73b74576f4d2e1f530203282cd68bbde.ppt
- Количество слайдов: 50
 Genregulation Physics of transcription control and expression analysis Systems biophysics 2010/05/11 Literature - Alberts/Lehninger - Kim Sneppen & G. Zocchi: Physics in Molecular Biology - E. Klipp et al. : Systems Biology in Practice
Genregulation Physics of transcription control and expression analysis Systems biophysics 2010/05/11 Literature - Alberts/Lehninger - Kim Sneppen & G. Zocchi: Physics in Molecular Biology - E. Klipp et al. : Systems Biology in Practice
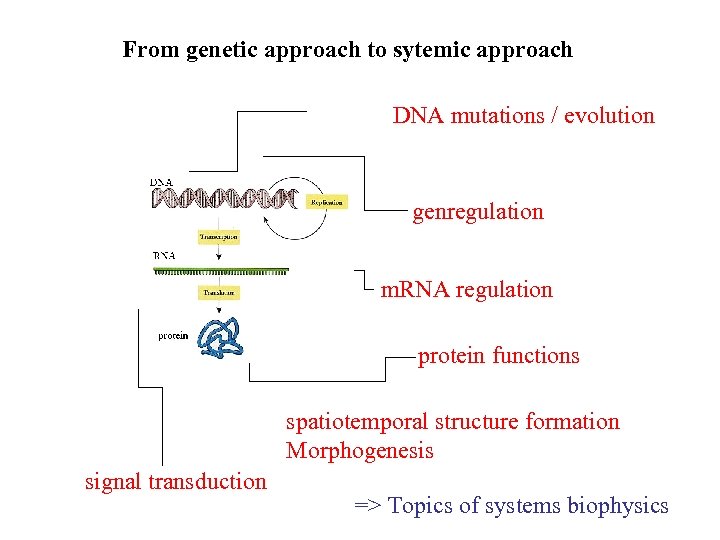 From genetic approach to sytemic approach DNA mutations / evolution genregulation m. RNA regulation protein functions spatiotemporal structure formation Morphogenesis signal transduction => Topics of systems biophysics
From genetic approach to sytemic approach DNA mutations / evolution genregulation m. RNA regulation protein functions spatiotemporal structure formation Morphogenesis signal transduction => Topics of systems biophysics
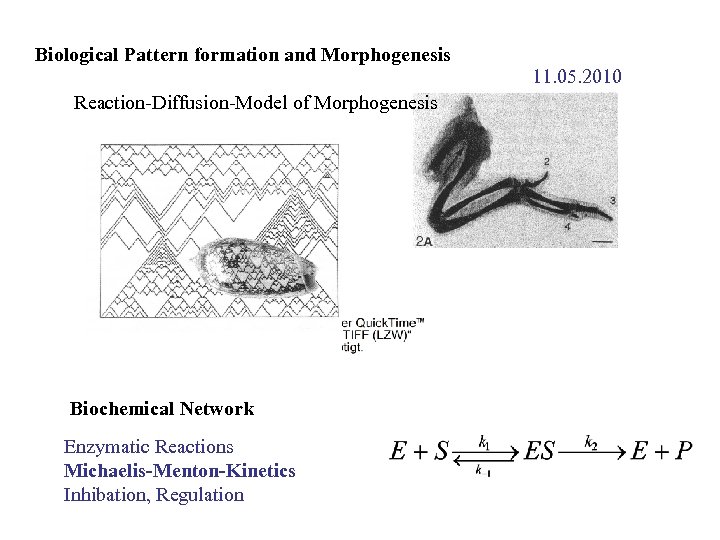 Biological Pattern formation and Morphogenesis Reaction-Diffusion-Model of Morphogenesis Biochemical Network Enzymatic Reactions Michaelis-Menton-Kinetics Inhibation, Regulation 11. 05. 2010
Biological Pattern formation and Morphogenesis Reaction-Diffusion-Model of Morphogenesis Biochemical Network Enzymatic Reactions Michaelis-Menton-Kinetics Inhibation, Regulation 11. 05. 2010
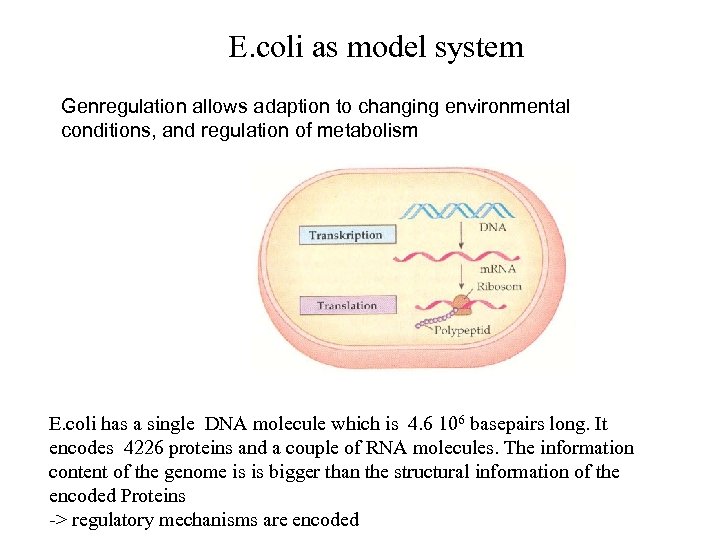 E. coli as model system Genregulation allows adaption to changing environmental conditions, and regulation of metabolism E. coli has a single DNA molecule which is 4. 6 106 basepairs long. It encodes 4226 proteins and a couple of RNA molecules. The information content of the genome is is bigger than the structural information of the encoded Proteins -> regulatory mechanisms are encoded
E. coli as model system Genregulation allows adaption to changing environmental conditions, and regulation of metabolism E. coli has a single DNA molecule which is 4. 6 106 basepairs long. It encodes 4226 proteins and a couple of RNA molecules. The information content of the genome is is bigger than the structural information of the encoded Proteins -> regulatory mechanisms are encoded
 Content of this lecture: Basics: Monod Model, Lac Operon Statistical Physics of DNA-binding Proteins Modelling of genregulatory Networks (ODE & Boolian Networks) Dynamics of Protein-DNA binding DNA looping Analysis of gene expression data Synthetic Networks
Content of this lecture: Basics: Monod Model, Lac Operon Statistical Physics of DNA-binding Proteins Modelling of genregulatory Networks (ODE & Boolian Networks) Dynamics of Protein-DNA binding DNA looping Analysis of gene expression data Synthetic Networks
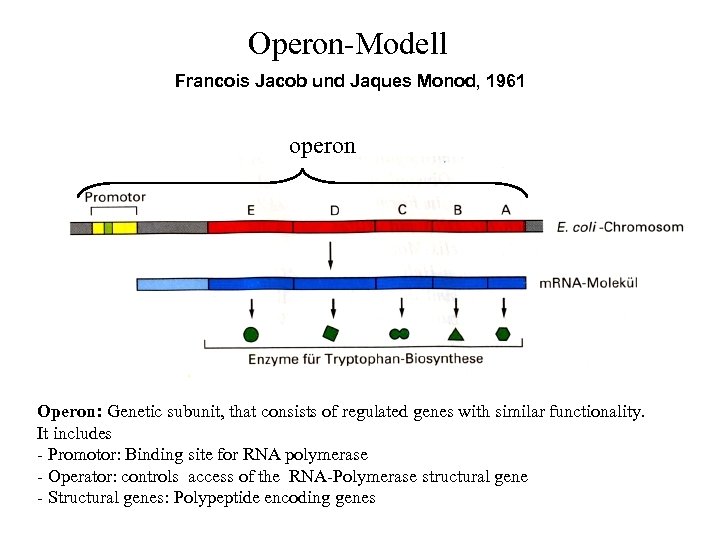 Operon-Modell Francois Jacob und Jaques Monod, 1961 operon Operon: Genetic subunit, that consists of regulated genes with similar functionality. It includes - Promotor: Binding site for RNA polymerase - Operator: controls access of the RNA-Polymerase structural gene - Structural genes: Polypeptide encoding genes
Operon-Modell Francois Jacob und Jaques Monod, 1961 operon Operon: Genetic subunit, that consists of regulated genes with similar functionality. It includes - Promotor: Binding site for RNA polymerase - Operator: controls access of the RNA-Polymerase structural gene - Structural genes: Polypeptide encoding genes
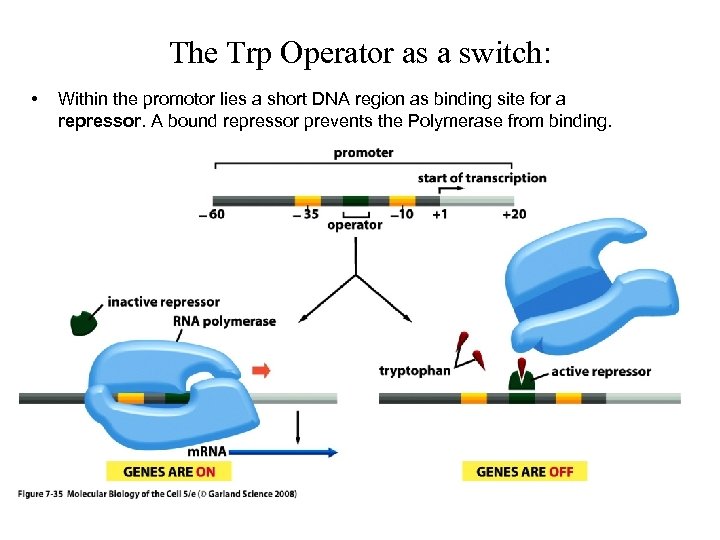 The Trp Operator as a switch: • Within the promotor lies a short DNA region as binding site for a repressor. A bound repressor prevents the Polymerase from binding.
The Trp Operator as a switch: • Within the promotor lies a short DNA region as binding site for a repressor. A bound repressor prevents the Polymerase from binding.
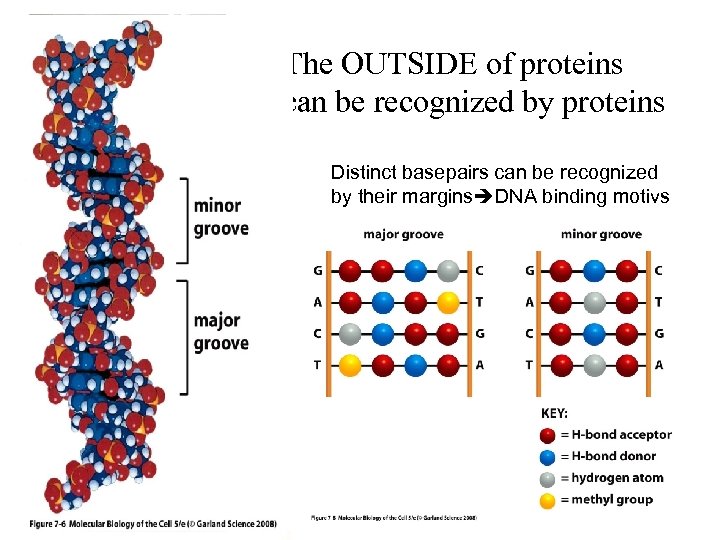 The OUTSIDE of proteins can be recognized by proteins Small channel Large channel Distinct basepairs can be recognized by their margins DNA binding motivs
The OUTSIDE of proteins can be recognized by proteins Small channel Large channel Distinct basepairs can be recognized by their margins DNA binding motivs
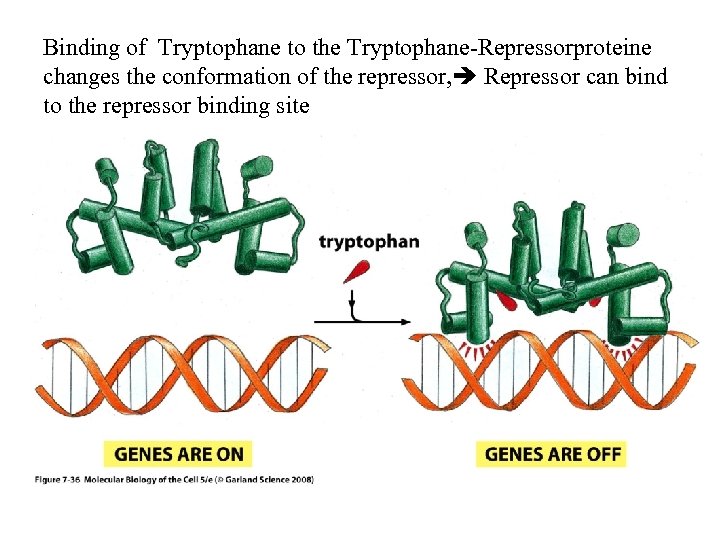 Binding of Tryptophane to the Tryptophane-Repressorproteine changes the conformation of the repressor, Repressor can bind to the repressor binding site
Binding of Tryptophane to the Tryptophane-Repressorproteine changes the conformation of the repressor, Repressor can bind to the repressor binding site
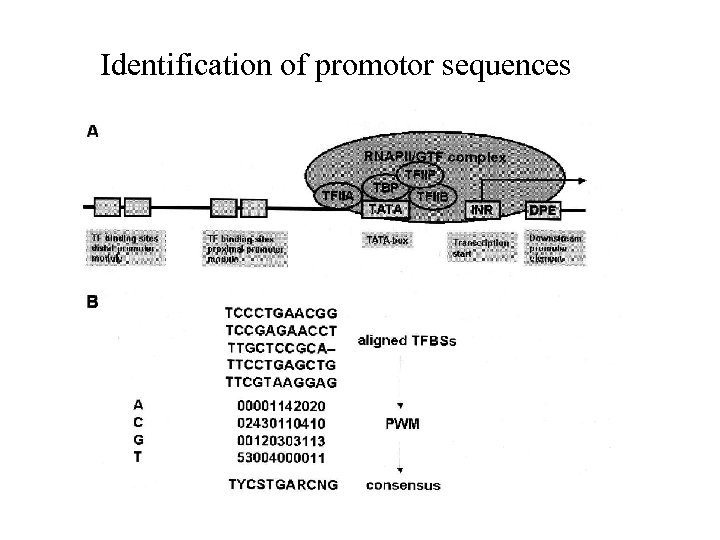 Identification of promotor sequences
Identification of promotor sequences
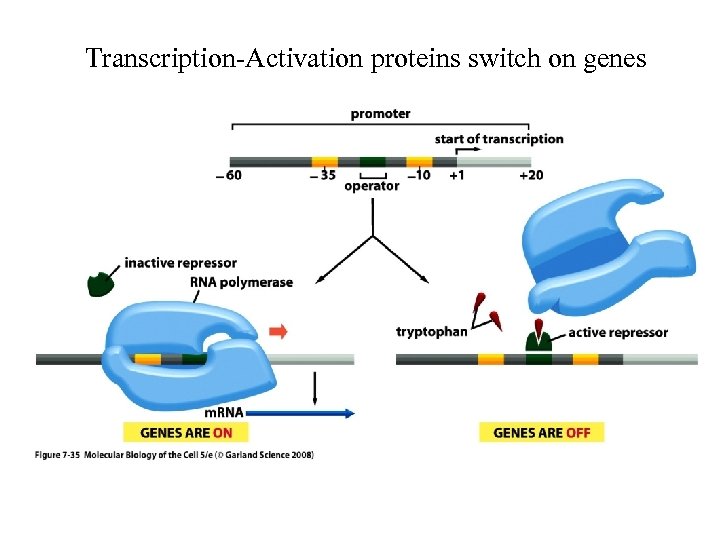 Transcription-Activation proteins switch on genes
Transcription-Activation proteins switch on genes
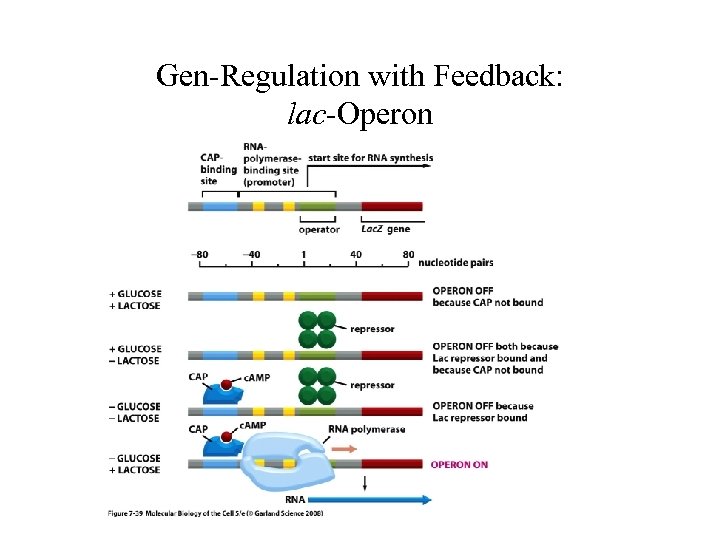 Gen-Regulation with Feedback: lac-Operon IPTG, TMG Lac. I
Gen-Regulation with Feedback: lac-Operon IPTG, TMG Lac. I
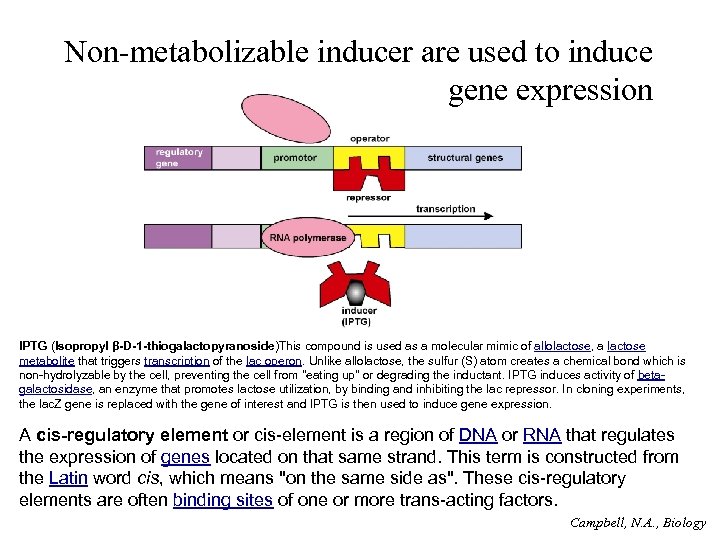 Non-metabolizable inducer are used to induce gene expression IPTG (Isopropyl β-D-1 -thiogalactopyranoside)This compound is used as a molecular mimic of allolactose, a lactose metabolite that triggers transcription of the lac operon. Unlike allolactose, the sulfur (S) atom creates a chemical bond which is non-hydrolyzable by the cell, preventing the cell from "eating up" or degrading the inductant. IPTG induces activity of betagalactosidase, an enzyme that promotes lactose utilization, by binding and inhibiting the lac repressor. In cloning experiments, the lac. Z gene is replaced with the gene of interest and IPTG is then used to induce gene expression. A cis-regulatory element or cis-element is a region of DNA or RNA that regulates the expression of genes located on that same strand. This term is constructed from the Latin word cis, which means "on the same side as". These cis-regulatory elements are often binding sites of one or more trans-acting factors. Campbell, N. A. , Biology
Non-metabolizable inducer are used to induce gene expression IPTG (Isopropyl β-D-1 -thiogalactopyranoside)This compound is used as a molecular mimic of allolactose, a lactose metabolite that triggers transcription of the lac operon. Unlike allolactose, the sulfur (S) atom creates a chemical bond which is non-hydrolyzable by the cell, preventing the cell from "eating up" or degrading the inductant. IPTG induces activity of betagalactosidase, an enzyme that promotes lactose utilization, by binding and inhibiting the lac repressor. In cloning experiments, the lac. Z gene is replaced with the gene of interest and IPTG is then used to induce gene expression. A cis-regulatory element or cis-element is a region of DNA or RNA that regulates the expression of genes located on that same strand. This term is constructed from the Latin word cis, which means "on the same side as". These cis-regulatory elements are often binding sites of one or more trans-acting factors. Campbell, N. A. , Biology
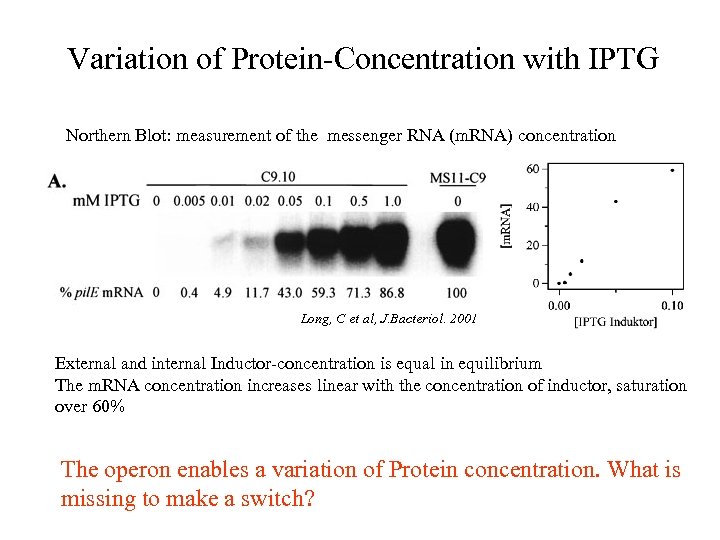 Variation of Protein-Concentration with IPTG Northern Blot: measurement of the messenger RNA (m. RNA) concentration Long, C et al, J. Bacteriol. 2001 External and internal Inductor-concentration is equal in equilibrium The m. RNA concentration increases linear with the concentration of inductor, saturation over 60% The operon enables a variation of Protein concentration. What is missing to make a switch?
Variation of Protein-Concentration with IPTG Northern Blot: measurement of the messenger RNA (m. RNA) concentration Long, C et al, J. Bacteriol. 2001 External and internal Inductor-concentration is equal in equilibrium The m. RNA concentration increases linear with the concentration of inductor, saturation over 60% The operon enables a variation of Protein concentration. What is missing to make a switch?
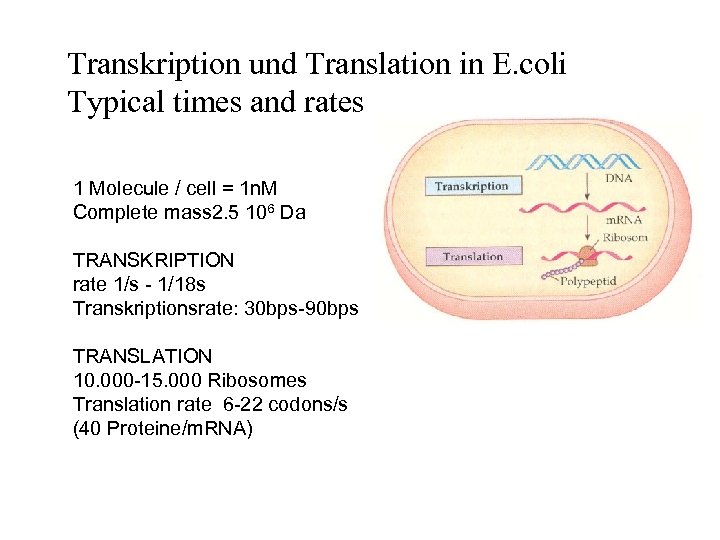 Transkription und Translation in E. coli Typical times and rates 1 Molecule / cell = 1 n. M Complete mass 2. 5 106 Da TRANSKRIPTION rate 1/s - 1/18 s Transkriptionsrate: 30 bps-90 bps TRANSLATION 10. 000 -15. 000 Ribosomes Translation rate 6 -22 codons/s (40 Proteine/m. RNA)
Transkription und Translation in E. coli Typical times and rates 1 Molecule / cell = 1 n. M Complete mass 2. 5 106 Da TRANSKRIPTION rate 1/s - 1/18 s Transkriptionsrate: 30 bps-90 bps TRANSLATION 10. 000 -15. 000 Ribosomes Translation rate 6 -22 codons/s (40 Proteine/m. RNA)
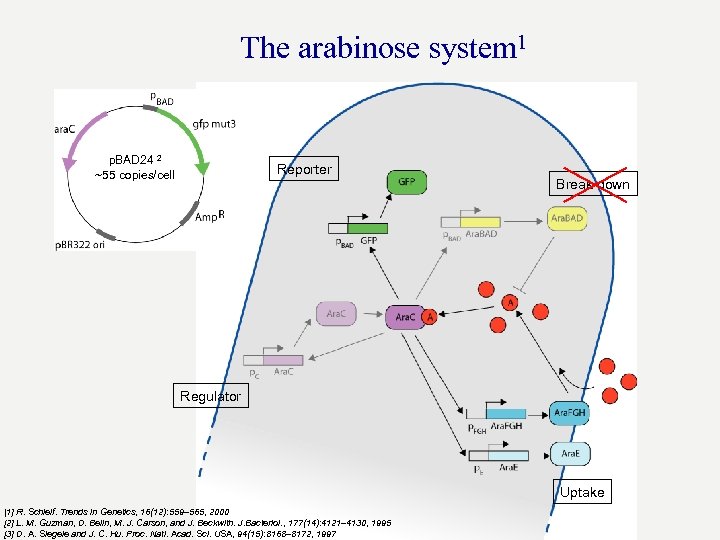 The arabinose system 1 p. BAD 24 2 ~55 copies/cell Reporter Break down Regulator Uptake [1] R. Schleif. Trends in Genetics, 16(12): 559– 565, 2000 [2] L. M. Guzman, D. Belin, M. J. Carson, and J. Beckwith. J. Bacteriol. , 177(14): 4121– 4130, 1995 [3] D. A. Siegele and J. C. Hu. Proc. Natl. Acad. Sci. USA, 94(15): 8168– 8172, 1997
The arabinose system 1 p. BAD 24 2 ~55 copies/cell Reporter Break down Regulator Uptake [1] R. Schleif. Trends in Genetics, 16(12): 559– 565, 2000 [2] L. M. Guzman, D. Belin, M. J. Carson, and J. Beckwith. J. Bacteriol. , 177(14): 4121– 4130, 1995 [3] D. A. Siegele and J. C. Hu. Proc. Natl. Acad. Sci. USA, 94(15): 8168– 8172, 1997
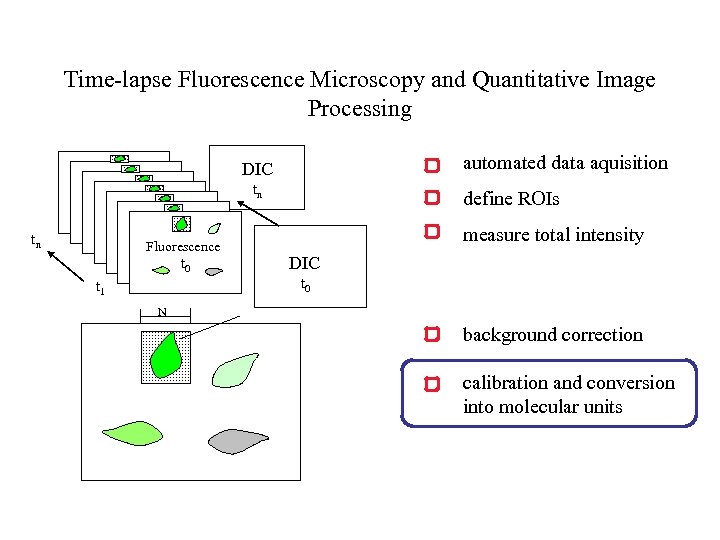 Time-lapse Fluorescence Microscopy and Quantitative Image Processing automated data aquisition DIC tn tn Fluorescence t 0 define ROIs measure total intensity DIC t 0 t 1 N background correction calibration and conversion into molecular units
Time-lapse Fluorescence Microscopy and Quantitative Image Processing automated data aquisition DIC tn tn Fluorescence t 0 define ROIs measure total intensity DIC t 0 t 1 N background correction calibration and conversion into molecular units
 Judith. Megerle@physik. lmu. de
Judith. Megerle@physik. lmu. de
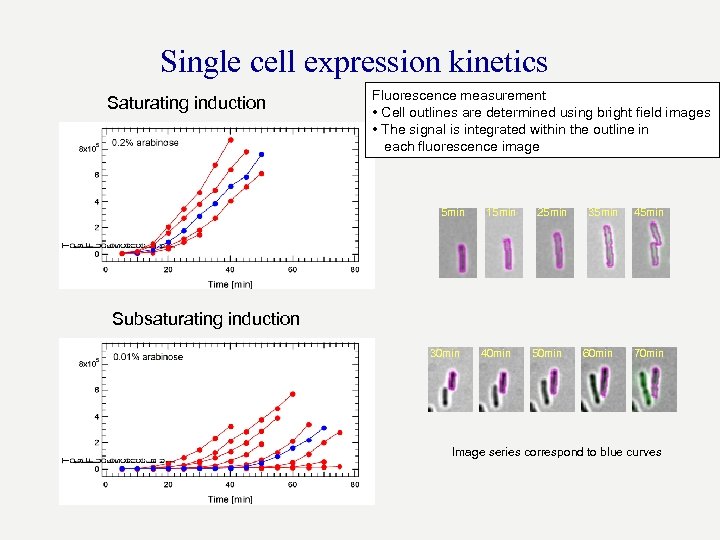 Single cell expression kinetics Saturating induction Fluorescence measurement • Cell outlines are determined using bright field images • The signal is integrated within the outline in each fluorescence image 5 min 15 min 25 min 35 min 45 min Subsaturating induction 30 min 40 min 50 min 60 min 70 min Image series correspond to blue curves
Single cell expression kinetics Saturating induction Fluorescence measurement • Cell outlines are determined using bright field images • The signal is integrated within the outline in each fluorescence image 5 min 15 min 25 min 35 min 45 min Subsaturating induction 30 min 40 min 50 min 60 min 70 min Image series correspond to blue curves
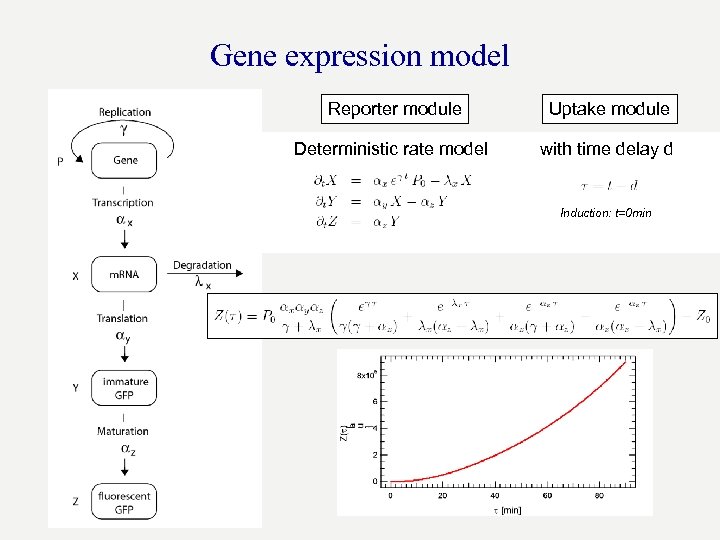 Gene expression model Reporter module Uptake module Deterministic rate model with time delay d Induction: t=0 min
Gene expression model Reporter module Uptake module Deterministic rate model with time delay d Induction: t=0 min
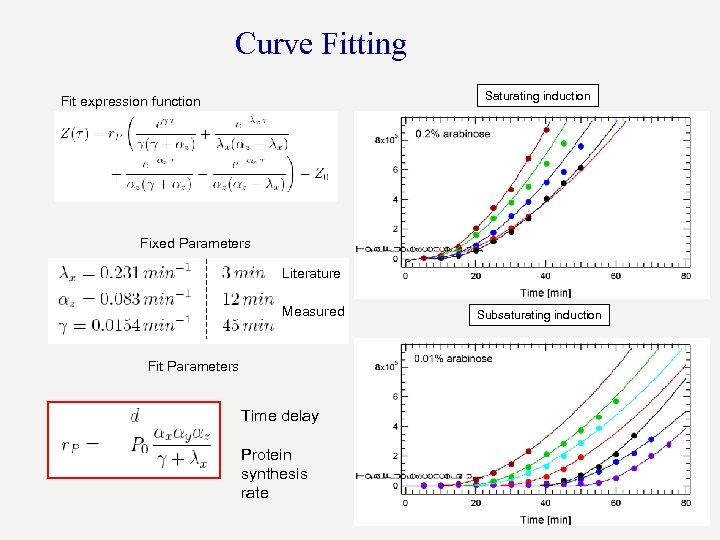 Curve Fitting Saturating induction Fit expression function Fixed Parameters Literature Measured Fit Parameters Time delay Protein synthesis rate Subsaturating induction
Curve Fitting Saturating induction Fit expression function Fixed Parameters Literature Measured Fit Parameters Time delay Protein synthesis rate Subsaturating induction
 Ohter example: Quorum Sensing
Ohter example: Quorum Sensing
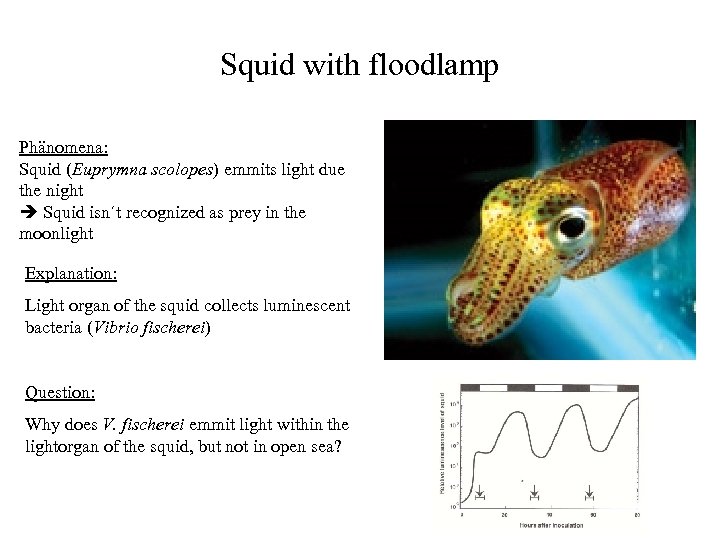 Squid with floodlamp Phänomena: Squid (Euprymna scolopes) emmits light due the night Squid isn´t recognized as prey in the moonlight Explanation: Light organ of the squid collects luminescent bacteria (Vibrio fischerei) Question: Why does V. fischerei emmit light within the lightorgan of the squid, but not in open sea?
Squid with floodlamp Phänomena: Squid (Euprymna scolopes) emmits light due the night Squid isn´t recognized as prey in the moonlight Explanation: Light organ of the squid collects luminescent bacteria (Vibrio fischerei) Question: Why does V. fischerei emmit light within the lightorgan of the squid, but not in open sea?
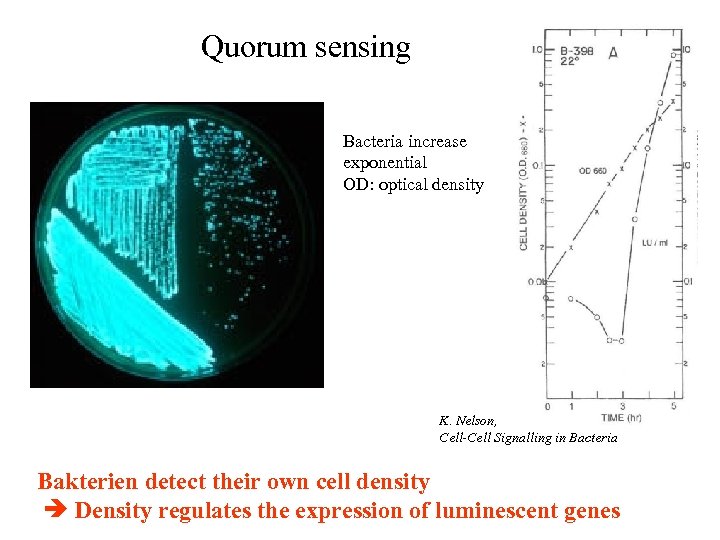 Quorum sensing Bacteria increase exponential OD: optical density K. Nelson, Cell-Cell Signalling in Bacteria Bakterien detect their own cell density Density regulates the expression of luminescent genes
Quorum sensing Bacteria increase exponential OD: optical density K. Nelson, Cell-Cell Signalling in Bacteria Bakterien detect their own cell density Density regulates the expression of luminescent genes
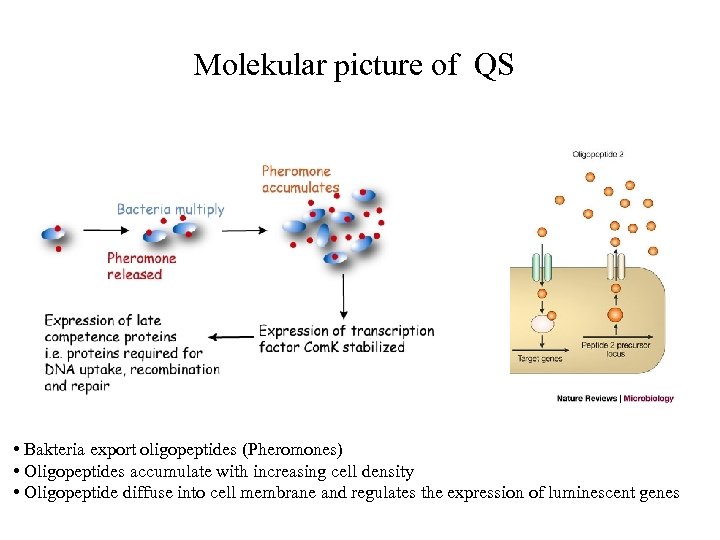 Molekular picture of QS • Bakteria export oligopeptides (Pheromones) • Oligopeptides accumulate with increasing cell density • Oligopeptide diffuse into cell membrane and regulates the expression of luminescent genes
Molekular picture of QS • Bakteria export oligopeptides (Pheromones) • Oligopeptides accumulate with increasing cell density • Oligopeptide diffuse into cell membrane and regulates the expression of luminescent genes
 Searching the binding site
Searching the binding site
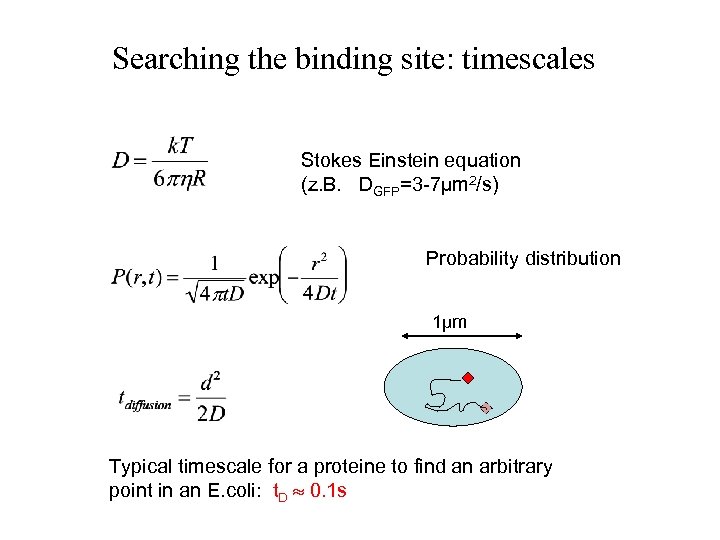 Searching the binding site: timescales Stokes Einstein equation (z. B. DGFP=3 -7µm 2/s) Probability distribution 1µm Typical timescale for a proteine to find an arbitrary point in an E. coli: t. D 0. 1 s
Searching the binding site: timescales Stokes Einstein equation (z. B. DGFP=3 -7µm 2/s) Probability distribution 1µm Typical timescale for a proteine to find an arbitrary point in an E. coli: t. D 0. 1 s
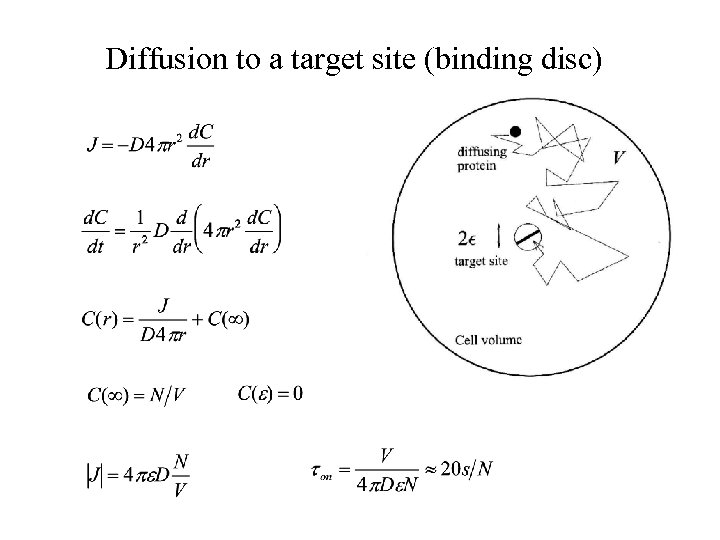 Diffusion to a target site (binding disc)
Diffusion to a target site (binding disc)
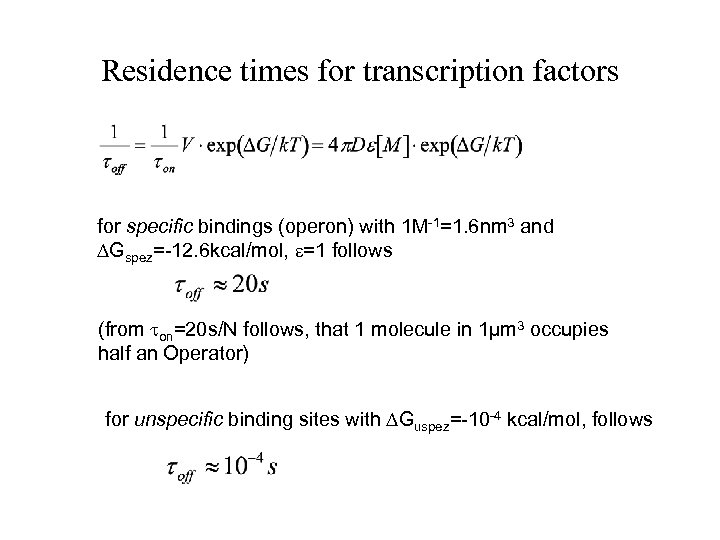 Residence times for transcription factors for specific bindings (operon) with 1 M-1=1. 6 nm 3 and Gspez=-12. 6 kcal/mol, =1 follows (from on=20 s/N follows, that 1 molecule in 1µm 3 occupies half an Operator) for unspecific binding sites with Guspez=-10 -4 kcal/mol, follows
Residence times for transcription factors for specific bindings (operon) with 1 M-1=1. 6 nm 3 and Gspez=-12. 6 kcal/mol, =1 follows (from on=20 s/N follows, that 1 molecule in 1µm 3 occupies half an Operator) for unspecific binding sites with Guspez=-10 -4 kcal/mol, follows
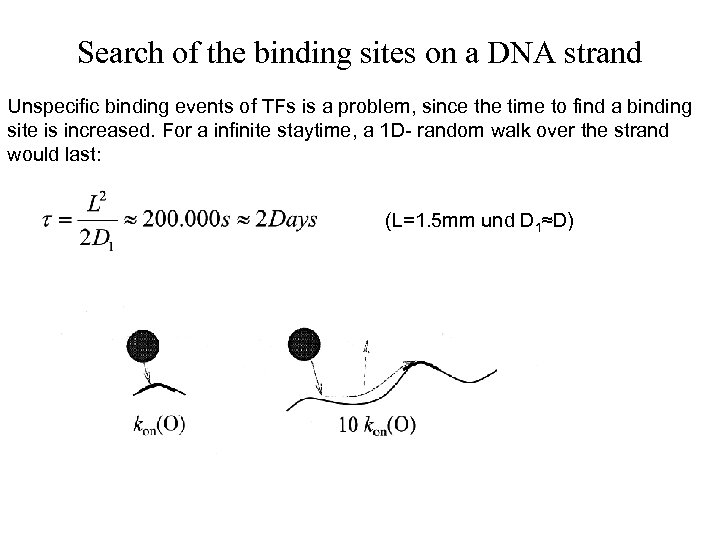 Search of the binding sites on a DNA strand Unspecific binding events of TFs is a problem, since the time to find a binding site is increased. For a infinite staytime, a 1 D- random walk over the strand would last: (L=1. 5 mm und D 1≈D)
Search of the binding sites on a DNA strand Unspecific binding events of TFs is a problem, since the time to find a binding site is increased. For a infinite staytime, a 1 D- random walk over the strand would last: (L=1. 5 mm und D 1≈D)
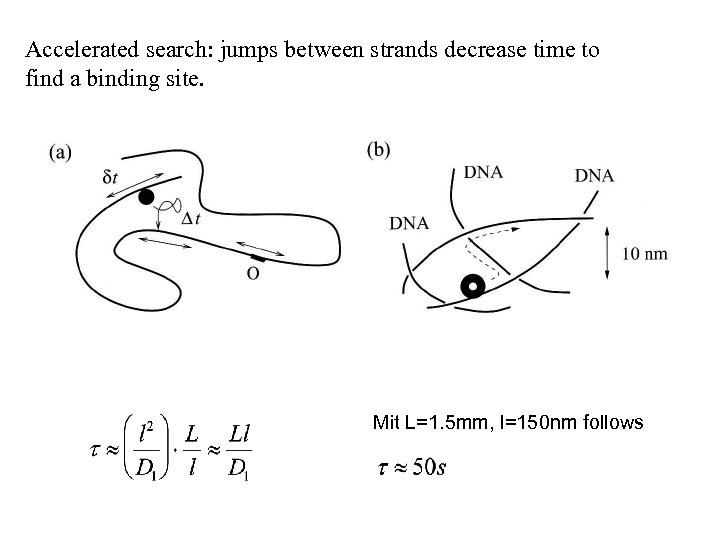 Accelerated search: jumps between strands decrease time to find a binding site. Mit L=1. 5 mm, l=150 nm follows
Accelerated search: jumps between strands decrease time to find a binding site. Mit L=1. 5 mm, l=150 nm follows
 Boolian Networks, or what cells and computers have in common.
Boolian Networks, or what cells and computers have in common.
 (Nature, Dec 99)
(Nature, Dec 99)
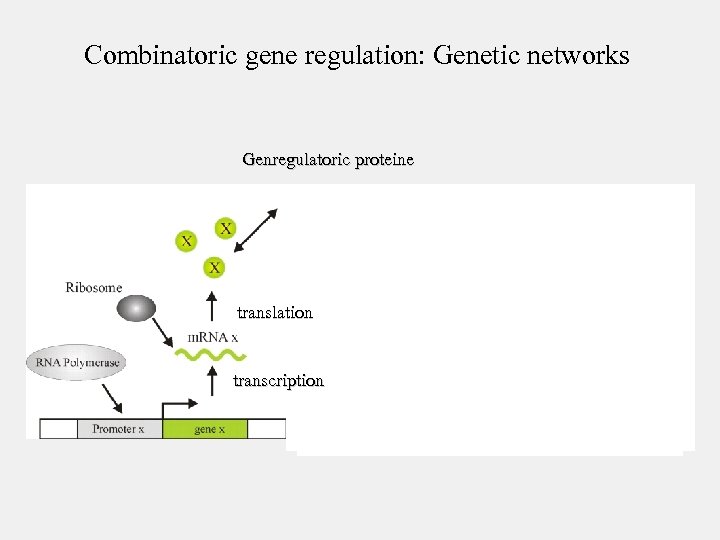 Combinatoric gene regulation: Genetic networks Genregulatoric proteine translation transcription
Combinatoric gene regulation: Genetic networks Genregulatoric proteine translation transcription
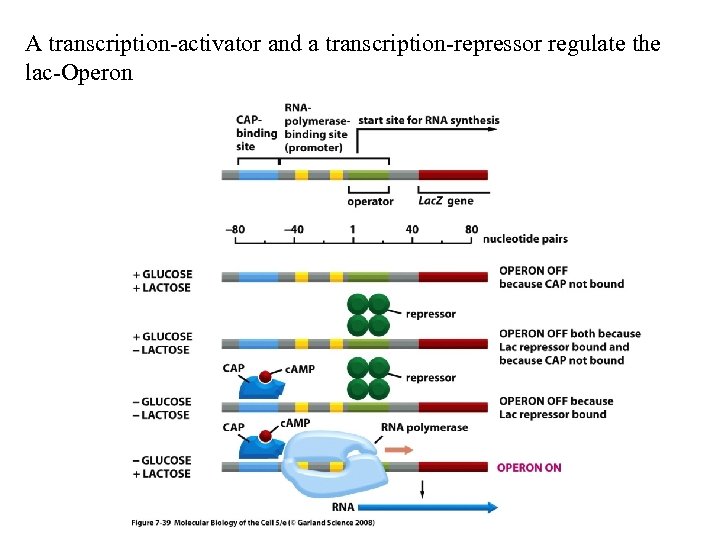 A transcription-activator and a transcription-repressor regulate the lac-Operon
A transcription-activator and a transcription-repressor regulate the lac-Operon
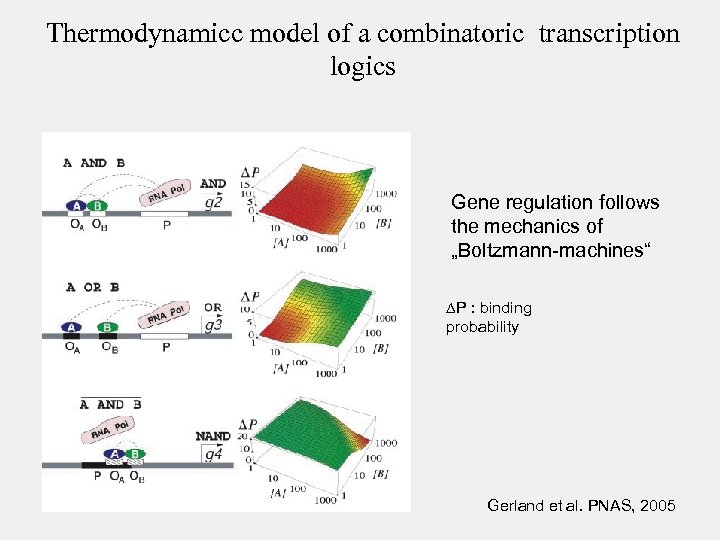 Thermodynamicc model of a combinatoric transcription logics Gene regulation follows the mechanics of „Boltzmann-machines“ P : binding probability Gerland et al. PNAS, 2005
Thermodynamicc model of a combinatoric transcription logics Gene regulation follows the mechanics of „Boltzmann-machines“ P : binding probability Gerland et al. PNAS, 2005
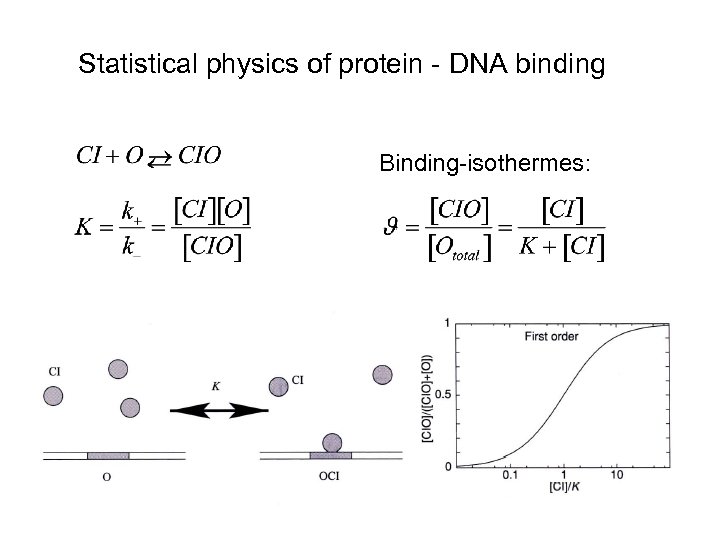 Statistical physics of protein - DNA binding Binding-isothermes:
Statistical physics of protein - DNA binding Binding-isothermes:
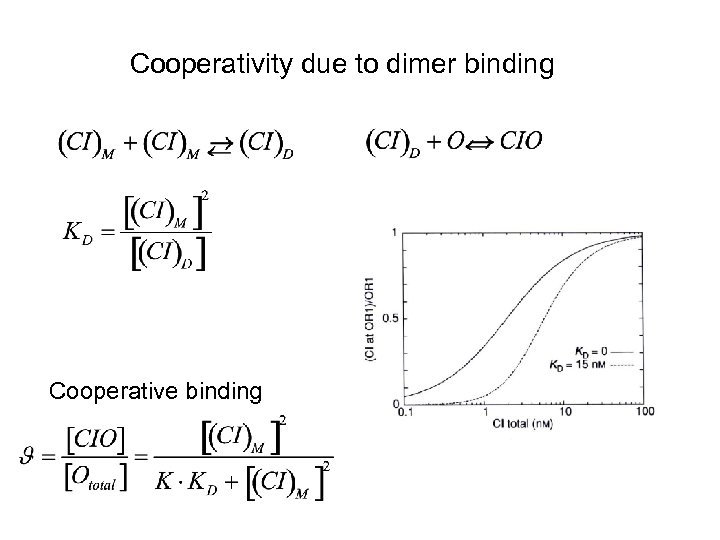 Cooperativity due to dimer binding Cooperative binding
Cooperativity due to dimer binding Cooperative binding
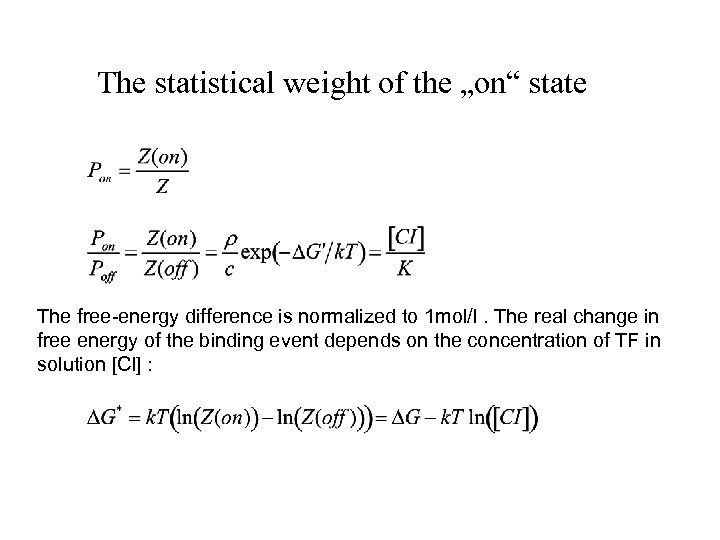 The statistical weight of the „on“ state The free-energy difference is normalized to 1 mol/l. The real change in free energy of the binding event depends on the concentration of TF in solution [Cl] :
The statistical weight of the „on“ state The free-energy difference is normalized to 1 mol/l. The real change in free energy of the binding event depends on the concentration of TF in solution [Cl] :
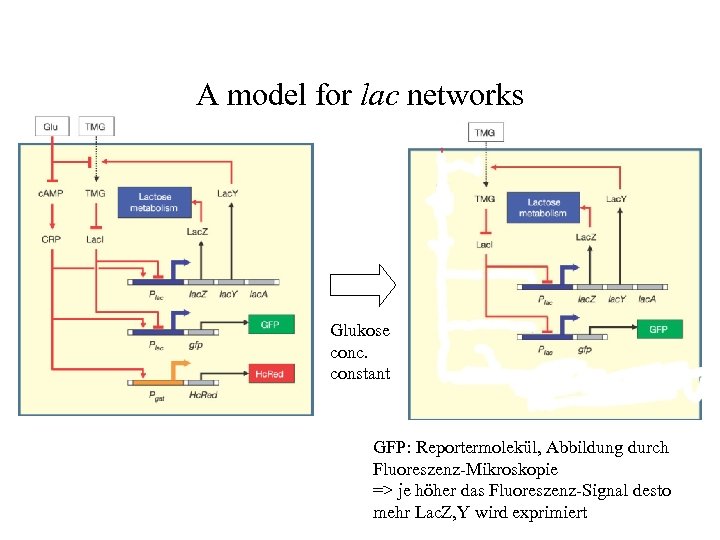 A model for lac networks Glukose conc. constant GFP: Reportermolekül, Abbildung durch Fluoreszenz-Mikroskopie => je höher das Fluoreszenz-Signal desto mehr Lac. Z, Y wird exprimiert
A model for lac networks Glukose conc. constant GFP: Reportermolekül, Abbildung durch Fluoreszenz-Mikroskopie => je höher das Fluoreszenz-Signal desto mehr Lac. Z, Y wird exprimiert
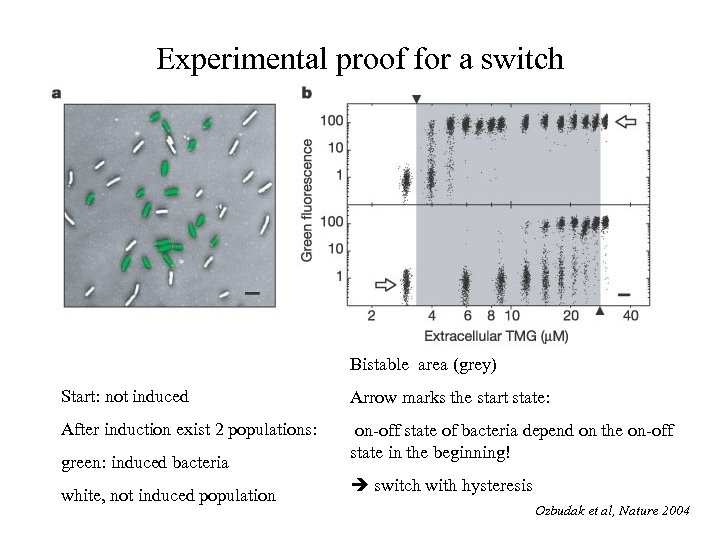 Experimental proof for a switch Bistable area (grey) Start: not induced Arrow marks the start state: After induction exist 2 populations: on-off state of bacteria depend on the on-off state in the beginning! green: induced bacteria white, not induced population switch with hysteresis Ozbudak et al, Nature 2004
Experimental proof for a switch Bistable area (grey) Start: not induced Arrow marks the start state: After induction exist 2 populations: on-off state of bacteria depend on the on-off state in the beginning! green: induced bacteria white, not induced population switch with hysteresis Ozbudak et al, Nature 2004
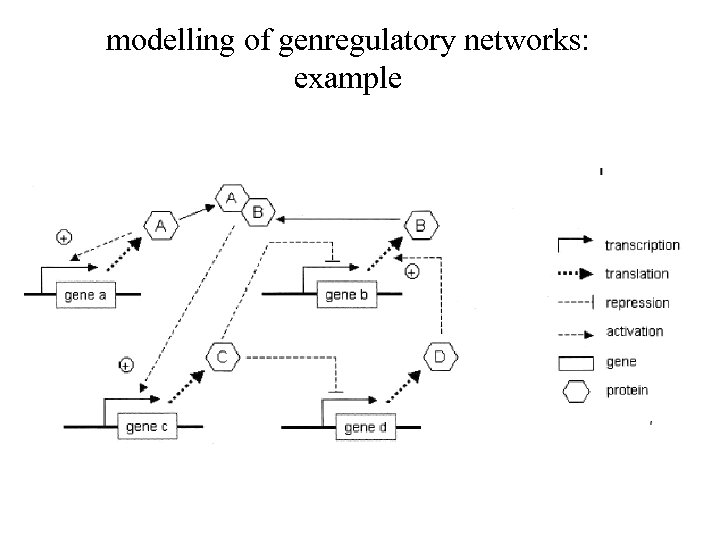 modelling of genregulatory networks: example
modelling of genregulatory networks: example
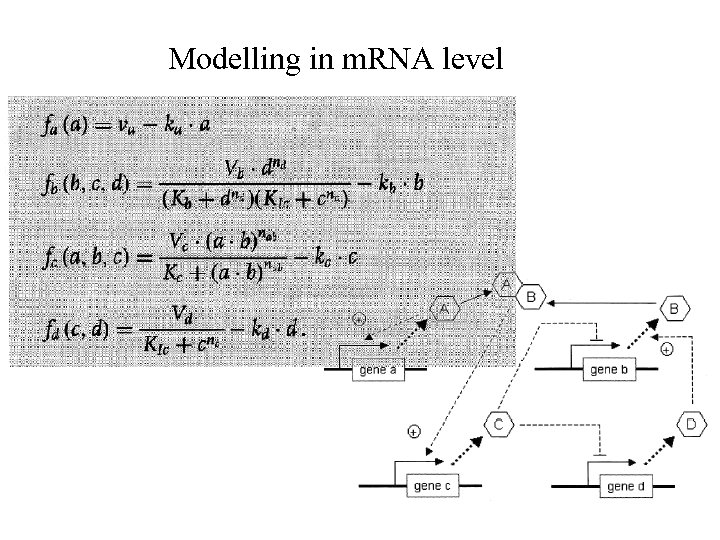 Modelling in m. RNA level
Modelling in m. RNA level
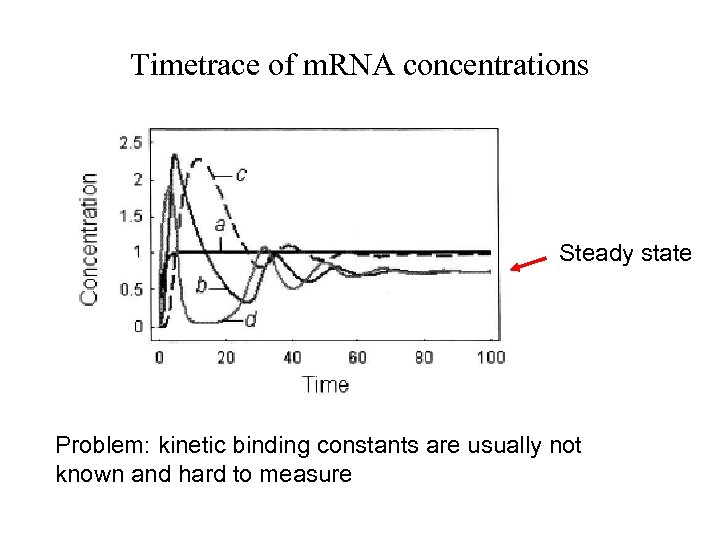 Timetrace of m. RNA concentrations Steady state Problem: kinetic binding constants are usually not known and hard to measure
Timetrace of m. RNA concentrations Steady state Problem: kinetic binding constants are usually not known and hard to measure
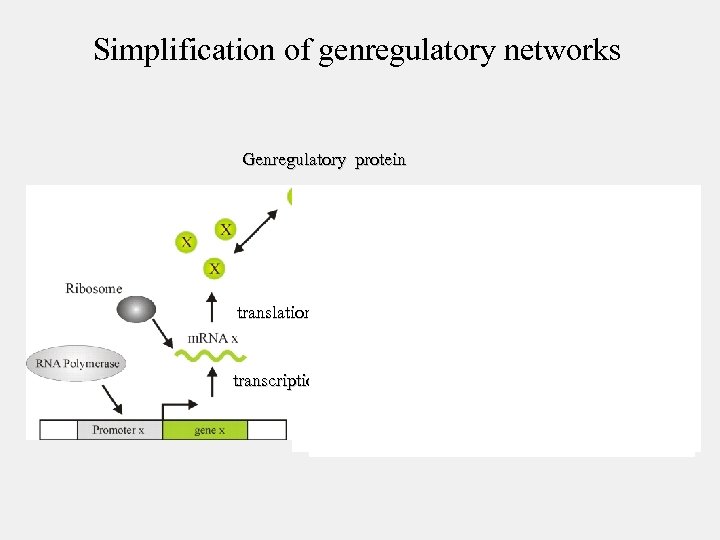 Simplification of genregulatory networks Genregulatory protein translation transcription
Simplification of genregulatory networks Genregulatory protein translation transcription
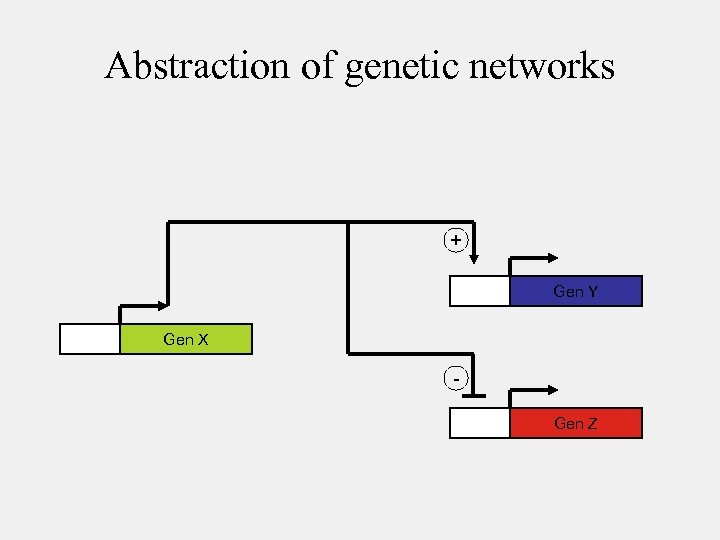 Abstraction of genetic networks + Gen Y Gen X Gen Z
Abstraction of genetic networks + Gen Y Gen X Gen Z
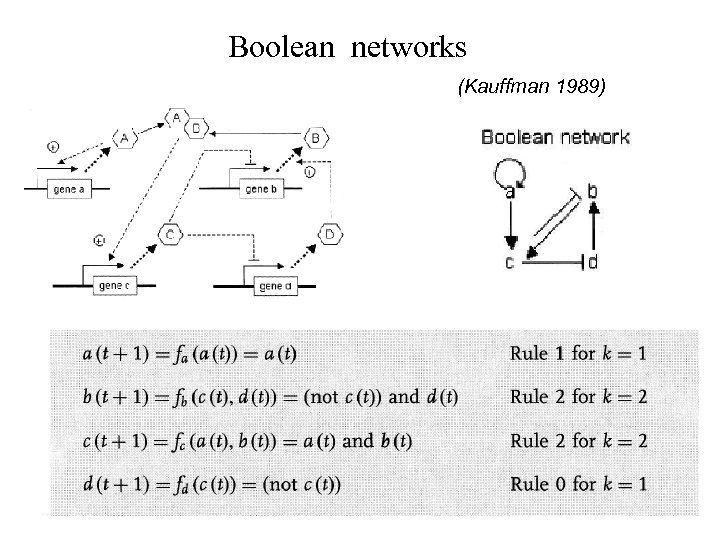 Boolean networks (Kauffman 1989)
Boolean networks (Kauffman 1989)
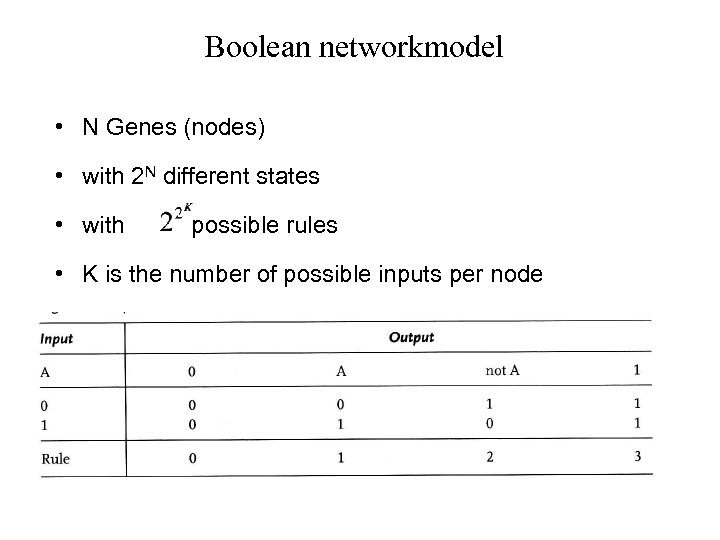 Boolean networkmodel • N Genes (nodes) • with 2 N different states • with possible rules • K is the number of possible inputs per node
Boolean networkmodel • N Genes (nodes) • with 2 N different states • with possible rules • K is the number of possible inputs per node
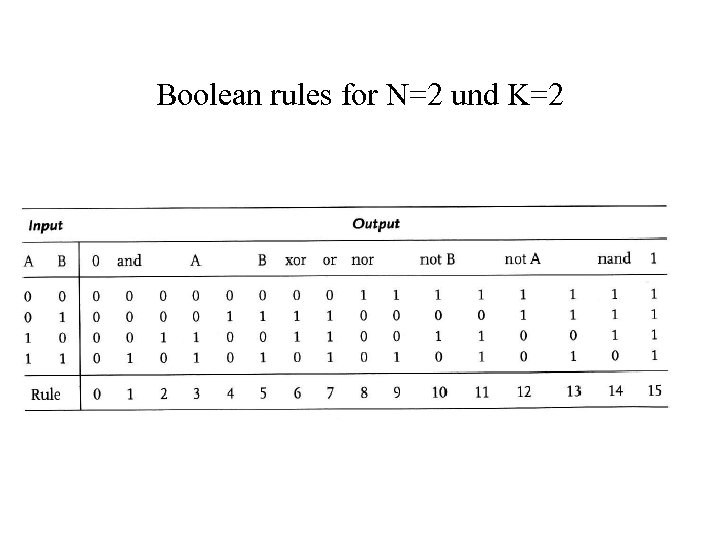 Boolean rules for N=2 und K=2
Boolean rules for N=2 und K=2
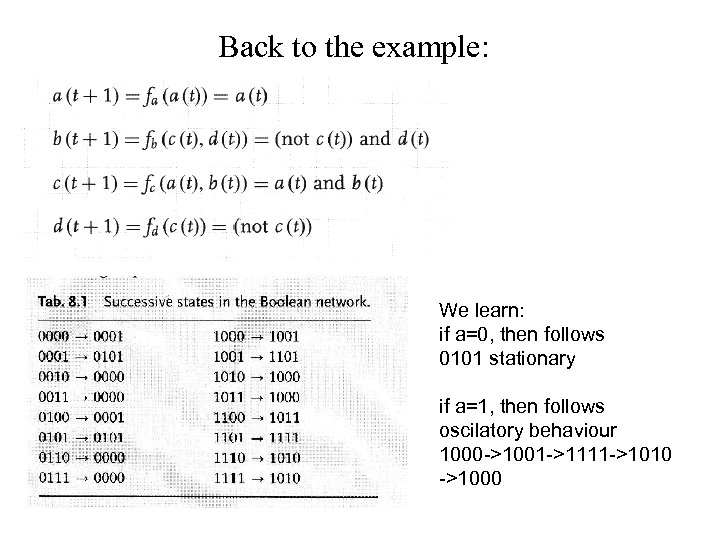 Back to the example: We learn: if a=0, then follows 0101 stationary if a=1, then follows oscilatory behaviour 1000 ->1001 ->1111 ->1010 ->1000
Back to the example: We learn: if a=0, then follows 0101 stationary if a=1, then follows oscilatory behaviour 1000 ->1001 ->1111 ->1010 ->1000


