5b1d3727e7553d11e7e3cacbbfb1606f.ppt
- Количество слайдов: 32
 GOSt a Gene Ontology mining tool Jüri Reimand
GOSt a Gene Ontology mining tool Jüri Reimand
 Overview • • • Introduction, bioinformatics Gene Ontology (GO) GOSt, a Gene Ontology mining tool Statistics and thresholds Ordered gene lists Extending GO
Overview • • • Introduction, bioinformatics Gene Ontology (GO) GOSt, a Gene Ontology mining tool Statistics and thresholds Ordered gene lists Extending GO
 Introduction • Bioinformatics – Analysis of experimental data • Genes encode proteins – Proteins : building blocks of living organisms – Gene expression : protein production from genetic code • Microarray experiments measure gene expression – Thousands of genes simultaneously – Expression levels over time – Different biological conditions – Comparison of healthy and diseased cells cluster similar profiles measures over time
Introduction • Bioinformatics – Analysis of experimental data • Genes encode proteins – Proteins : building blocks of living organisms – Gene expression : protein production from genetic code • Microarray experiments measure gene expression – Thousands of genes simultaneously – Expression levels over time – Different biological conditions – Comparison of healthy and diseased cells cluster similar profiles measures over time
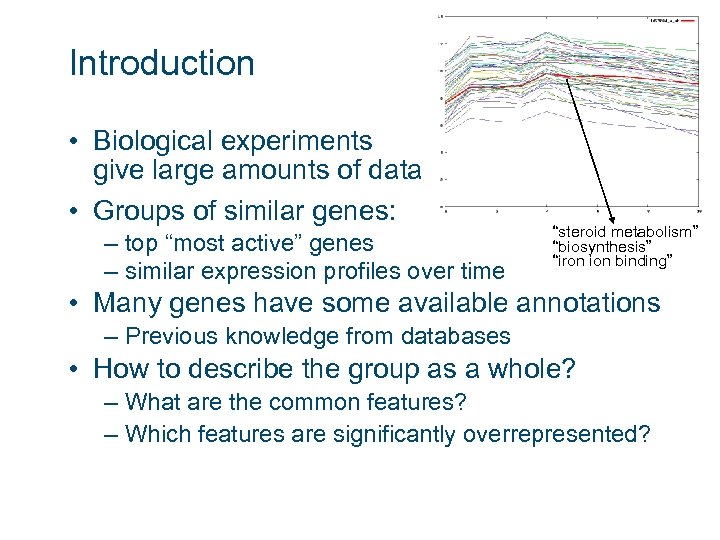 Introduction • Biological experiments give large amounts of data • Groups of similar genes: – top “most active” genes – similar expression profiles over time “steroid metabolism” “biosynthesis” “iron ion binding” • Many genes have some available annotations – Previous knowledge from databases • How to describe the group as a whole? – What are the common features? – Which features are significantly overrepresented?
Introduction • Biological experiments give large amounts of data • Groups of similar genes: – top “most active” genes – similar expression profiles over time “steroid metabolism” “biosynthesis” “iron ion binding” • Many genes have some available annotations – Previous knowledge from databases • How to describe the group as a whole? – What are the common features? – Which features are significantly overrepresented?
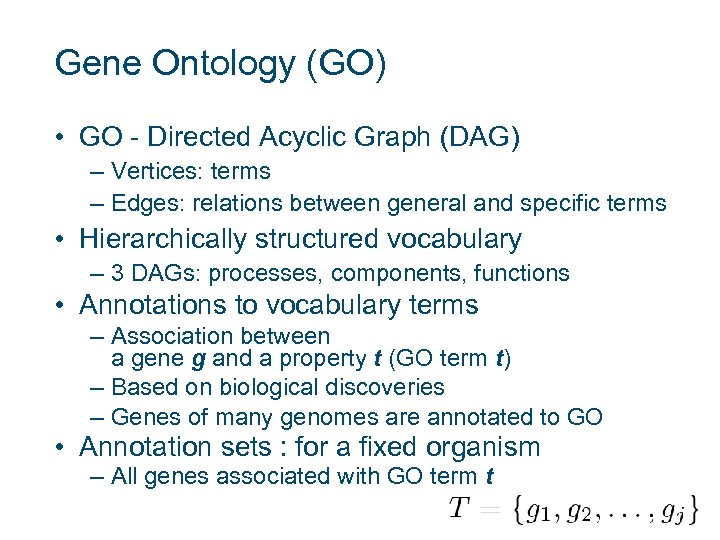 Gene Ontology (GO) • GO - Directed Acyclic Graph (DAG) – Vertices: terms – Edges: relations between general and specific terms • Hierarchically structured vocabulary – 3 DAGs: processes, components, functions • Annotations to vocabulary terms – Association between a gene g and a property t (GO term t) – Based on biological discoveries – Genes of many genomes are annotated to GO • Annotation sets : for a fixed organism – All genes associated with GO term t
Gene Ontology (GO) • GO - Directed Acyclic Graph (DAG) – Vertices: terms – Edges: relations between general and specific terms • Hierarchically structured vocabulary – 3 DAGs: processes, components, functions • Annotations to vocabulary terms – Association between a gene g and a property t (GO term t) – Based on biological discoveries – Genes of many genomes are annotated to GO • Annotation sets : for a fixed organism – All genes associated with GO term t
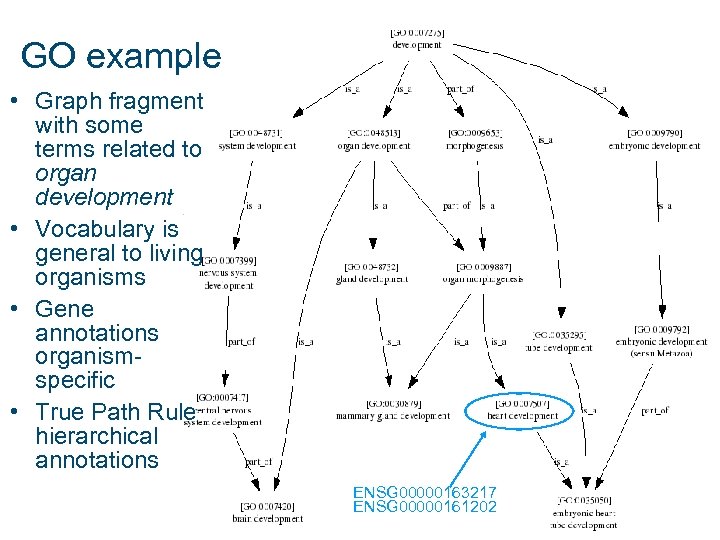 GO example • Graph fragment with some terms related to organ development • Vocabulary is general to living organisms • Gene annotations organismspecific • True Path Rule hierarchical annotations ENSG 00000163217 ENSG 00000161202
GO example • Graph fragment with some terms related to organ development • Vocabulary is general to living organisms • Gene annotations organismspecific • True Path Rule hierarchical annotations ENSG 00000163217 ENSG 00000161202
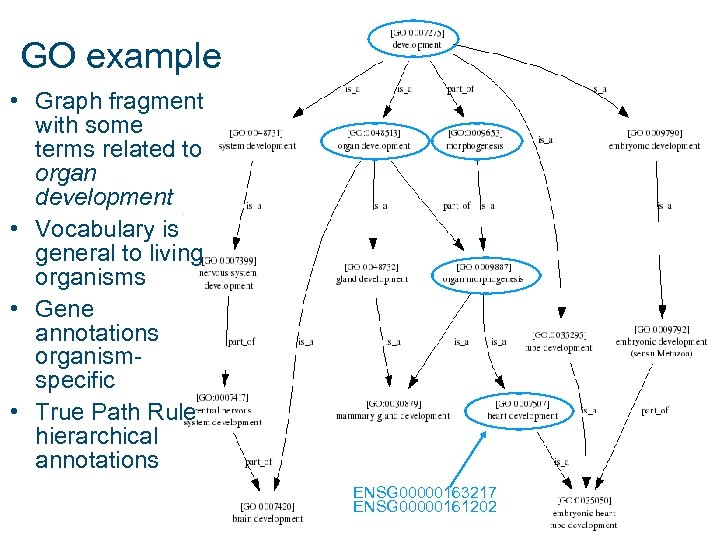 GO example • Graph fragment with some terms related to organ development • Vocabulary is general to living organisms • Gene annotations organismspecific • True Path Rule hierarchical annotations ENSG 00000163217 ENSG 00000161202
GO example • Graph fragment with some terms related to organ development • Vocabulary is general to living organisms • Gene annotations organismspecific • True Path Rule hierarchical annotations ENSG 00000163217 ENSG 00000161202
 GOSt – Gene Ontology Statistics • • • GO annotations to groups of genes Statistical significance of results Thresholds for distinguishing significant results Analysing ordered lists of genes Visualisation methods, WWW interface Command line toolset for large-scale analysis
GOSt – Gene Ontology Statistics • • • GO annotations to groups of genes Statistical significance of results Thresholds for distinguishing significant results Analysing ordered lists of genes Visualisation methods, WWW interface Command line toolset for large-scale analysis
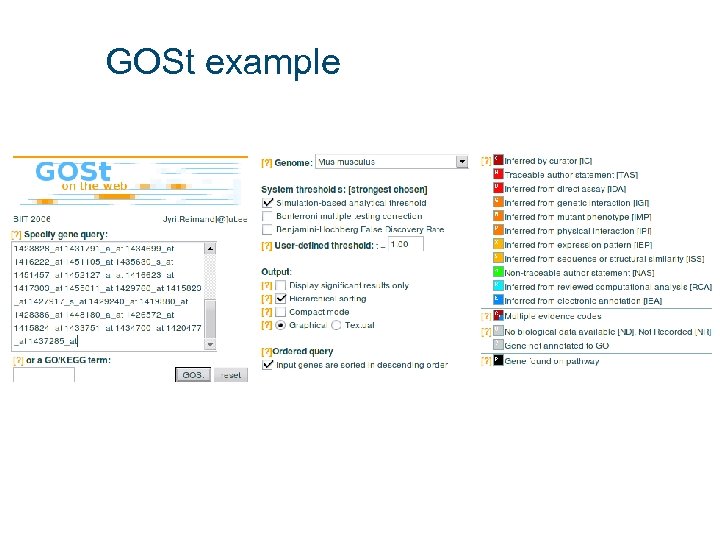 GOSt example
GOSt example
 45 mouse genes 338 GO
45 mouse genes 338 GO
 Evidence codes Genes P-value GO terms
Evidence codes Genes P-value GO terms
 Annotations to gene groups Gq Query GO Term e. g. Gt heart development • Result: term t matches query Q
Annotations to gene groups Gq Query GO Term e. g. Gt heart development • Result: term t matches query Q
 Statistical significance • Is intersection Q∩ significant? T • Fisher's one-tailed test – Cumulative hypergeometric probability – Get observed or more genes in intersection Q∩T – P ( pick k white balls out of K white and N-K black balls ) • Multiple testing – Every query results in a number of p-values – Matching GO terms are not independent – Increased rate of false positive matches • Which p-values are significant?
Statistical significance • Is intersection Q∩ significant? T • Fisher's one-tailed test – Cumulative hypergeometric probability – Get observed or more genes in intersection Q∩T – P ( pick k white balls out of K white and N-K black balls ) • Multiple testing – Every query results in a number of p-values – Matching GO terms are not independent – Increased rate of false positive matches • Which p-values are significant?
 Experimental thresholds • Simulation experiment – Fix some gene query size k – Repeat 1000 times: • Generate synthetic query Q with k elements : random subset of organism's genes • Observe best p-value p for query Q • Store p-value, p --> P – Choose p', 50 th smallest p-value from P – Threshold p' – top 5% of p-values for random queries of size k • Calculate for query lengths k = [1, 1000] • Compare with standard multiple testing corrections – Bonferroni (1936), Benjamini-Hochberg (1995)
Experimental thresholds • Simulation experiment – Fix some gene query size k – Repeat 1000 times: • Generate synthetic query Q with k elements : random subset of organism's genes • Observe best p-value p for query Q • Store p-value, p --> P – Choose p', 50 th smallest p-value from P – Threshold p' – top 5% of p-values for random queries of size k • Calculate for query lengths k = [1, 1000] • Compare with standard multiple testing corrections – Bonferroni (1936), Benjamini-Hochberg (1995)
 Analytical thresholds • Analytical approach to simulated thresholds – Fix gene query size k – Observe all sizes and frequencies of GO annotation sets T – Presume events with different T independent – Observe possible p-values p with query of k elements – Always correct p by constant c=0. 97 (set dependencies!) – Find such threshold p', that gives p ~= 0. 95 • Repeat for query lengths k = [1, 1000]
Analytical thresholds • Analytical approach to simulated thresholds – Fix gene query size k – Observe all sizes and frequencies of GO annotation sets T – Presume events with different T independent – Observe possible p-values p with query of k elements – Always correct p by constant c=0. 97 (set dependencies!) – Find such threshold p', that gives p ~= 0. 95 • Repeat for query lengths k = [1, 1000]
 Significance thresholds
Significance thresholds
 Significance thresholds
Significance thresholds
 Significance thresholds
Significance thresholds
 Significance thresholds
Significance thresholds
 Ordered lists of genes • Gene groups may be ordered – Interesting gene and few most similar genes – Top “most active” genes – Increasing distance from cluster centre • Top of the list, but how many? – Compare list with GO term – Which portion gives best p-value? – Peak significance of ordered query
Ordered lists of genes • Gene groups may be ordered – Interesting gene and few most similar genes – Top “most active” genes – Increasing distance from cluster centre • Top of the list, but how many? – Compare list with GO term – Which portion gives best p-value? – Peak significance of ordered query
 GOSt algorithms • Unordered query – Intersections with all annotation sets T • Exhaustive algorithm for ordered queries: – intersections with all Qi and annotation sets T • Approximate algorithm for ordered queries: – for every annotation set T, view only list portions that give local p-value extremes • local best p : list ends with matching gene • local worst p : list ends just before matching gene
GOSt algorithms • Unordered query – Intersections with all annotation sets T • Exhaustive algorithm for ordered queries: – intersections with all Qi and annotation sets T • Approximate algorithm for ordered queries: – for every annotation set T, view only list portions that give local p-value extremes • local best p : list ends with matching gene • local worst p : list ends just before matching gene
 Example: Ordered list analysis Peak significance at ordered list of 28 genes p-value query length List of genes, and matches for “Biosynthesis of steroids”
Example: Ordered list analysis Peak significance at ordered list of 28 genes p-value query length List of genes, and matches for “Biosynthesis of steroids”
 Evidence codes Genes P-value Ordered list query GO categories
Evidence codes Genes P-value Ordered list query GO categories
 Algorithm speed comparison 24 sec 2. 8 sec
Algorithm speed comparison 24 sec 2. 8 sec
 GOSt features • Command line interface (C/C++ and Perl) • Graphical user interface in web http: //bioinf. ebc. ee/GOST – SWOG (Graphics language, Jaanus Hansen 2005) • Data for multiple organisms – yeast, chicken, cow, mouse, rat, human. . . • Wrappers for parallel applications (GRID, MPI) • Pipelines for gene expression data analysis
GOSt features • Command line interface (C/C++ and Perl) • Graphical user interface in web http: //bioinf. ebc. ee/GOST – SWOG (Graphics language, Jaanus Hansen 2005) • Data for multiple organisms – yeast, chicken, cow, mouse, rat, human. . . • Wrappers for parallel applications (GRID, MPI) • Pipelines for gene expression data analysis
 Extending GO ( i ) • Pathway – a network of interacting genes and proteins – metabolism pathways, disease pathways, . . • Include pathway data to GO vocabulary – KEGG Pathway database – pathways as vocabulary terms – related genes as annotations to terms • KEGG terms independent of GO vocabulary GO: 0003674 GO molecular_function GO: 0005575 cellular_component GO: 0008150 biological_process KEGG: 00000 KEGG pathways
Extending GO ( i ) • Pathway – a network of interacting genes and proteins – metabolism pathways, disease pathways, . . • Include pathway data to GO vocabulary – KEGG Pathway database – pathways as vocabulary terms – related genes as annotations to terms • KEGG terms independent of GO vocabulary GO: 0003674 GO molecular_function GO: 0005575 cellular_component GO: 0008150 biological_process KEGG: 00000 KEGG pathways
 KEGG: 05010 - Alzheimer's disease
KEGG: 05010 - Alzheimer's disease
 Extending GO ( ii ) • Gene expression started by transcription factors (TF) • TFs bind to certain patterns in DNA – Transcription Factor Binding Sites (TFBS) – Often found in regions close to gene (1 k bp) • Include TFBS data from TRANSFAC – Patterns (putative TFBS) as vocabulary terms – annotations to genes near patterns Transcription factor ATATAATAAAGATGAGGCGAATATACCGGCCCTTAGCGCGAAGCAATTCATCATATAAGCGAGAGAGGCCAATATGCAATCTTCGACAGCAT TF binding site gene
Extending GO ( ii ) • Gene expression started by transcription factors (TF) • TFs bind to certain patterns in DNA – Transcription Factor Binding Sites (TFBS) – Often found in regions close to gene (1 k bp) • Include TFBS data from TRANSFAC – Patterns (putative TFBS) as vocabulary terms – annotations to genes near patterns Transcription factor ATATAATAAAGATGAGGCGAATATACCGGCCCTTAGCGCGAAGCAATTCATCATATAAGCGAGAGAGGCCAATATGCAATCTTCGACAGCAT TF binding site gene
 TRANSFAC motifs • Motifs added in a hierarchy – according to PWM score – 5 levels: • near_threshold • . . . • near_MAX_score depth in hierarchy • Work in progress TF: M 00431_4 TF: M 00431_3 TF: M 00431_2 TF: M 00431_1 TF: M 00431_0 TF: M 00328_4 TF: M 00328_3 TF: M 00328_2 TF: M 00000 – Hedi Peterson TTTSGCGS: 4 TTTSGCGS: 3 TTTSGCGS: 2 TTTSGCGS: 1 TTTSGCGS: 0 NCNNTNNTGCRTGANNNN: 4 NCNNTNNTGCRTGANNNN: 3 NCNNTNNTGCRTGANNNN: 2 TRANSFAC motifs GO: 0003674 GO molecular_function GO: 0005575 cellular_component GO: 0008150 biological_process KEGG: 00000 KEGG pathways
TRANSFAC motifs • Motifs added in a hierarchy – according to PWM score – 5 levels: • near_threshold • . . . • near_MAX_score depth in hierarchy • Work in progress TF: M 00431_4 TF: M 00431_3 TF: M 00431_2 TF: M 00431_1 TF: M 00431_0 TF: M 00328_4 TF: M 00328_3 TF: M 00328_2 TF: M 00000 – Hedi Peterson TTTSGCGS: 4 TTTSGCGS: 3 TTTSGCGS: 2 TTTSGCGS: 1 TTTSGCGS: 0 NCNNTNNTGCRTGANNNN: 4 NCNNTNNTGCRTGANNNN: 3 NCNNTNNTGCRTGANNNN: 2 TRANSFAC motifs GO: 0003674 GO molecular_function GO: 0005575 cellular_component GO: 0008150 biological_process KEGG: 00000 KEGG pathways
 Summary • We investigated means for finding GO annotations to groups of genes, and statistical methods for determining significance of results. • We combined GO vocabulary with various types of biological data, such as KEGG pathways and TRANSFAC regulatory elements. • We proposed analytical thresholds for distinguishing significant results from structured and partly dependent GO annotations, and verified thresholds with simulation experiments. • We proposed a novel concept of analyzing GO annotations for ordered lists of genes, and implemented fast algorithms for the purpose. • The practical result of our work is GOSt, a GO mining tool. Command line interface is suitable for large-scale automatic analysis, while graphical web interface enables highly visualized and interactive analysis.
Summary • We investigated means for finding GO annotations to groups of genes, and statistical methods for determining significance of results. • We combined GO vocabulary with various types of biological data, such as KEGG pathways and TRANSFAC regulatory elements. • We proposed analytical thresholds for distinguishing significant results from structured and partly dependent GO annotations, and verified thresholds with simulation experiments. • We proposed a novel concept of analyzing GO annotations for ordered lists of genes, and implemented fast algorithms for the purpose. • The practical result of our work is GOSt, a GO mining tool. Command line interface is suitable for large-scale automatic analysis, while graphical web interface enables highly visualized and interactive analysis.
 Sneak preview • GO analysis of hierarchical clustering tree – Cluster genes according to expression similarity and. . –. . “Wrap up” nodes that show no significant annotations in GO • Work in progress – Meelis Kull – Darja Krushevskaja
Sneak preview • GO analysis of hierarchical clustering tree – Cluster genes according to expression similarity and. . –. . “Wrap up” nodes that show no significant annotations in GO • Work in progress – Meelis Kull – Darja Krushevskaja
 Acknowledgments Jaak Vilo BIIT group Hedi Peterson Meelis Kull Jaanus Hansen Priit Adler Ilja Livenson Raivo Kolde Konstantin Tretjakov Pavlos Pavlidis Asko Tiidumaa Darja Krushevskaja Fun. Gen. ES Consortium
Acknowledgments Jaak Vilo BIIT group Hedi Peterson Meelis Kull Jaanus Hansen Priit Adler Ilja Livenson Raivo Kolde Konstantin Tretjakov Pavlos Pavlidis Asko Tiidumaa Darja Krushevskaja Fun. Gen. ES Consortium


