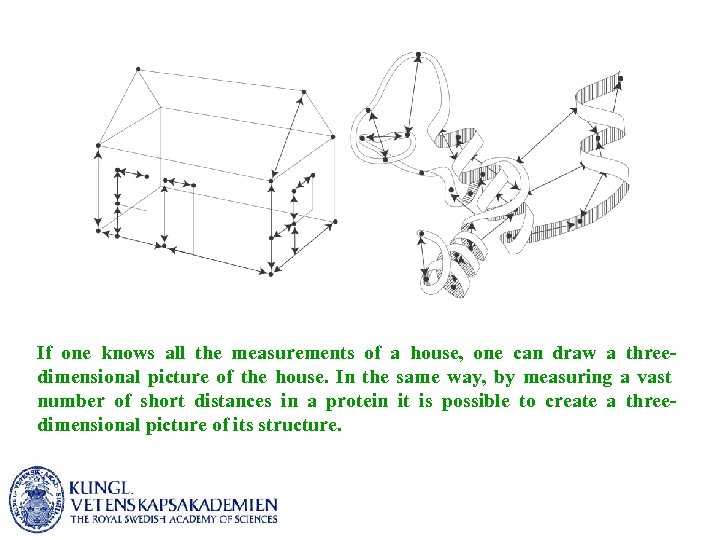 If one knows all the measurements of a house, one can draw a threedimensional picture of the house. In the same way, by measuring a vast number of short distances in a protein it is possible to create a threedimensional picture of its structure.
If one knows all the measurements of a house, one can draw a threedimensional picture of the house. In the same way, by measuring a vast number of short distances in a protein it is possible to create a threedimensional picture of its structure.
 NMR Spectroscopy Most NMR spectroscopists use magnets that are 500 megahertz to 800 megahertz. This magnet is 900 megahertz—the strongest one available. National Institutes of Health, USA
NMR Spectroscopy Most NMR spectroscopists use magnets that are 500 megahertz to 800 megahertz. This magnet is 900 megahertz—the strongest one available. National Institutes of Health, USA
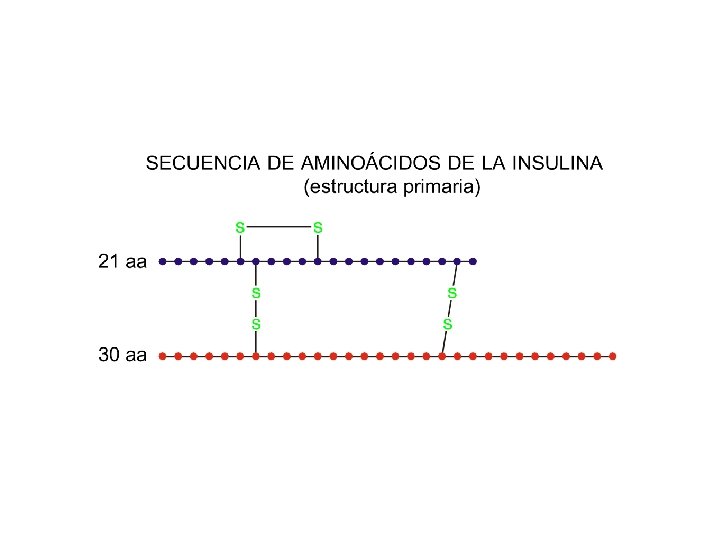
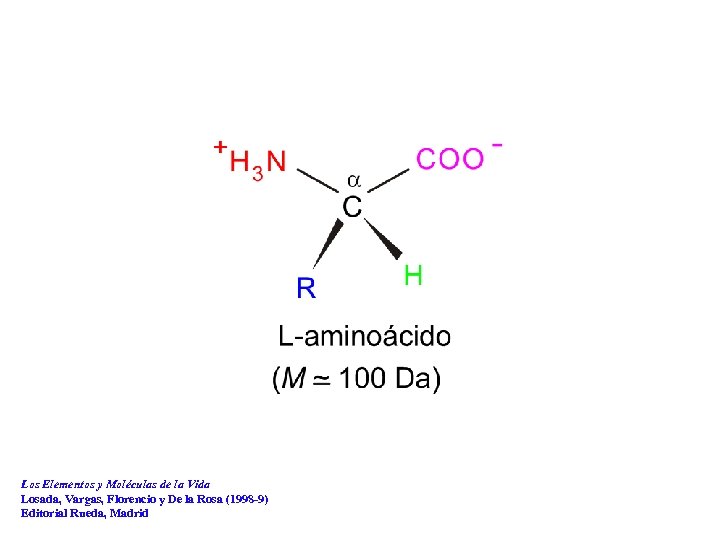 Los Elementos y Moléculas de la Vida Losada, Vargas, Florencio y De la Rosa (1998 -9) Editorial Rueda, Madrid
Los Elementos y Moléculas de la Vida Losada, Vargas, Florencio y De la Rosa (1998 -9) Editorial Rueda, Madrid
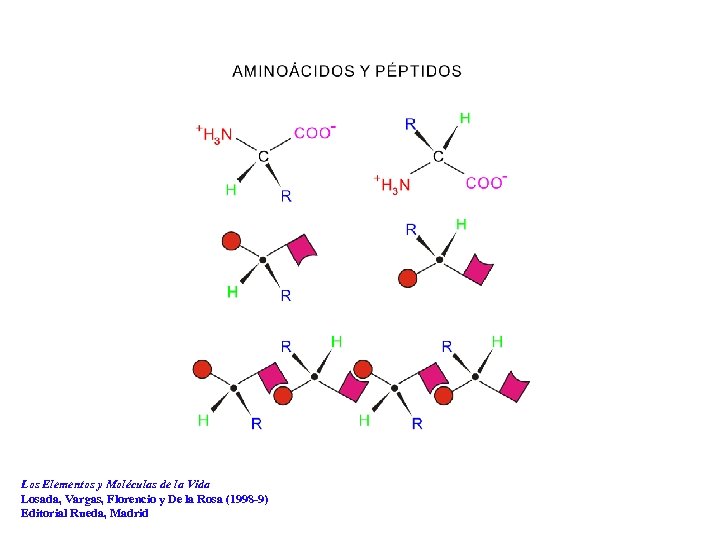 Los Elementos y Moléculas de la Vida Losada, Vargas, Florencio y De la Rosa (1998 -9) Editorial Rueda, Madrid
Los Elementos y Moléculas de la Vida Losada, Vargas, Florencio y De la Rosa (1998 -9) Editorial Rueda, Madrid
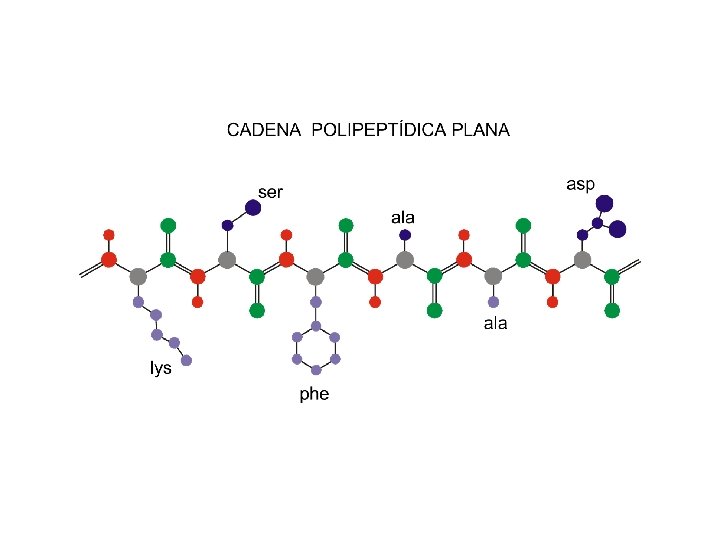
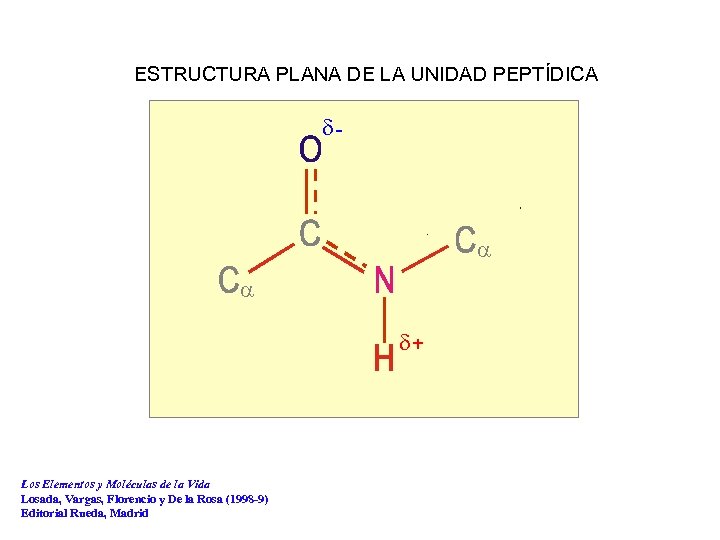 ESTRUCTURA PLANA DE LA UNIDAD PEPTÍDICA d- a a d+ Los Elementos y Moléculas de la Vida Losada, Vargas, Florencio y De la Rosa (1998 -9) Editorial Rueda, Madrid
ESTRUCTURA PLANA DE LA UNIDAD PEPTÍDICA d- a a d+ Los Elementos y Moléculas de la Vida Losada, Vargas, Florencio y De la Rosa (1998 -9) Editorial Rueda, Madrid
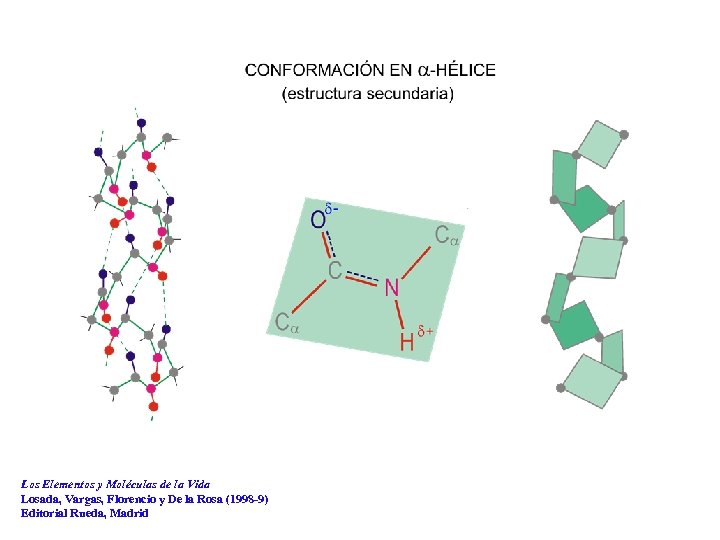 Los Elementos y Moléculas de la Vida Losada, Vargas, Florencio y De la Rosa (1998 -9) Editorial Rueda, Madrid
Los Elementos y Moléculas de la Vida Losada, Vargas, Florencio y De la Rosa (1998 -9) Editorial Rueda, Madrid
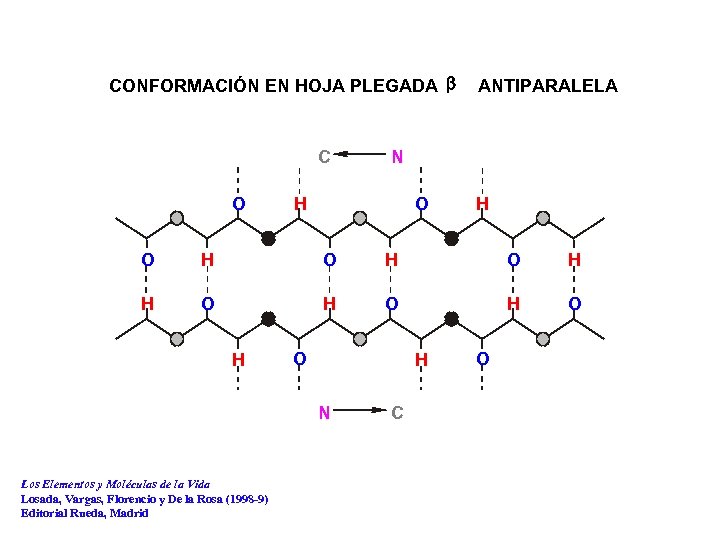 CONFORMACIÓN EN HOJA PLEGADA b C O ANTIPARALELA N O H H O H O H O H N Los Elementos y Moléculas de la Vida Losada, Vargas, Florencio y De la Rosa (1998 -9) Editorial Rueda, Madrid C O
CONFORMACIÓN EN HOJA PLEGADA b C O ANTIPARALELA N O H H O H O H O H N Los Elementos y Moléculas de la Vida Losada, Vargas, Florencio y De la Rosa (1998 -9) Editorial Rueda, Madrid C O
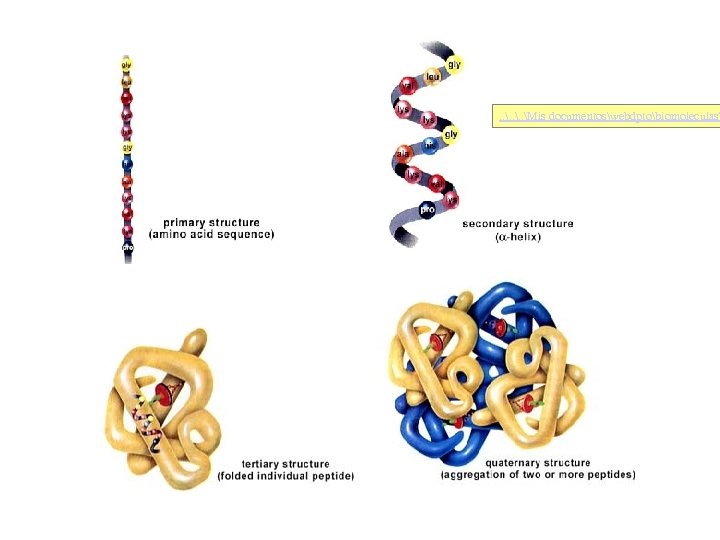 . . Mis documentoswebdptobiomoleculas
. . Mis documentoswebdptobiomoleculas
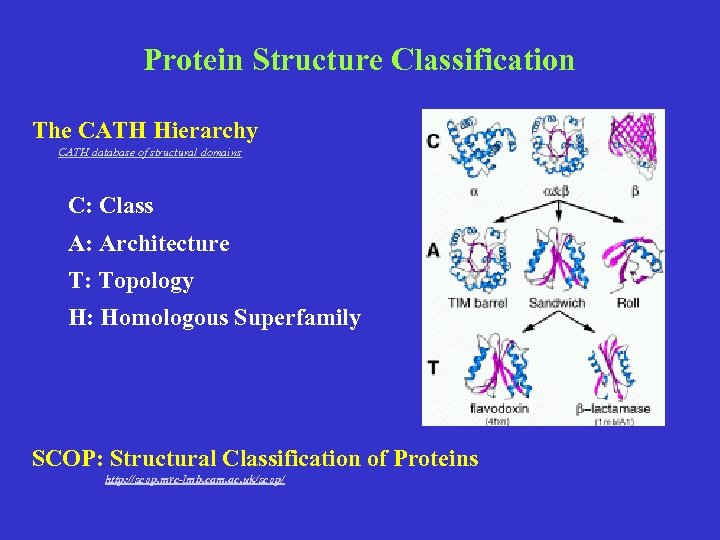 Protein Structure Classification The CATH Hierarchy CATH database of structural domains C: Class A: Architecture T: Topology H: Homologous Superfamily SCOP: Structural Classification of Proteins http: //scop. mrc-lmb. cam. ac. uk/scop/
Protein Structure Classification The CATH Hierarchy CATH database of structural domains C: Class A: Architecture T: Topology H: Homologous Superfamily SCOP: Structural Classification of Proteins http: //scop. mrc-lmb. cam. ac. uk/scop/
 Proteínas: Evolución a nivel molecular
Proteínas: Evolución a nivel molecular
 A strict principle of economy: The same economy that reuses a few motifs to subserve different functions (divergent evolution) increased the chance of different biological systems coming up with different solutions to the same problem (convergent evolution).
A strict principle of economy: The same economy that reuses a few motifs to subserve different functions (divergent evolution) increased the chance of different biological systems coming up with different solutions to the same problem (convergent evolution).
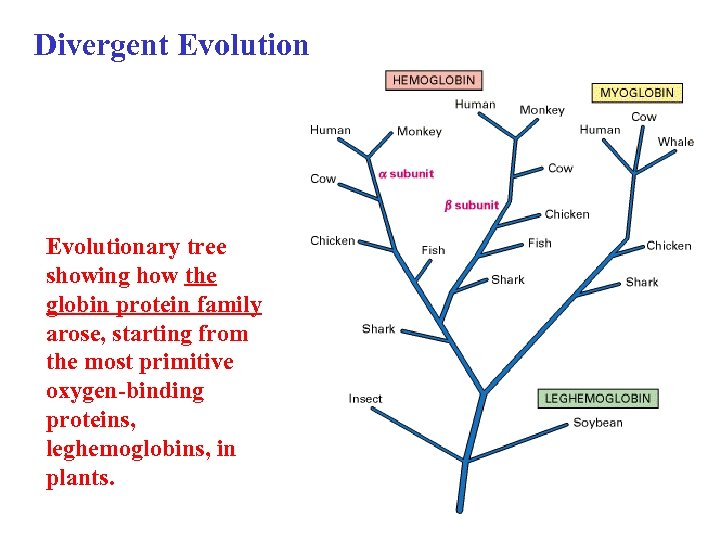 Divergent Evolutionary tree showing how the globin protein family arose, starting from the most primitive oxygen-binding proteins, leghemoglobins, in plants.
Divergent Evolutionary tree showing how the globin protein family arose, starting from the most primitive oxygen-binding proteins, leghemoglobins, in plants.
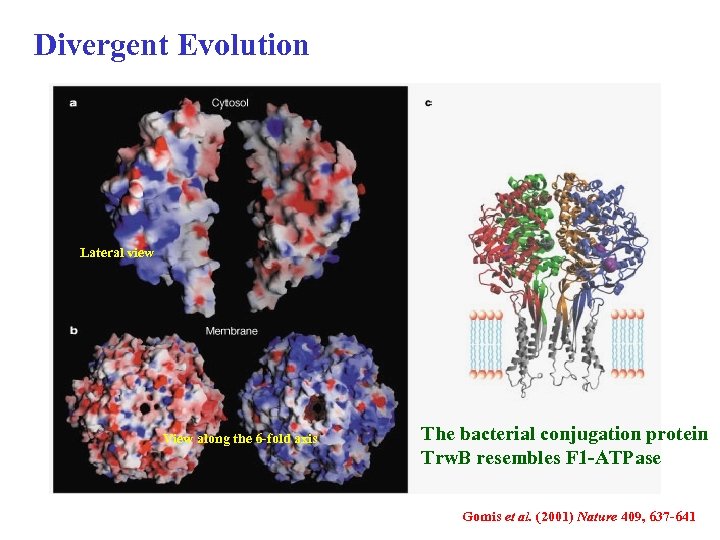 Divergent Evolution Lateral view View along the 6 -fold axis The bacterial conjugation protein Trw. B resembles F 1 -ATPase Gomis et al. (2001) Nature 409, 637 -641
Divergent Evolution Lateral view View along the 6 -fold axis The bacterial conjugation protein Trw. B resembles F 1 -ATPase Gomis et al. (2001) Nature 409, 637 -641
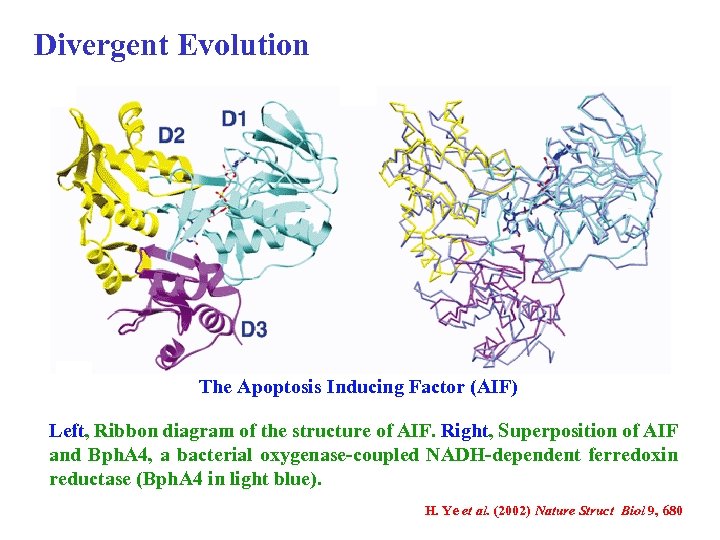 Divergent Evolution The Apoptosis Inducing Factor (AIF) Left, Ribbon diagram of the structure of AIF. Right, Superposition of AIF and Bph. A 4, a bacterial oxygenase-coupled NADH-dependent ferredoxin reductase (Bph. A 4 in light blue). H. Ye et al. (2002) Nature Struct Biol 9, 680
Divergent Evolution The Apoptosis Inducing Factor (AIF) Left, Ribbon diagram of the structure of AIF. Right, Superposition of AIF and Bph. A 4, a bacterial oxygenase-coupled NADH-dependent ferredoxin reductase (Bph. A 4 in light blue). H. Ye et al. (2002) Nature Struct Biol 9, 680
 Apoptosis (or programmed cell death, PCD) is a highly organized multistep process, with the induction of mitochondrial membrane permeabilization as a decisive event in the commitment to cell death. The execution of apoptosis comprises both caspase-dependent and caspase-independent processes. The Apoptosis Inducing Factor (AIF), a resident protein of the intermitochondrial space, has been implicated as a crucial early effector of caspase-independent apoptosis, acting before or in parallel with the onset of caspase-dependent processes. The ectopic presence of AIF in the extra-mitochondrial compartment suffices to kill cells. H. Ye et al. (2002) Nature Struct Biol 9, 680
Apoptosis (or programmed cell death, PCD) is a highly organized multistep process, with the induction of mitochondrial membrane permeabilization as a decisive event in the commitment to cell death. The execution of apoptosis comprises both caspase-dependent and caspase-independent processes. The Apoptosis Inducing Factor (AIF), a resident protein of the intermitochondrial space, has been implicated as a crucial early effector of caspase-independent apoptosis, acting before or in parallel with the onset of caspase-dependent processes. The ectopic presence of AIF in the extra-mitochondrial compartment suffices to kill cells. H. Ye et al. (2002) Nature Struct Biol 9, 680
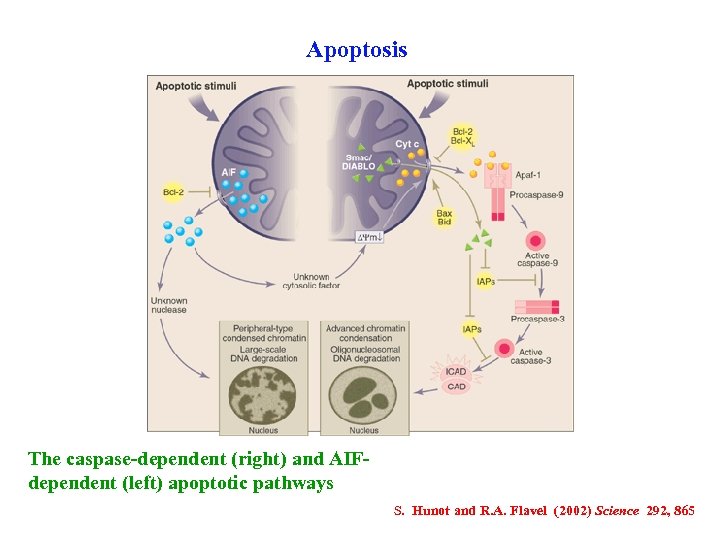 Apoptosis The caspase-dependent (right) and AIFdependent (left) apoptotic pathways S. Hunot and R. A. Flavel (2002) Science 292, 865
Apoptosis The caspase-dependent (right) and AIFdependent (left) apoptotic pathways S. Hunot and R. A. Flavel (2002) Science 292, 865
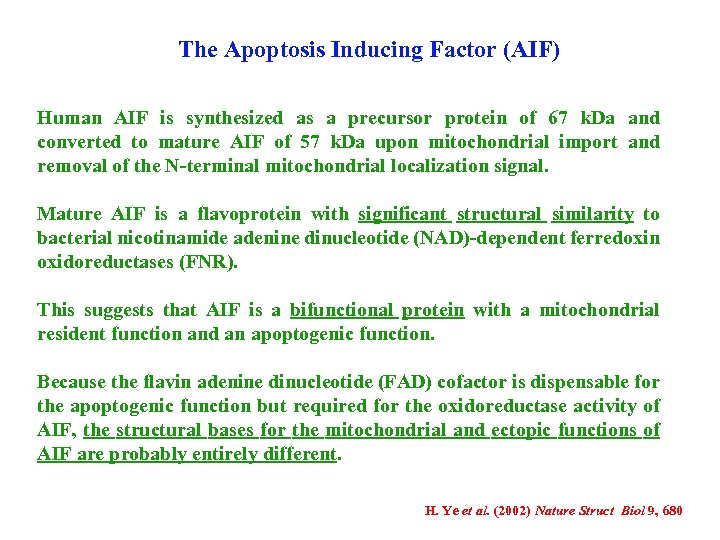 The Apoptosis Inducing Factor (AIF) Human AIF is synthesized as a precursor protein of 67 k. Da and converted to mature AIF of 57 k. Da upon mitochondrial import and removal of the N-terminal mitochondrial localization signal. Mature AIF is a flavoprotein with significant structural similarity to bacterial nicotinamide adenine dinucleotide (NAD)-dependent ferredoxin oxidoreductases (FNR). This suggests that AIF is a bifunctional protein with a mitochondrial resident function and an apoptogenic function. Because the flavin adenine dinucleotide (FAD) cofactor is dispensable for the apoptogenic function but required for the oxidoreductase activity of AIF, the structural bases for the mitochondrial and ectopic functions of AIF are probably entirely different. H. Ye et al. (2002) Nature Struct Biol 9, 680
The Apoptosis Inducing Factor (AIF) Human AIF is synthesized as a precursor protein of 67 k. Da and converted to mature AIF of 57 k. Da upon mitochondrial import and removal of the N-terminal mitochondrial localization signal. Mature AIF is a flavoprotein with significant structural similarity to bacterial nicotinamide adenine dinucleotide (NAD)-dependent ferredoxin oxidoreductases (FNR). This suggests that AIF is a bifunctional protein with a mitochondrial resident function and an apoptogenic function. Because the flavin adenine dinucleotide (FAD) cofactor is dispensable for the apoptogenic function but required for the oxidoreductase activity of AIF, the structural bases for the mitochondrial and ectopic functions of AIF are probably entirely different. H. Ye et al. (2002) Nature Struct Biol 9, 680
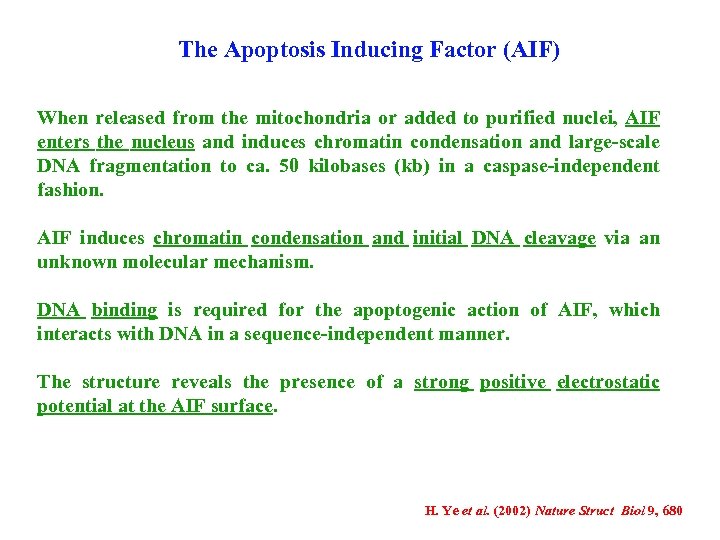 The Apoptosis Inducing Factor (AIF) When released from the mitochondria or added to purified nuclei, AIF enters the nucleus and induces chromatin condensation and large-scale DNA fragmentation to ca. 50 kilobases (kb) in a caspase-independent fashion. AIF induces chromatin condensation and initial DNA cleavage via an unknown molecular mechanism. DNA binding is required for the apoptogenic action of AIF, which interacts with DNA in a sequence-independent manner. The structure reveals the presence of a strong positive electrostatic potential at the AIF surface. H. Ye et al. (2002) Nature Struct Biol 9, 680
The Apoptosis Inducing Factor (AIF) When released from the mitochondria or added to purified nuclei, AIF enters the nucleus and induces chromatin condensation and large-scale DNA fragmentation to ca. 50 kilobases (kb) in a caspase-independent fashion. AIF induces chromatin condensation and initial DNA cleavage via an unknown molecular mechanism. DNA binding is required for the apoptogenic action of AIF, which interacts with DNA in a sequence-independent manner. The structure reveals the presence of a strong positive electrostatic potential at the AIF surface. H. Ye et al. (2002) Nature Struct Biol 9, 680
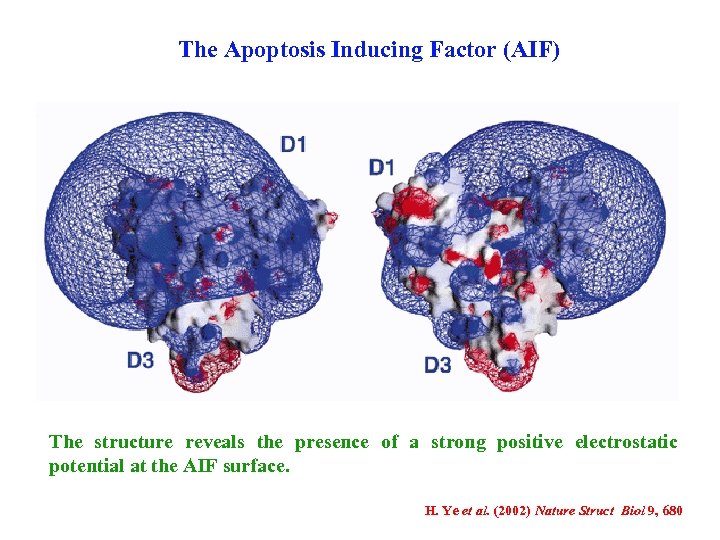 The Apoptosis Inducing Factor (AIF) The structure reveals the presence of a strong positive electrostatic potential at the AIF surface. H. Ye et al. (2002) Nature Struct Biol 9, 680
The Apoptosis Inducing Factor (AIF) The structure reveals the presence of a strong positive electrostatic potential at the AIF surface. H. Ye et al. (2002) Nature Struct Biol 9, 680
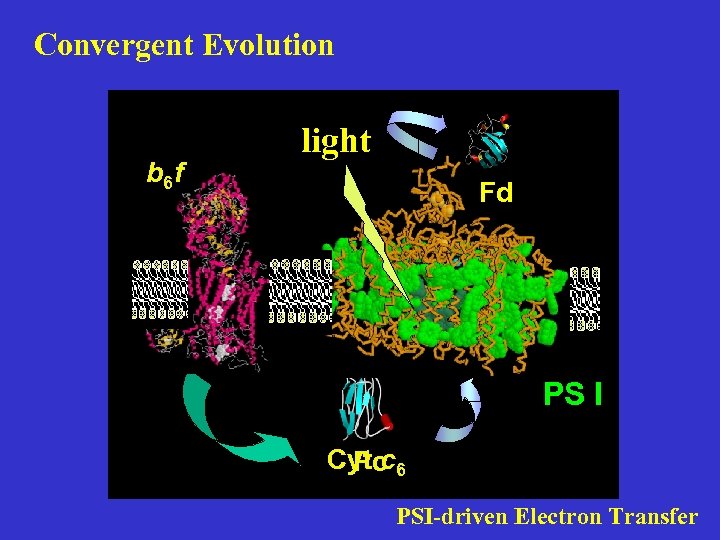 Convergent Evolution b 6 f light Fd PS I Cyt c 6 Pc PSI-driven Electron Transfer
Convergent Evolution b 6 f light Fd PS I Cyt c 6 Pc PSI-driven Electron Transfer
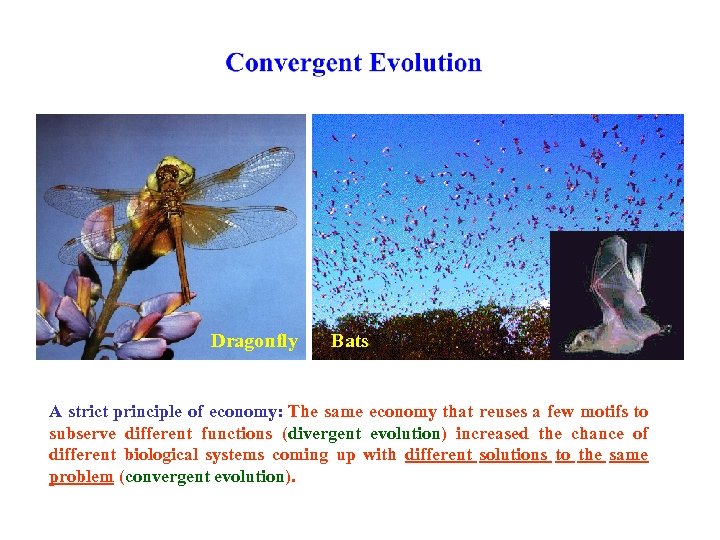 Dragonfly Bats A strict principle of economy: The same economy that reuses a few motifs to subserve different functions (divergent evolution) increased the chance of different biological systems coming up with different solutions to the same problem (convergent evolution).
Dragonfly Bats A strict principle of economy: The same economy that reuses a few motifs to subserve different functions (divergent evolution) increased the chance of different biological systems coming up with different solutions to the same problem (convergent evolution).
 The Three Solutions to Vertebrate Flight Pterosauria (pterosaurs) Aves (birds) Chiroptera (bats)
The Three Solutions to Vertebrate Flight Pterosauria (pterosaurs) Aves (birds) Chiroptera (bats)
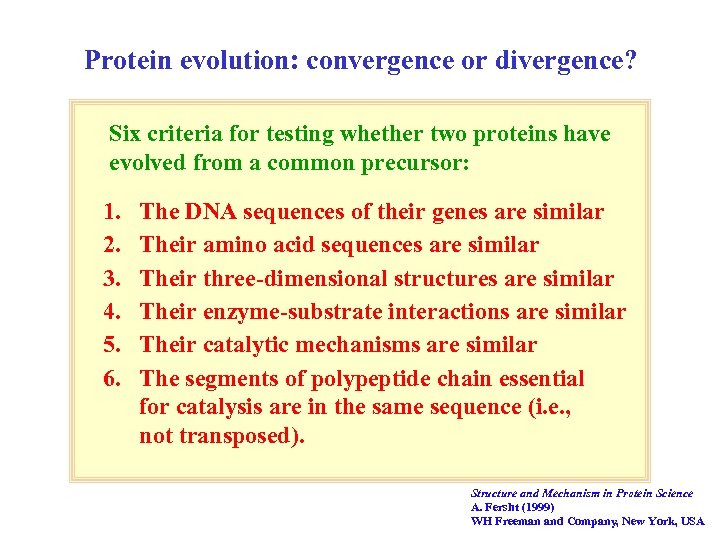 Protein evolution: convergence or divergence? Six criteria for testing whether two proteins have evolved from a common precursor: 1. 2. 3. 4. 5. 6. The DNA sequences of their genes are similar Their amino acid sequences are similar Their three-dimensional structures are similar Their enzyme-substrate interactions are similar Their catalytic mechanisms are similar The segments of polypeptide chain essential for catalysis are in the same sequence (i. e. , not transposed). Structure and Mechanism in Protein Science A. Fersht (1999) WH Freeman and Company, New York, USA
Protein evolution: convergence or divergence? Six criteria for testing whether two proteins have evolved from a common precursor: 1. 2. 3. 4. 5. 6. The DNA sequences of their genes are similar Their amino acid sequences are similar Their three-dimensional structures are similar Their enzyme-substrate interactions are similar Their catalytic mechanisms are similar The segments of polypeptide chain essential for catalysis are in the same sequence (i. e. , not transposed). Structure and Mechanism in Protein Science A. Fersht (1999) WH Freeman and Company, New York, USA
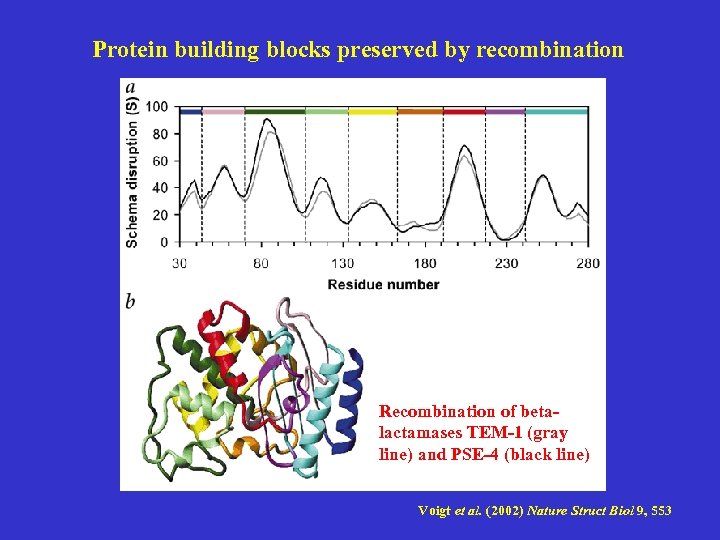 Protein building blocks preserved by recombination Recombination of betalactamases TEM-1 (gray line) and PSE-4 (black line) Voigt et al. (2002) Nature Struct Biol 9, 553
Protein building blocks preserved by recombination Recombination of betalactamases TEM-1 (gray line) and PSE-4 (black line) Voigt et al. (2002) Nature Struct Biol 9, 553
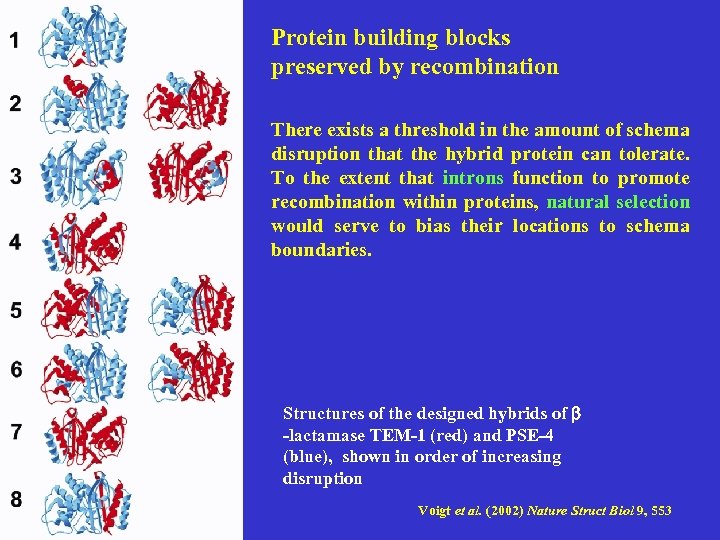 Protein building blocks preserved by recombination There exists a threshold in the amount of schema disruption that the hybrid protein can tolerate. To the extent that introns function to promote recombination within proteins, natural selection would serve to bias their locations to schema boundaries. Structures of the designed hybrids of b -lactamase TEM-1 (red) and PSE-4 (blue), shown in order of increasing disruption Voigt et al. (2002) Nature Struct Biol 9, 553
Protein building blocks preserved by recombination There exists a threshold in the amount of schema disruption that the hybrid protein can tolerate. To the extent that introns function to promote recombination within proteins, natural selection would serve to bias their locations to schema boundaries. Structures of the designed hybrids of b -lactamase TEM-1 (red) and PSE-4 (blue), shown in order of increasing disruption Voigt et al. (2002) Nature Struct Biol 9, 553
 Dinámica Molecular de las Estructuras Proteicas La biología es inconcebible sin movimiento
Dinámica Molecular de las Estructuras Proteicas La biología es inconcebible sin movimiento
 ATP synthase Animation of the complete mechanism Lecture 10, ATP synthase http: //www. life. uiuc. edu/crofts/bioph 354/lect 10. html
ATP synthase Animation of the complete mechanism Lecture 10, ATP synthase http: //www. life. uiuc. edu/crofts/bioph 354/lect 10. html
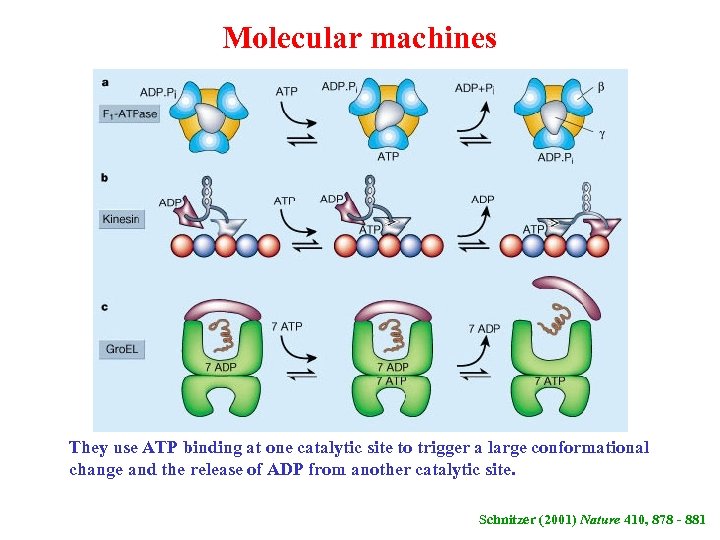 Molecular machines They use ATP binding at one catalytic site to trigger a large conformational change and the release of ADP from another catalytic site. Schnitzer (2001) Nature 410, 878 - 881
Molecular machines They use ATP binding at one catalytic site to trigger a large conformational change and the release of ADP from another catalytic site. Schnitzer (2001) Nature 410, 878 - 881
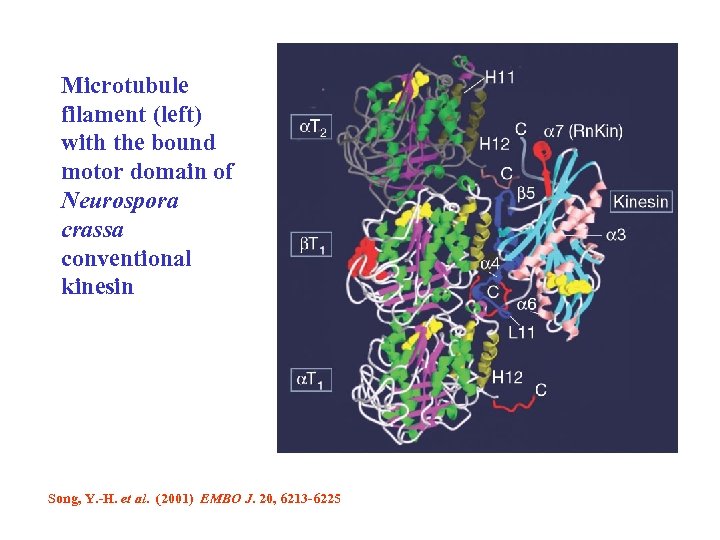 Microtubule filament (left) with the bound motor domain of Neurospora crassa conventional kinesin Song, Y. -H. et al. (2001) EMBO J. 20, 6213 -6225
Microtubule filament (left) with the bound motor domain of Neurospora crassa conventional kinesin Song, Y. -H. et al. (2001) EMBO J. 20, 6213 -6225
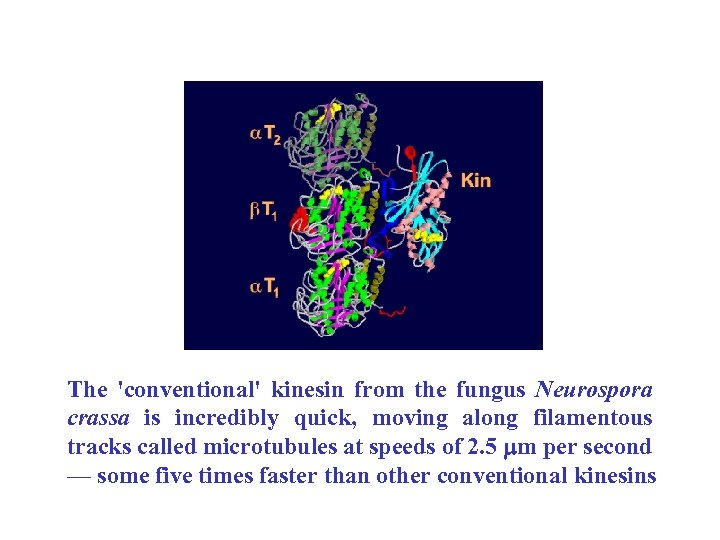 The 'conventional' kinesin from the fungus Neurospora crassa is incredibly quick, moving along filamentous tracks called microtubules at speeds of 2. 5 mm per second — some five times faster than other conventional kinesins
The 'conventional' kinesin from the fungus Neurospora crassa is incredibly quick, moving along filamentous tracks called microtubules at speeds of 2. 5 mm per second — some five times faster than other conventional kinesins
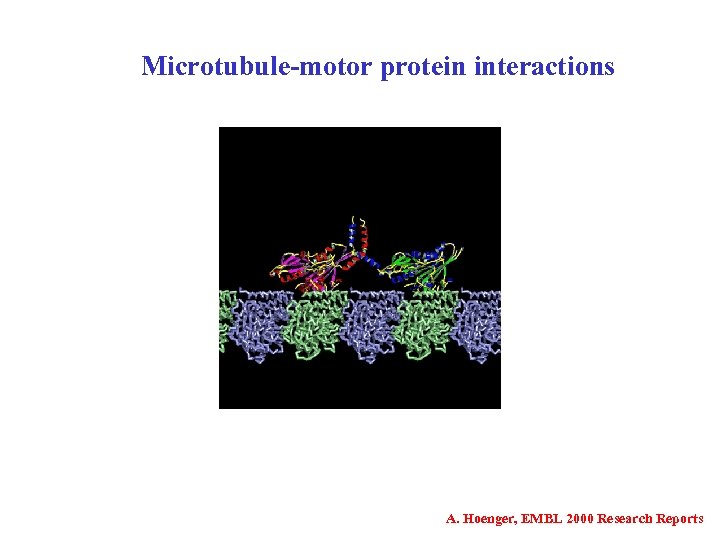 Microtubule-motor protein interactions A. Hoenger, EMBL 2000 Research Reports
Microtubule-motor protein interactions A. Hoenger, EMBL 2000 Research Reports
 Myosin, a cellular motor protein It takes 37 -nm steps by placing one “foot” after the other Cover - Science 27 June 2003
Myosin, a cellular motor protein It takes 37 -nm steps by placing one “foot” after the other Cover - Science 27 June 2003
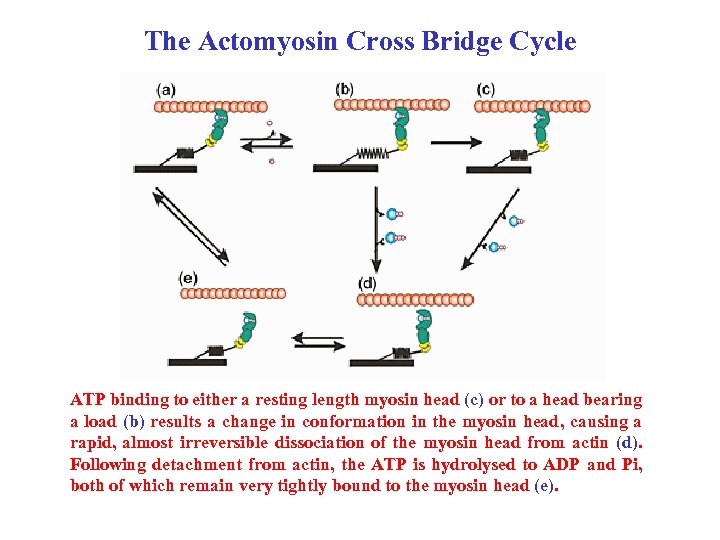 The Actomyosin Cross Bridge Cycle ATP binding to either a resting length myosin head (c) or to a head bearing a load (b) results a change in conformation in the myosin head, causing a rapid, almost irreversible dissociation of the myosin head from actin (d). Following detachment from actin, the ATP is hydrolysed to ADP and Pi, both of which remain very tightly bound to the myosin head (e).
The Actomyosin Cross Bridge Cycle ATP binding to either a resting length myosin head (c) or to a head bearing a load (b) results a change in conformation in the myosin head, causing a rapid, almost irreversible dissociation of the myosin head from actin (d). Following detachment from actin, the ATP is hydrolysed to ADP and Pi, both of which remain very tightly bound to the myosin head (e).
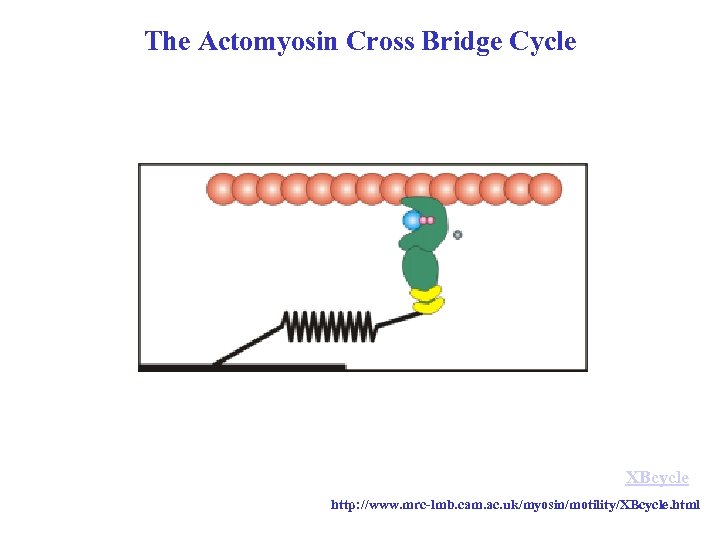 The Actomyosin Cross Bridge Cycle XBcycle http: //www. mrc-lmb. cam. ac. uk/myosin/motility/XBcycle. html
The Actomyosin Cross Bridge Cycle XBcycle http: //www. mrc-lmb. cam. ac. uk/myosin/motility/XBcycle. html
 Myosin http: //molmovdb. mbb. yale. edu/Mol. Mov. DB/cgi-bin/morph. cgi? ID=12221 -32592
Myosin http: //molmovdb. mbb. yale. edu/Mol. Mov. DB/cgi-bin/morph. cgi? ID=12221 -32592
 Protein mobility and enzyme mechanism A major question is: Are the modes of mobility observed in enzymes just incidental (. . . ) or are they essential for catalysis? Flexibility could be useful in aiding the access of ligands to active sites. Structure and Mechanism in Protein Science A. Fersht (1999) WH Freeman and Company, New York, USA
Protein mobility and enzyme mechanism A major question is: Are the modes of mobility observed in enzymes just incidental (. . . ) or are they essential for catalysis? Flexibility could be useful in aiding the access of ligands to active sites. Structure and Mechanism in Protein Science A. Fersht (1999) WH Freeman and Company, New York, USA
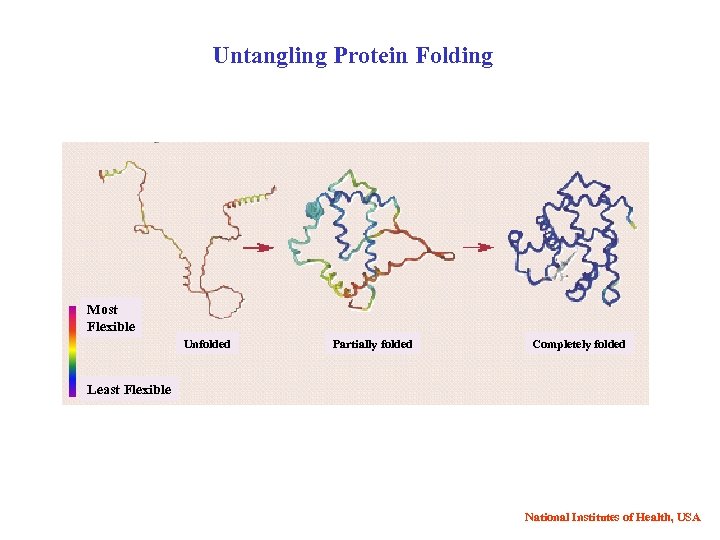 Untangling Protein Folding Most Flexible Unfolded Partially folded Completely folded Least Flexible National Institutes of Health, USA
Untangling Protein Folding Most Flexible Unfolded Partially folded Completely folded Least Flexible National Institutes of Health, USA
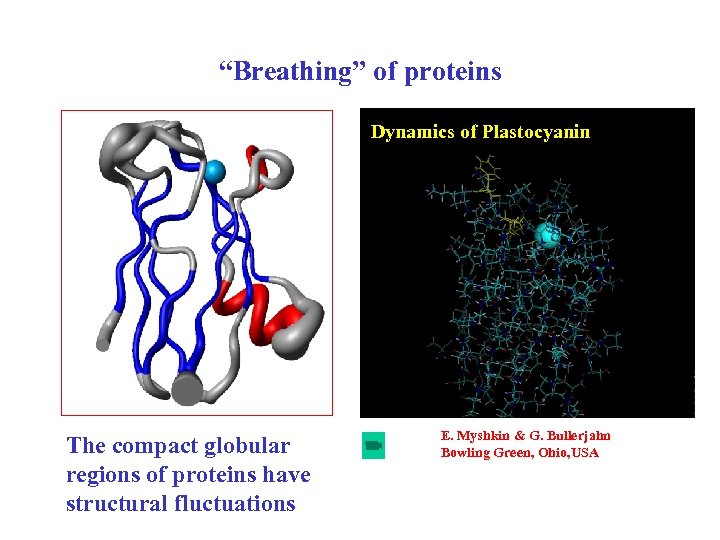 “Breathing” of proteins Dynamics of Plastocyanin The compact globular regions of proteins have structural fluctuations E. Myshkin & G. Bullerjahn Bowling Green, Ohio, USA
“Breathing” of proteins Dynamics of Plastocyanin The compact globular regions of proteins have structural fluctuations E. Myshkin & G. Bullerjahn Bowling Green, Ohio, USA
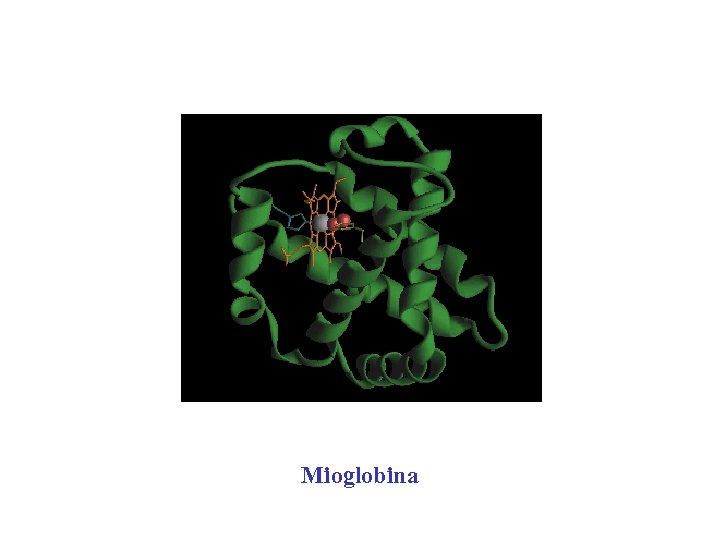 Mioglobina
Mioglobina
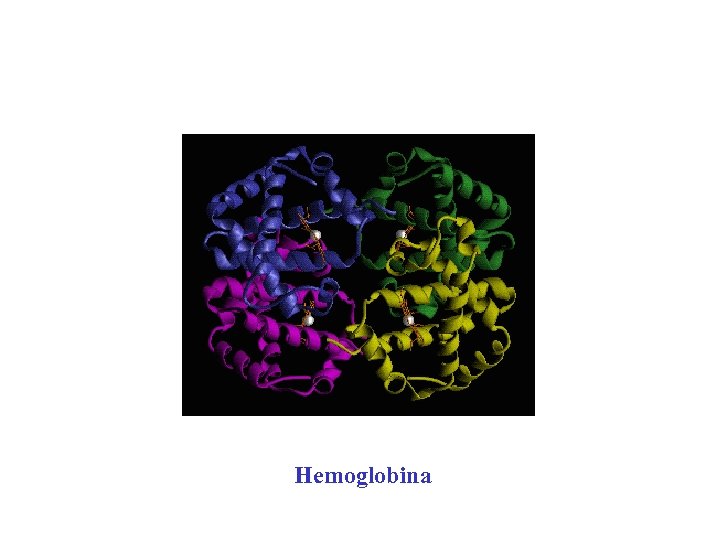 Hemoglobina
Hemoglobina
 Domain movements: segment flexibility All domains movements may be constructucted from a combination of hinge and shear motions: Hinge motions, in which two elements of structure open and close as if connected by a hinge. Shear motions, in which one element of structure slides relative to the other. Structure and Mechanism in Protein Science A. Fersht (1999) WH Freeman and Company, New York, USA
Domain movements: segment flexibility All domains movements may be constructucted from a combination of hinge and shear motions: Hinge motions, in which two elements of structure open and close as if connected by a hinge. Shear motions, in which one element of structure slides relative to the other. Structure and Mechanism in Protein Science A. Fersht (1999) WH Freeman and Company, New York, USA
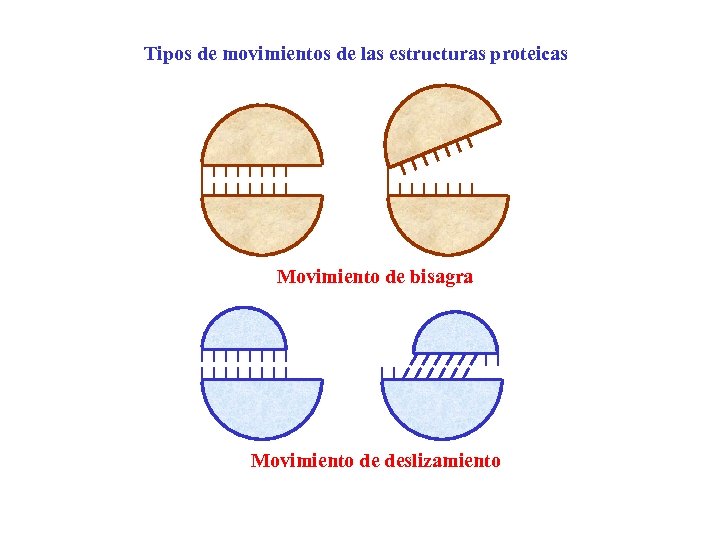 Tipos de movimientos de las estructuras proteicas Movimiento de bisagra Movimiento de deslizamiento
Tipos de movimientos de las estructuras proteicas Movimiento de bisagra Movimiento de deslizamiento
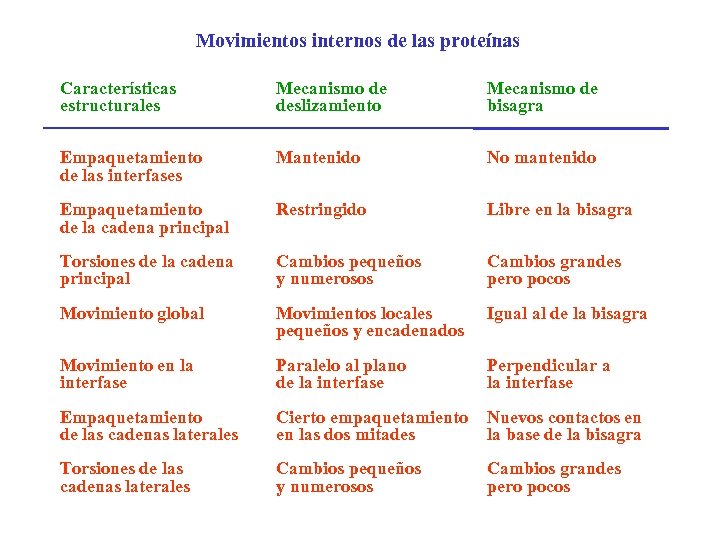 Movimientos internos de las proteínas Características estructurales Mecanismo de deslizamiento Mecanismo de bisagra Empaquetamiento de las interfases Mantenido No mantenido Empaquetamiento de la cadena principal Restringido Libre en la bisagra Torsiones de la cadena principal Cambios pequeños y numerosos Cambios grandes pero pocos Movimiento global Movimientos locales pequeños y encadenados Igual al de la bisagra Movimiento en la interfase Paralelo al plano de la interfase Perpendicular a la interfase Empaquetamiento de las cadenas laterales Cierto empaquetamiento en las dos mitades Nuevos contactos en la base de la bisagra Torsiones de las cadenas laterales Cambios pequeños y numerosos Cambios grandes pero pocos
Movimientos internos de las proteínas Características estructurales Mecanismo de deslizamiento Mecanismo de bisagra Empaquetamiento de las interfases Mantenido No mantenido Empaquetamiento de la cadena principal Restringido Libre en la bisagra Torsiones de la cadena principal Cambios pequeños y numerosos Cambios grandes pero pocos Movimiento global Movimientos locales pequeños y encadenados Igual al de la bisagra Movimiento en la interfase Paralelo al plano de la interfase Perpendicular a la interfase Empaquetamiento de las cadenas laterales Cierto empaquetamiento en las dos mitades Nuevos contactos en la base de la bisagra Torsiones de las cadenas laterales Cambios pequeños y numerosos Cambios grandes pero pocos
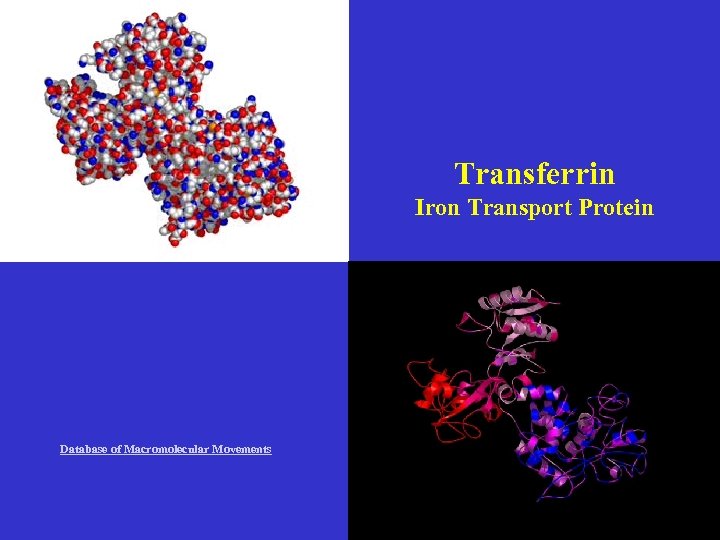 Transferrin Iron Transport Protein Database of Macromolecular Movements
Transferrin Iron Transport Protein Database of Macromolecular Movements
 Database of Macromolecular Movements Ca - ATPase
Database of Macromolecular Movements Ca - ATPase

 I. Grupos R no polares (a) L-Glicina (Gly) : H L-Alanina (Ala) : CH 3 L-Valina (Val) : CH 3 CH CH 3 L-Leucina (Leu) : CH 3 C H - CH 2 C H 3 L-Isoleucina (Ileu) : CH 3 - CH 2 CH C H 3 L-Metionina (Met) : CH 3 - S - CH 2 -
I. Grupos R no polares (a) L-Glicina (Gly) : H L-Alanina (Ala) : CH 3 L-Valina (Val) : CH 3 CH CH 3 L-Leucina (Leu) : CH 3 C H - CH 2 C H 3 L-Isoleucina (Ileu) : CH 3 - CH 2 CH C H 3 L-Metionina (Met) : CH 3 - S - CH 2 -
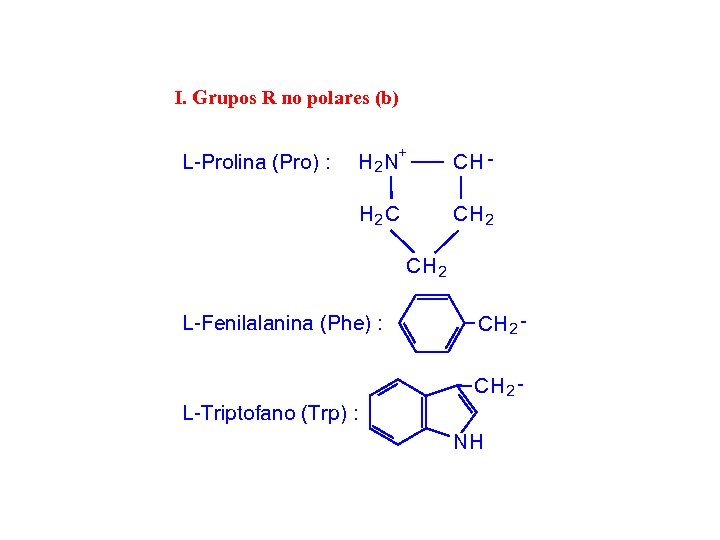 I. Grupos R no polares (b) H 2 N+ CH - H 2 C L-Prolina (Pro) : CH 2 C H 2 L-Fenilalanina (Phe) : CH 2 - L-Triptofano (Trp) : NH
I. Grupos R no polares (b) H 2 N+ CH - H 2 C L-Prolina (Pro) : CH 2 C H 2 L-Fenilalanina (Phe) : CH 2 - L-Triptofano (Trp) : NH
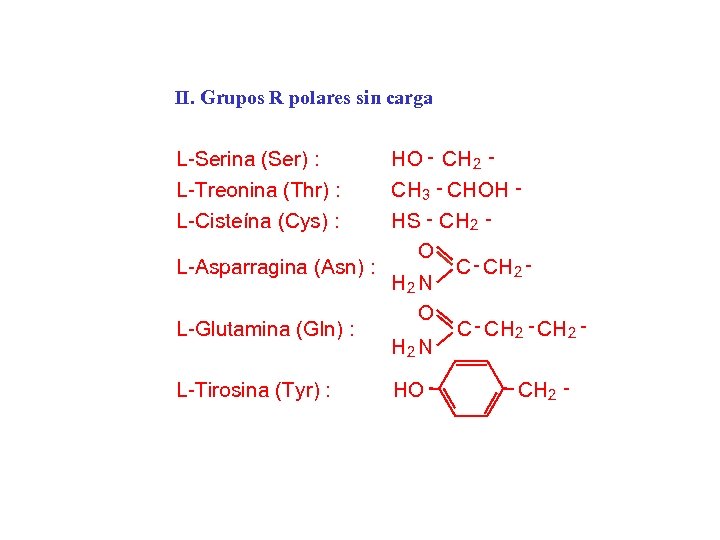 II. Grupos R polares sin carga HO - CH 2 CH 3 - CH OH H S - CH 2 O L-Asparragina (Asn) : C - CH 2 N O L-Glutamina (Gln) : C - C H 2 - CH 2 N L-Serina (Ser) : L-Treonina (Thr) : L-Cisteína (Cys) : L-Tirosina (Tyr) : HO CH 2 -
II. Grupos R polares sin carga HO - CH 2 CH 3 - CH OH H S - CH 2 O L-Asparragina (Asn) : C - CH 2 N O L-Glutamina (Gln) : C - C H 2 - CH 2 N L-Serina (Ser) : L-Treonina (Thr) : L-Cisteína (Cys) : L-Tirosina (Tyr) : HO CH 2 -
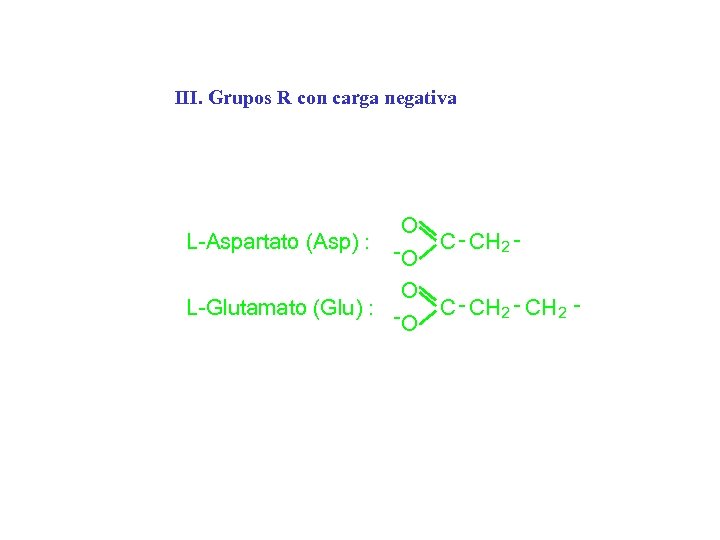 III. Grupos R con carga negativa O L-Aspartato (Asp) : C - C H 2 O O L-Glutamato (Glu) : C - C H 2 - CH 2 O
III. Grupos R con carga negativa O L-Aspartato (Asp) : C - C H 2 O O L-Glutamato (Glu) : C - C H 2 - CH 2 O
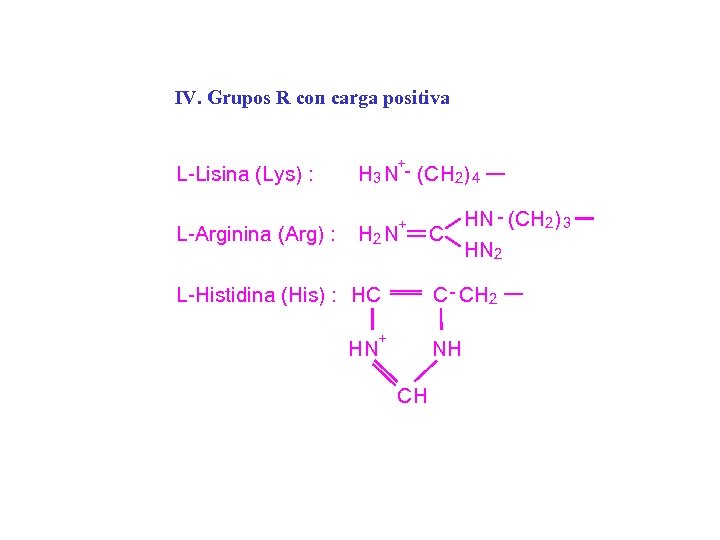 IV. Grupos R con carga positiva L-Lisina (Lys) : L-Arginina (Arg) : + H 3 N - (CH 2) 4 + H 2 N HN - (CH 2) 3 C HN 2 C - CH 2 L-Histidina (His) : HC + HN NH CH
IV. Grupos R con carga positiva L-Lisina (Lys) : L-Arginina (Arg) : + H 3 N - (CH 2) 4 + H 2 N HN - (CH 2) 3 C HN 2 C - CH 2 L-Histidina (His) : HC + HN NH CH