0b5d7092455af5a7b4dbb266cdee077c.ppt
- Количество слайдов: 33
 Herpesviral Protein Networks and Their Interaction with the Human Proteome Presentation by: Kyle Borge, David Byon, & Jim Hall reconstruction of a Herpes Virus capsid Presentation by: Kyle Borge, David Byon, & Jim Hall
Herpesviral Protein Networks and Their Interaction with the Human Proteome Presentation by: Kyle Borge, David Byon, & Jim Hall reconstruction of a Herpes Virus capsid Presentation by: Kyle Borge, David Byon, & Jim Hall
 Introduction to the Herpesvirus • Large double-stranded DNA genomes • Eight different strains • Causes diseases ranging from cold sores to shingles • Vaccine available for Varicella-Zoster Virus (VZV) • Little known about protein interactions
Introduction to the Herpesvirus • Large double-stranded DNA genomes • Eight different strains • Causes diseases ranging from cold sores to shingles • Vaccine available for Varicella-Zoster Virus (VZV) • Little known about protein interactions
 Types of Herpesviruses Investigated • Kaposi’s Sarcoma-associated Herpesvirus (KSHV) – – – In the gamma (γ) herpes virus phylogenetic class Causes cancerous tumors Mostly associated with HIV patients Sequenced in 1996 Genome is roughly 165 kbs 89 open reading frames (ORFs) • 113 ORFs used in experiment (included 15 cytoplasmic and 5 external domains derived from transmembrane proteins) • Varicella-Zoster Virus (VZV) , in the alpha (α) herpesvirus phylogenetic class – Causes chicken pox in children and shingles in adults – Sequenced in 1986 – Genome is roughly 125 kbs – 69 open reading frames (ORFs) • 96 ORFs used in experiment (Included 13 cytoplasmic and 10 external domains derived from transmembrane proteins)
Types of Herpesviruses Investigated • Kaposi’s Sarcoma-associated Herpesvirus (KSHV) – – – In the gamma (γ) herpes virus phylogenetic class Causes cancerous tumors Mostly associated with HIV patients Sequenced in 1996 Genome is roughly 165 kbs 89 open reading frames (ORFs) • 113 ORFs used in experiment (included 15 cytoplasmic and 5 external domains derived from transmembrane proteins) • Varicella-Zoster Virus (VZV) , in the alpha (α) herpesvirus phylogenetic class – Causes chicken pox in children and shingles in adults – Sequenced in 1986 – Genome is roughly 125 kbs – 69 open reading frames (ORFs) • 96 ORFs used in experiment (Included 13 cytoplasmic and 10 external domains derived from transmembrane proteins)
 Methods of Investigating Protein Interactions (PPI) • Many Methods • The Y 2 H technique is one of the top techniques for detecting protein-protein interactions • This article used Y 2 H to investigate protein interactions
Methods of Investigating Protein Interactions (PPI) • Many Methods • The Y 2 H technique is one of the top techniques for detecting protein-protein interactions • This article used Y 2 H to investigate protein interactions
 Y 2 H Advantages • • Relatively simple (automated) • Quick • Inexpensive http: //www. dnatube. com/video/993/Plasmid • Only Cloningneed the sequenced genome (or sequence of interest) • Scalable, its possible to screen for interactions among many proteins creating a more high-throughput screen (ex. viral genome) • Protein/polypeptides can be from various sources; eukaryotes, prokaryotes, viruses and even artificial sequences…allows the comparison of interactomes w/in and between different species…in this paper, eukaryote (human) interactome vs. viral interactome
Y 2 H Advantages • • Relatively simple (automated) • Quick • Inexpensive http: //www. dnatube. com/video/993/Plasmid • Only Cloningneed the sequenced genome (or sequence of interest) • Scalable, its possible to screen for interactions among many proteins creating a more high-throughput screen (ex. viral genome) • Protein/polypeptides can be from various sources; eukaryotes, prokaryotes, viruses and even artificial sequences…allows the comparison of interactomes w/in and between different species…in this paper, eukaryote (human) interactome vs. viral interactome
 Y 2 H Limitations • The Y 2 H system cant analyze some classes of proteins • Transmembrane proteins, specifically their hydrophobic regions which may prevent the protein from reaching the nucleus • http: //www. dnatube. com/video/993/Plasmid • Transcriptional activators; may activate transcription w/out any Cloning interaction • False-negatives • Y 2 H screen fails to detect a protein-protein interactions • False-positives • Y 2 H screen produces a positive result (characterized by reporter gene activity) where no protein-protein interaction took place Ex. bait proteins activate, transcribing the reporter gene, w/out the binding of the AD (bait proteins act as transcriptional activators)
Y 2 H Limitations • The Y 2 H system cant analyze some classes of proteins • Transmembrane proteins, specifically their hydrophobic regions which may prevent the protein from reaching the nucleus • http: //www. dnatube. com/video/993/Plasmid • Transcriptional activators; may activate transcription w/out any Cloning interaction • False-negatives • Y 2 H screen fails to detect a protein-protein interactions • False-positives • Y 2 H screen produces a positive result (characterized by reporter gene activity) where no protein-protein interaction took place Ex. bait proteins activate, transcribing the reporter gene, w/out the binding of the AD (bait proteins act as transcriptional activators)
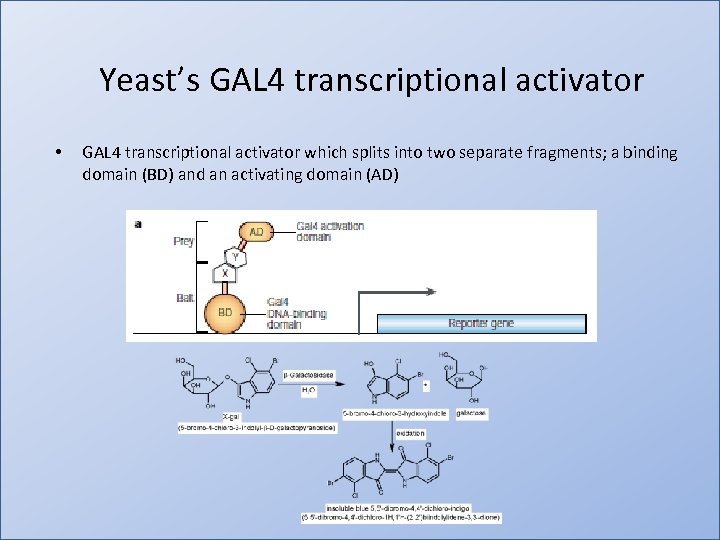 Yeast’s GAL 4 transcriptional activator • GAL 4 transcriptional activator which splits into two separate fragments; a binding domain (BD) and an activating domain (AD)
Yeast’s GAL 4 transcriptional activator • GAL 4 transcriptional activator which splits into two separate fragments; a binding domain (BD) and an activating domain (AD)
 Y 2 H Method • ORFs selected from published sequences • Amplified by nested PCR – Made primer sets of ends of ORFs • Y 2 H bait and prey vectors • Vectors transformed into Y 187 and AH 109 haploid yeast cells creating pools; a bait pool and a prey pool • Bait and prey mated in quadruplicates • Positive diploid yeasts are selected
Y 2 H Method • ORFs selected from published sequences • Amplified by nested PCR – Made primer sets of ends of ORFs • Y 2 H bait and prey vectors • Vectors transformed into Y 187 and AH 109 haploid yeast cells creating pools; a bait pool and a prey pool • Bait and prey mated in quadruplicates • Positive diploid yeasts are selected
 Open Reading Frames (ORFs) • Every ORFs of both KSHV & VZV were cloned & ligated into both a bait and prey GAL 4 vector • Bait – protein of interest – the protein is fused to the yeast Gal 4 DNA-binding domain (DBD) • Prey – a protein/ORF fused to the Gal 4 transcriptional activation domain (AD) – interacting protein • Physical interaction between the bait and prey brings the DNA-BD and an AD of Gal 4 together, thus re-creating a transcriptionally active Gal 4 hybrid • Gal 4 activity can be assayed by the expression of reporter genes and selectable markers
Open Reading Frames (ORFs) • Every ORFs of both KSHV & VZV were cloned & ligated into both a bait and prey GAL 4 vector • Bait – protein of interest – the protein is fused to the yeast Gal 4 DNA-binding domain (DBD) • Prey – a protein/ORF fused to the Gal 4 transcriptional activation domain (AD) – interacting protein • Physical interaction between the bait and prey brings the DNA-BD and an AD of Gal 4 together, thus re-creating a transcriptionally active Gal 4 hybrid • Gal 4 activity can be assayed by the expression of reporter genes and selectable markers
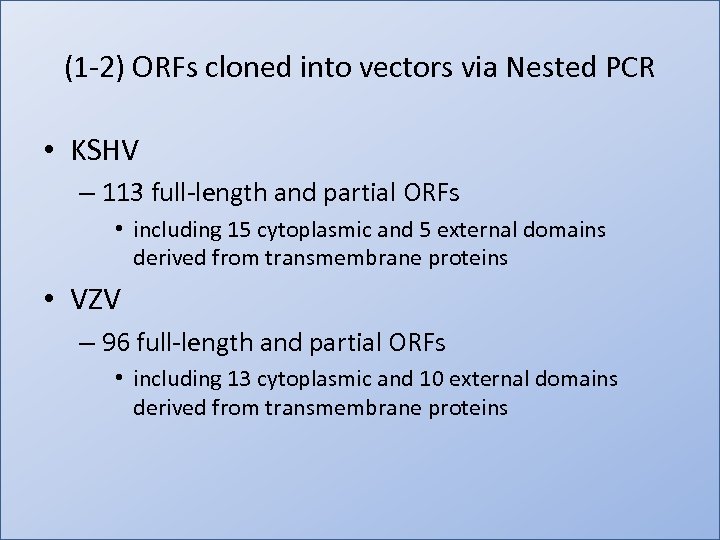 (1 -2) ORFs cloned into vectors via Nested PCR • KSHV – 113 full-length and partial ORFs • including 15 cytoplasmic and 5 external domains derived from transmembrane proteins • VZV – 96 full-length and partial ORFs • including 13 cytoplasmic and 10 external domains derived from transmembrane proteins
(1 -2) ORFs cloned into vectors via Nested PCR • KSHV – 113 full-length and partial ORFs • including 15 cytoplasmic and 5 external domains derived from transmembrane proteins • VZV – 96 full-length and partial ORFs • including 13 cytoplasmic and 10 external domains derived from transmembrane proteins
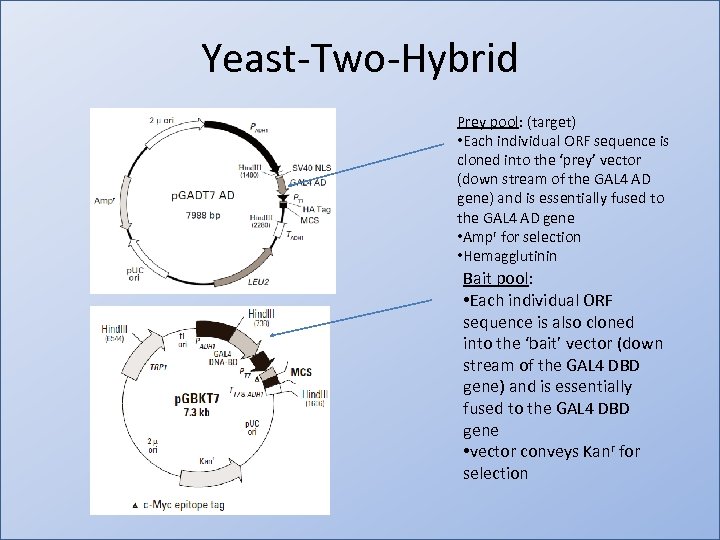 Yeast-Two-Hybrid Prey pool: (target) • Each individual ORF sequence is cloned into the ‘prey’ vector (down stream of the GAL 4 AD gene) and is essentially fused to the GAL 4 AD gene • Ampr for selection • Hemagglutinin Bait pool: • Each individual ORF sequence is also cloned into the ‘bait’ vector (down stream of the GAL 4 DBD gene) and is essentially fused to the GAL 4 DBD gene • vector conveys Kanr for selection
Yeast-Two-Hybrid Prey pool: (target) • Each individual ORF sequence is cloned into the ‘prey’ vector (down stream of the GAL 4 AD gene) and is essentially fused to the GAL 4 AD gene • Ampr for selection • Hemagglutinin Bait pool: • Each individual ORF sequence is also cloned into the ‘bait’ vector (down stream of the GAL 4 DBD gene) and is essentially fused to the GAL 4 DBD gene • vector conveys Kanr for selection
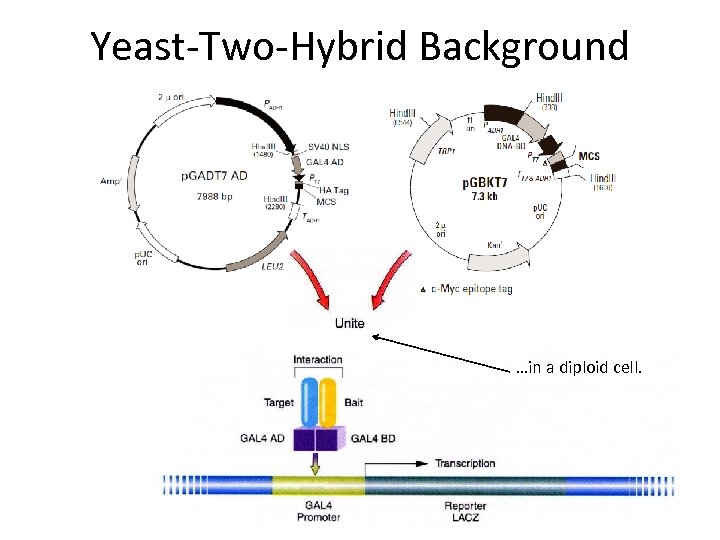 Yeast-Two-Hybrid Background …in a diploid cell.
Yeast-Two-Hybrid Background …in a diploid cell.
 Viral Protein Interactions in KSHV • 12, 000 Viral Protein Interactions tested • Identified 123 nonredundant interacting protein pairs • 118/123 were novel • 7/123 were previously reported • Screen captures 5/7 (71%) of previously reported interactions • 50% of Y 2 H interactions confirmed by coimmunoprecipitation (Co. IP)
Viral Protein Interactions in KSHV • 12, 000 Viral Protein Interactions tested • Identified 123 nonredundant interacting protein pairs • 118/123 were novel • 7/123 were previously reported • Screen captures 5/7 (71%) of previously reported interactions • 50% of Y 2 H interactions confirmed by coimmunoprecipitation (Co. IP)
 Viral Protein Interactions in KSHV
Viral Protein Interactions in KSHV
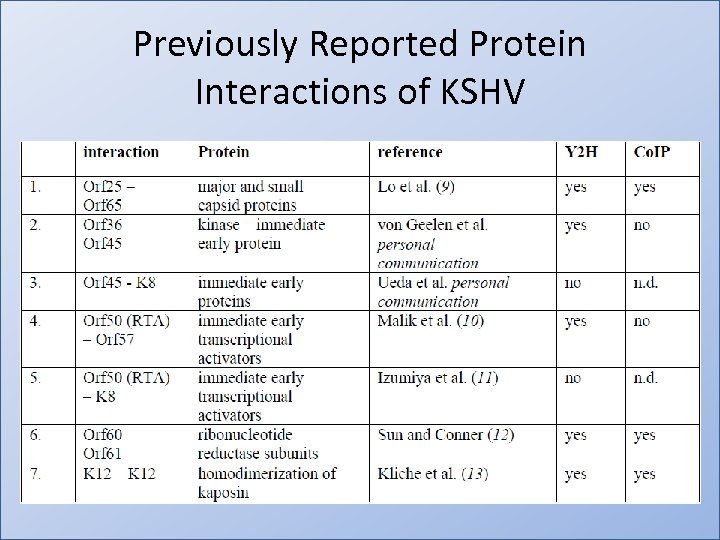 Previously Reported Protein Interactions of KSHV
Previously Reported Protein Interactions of KSHV
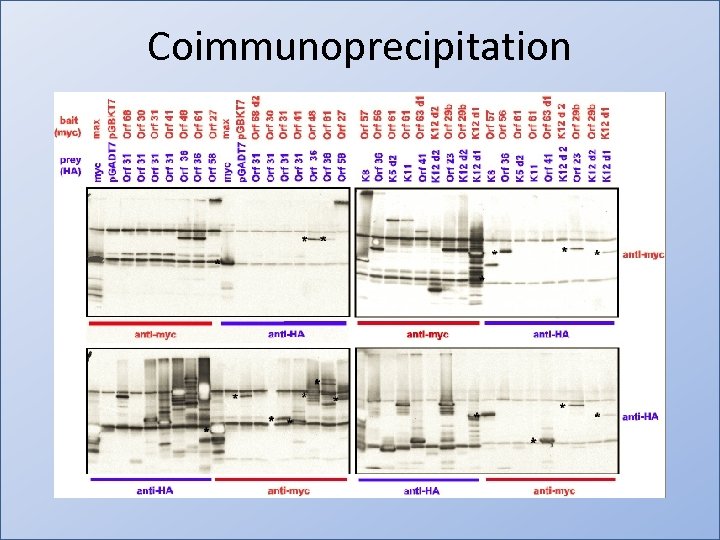 Coimmunoprecipitation
Coimmunoprecipitation
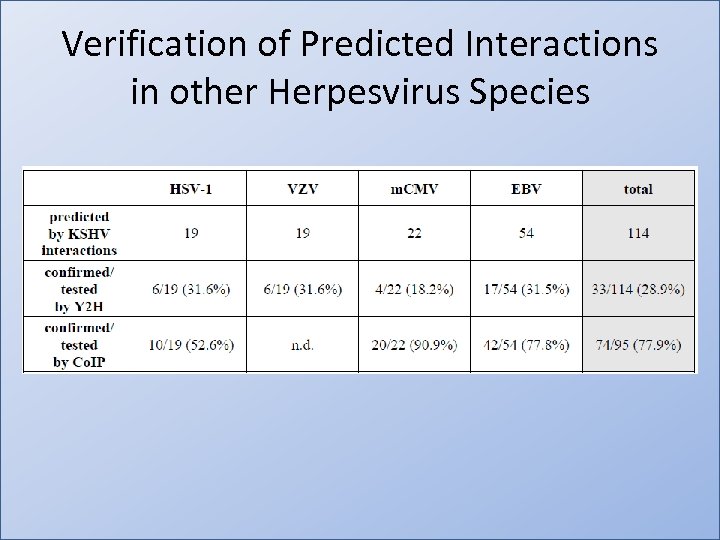 Verification of Predicted Interactions in other Herpesvirus Species
Verification of Predicted Interactions in other Herpesvirus Species
![Correlation Between Viral Protein Interaction and Expression Profile • Average expression correlation [AEC]was calculated Correlation Between Viral Protein Interaction and Expression Profile • Average expression correlation [AEC]was calculated](https://present5.com/presentation/0b5d7092455af5a7b4dbb266cdee077c/image-18.jpg) Correlation Between Viral Protein Interaction and Expression Profile • Average expression correlation [AEC]was calculated • For random pairs of ORFs: 0. 804 • For interacting pairs of ORFs: 0. 839 • Correlation between AEC and clustering coefficient • Used to propose static or dynamic interaction for viral hubs
Correlation Between Viral Protein Interaction and Expression Profile • Average expression correlation [AEC]was calculated • For random pairs of ORFs: 0. 804 • For interacting pairs of ORFs: 0. 839 • Correlation between AEC and clustering coefficient • Used to propose static or dynamic interaction for viral hubs
 Protein Interaction Networks
Protein Interaction Networks
 Network Terminology • • • Node – represents a protein Edge – represents interaction between two nodes Average (node) degree – the average number of neighbors or connections that any given node has Power coefficient (g) – derived from an approximate power law degree distribution plotted on a bilogarithmic scale and fitted by linear regression P value - (significance under linear regression) as fitted by a power-law degree distribution (‘‘scale-free’’ property) Characteristic path length – the distance between two nodes Diameter (d) - describes the interconnectedness of a network; defined as the average length of the shortest paths between any two nodes in the network Clustering coefficient – A value given to depict the number of fold enrichment over comparable random networks (‘‘small-world’’ property) Small world property/network – Any network that has characteristics of a relatively short path and dense cluster (high cluster coefficient)
Network Terminology • • • Node – represents a protein Edge – represents interaction between two nodes Average (node) degree – the average number of neighbors or connections that any given node has Power coefficient (g) – derived from an approximate power law degree distribution plotted on a bilogarithmic scale and fitted by linear regression P value - (significance under linear regression) as fitted by a power-law degree distribution (‘‘scale-free’’ property) Characteristic path length – the distance between two nodes Diameter (d) - describes the interconnectedness of a network; defined as the average length of the shortest paths between any two nodes in the network Clustering coefficient – A value given to depict the number of fold enrichment over comparable random networks (‘‘small-world’’ property) Small world property/network – Any network that has characteristics of a relatively short path and dense cluster (high cluster coefficient)
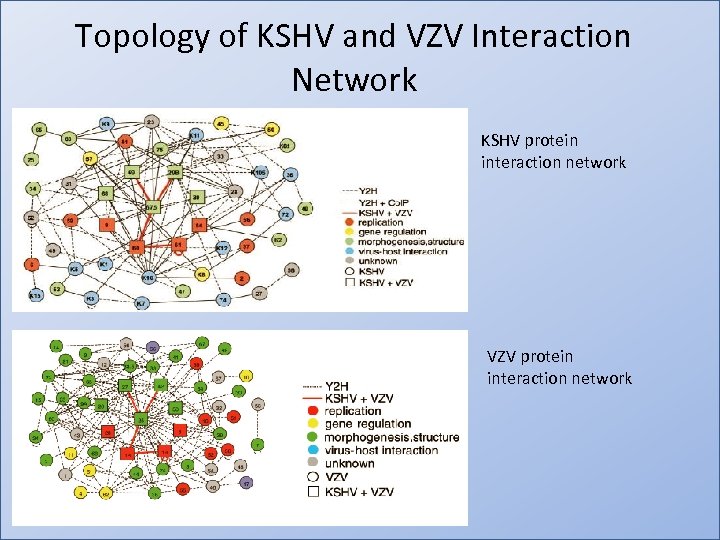 Topology of KSHV and VZV Interaction Network KSHV protein interaction network VZV protein interaction network
Topology of KSHV and VZV Interaction Network KSHV protein interaction network VZV protein interaction network
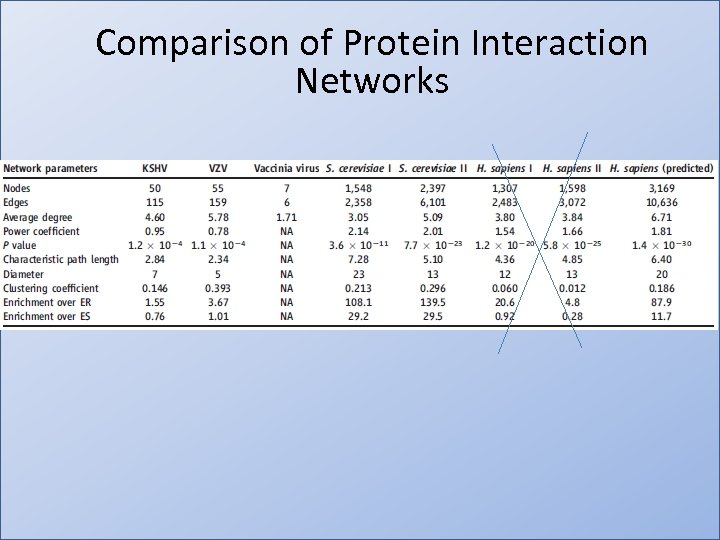 Comparison of Protein Interaction Networks
Comparison of Protein Interaction Networks
 Power Law Distribution Comparison • http: //www. dnatube. com/video/993/Plasmid. Cloning
Power Law Distribution Comparison • http: //www. dnatube. com/video/993/Plasmid. Cloning
 Removal of Nodes in KSHV Network
Removal of Nodes in KSHV Network
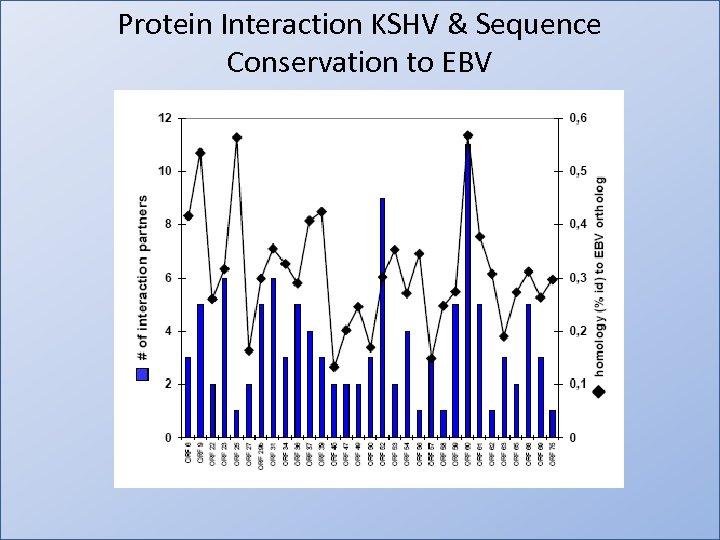 Protein Interaction KSHV & Sequence Conservation to EBV
Protein Interaction KSHV & Sequence Conservation to EBV
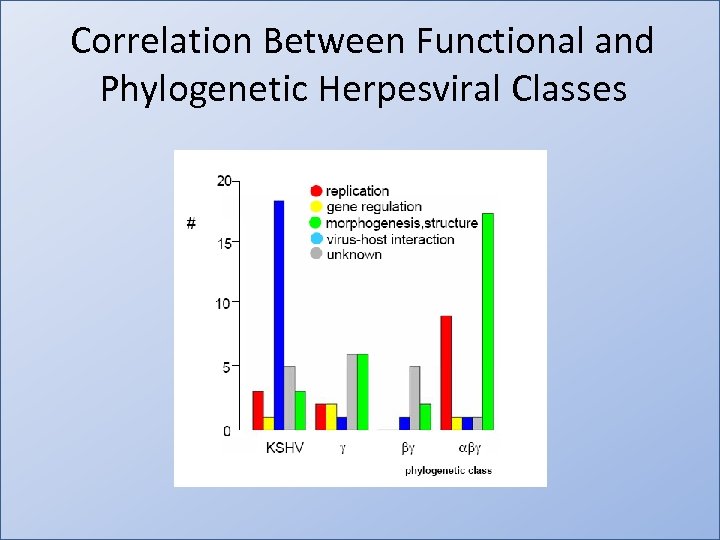 Correlation Between Functional and Phylogenetic Herpesviral Classes
Correlation Between Functional and Phylogenetic Herpesviral Classes
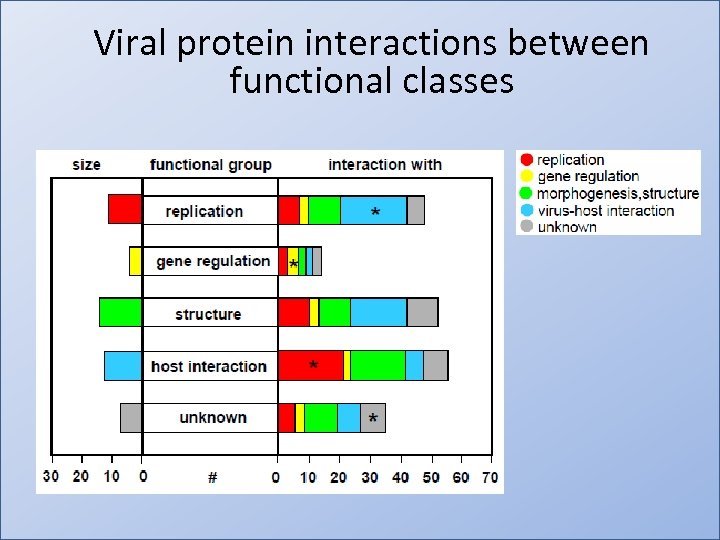 Viral protein interactions between functional classes • http: //www. dnatube. com/video/993/Plasmid. Cloning
Viral protein interactions between functional classes • http: //www. dnatube. com/video/993/Plasmid. Cloning
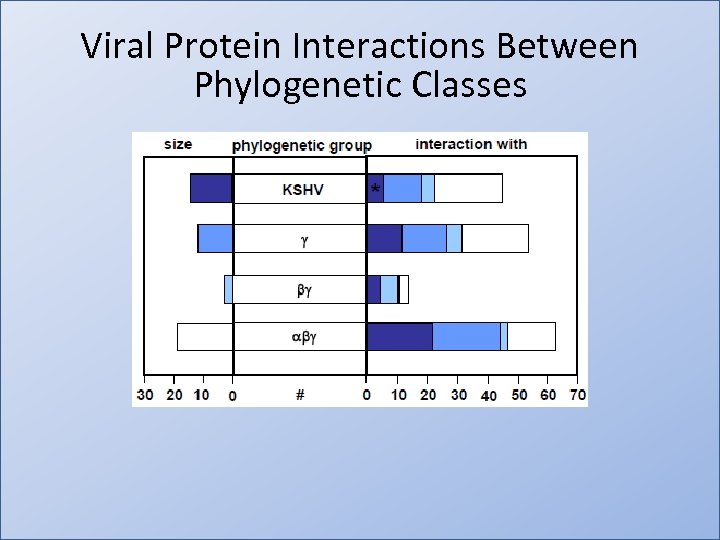 Viral Protein Interactions Between Phylogenetic Classes
Viral Protein Interactions Between Phylogenetic Classes
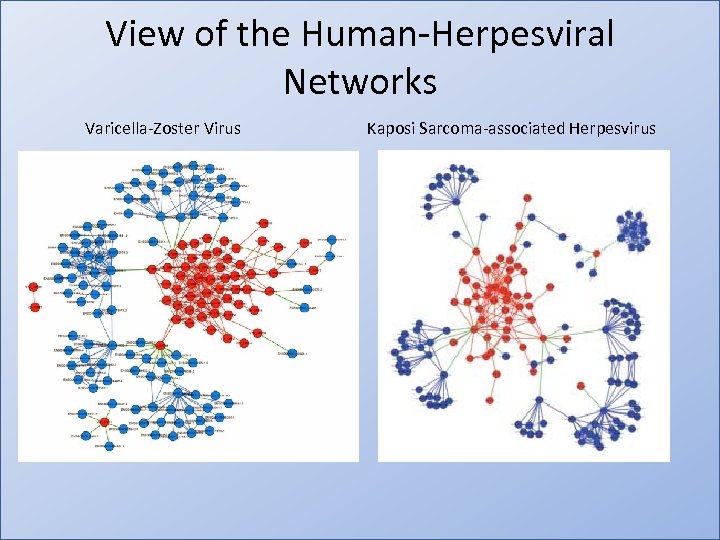 View of the Human-Herpesviral Networks Varicella-Zoster Virus Kaposi Sarcoma-associated Herpesvirus
View of the Human-Herpesviral Networks Varicella-Zoster Virus Kaposi Sarcoma-associated Herpesvirus
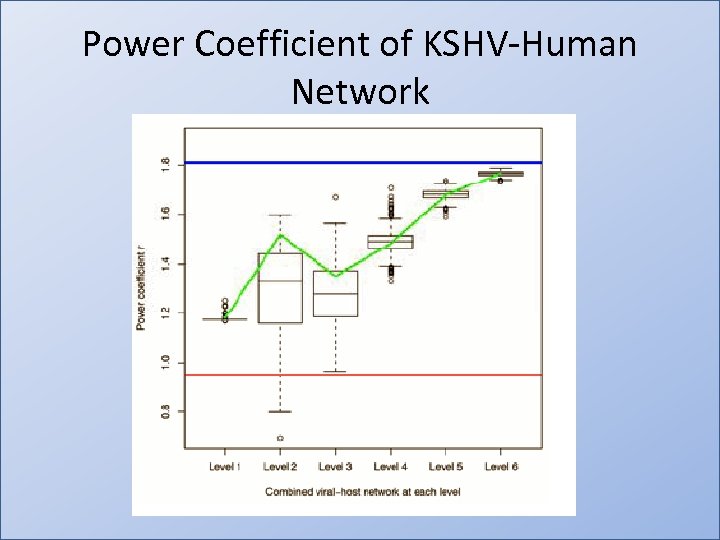 Power Coefficient of KSHV-Human Network
Power Coefficient of KSHV-Human Network
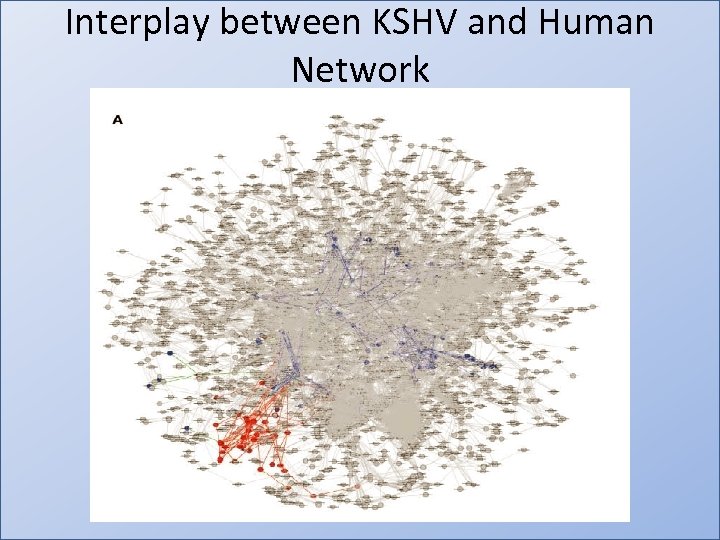 Interplay between KSHV and Human Network
Interplay between KSHV and Human Network
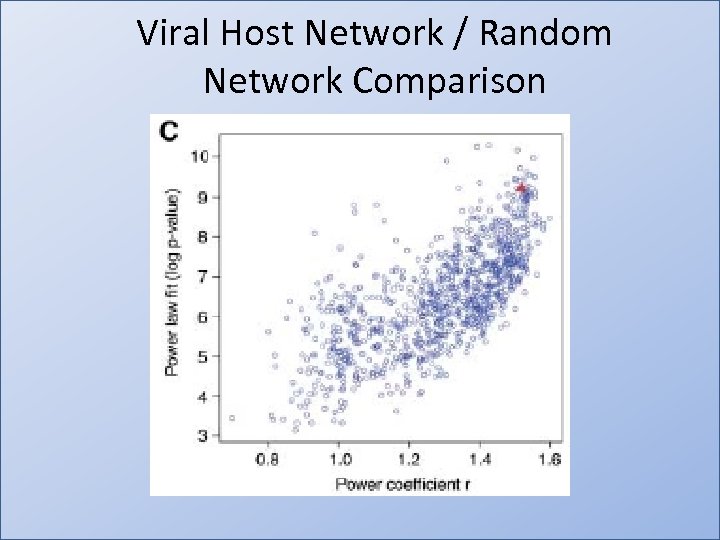 Viral Host Network / Random Network Comparison
Viral Host Network / Random Network Comparison
 Conclusions • Virus and host interactomes possess distinct network topologies • Integration of viral and host protein network may lead to better understanding of viral pathogenicity • Future interactome data from other viruses may improve understanding of functions of viral proteins and their phylogeny • Understanding networks may help to develop future therapies
Conclusions • Virus and host interactomes possess distinct network topologies • Integration of viral and host protein network may lead to better understanding of viral pathogenicity • Future interactome data from other viruses may improve understanding of functions of viral proteins and their phylogeny • Understanding networks may help to develop future therapies


