bd378d86b4c3a55dd36b1600df54542b.ppt
- Количество слайдов: 30

GUS 3. 0: Web Sites and Tools June 20, 2002 Jonathan Crabtree crabtree@pcbi. upenn. edu

Outline " Current web interfaces examples: Java Servlet, CGI-based reusable " allgenes. org, Plasmo. DB. org Java and Perl code, install scripts The future? PHP and JSP "GUSWWW" schema redesign
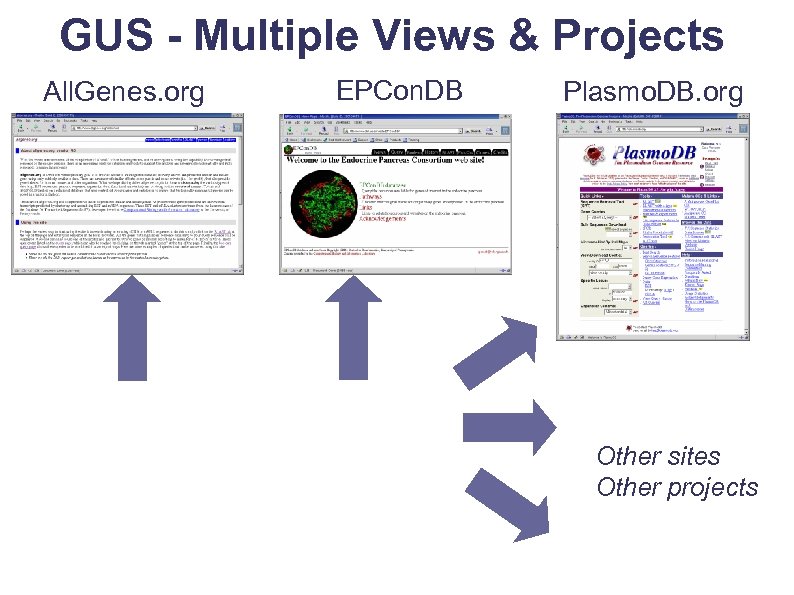
GUS - Multiple Views & Projects All. Genes. org EPCon. DB Plasmo. DB. org Other sites Other projects

allgenes. org query: "Is my c. DNA similar to any mouse genes that are predicted to encode transcription factors and have been localized to mouse chromosome 5? "
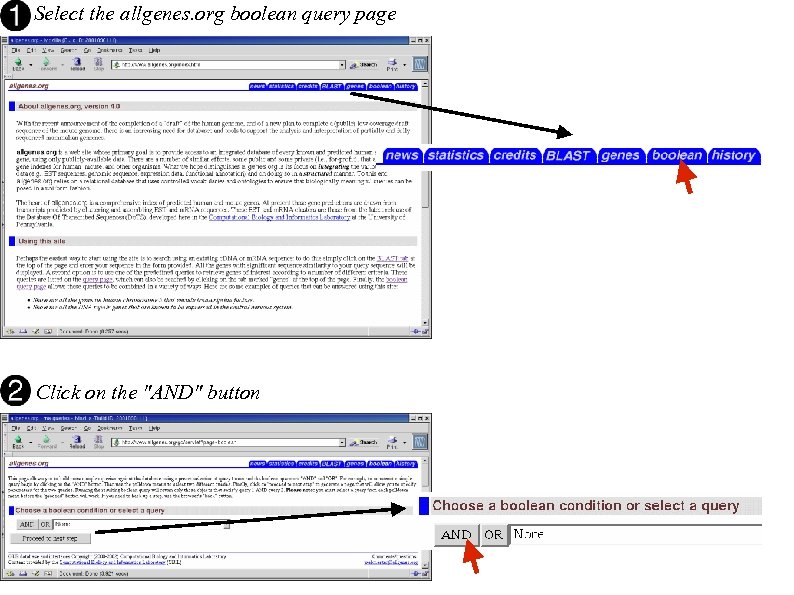
Select the allgenes. org boolean query page Click on the "AND" button
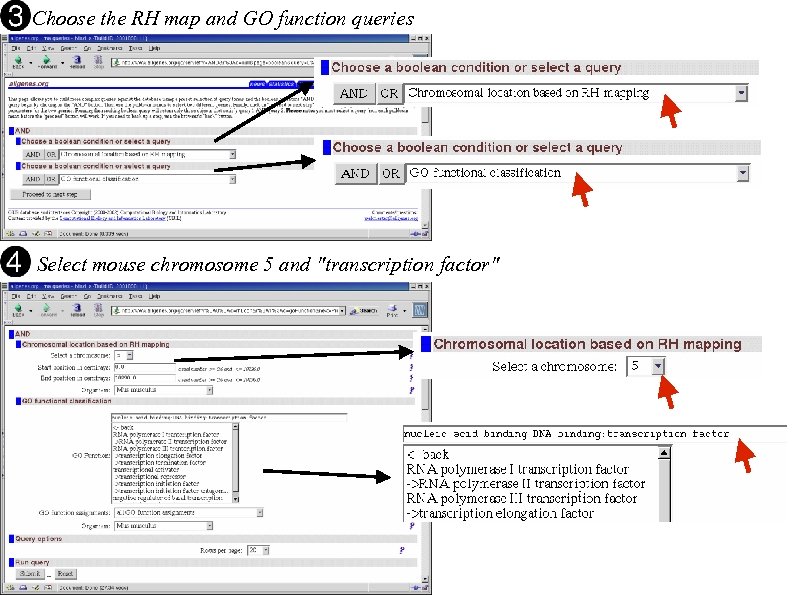
Choose the RH map and GO function queries Select mouse chromosome 5 and "transcription factor"
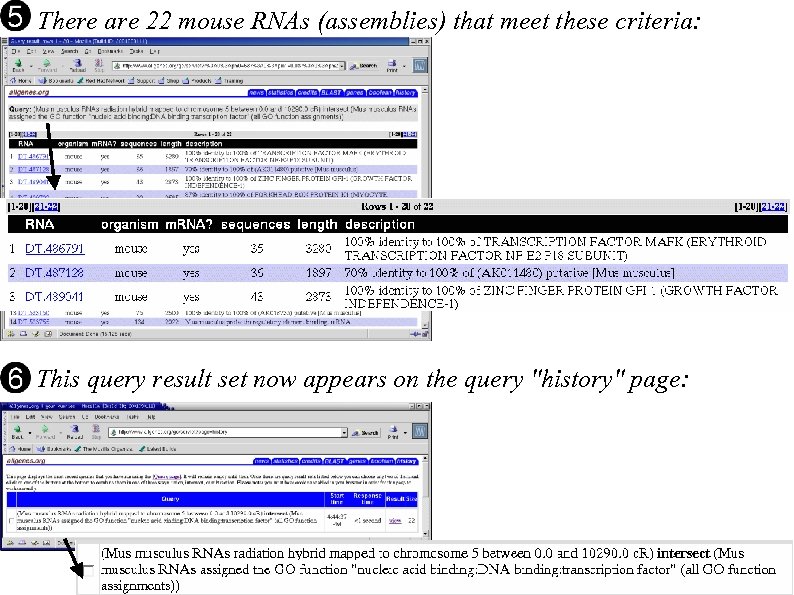
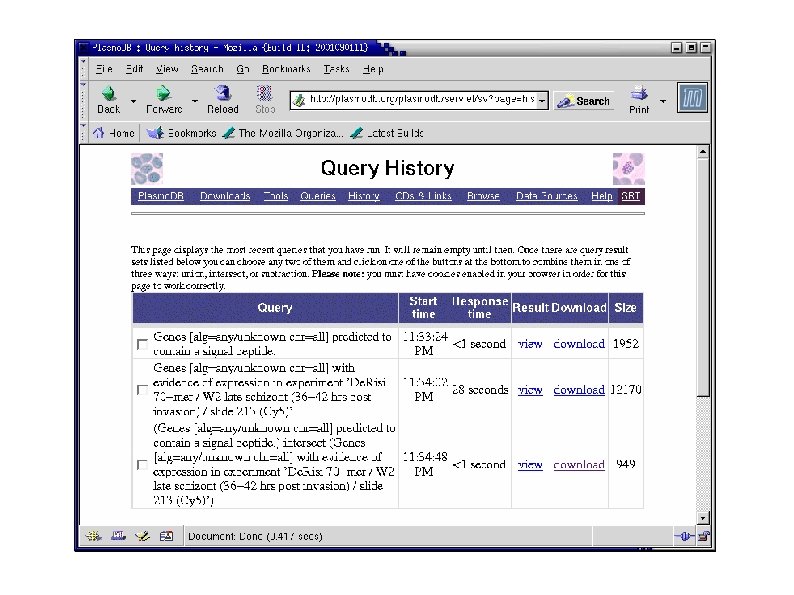
There are 22 mouse RNAs (assemblies) that meet these criteria: This query result set now appears on the query "history" page:
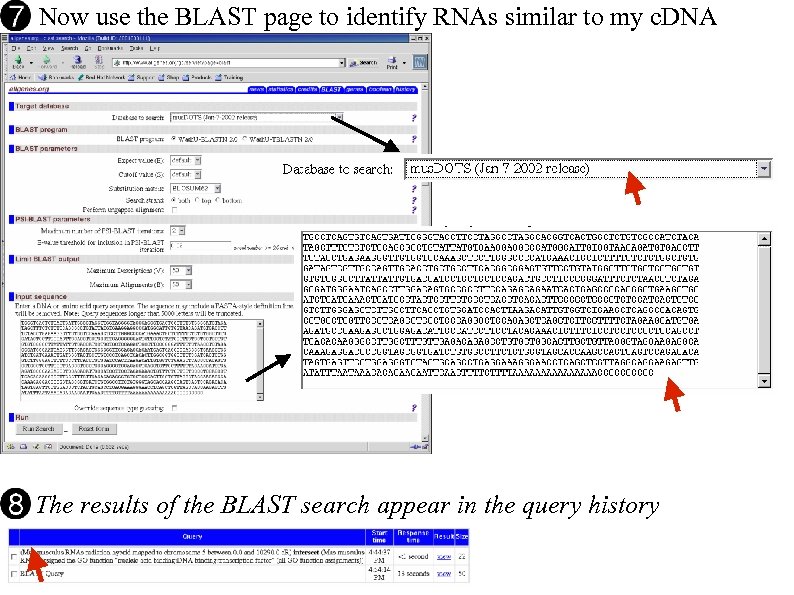
Now use the BLAST page to identify RNAs similar to my c. DNA The results of the BLAST search appear in the query history
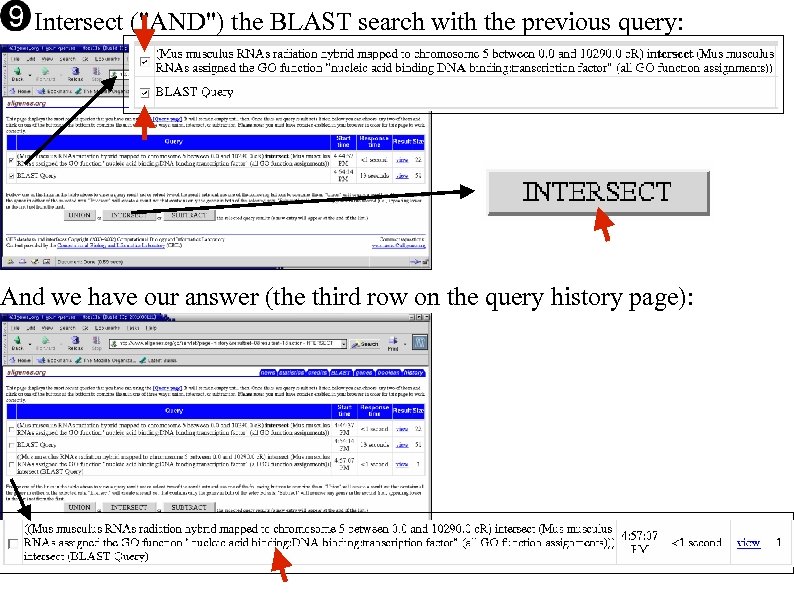
Intersect ("AND") the BLAST search with the previous query: And we have our answer (the third row on the query history page):
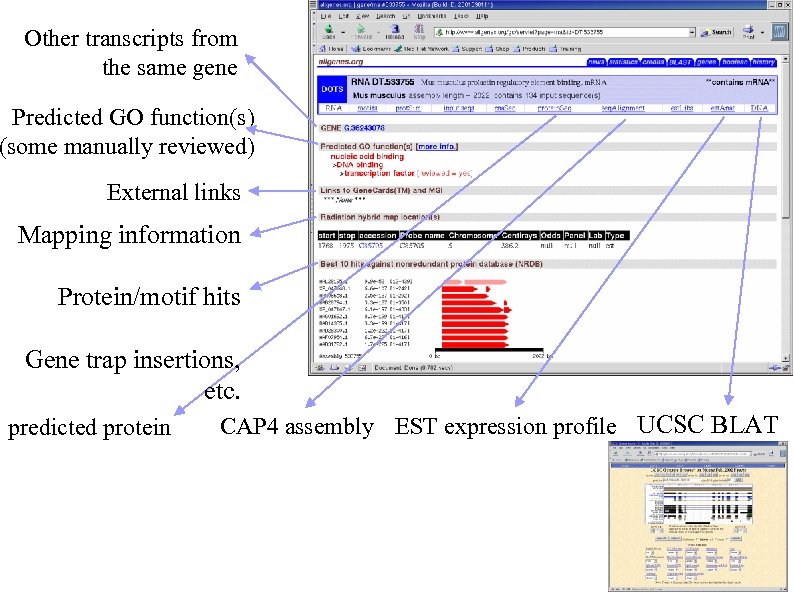
Other transcripts from the same gene Predicted GO function(s) (some manually reviewed) External links Mapping information Protein/motif hits Gene trap insertions, etc. predicted protein CAP 4 assembly EST expression profile UCSC BLAT

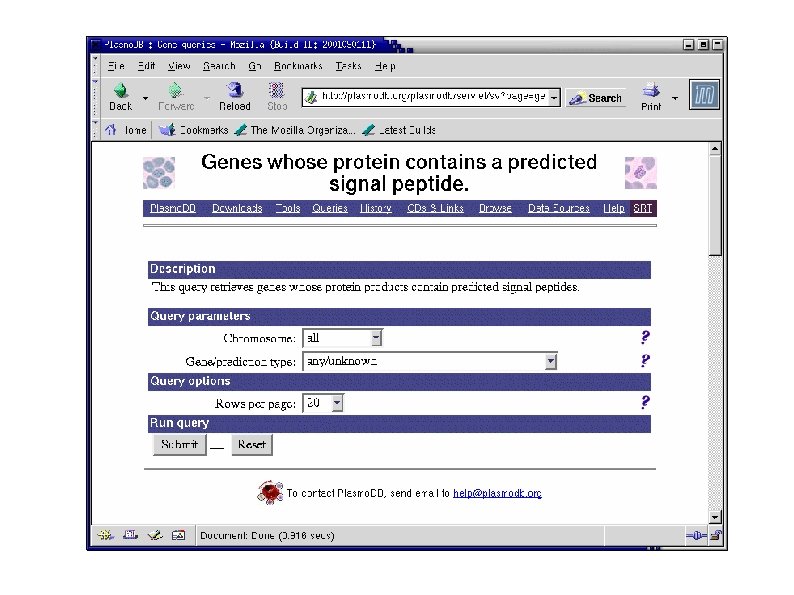
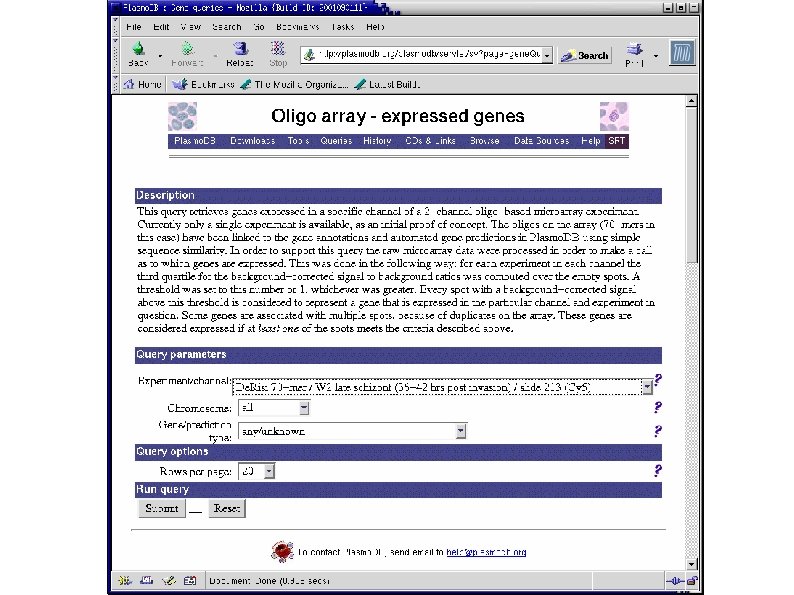
Plasmo. DB: Combining Expression and Sequence Data "List all genes whose proteins are predicted to contain a signal peptide and for which there is evidence that they are expressed in Plasmodium falciparum's late schizont stage. "

Web Interface Components GUS/www/allgenes/htdocs/index. html. in. . . GUS/www/allgenes/cgi-bin/rna. Prot. Sim. Png. pl. in. . . GUS/java/cbil/gus/servlet/Site. Servlet. java. . . GUS/www/install/allgenes-config. in GUS/www/install. Servlet. pl GUS/perl/servlet/allgenes/rna. Prot. Sim. pl. in. . .
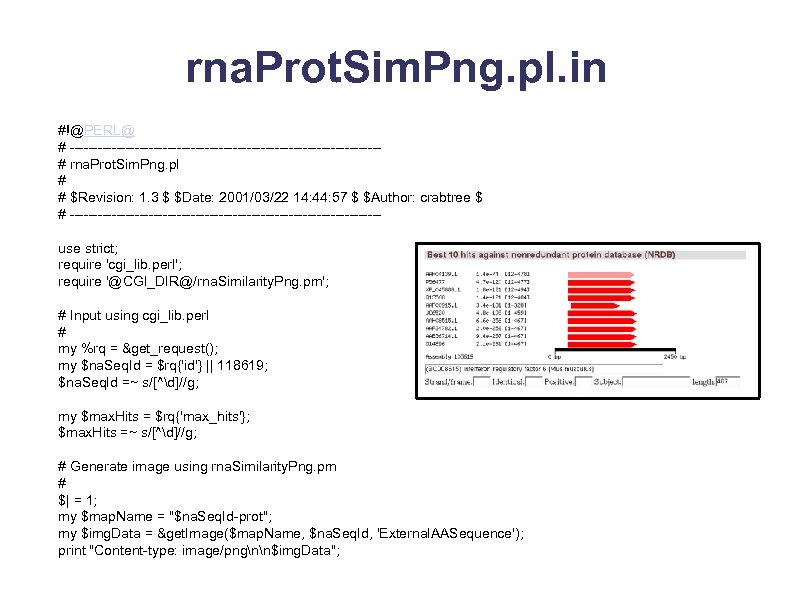
rna. Prot. Sim. Png. pl. in #!@PERL@ # ---------------------------------# rna. Prot. Sim. Png. pl # # $Revision: 1. 3 $ $Date: 2001/03/22 14: 44: 57 $ $Author: crabtree $ # ---------------------------------use strict; require 'cgi_lib. perl'; require '@CGI_DIR@/rna. Similarity. Png. pm'; # Input using cgi_lib. perl # my %rq = &get_request(); my $na. Seq. Id = $rq{'id'} || 118619; $na. Seq. Id =~ s/[^d]//g; my $max. Hits = $rq{'max_hits'}; $max. Hits =~ s/[^d]//g; # Generate image using rna. Similarity. Png. pm # $| = 1; my $map. Name = "$na. Seq. Id-prot"; my $img. Data = &get. Image($map. Name, $na. Seq. Id, 'External. AASequence'); print "Content-type: image/pngnn$img. Data";

cbil. gus. servlet. Site. Servlet " extends javax. servlet. http. Http. Servlet and " is the only actual servlet in our Java code reads a configuration file and instantiates the set of Java. Beans defined therein: instances of Page. Generator. I - content generators Sql. Query - parameterized SQL queries "Param" " and "Formatter" classes implements logging, dispatches requests

allgenes-config. in # Oracle-specific routines # gus. Ora. Sql. class=cbil. gus. servlet. db. oracle. SQL # Set of logins to GUS or GUSdev # gus. Login. class=cbil. gus. servlet. db. Connection. Pool gus. Login=@ORACLE_LOGIN@ gus. Login. Password=@ORACLE_PASSWORD@ gus. Login. DBUrl=@ORACLE_JDBC_URL@ gus. Login. Num. Connections=6 gus. Login. Max. Query. Time=120 gus. Login. Check. Interval=30 gus. Login. JDBCDrivers=oracle. jdbc. driver. Oracle. Driver gus. Login. Sql=gus. Ora. Sql gus. Login. Print. Status. Messages=true. . .
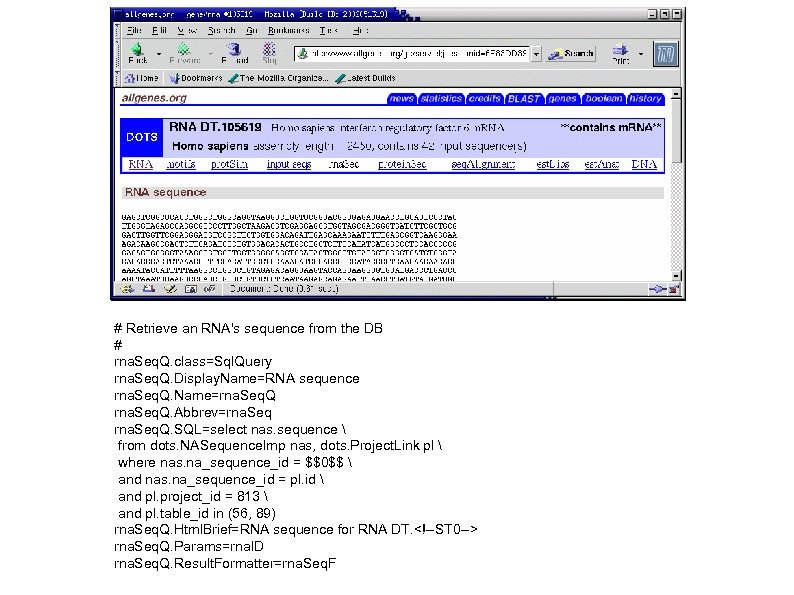
# Retrieve an RNA's sequence from the DB # rna. Seq. Q. class=Sql. Query rna. Seq. Q. Display. Name=RNA sequence rna. Seq. Q. Name=rna. Seq. Q. Abbrev=rna. Seq. Q. SQL=select nas. sequence from dots. NASequence. Imp nas, dots. Project. Link pl where nas. na_sequence_id = $$0$$ and nas. na_sequence_id = pl. id and pl. project_id = 813 and pl. table_id in (56, 89) rna. Seq. Q. Html. Brief=RNA sequence for RNA DT. <!--ST 0 --> rna. Seq. Q. Params=rna. ID rna. Seq. Q. Result. Formatter=rna. Seq. F
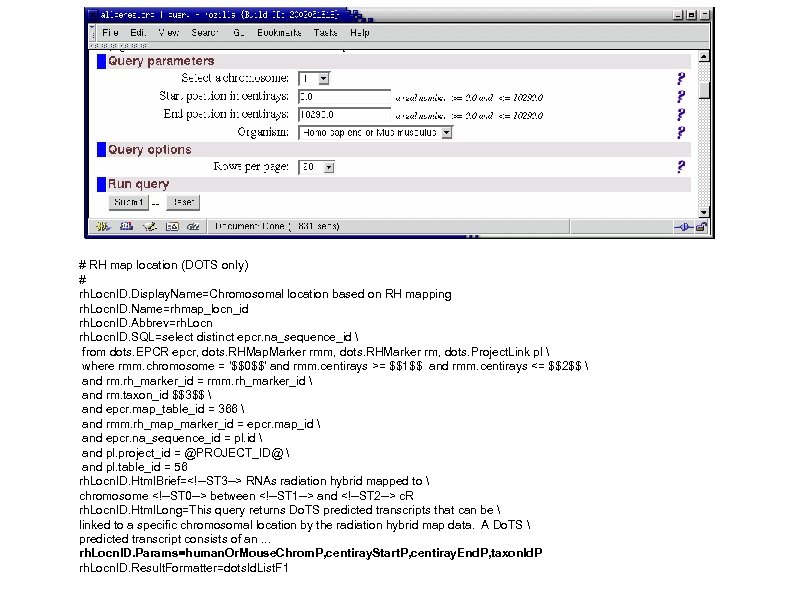
# RH map location (DOTS only) # rh. Locn. ID. Display. Name=Chromosomal location based on RH mapping rh. Locn. ID. Name=rhmap_locn_id rh. Locn. ID. Abbrev=rh. Locn. ID. SQL=select distinct epcr. na_sequence_id from dots. EPCR epcr, dots. RHMap. Marker rmm, dots. RHMarker rm, dots. Project. Link pl where rmm. chromosome = '$$0$$' and rmm. centirays >= $$1$$ and rmm. centirays <= $$2$$ and rm. rh_marker_id = rmm. rh_marker_id and rm. taxon_id $$3$$ and epcr. map_table_id = 366 and rmm. rh_map_marker_id = epcr. map_id and epcr. na_sequence_id = pl. id and pl. project_id = @PROJECT_ID@ and pl. table_id = 56 rh. Locn. ID. Html. Brief=<!--ST 3 --> RNAs radiation hybrid mapped to chromosome <!--ST 0 --> between <!--ST 1 --> and <!--ST 2 --> c. R rh. Locn. ID. Html. Long=This query returns Do. TS predicted transcripts that can be linked to a specific chromosomal location by the radiation hybrid map data. A Do. TS predicted transcript consists of an. . . rh. Locn. ID. Params=human. Or. Mouse. Chrom. P, centiray. Start. P, centiray. End. P, taxon. Id. P rh. Locn. ID. Result. Formatter=dots. Id. List. F 1
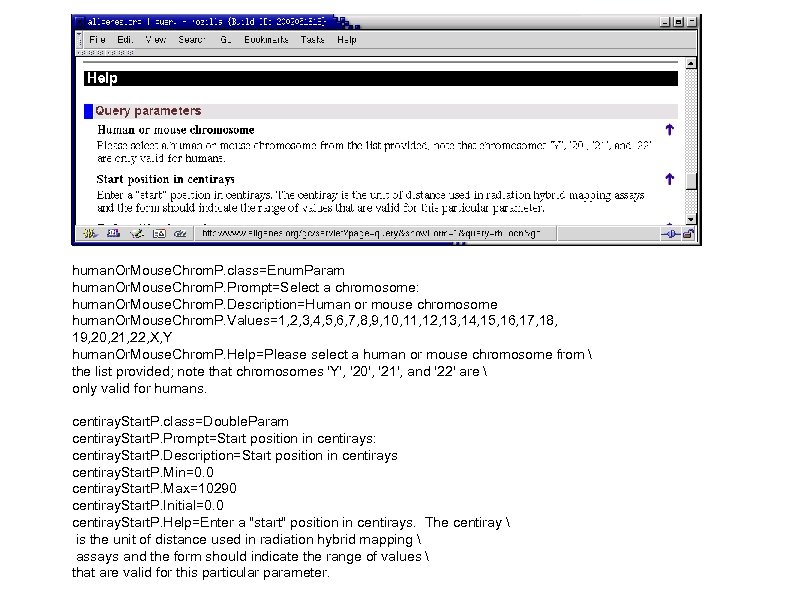
human. Or. Mouse. Chrom. P. class=Enum. Param human. Or. Mouse. Chrom. P. Prompt=Select a chromosome: human. Or. Mouse. Chrom. P. Description=Human or mouse chromosome human. Or. Mouse. Chrom. P. Values=1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, X, Y human. Or. Mouse. Chrom. P. Help=Please select a human or mouse chromosome from the list provided; note that chromosomes 'Y', '20', '21', and '22' are only valid for humans. centiray. Start. P. class=Double. Param centiray. Start. P. Prompt=Start position in centirays: centiray. Start. P. Description=Start position in centirays centiray. Start. P. Min=0. 0 centiray. Start. P. Max=10290 centiray. Start. P. Initial=0. 0 centiray. Start. P. Help=Enter a "start" position in centirays. The centiray is the unit of distance used in radiation hybrid mapping assays and the form should indicate the range of values that are valid for this particular parameter.
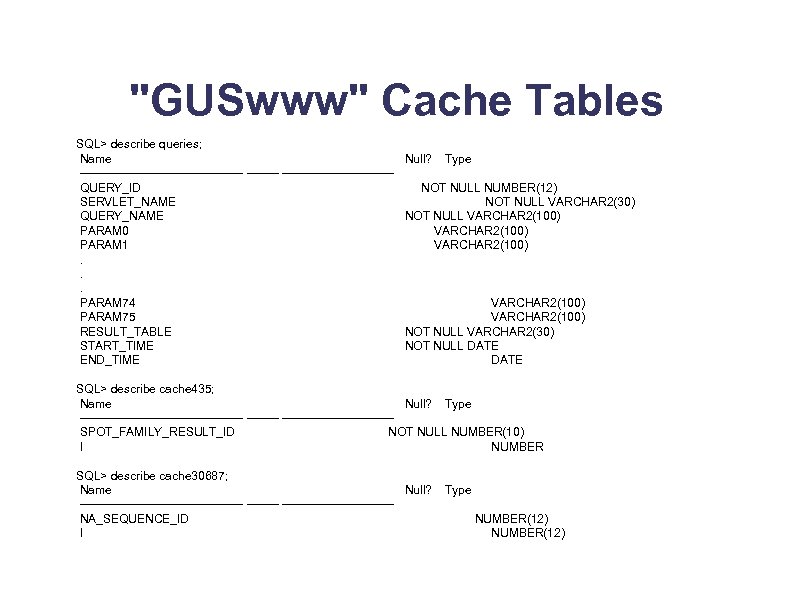
"GUSwww" Cache Tables SQL> describe queries; Name ---------------------QUERY_ID SERVLET_NAME QUERY_NAME PARAM 0 PARAM 1. . . PARAM 74 PARAM 75 RESULT_TABLE START_TIME END_TIME Null? Type NOT NULL NUMBER(12) NOT NULL VARCHAR 2(30) NOT NULL VARCHAR 2(100) VARCHAR 2(100) NOT NULL VARCHAR 2(30) NOT NULL DATE SQL> describe cache 435; Name Null? Type ---------------------SPOT_FAMILY_RESULT_ID NOT NULL NUMBER(10) I NUMBER SQL> describe cache 30687; Name Null? ---------------------NA_SEQUENCE_ID I Type NUMBER(12)
![install. Servlet. pl [crabtree@zeus install]$. /install. Servlet. pl --port=9000 --cgi. Dir=/world/www. allgenes/cgi-bin/ install. Servlet. pl [crabtree@zeus install]$. /install. Servlet. pl --port=9000 --cgi. Dir=/world/www. allgenes/cgi-bin/](https://present5.com/presentation/bd378d86b4c3a55dd36b1600df54542b/image-23.jpg)
install. Servlet. pl [crabtree@zeus install]$. /install. Servlet. pl --port=9000 --cgi. Dir=/world/www. allgenes/cgi-bin/ --htdocs. Dir=/world/www. allgenes/htdocs --cgi. URL=http: //www. allgenes. org/cgi-bin --htdocs. URL=http: //www. allgenes. org --install. Dir=/world/www. allgenes/servlet --servlet. Name=allgenes-zeus --servlet. File. Prefix=allgenes --servlet. Config=allgenes-zeus --production --servlet. URL=http: //www. allgenes. org/gc/servlet -install htdocs and cgi-bin files perform substitutions defined by 'allgenes-zeus' (e. g. ORA_LOGIN, ORA_PASSWORD, PROJECT_ID) -compile Java code, create. jar file and install -install servlet configuration file
![Features of Current [Servlet] Implementation " Automatic generation of HTML FORMs Automated Integrated INPUT Features of Current [Servlet] Implementation " Automatic generation of HTML FORMs Automated Integrated INPUT](https://present5.com/presentation/bd378d86b4c3a55dd36b1600df54542b/image-24.jpg)
Features of Current [Servlet] Implementation " Automatic generation of HTML FORMs Automated Integrated INPUT input checking help features elements populated from the database " Query history facility " Boolean queries (AND, OR, SUBTRACT) " Declarative configuration file " Base system is relatively independent of GUS
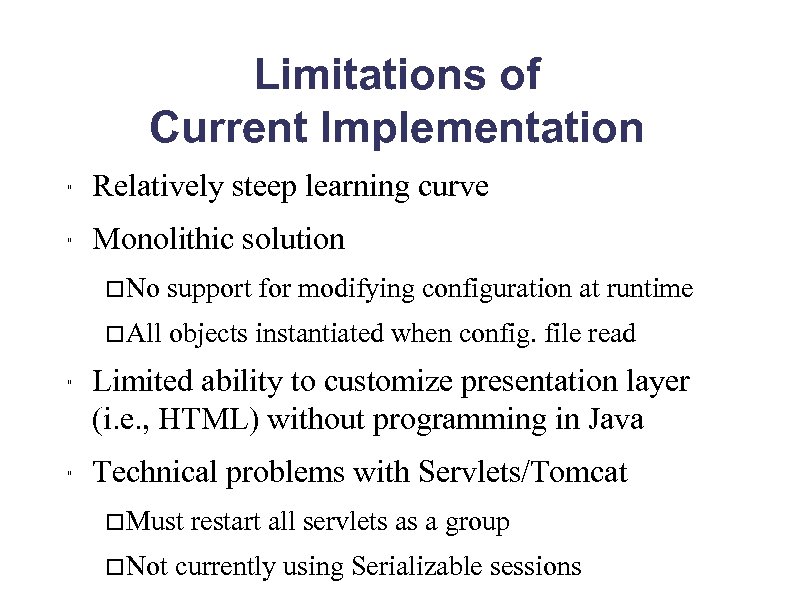
Limitations of Current Implementation " Relatively steep learning curve " Monolithic solution No All " " support for modifying configuration at runtime objects instantiated when config. file read Limited ability to customize presentation layer (i. e. , HTML) without programming in Java Technical problems with Servlets/Tomcat Must Not restart all servlets as a group currently using Serializable sessions
Dynamic Web Content " HTML fragments embedded in a program: CGI Java " programs (e. g. Perl - interpreted) Servlets (compiled) Program fragments embedded in HTML: PHP JSP " (interpreted) (compiled; once, as needed) Another axis: persistent vs. not (CGI/Fast. CGI)
Program Fragments in HTML " Advantages: faster development cycle; can edit in place easier to see/validate structure of HTML pages HTML " Disadvantages: must " has no functions, Java and PHP do take care to manage complexity of application Recommendations: move towards adopting this approach move all persistent state into the database
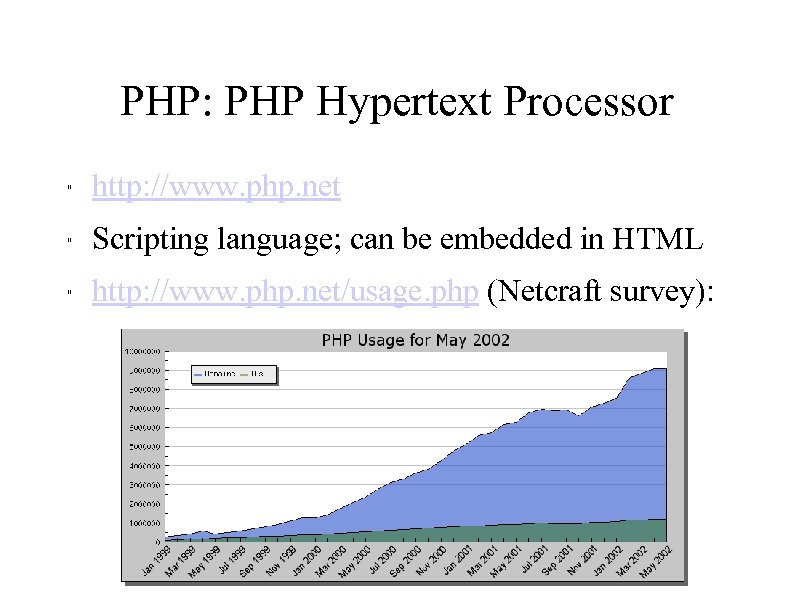
PHP: PHP Hypertext Processor " http: //www. php. net " Scripting language; can be embedded in HTML " http: //www. php. net/usage. php (Netcraft survey):

JSP - Java Server Pages " Based on and can interact with Java Servlets " Essentially Java embedded in HTML " XML-based tags, scriptlets, and Java. Bean calls " Standard tag libraries available " Pages typically compiled on demand " Multiple implementations? (vs. single for PHP)

Next steps " Agree on desired user interface functionality saving queries for Plasmo. DB persistent preferences for genome browser " Design parts of the schema to support it " Migrate old code/write new code " Easier to migrate existing code with JSP
bd378d86b4c3a55dd36b1600df54542b.ppt