22ba76b2a04d9336215cb689790fc440.ppt
- Количество слайдов: 20
Glycoprotein Microheterogeneity via N-Glycopeptide Identification Kevin Brown Chandler, Petr Pompach, Radoslav Goldman, Nathan Edwards Georgetown University Medical Center
The challenge l Identify glycopeptides in large-scale tandem mass-spectrometry datasets l l l Good, but not great, instrumentation l l Many glycopeptide enriched fractions Many tandem mass-spectra / fraction QStar Elite – CID, good MS 1/MS 2 resolution Strive for hypothesis-generating analysis l l Site-specific glycopeptide characterization Glycoform occupancy in differentiated samples 2

Observations l Oxonium ions (204, 366) help distinguish glycopeptides from peptides… l l Few peptide b/y-ions to identify peptides… l l …but do little to identify the glycopeptide …but intact peptide fragments are common If the peptide can be guessed, then… l …the glycan's mass can be determined 3
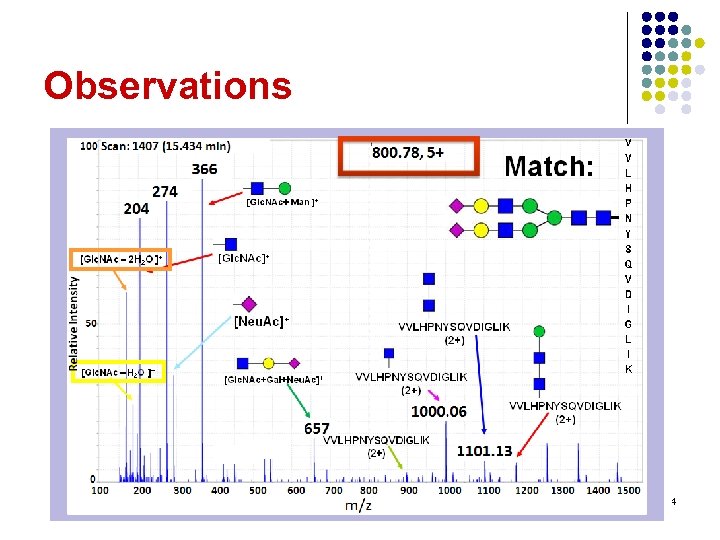
Observations 4
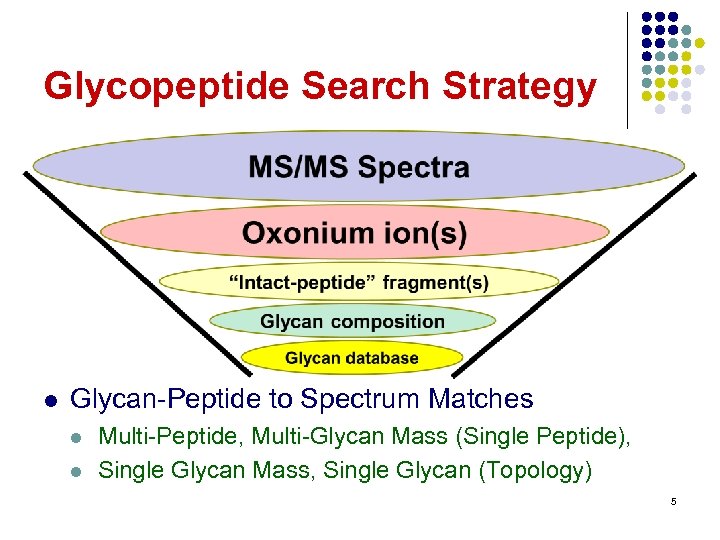
Glycopeptide Search Strategy l Glycan-Peptide to Spectrum Matches l l Multi-Peptide, Multi-Glycan Mass (Single Peptide), Single Glycan Mass, Single Glycan (Topology) 5

Compromises l Single protein / Simple protein mixture l l Single N-glycan per peptide l l Subtraction from precursor Digest may not resolve site l l Few peptides to distinguish Need peptide/glycan fragments to distinguish Isobaric peptide-glycan pairs are not resolved l Need peptide/glycan fragments to distinguish 6

Glycan Databases l Link putative glycan masses to N-linked glycan structures (and organism, etc. ): l l l Human N-linked Glycome. DB Cartoonist structure enumeration CFG Mammalian Array (v 5. 0) In-house database (Oxford notation) Database(s) provide "biased" search space: l l Coverage vs. "Reasonableness" Trade off: Time, Specificity, Biology 7
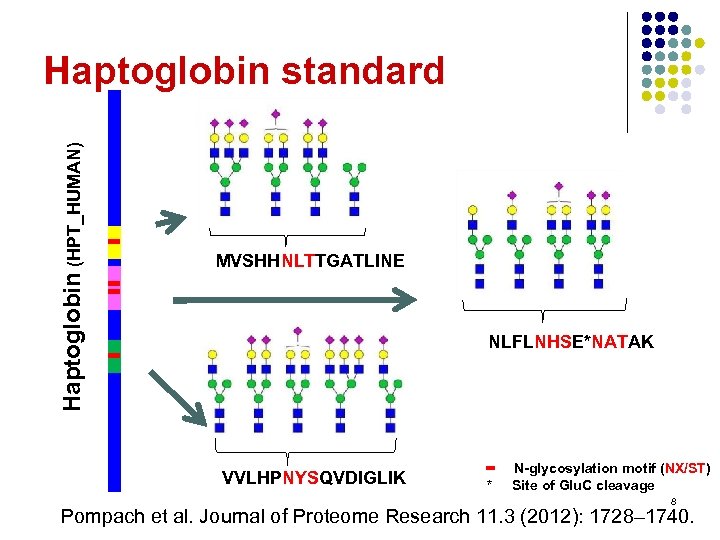
Haptoglobin (HPT_HUMAN) Haptoglobin standard MVSHHNLTTGATLINE NLFLNHSE*NATAK VVLHPNYSQVDIGLIK • * N-glycosylation motif (NX/ST) Site of Glu. C cleavage 8 Pompach et al. Journal of Proteome Research 11. 3 (2012): 1728– 1740.
Haptoglobin standard l 11 HILIC fractions enriched for glycopeptides l l 263 spectra matched to peptide-glycan pairs l l l 11 x LC-MS/MS acquisitions (≥ 15 k spectra) 2887/3288 MS/MS spectra have oxonium ion(s) 317 have "intact-peptide" fragment ions 52% matched single-glycan 8% matched multi-peptide 27 distinct (mass) glycans on 11 peptides l Glycans identified on all 4 haptoglobin sites 9
Algorithms & Infrastructure l Glycan databases indexed by composition, mass, N-linked, and motif/type l l l Monosaccharide decomposition of glycan mass l l Formats: IUPAC, Linear Code, Glyco. CT_condensed Implemented: Glycome. DB, Cartoonist, CFG Array Böcker et al. Efficient mass decomposition (2005) χ2 Goodness-of-fit test for precursor cluster l l Theoretical isotope cluster from composition. ICScore based on χ2 -test p-value. 10

False Discovery Rate (FDR) l How confident can we be in these massmatches? 11

False Discovery Rate (FDR) l How confident can we be in these massmatches? FDR: 3. 9% [ ~ 10 / 263 spectra ] 12
False Discovery Rate (FDR) l How confident can we be in these massmatches? FDR: 3. 9% [ ~ 10 / 263 spectra ] l Estimate the number of errors by searching with non-N-linked motif (decoy) peptides too. l l Count spectra matched to decoy peptide-glycan pairs. Rescale decoy counts to balance the number of motif and non-motif peptides. 13
Tuning the filters… l Adjusting thresholds and parameters to l l Increase specificity (lower FDR, fewer spectra), or Increase sensitivity (more spectra, higher FDR) 14
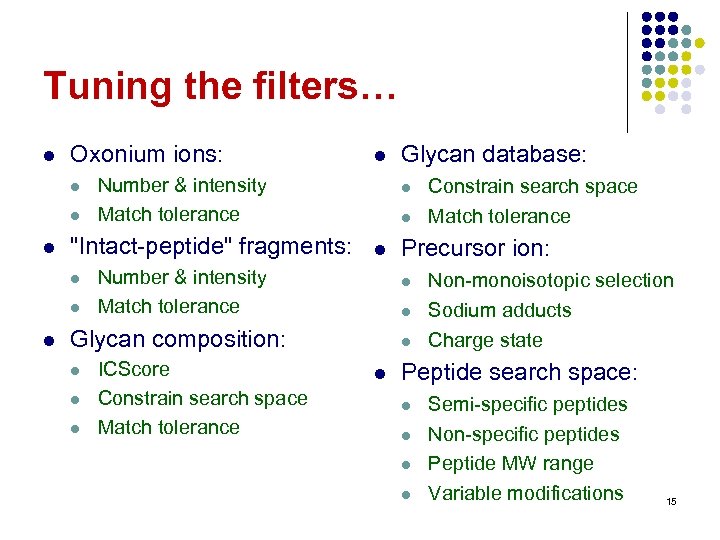
Tuning the filters… l Oxonium ions: l l l Number & intensity Match tolerance "Intact-peptide" fragments: l l l Number & intensity Match tolerance l l ICScore Constrain search space Match tolerance l l l Constrain search space Match tolerance Precursor ion: l Glycan composition: l Glycan database: Non-monoisotopic selection Sodium adducts Charge state Peptide search space: l l Semi-specific peptides Non-specific peptides Peptide MW range Variable modifications 15
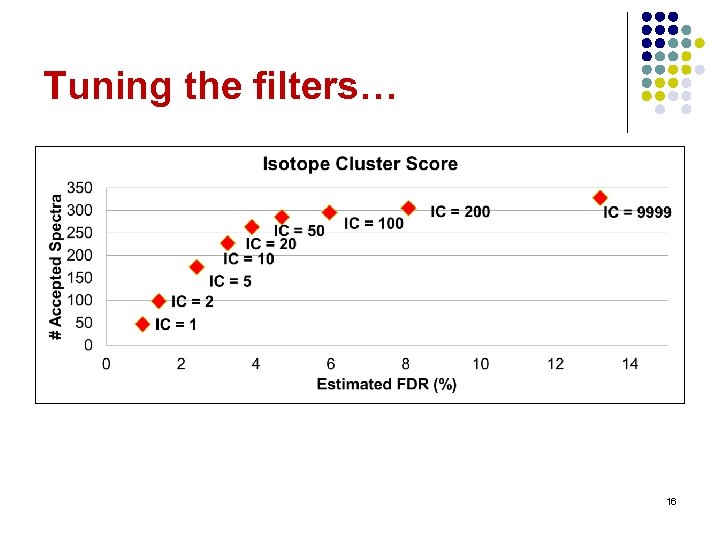
Tuning the filters… 16
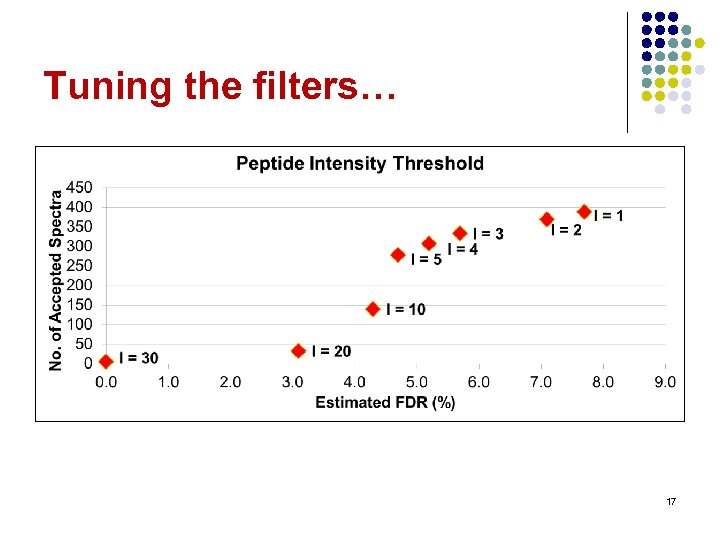
Tuning the filters… 17

Glyco. Peptide. Search (GPS) 1. 3 l Freely available implementation l l l Windows, Linux Reads open-format spectra (mz. XML, MGF) Pre-indexed Glycan databases l l l Human & Mammalian Glycome. DB Mammalian CFG Array (v 5. 0) User-Named (Oxford notation) l In silico digest and N-linked motif identification l Automatic target/decoy analysis for FDR http: //edwardslab. bmcb. georgetown. edu/GPS l 18

Where to from here? l l Demonstrate utility on new instrument platforms, proteins, samples Develop a scoring model for fragments Re-implement Cartoonist demerits Exploit relationships between l l MS 2 spectra, MSn spectra Explore application to l O-glycopeptides, N-glycans, O-glycans 19
Acknowledgements l Edwards Lab (Georgetown) l l Goldman Lab (Georgetown) l l Radoslav Goldman (Poster 6) Petr Pompach Miloslav Sanda (Poster 23) Marshal Bern (Xerox PARC) l l Kevin Brown Chandler [NSF] (Poster 32) Cartoonist, Peptoonist Rene Ranzinger (CCRC) l Glycome. DB 20
22ba76b2a04d9336215cb689790fc440.ppt