c30c67af9992beebb2f01189c4232afe.ppt
- Количество слайдов: 54
 Genomic Evaluations G. R. Wiggans Animal Improvement Programs Laboratory Agricultural Research Service, USDA Beltsville, MD george. wiggans@ars. usda. gov Alta Genetics meeting May 2010 ( ) 1 G. R. Wiggans
Genomic Evaluations G. R. Wiggans Animal Improvement Programs Laboratory Agricultural Research Service, USDA Beltsville, MD george. wiggans@ars. usda. gov Alta Genetics meeting May 2010 ( ) 1 G. R. Wiggans
 How the system works l l l Studs and breeds nominate animals through AIPL web site Hair, blood, semen, or extracted DNA sent to 1 of 4 Labs Genotypes sent to AIPL monthly Monthly evaluation updates released on the first Tuesday of most months Official evaluations updated only at tri-annual traditional runs (except for C & P bulls) G. R. Wiggans
How the system works l l l Studs and breeds nominate animals through AIPL web site Hair, blood, semen, or extracted DNA sent to 1 of 4 Labs Genotypes sent to AIPL monthly Monthly evaluation updates released on the first Tuesday of most months Official evaluations updated only at tri-annual traditional runs (except for C & P bulls) G. R. Wiggans
 Pedigree & Nomination l l Studs may submit pedigree and nominate in batch files Pedigree for CAN, AUS, GBR automatically collected from web sites Nomination expected by the time sample arrives at lab Sample ID reported at nomination must match ID on sample at lab G. R. Wiggans
Pedigree & Nomination l l Studs may submit pedigree and nominate in batch files Pedigree for CAN, AUS, GBR automatically collected from web sites Nomination expected by the time sample arrives at lab Sample ID reported at nomination must match ID on sample at lab G. R. Wiggans
 Conflict processing l Parent-progeny conflicts detected l Sex and breed checked l l l Conflicts reported to lab and requester for resolution Pedigree changes automatically update genotype usability Foreign pedigree updates not automatic G. R. Wiggans
Conflict processing l Parent-progeny conflicts detected l Sex and breed checked l l l Conflicts reported to lab and requester for resolution Pedigree changes automatically update genotype usability Foreign pedigree updates not automatic G. R. Wiggans
 Changes in April l Deviations of predictor cows adjusted to be like bulls with similar reliability to improve their contribution to accuracy Genotypes of dams of genotyped animals imputed to add predictor animals Sum of genomic relationships of each animal with the predictor animals used to improve estimation of Reliability G. R. Wiggans
Changes in April l Deviations of predictor cows adjusted to be like bulls with similar reliability to improve their contribution to accuracy Genotypes of dams of genotyped animals imputed to add predictor animals Sum of genomic relationships of each animal with the predictor animals used to improve estimation of Reliability G. R. Wiggans
 Imputation l l Determine an animal’s genotype from genotypes of its parents and progeny Genotype separated into sire and dam contributions. Identifies the allele on each member of a chromosome pair G. R. Wiggans
Imputation l l Determine an animal’s genotype from genotypes of its parents and progeny Genotype separated into sire and dam contributions. Identifies the allele on each member of a chromosome pair G. R. Wiggans
 O-Style Haplotypes chromosome 15 G. R. Wiggans
O-Style Haplotypes chromosome 15 G. R. Wiggans
 Imputation (cont. ) l l l Inheritance of haplotypes tracked Accuracy of imputation improves with number of progeny Crossovers during meiosis contribute to uncertainty G. R. Wiggans
Imputation (cont. ) l l l Inheritance of haplotypes tracked Accuracy of imputation improves with number of progeny Crossovers during meiosis contribute to uncertainty G. R. Wiggans
 Imputation Plans l l l Add separate genomic indicator code (probably 3) to cow format 105 to identify imputed cows. Already are identified in XML files. 3 K genotypes will be imputed to 50 K, chip type code to be added to XML No authorization to release evaluations on imputed bulls. 15 HO bulls have 5+ genotyped progeny whose dams are genotyped. G. R. Wiggans
Imputation Plans l l l Add separate genomic indicator code (probably 3) to cow format 105 to identify imputed cows. Already are identified in XML files. 3 K genotypes will be imputed to 50 K, chip type code to be added to XML No authorization to release evaluations on imputed bulls. 15 HO bulls have 5+ genotyped progeny whose dams are genotyped. G. R. Wiggans
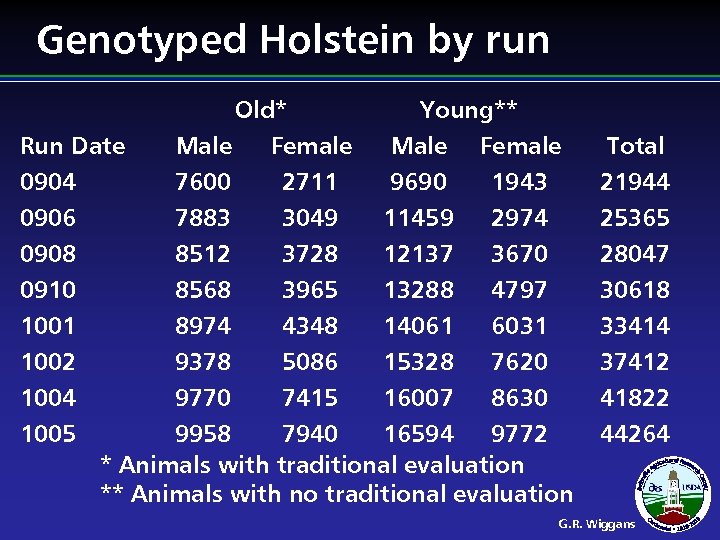 Genotyped Holstein by run Run Date 0904 0906 0908 0910 1001 1002 1004 1005 Old* Male Female 7600 2711 7883 3049 Young** Male Female 9690 1943 11459 2974 8512 3728 12137 3670 8568 3965 13288 4797 8974 4348 14061 6031 9378 5086 15328 7620 9770 7415 16007 8630 9958 7940 16594 9772 * Animals with traditional evaluation ** Animals with no traditional evaluation Total 21944 25365 28047 30618 33414 37412 41822 44264 G. R. Wiggans
Genotyped Holstein by run Run Date 0904 0906 0908 0910 1001 1002 1004 1005 Old* Male Female 7600 2711 7883 3049 Young** Male Female 9690 1943 11459 2974 8512 3728 12137 3670 8568 3965 13288 4797 8974 4348 14061 6031 9378 5086 15328 7620 9770 7415 16007 8630 9958 7940 16594 9772 * Animals with traditional evaluation ** Animals with no traditional evaluation Total 21944 25365 28047 30618 33414 37412 41822 44264 G. R. Wiggans
 Cow Adjustment l l Evaluations of elite cows biased upward Cutoff studies showed little benefit from including cows as predictors Reducing heritability would reduce the problem but industry is reluctant to do so Adjustment of cow evaluations implemented G. R. Wiggans
Cow Adjustment l l Evaluations of elite cows biased upward Cutoff studies showed little benefit from including cows as predictors Reducing heritability would reduce the problem but industry is reluctant to do so Adjustment of cow evaluations implemented G. R. Wiggans
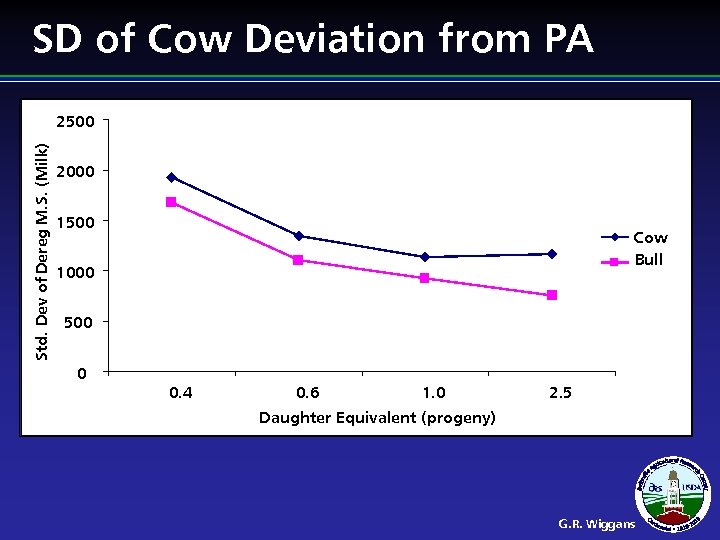 SD of Cow Deviation from PA Std. Dev of Dereg M. S. (Milk) 2500 2000 1500 Cow Bull 1000 500 0 0. 4 0. 6 1. 0 2. 5 Daughter Equivalent (progeny) G. R. Wiggans
SD of Cow Deviation from PA Std. Dev of Dereg M. S. (Milk) 2500 2000 1500 Cow Bull 1000 500 0 0. 4 0. 6 1. 0 2. 5 Daughter Equivalent (progeny) G. R. Wiggans
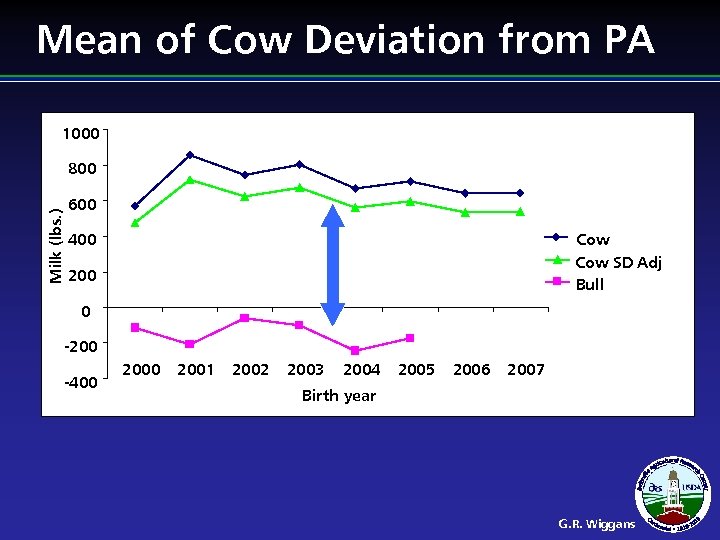 Mean of Cow Deviation from PA 1000 Milk (lbs. ) 800 600 400 Cow SD Adj Bull 200 0 -200 -400 2001 2002 2003 2004 2005 2006 2007 Birth year G. R. Wiggans
Mean of Cow Deviation from PA 1000 Milk (lbs. ) 800 600 400 Cow SD Adj Bull 200 0 -200 -400 2001 2002 2003 2004 2005 2006 2007 Birth year G. R. Wiggans
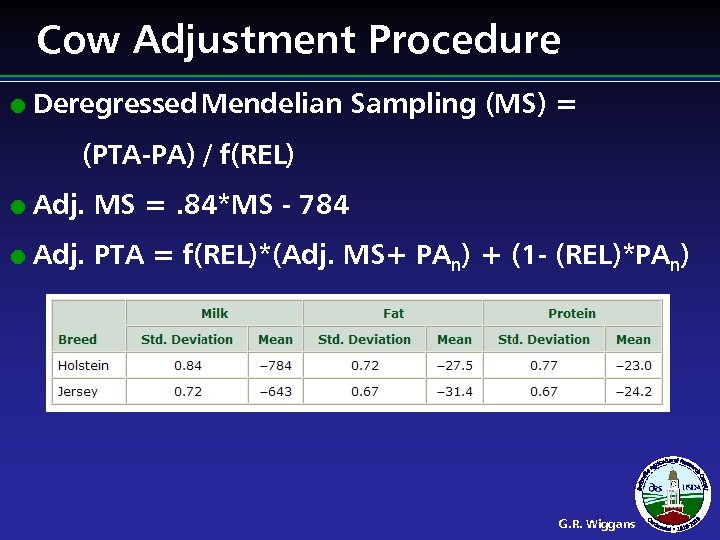 Cow Adjustment Procedure l Deregressed Mendelian Sampling (MS) = (PTA-PA) / f(REL) l Adj. MS =. 84*MS - 784 l Adj. PTA = f(REL)*(Adj. MS+ PAn) + (1 - (REL)*PAn) G. R. Wiggans
Cow Adjustment Procedure l Deregressed Mendelian Sampling (MS) = (PTA-PA) / f(REL) l Adj. MS =. 84*MS - 784 l Adj. PTA = f(REL)*(Adj. MS+ PAn) + (1 - (REL)*PAn) G. R. Wiggans
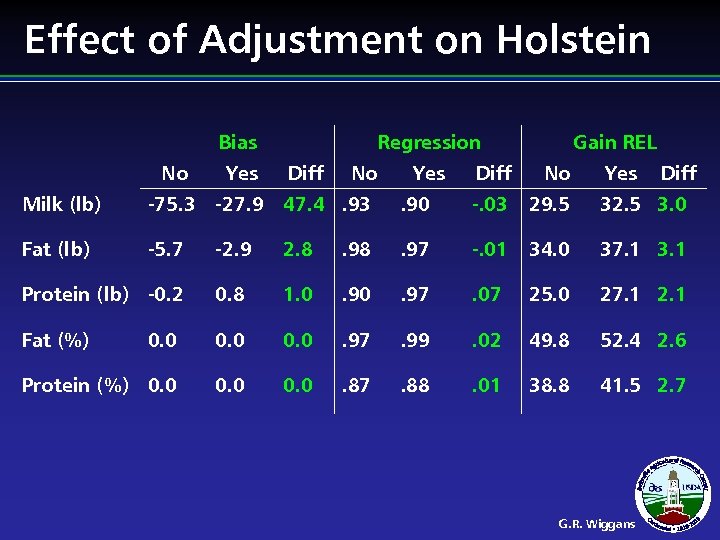 Effect of Adjustment on Holstein Bias No Yes Regression Diff No Yes Gain REL Diff No Yes Diff Milk (lb) -75. 3 -27. 9 47. 4. 93 . 90 -. 03 29. 5 32. 5 3. 0 Fat (lb) -5. 7 -2. 9 2. 8 . 97 -. 01 34. 0 37. 1 3. 1 Protein (lb) -0. 2 0. 8 1. 0 . 97 . 07 25. 0 27. 1 2. 1 Fat (%) 0. 0 . 97 . 99 . 02 49. 8 52. 4 2. 6 Protein (%) 0. 0 . 87 . 88 . 01 38. 8 41. 5 2. 7 G. R. Wiggans
Effect of Adjustment on Holstein Bias No Yes Regression Diff No Yes Gain REL Diff No Yes Diff Milk (lb) -75. 3 -27. 9 47. 4. 93 . 90 -. 03 29. 5 32. 5 3. 0 Fat (lb) -5. 7 -2. 9 2. 8 . 97 -. 01 34. 0 37. 1 3. 1 Protein (lb) -0. 2 0. 8 1. 0 . 97 . 07 25. 0 27. 1 2. 1 Fat (%) 0. 0 . 97 . 99 . 02 49. 8 52. 4 2. 6 Protein (%) 0. 0 . 87 . 88 . 01 38. 8 41. 5 2. 7 G. R. Wiggans
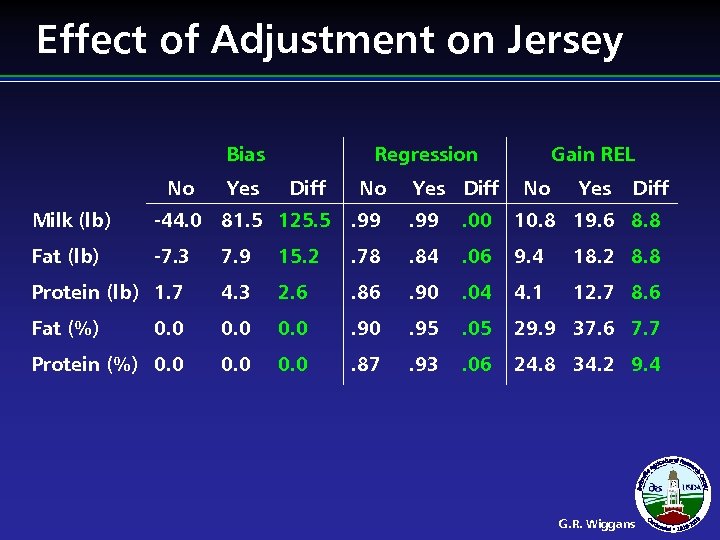 Effect of Adjustment on Jersey Bias No Yes Regression Diff No Yes Diff Gain REL No Yes Diff Milk (lb) -44. 0 81. 5 125. 5. 99 . 00 10. 8 19. 6 8. 8 Fat (lb) -7. 3 7. 9 15. 2 . 78 . 84 . 06 9. 4 18. 2 8. 8 Protein (lb) 1. 7 4. 3 2. 6 . 86 . 90 . 04 4. 1 12. 7 8. 6 Fat (%) 0. 0 . 95 . 05 29. 9 37. 6 7. 7 Protein (%) 0. 0 . 87 . 93 . 06 24. 8 34. 2 9. 4 G. R. Wiggans
Effect of Adjustment on Jersey Bias No Yes Regression Diff No Yes Diff Gain REL No Yes Diff Milk (lb) -44. 0 81. 5 125. 5. 99 . 00 10. 8 19. 6 8. 8 Fat (lb) -7. 3 7. 9 15. 2 . 78 . 84 . 06 9. 4 18. 2 8. 8 Protein (lb) 1. 7 4. 3 2. 6 . 86 . 90 . 04 4. 1 12. 7 8. 6 Fat (%) 0. 0 . 95 . 05 29. 9 37. 6 7. 7 Protein (%) 0. 0 . 87 . 93 . 06 24. 8 34. 2 9. 4 G. R. Wiggans
 Cow Adjustment Summary l l Increased reliability of genomic predictions Genomic evaluations of the top cows, top young bulls, and top heifers decreased Among bulls, foreign bulls with a high proportion of genotyped daughters had largest changes Adjusted PTA reported in XML traditional fields G. R. Wiggans
Cow Adjustment Summary l l Increased reliability of genomic predictions Genomic evaluations of the top cows, top young bulls, and top heifers decreased Among bulls, foreign bulls with a high proportion of genotyped daughters had largest changes Adjusted PTA reported in XML traditional fields G. R. Wiggans
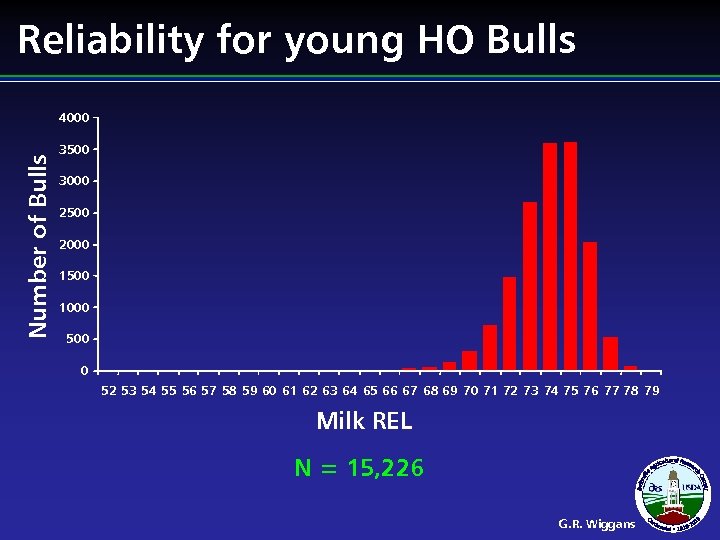 Reliability for young HO Bulls Number of Bulls 4000 3500 3000 2500 2000 1500 1000 500 0 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 Milk REL N = 15, 226 G. R. Wiggans
Reliability for young HO Bulls Number of Bulls 4000 3500 3000 2500 2000 1500 1000 500 0 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 Milk REL N = 15, 226 G. R. Wiggans
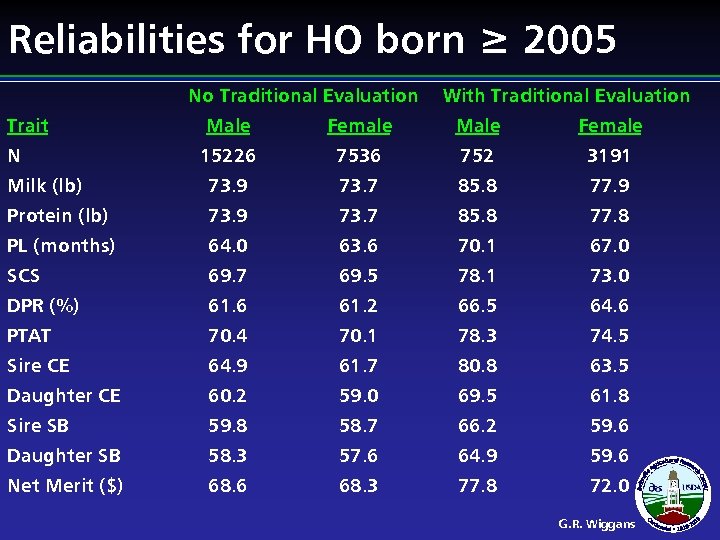 Reliabilities for HO born ≥ 2005 No Traditional Evaluation Trait With Traditional Evaluation Male Female 15226 7536 752 3191 Milk (lb) 73. 9 73. 7 85. 8 77. 9 Protein (lb) 73. 9 73. 7 85. 8 77. 8 PL (months) 64. 0 63. 6 70. 1 67. 0 SCS 69. 7 69. 5 78. 1 73. 0 DPR (%) 61. 6 61. 2 66. 5 64. 6 PTAT 70. 4 70. 1 78. 3 74. 5 Sire CE 64. 9 61. 7 80. 8 63. 5 Daughter CE 60. 2 59. 0 69. 5 61. 8 Sire SB 59. 8 58. 7 66. 2 59. 6 Daughter SB 58. 3 57. 6 64. 9 59. 6 Net Merit ($) 68. 6 68. 3 77. 8 72. 0 N G. R. Wiggans
Reliabilities for HO born ≥ 2005 No Traditional Evaluation Trait With Traditional Evaluation Male Female 15226 7536 752 3191 Milk (lb) 73. 9 73. 7 85. 8 77. 9 Protein (lb) 73. 9 73. 7 85. 8 77. 8 PL (months) 64. 0 63. 6 70. 1 67. 0 SCS 69. 7 69. 5 78. 1 73. 0 DPR (%) 61. 6 61. 2 66. 5 64. 6 PTAT 70. 4 70. 1 78. 3 74. 5 Sire CE 64. 9 61. 7 80. 8 63. 5 Daughter CE 60. 2 59. 0 69. 5 61. 8 Sire SB 59. 8 58. 7 66. 2 59. 6 Daughter SB 58. 3 57. 6 64. 9 59. 6 Net Merit ($) 68. 6 68. 3 77. 8 72. 0 N G. R. Wiggans
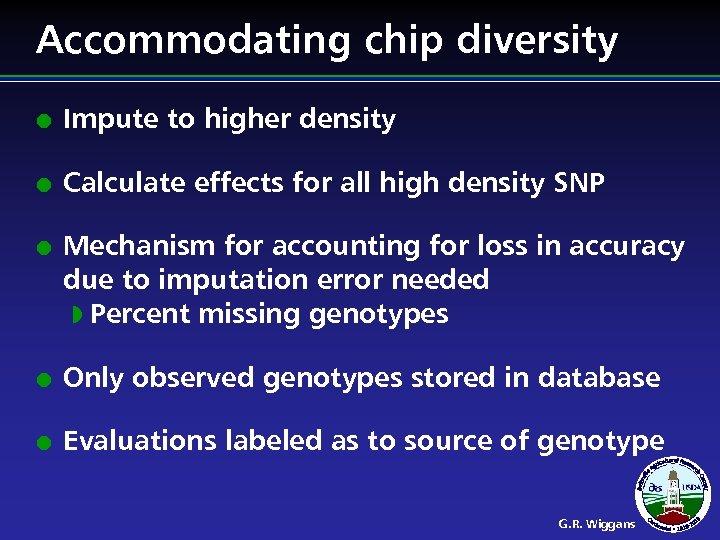 Accommodating chip diversity l Impute to higher density l Calculate effects for all high density SNP l Mechanism for accounting for loss in accuracy due to imputation error needed w Percent missing genotypes l Only observed genotypes stored in database l Evaluations labeled as to source of genotype G. R. Wiggans
Accommodating chip diversity l Impute to higher density l Calculate effects for all high density SNP l Mechanism for accounting for loss in accuracy due to imputation error needed w Percent missing genotypes l Only observed genotypes stored in database l Evaluations labeled as to source of genotype G. R. Wiggans
 Illumina 3 K chip l l SNP chosen w 3072, evenly spaced w Some Y specific SNP w 90 SNP for breed determination Expect to impute genotypes for 43, 382 SNP with high accuracy Expect breeds to use 3 K chip to replace microsatellites for parentage verification Breeds allowed to genotype bulls for parentage only G. R. Wiggans
Illumina 3 K chip l l SNP chosen w 3072, evenly spaced w Some Y specific SNP w 90 SNP for breed determination Expect to impute genotypes for 43, 382 SNP with high accuracy Expect breeds to use 3 K chip to replace microsatellites for parentage verification Breeds allowed to genotype bulls for parentage only G. R. Wiggans
 Proposed stud use of 3 K chip l Accuracy adequate for first stage screening l HD genotyping reserved for bulls acquired. Confirm ID w Second stage selection w l l Genotyping of more candidates Genotype remaining CDDR predictor bulls to meet or exceed Euro. Genomics reliabilities G. R. Wiggans
Proposed stud use of 3 K chip l Accuracy adequate for first stage screening l HD genotyping reserved for bulls acquired. Confirm ID w Second stage selection w l l Genotyping of more candidates Genotype remaining CDDR predictor bulls to meet or exceed Euro. Genomics reliabilities G. R. Wiggans
 HD chip l l Proposed 860 K SNP include current 43, 382 so can replace 50 K chip in current evaluations 3, 000+ genotypes at HD may be adequate to support imputation of HD from current 50 K SNP Expected gain in Rel < 2 May allow HO genotypes to contribute to accuracy of JE & BS genomic evaluations G. R. Wiggans
HD chip l l Proposed 860 K SNP include current 43, 382 so can replace 50 K chip in current evaluations 3, 000+ genotypes at HD may be adequate to support imputation of HD from current 50 K SNP Expected gain in Rel < 2 May allow HO genotypes to contribute to accuracy of JE & BS genomic evaluations G. R. Wiggans
 HD chip (Cont. ) l l Could share cost of HD genotyping with Europe to get more animals to improve accuracy of imputation Trend is toward higher densities Continued genotyping at 50 K may be shortsighted May allow reduction in polygenic effect giving increased accuracy G. R. Wiggans
HD chip (Cont. ) l l Could share cost of HD genotyping with Europe to get more animals to improve accuracy of imputation Trend is toward higher densities Continued genotyping at 50 K may be shortsighted May allow reduction in polygenic effect giving increased accuracy G. R. Wiggans
 Will data recording survive? l l Progeny test no longer required to market bulls In 2013, new entrants may have no data collection expense Loss in accuracy of SNP effect estimates occurs over time How much data is needed? G. R. Wiggans
Will data recording survive? l l Progeny test no longer required to market bulls In 2013, new entrants may have no data collection expense Loss in accuracy of SNP effect estimates occurs over time How much data is needed? G. R. Wiggans
 What replaces the PT program? l l G bulls will have thousands of daughters in their early traditional evaluations Milk recording is justified for management information Type data may come from breeder herds because they use G bulls Data on new traits will have to be paid for G. R. Wiggans
What replaces the PT program? l l G bulls will have thousands of daughters in their early traditional evaluations Milk recording is justified for management information Type data may come from breeder herds because they use G bulls Data on new traits will have to be paid for G. R. Wiggans
 Data into National Evaluations l l Progeny test herds could become data supply herds Data acquisition could be supported by a fee based on animals receiving a genomic evaluation Plan must be perceived as fair by all industry players Quality certification model could apply G. R. Wiggans
Data into National Evaluations l l Progeny test herds could become data supply herds Data acquisition could be supported by a fee based on animals receiving a genomic evaluation Plan must be perceived as fair by all industry players Quality certification model could apply G. R. Wiggans
 Questions l l l How can accuracy of evaluations from Euro. Genomics be exceeded? Should young bull purchases be based on 3 K genotypes? How will continued flow of data into genetic evaluations be assured? G. R. Wiggans
Questions l l l How can accuracy of evaluations from Euro. Genomics be exceeded? Should young bull purchases be based on 3 K genotypes? How will continued flow of data into genetic evaluations be assured? G. R. Wiggans
 Questions from Bob G. R. Wiggans
Questions from Bob G. R. Wiggans
 Will there be a code on. GPTA’s to distinguish between genotyped and imputed animals? l l Genomic indicator code of 3 planned for imputed cows in format 105 in August Already designated in XML files G. R. Wiggans
Will there be a code on. GPTA’s to distinguish between genotyped and imputed animals? l l Genomic indicator code of 3 planned for imputed cows in format 105 in August Already designated in XML files G. R. Wiggans
 From which EU countries are cow proofs used in genomics? (All, health, type? ) l Cow evaluations for milk, fat, and protein are collected from: NLD w DEU w FRA w GBR w ITA w DNK w G. R. Wiggans
From which EU countries are cow proofs used in genomics? (All, health, type? ) l Cow evaluations for milk, fat, and protein are collected from: NLD w DEU w FRA w GBR w ITA w DNK w G. R. Wiggans
 What caused the DPR changes from Dec – Feb – April? Can it happen again? l l l The traditional DPR PAs for some foreign bulls were incorrect in Feb May have been due to missing the dam or MGS We have increased checking for missing pedigree G. R. Wiggans
What caused the DPR changes from Dec – Feb – April? Can it happen again? l l l The traditional DPR PAs for some foreign bulls were incorrect in Feb May have been due to missing the dam or MGS We have increased checking for missing pedigree G. R. Wiggans
 What is the difference between selection index (US) and blending of proofs (CAN)? l Selection Index combines w w w l l l Genomic Traditional computed using only genotyped animals Theoretically justified DGV includes all information when both parents genotyped Used in most countries G. R. Wiggans
What is the difference between selection index (US) and blending of proofs (CAN)? l Selection Index combines w w w l l l Genomic Traditional computed using only genotyped animals Theoretically justified DGV includes all information when both parents genotyped Used in most countries G. R. Wiggans
 What is the difference between Selection Index (US) and. Blending of proofs (CAN)? (Cont. ) l Blending combines w Genomic w Traditional l Weighted by reliability l Simple to explain G. R. Wiggans
What is the difference between Selection Index (US) and. Blending of proofs (CAN)? (Cont. ) l Blending combines w Genomic w Traditional l Weighted by reliability l Simple to explain G. R. Wiggans
 What are the criteria for an animal to be included in the reference population for genomics? l Traditional Rel > PA Rel G. R. Wiggans
What are the criteria for an animal to be included in the reference population for genomics? l Traditional Rel > PA Rel G. R. Wiggans
 What other factors can change SNP effect estimates, beyond adding new animals to the reference population? l New traditional evaluations – tri-annual runs l Insufficient iterations in previous run l Change in SNP used G. R. Wiggans
What other factors can change SNP effect estimates, beyond adding new animals to the reference population? l New traditional evaluations – tri-annual runs l Insufficient iterations in previous run l Change in SNP used G. R. Wiggans
 Why do genomic evaluations change? l l l Reference population animals are added Changes in traditional PTA cause genomic evaluation to change particularly for high reliability bulls Small changes due to filling in missing SNP genotypes when possible G. R. Wiggans
Why do genomic evaluations change? l l l Reference population animals are added Changes in traditional PTA cause genomic evaluation to change particularly for high reliability bulls Small changes due to filling in missing SNP genotypes when possible G. R. Wiggans
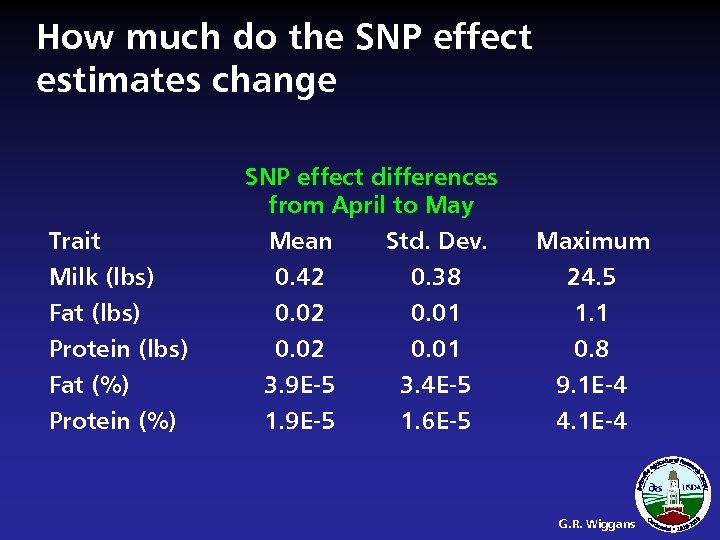 How much do the SNP effect estimates change SNP effect differences from April to May Trait Milk (lbs) Mean 0. 42 Std. Dev. 0. 38 Maximum 24. 5 Fat (lbs) Protein (lbs) Fat (%) Protein (%) 0. 02 3. 9 E-5 1. 9 E-5 0. 01 3. 4 E-5 1. 6 E-5 1. 1 0. 8 9. 1 E-4 4. 1 E-4 G. R. Wiggans
How much do the SNP effect estimates change SNP effect differences from April to May Trait Milk (lbs) Mean 0. 42 Std. Dev. 0. 38 Maximum 24. 5 Fat (lbs) Protein (lbs) Fat (%) Protein (%) 0. 02 3. 9 E-5 1. 9 E-5 0. 01 3. 4 E-5 1. 6 E-5 1. 1 0. 8 9. 1 E-4 4. 1 E-4 G. R. Wiggans
 Are reliability calculations different in the US vs. EU? And, why are reliability values similar with a large discrepancy in the number of predictor bulls? (9, 000 vs. 17, 000) l l ~8, 000 cows also contribute to accuracy US Rel is adjusted to reflect gains from cutoff studies G. R. Wiggans
Are reliability calculations different in the US vs. EU? And, why are reliability values similar with a large discrepancy in the number of predictor bulls? (9, 000 vs. 17, 000) l l ~8, 000 cows also contribute to accuracy US Rel is adjusted to reflect gains from cutoff studies G. R. Wiggans
 Can sire proofs be imputed? And, is this likely to happen? l l l Genotypes of bulls can be imputed Only 15 non-genotyped HO bulls with 5+ genotyped progeny with genotyped dams May be approved for bulls controlled by participating studs G. R. Wiggans
Can sire proofs be imputed? And, is this likely to happen? l l l Genotypes of bulls can be imputed Only 15 non-genotyped HO bulls with 5+ genotyped progeny with genotyped dams May be approved for bulls controlled by participating studs G. R. Wiggans
 Will there be an adjustment made to type in August? l No G. R. Wiggans
Will there be an adjustment made to type in August? l No G. R. Wiggans
 Is it possible to estimate the variation in the offspring for $NM when two genotyped animals are mated? l Yes, sum absolute value of SNP effects weighed as: w w Parents 0 and 2, weight 1 w l Parents both 0 or 2, weight 0 1 or both parents 1, weight 2 Weights are max difference in progeny genotypes G. R. Wiggans
Is it possible to estimate the variation in the offspring for $NM when two genotyped animals are mated? l Yes, sum absolute value of SNP effects weighed as: w w Parents 0 and 2, weight 1 w l Parents both 0 or 2, weight 0 1 or both parents 1, weight 2 Weights are max difference in progeny genotypes G. R. Wiggans
 Making genotyped and nongenotyped cows more comparable l High priority research area w Reduce h 2 w Add herd x dam interaction w Differential adjustment by herd G. R. Wiggans
Making genotyped and nongenotyped cows more comparable l High priority research area w Reduce h 2 w Add herd x dam interaction w Differential adjustment by herd G. R. Wiggans
 Will AIPL continue to impute cows during each genomic evaluation? l Yes G. R. Wiggans
Will AIPL continue to impute cows during each genomic evaluation? l Yes G. R. Wiggans
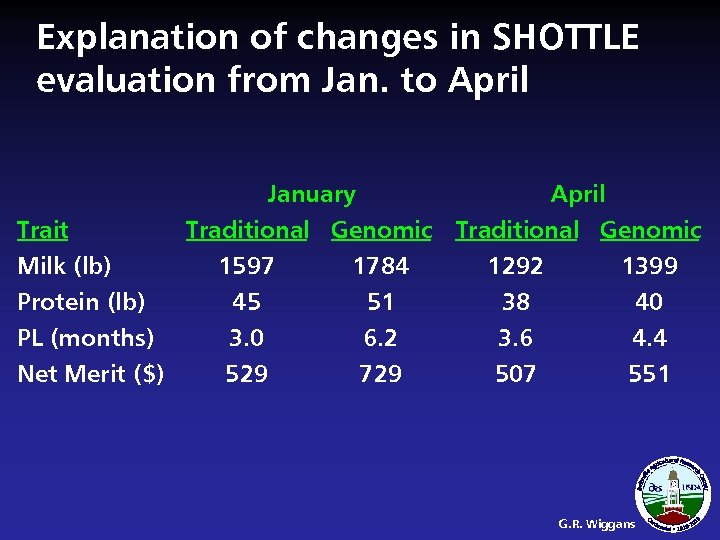 Explanation of changes in SHOTTLE evaluation from Jan. to April Trait Milk (lb) Protein (lb) PL (months) Net Merit ($) January April Traditional Genomic 1597 1784 1292 1399 45 51 38 40 3. 0 529 6. 2 729 3. 6 507 4. 4 551 G. R. Wiggans
Explanation of changes in SHOTTLE evaluation from Jan. to April Trait Milk (lb) Protein (lb) PL (months) Net Merit ($) January April Traditional Genomic 1597 1784 1292 1399 45 51 38 40 3. 0 529 6. 2 729 3. 6 507 4. 4 551 G. R. Wiggans
 Why were cows in Advantage herds with no preferential treatment adjusted? l l l All genotyped cows were adjusted in the same way The maternal component of PA was adjusted Investigating if accuracy can be improved by adjusting each herd based on its own average PTA-PA G. R. Wiggans
Why were cows in Advantage herds with no preferential treatment adjusted? l l l All genotyped cows were adjusted in the same way The maternal component of PA was adjusted Investigating if accuracy can be improved by adjusting each herd based on its own average PTA-PA G. R. Wiggans
 What changes to the imputation process were made in May? l l Maternal grandparents were checked for haplotypes where parents were not available Current allele frequencies replaced base population frequencies for unknown genotypes G. R. Wiggans
What changes to the imputation process were made in May? l l Maternal grandparents were checked for haplotypes where parents were not available Current allele frequencies replaced base population frequencies for unknown genotypes G. R. Wiggans
 How do you incorporate various chip sets (ex. 3 K, 50 K, 700 K, 850 K) into a single genomic evaluation? And, what level of imputing will take place? l l Lower densities will be imputed to highest density If the larger HD chip does not include all the SNP of the smaller one, then combined set must be imputed or some SNP ignored G. R. Wiggans
How do you incorporate various chip sets (ex. 3 K, 50 K, 700 K, 850 K) into a single genomic evaluation? And, what level of imputing will take place? l l Lower densities will be imputed to highest density If the larger HD chip does not include all the SNP of the smaller one, then combined set must be imputed or some SNP ignored G. R. Wiggans
 What will be the gain in accuracy from going from 50 K to 850 K? l <2 increase in reliability G. R. Wiggans
What will be the gain in accuracy from going from 50 K to 850 K? l <2 increase in reliability G. R. Wiggans
 What are the biggest changes and challenges after March 2013 when anyone can get a genomic evaluation of a bull? l Maintaining support for data collection for genetic evaluations G. R. Wiggans
What are the biggest changes and challenges after March 2013 when anyone can get a genomic evaluation of a bull? l Maintaining support for data collection for genetic evaluations G. R. Wiggans
 What are the relative weights given to SNP information for the. GPTAs of 1 st lactation cows, 1 st crop bulls, and 2 nd crop bulls? l Proportional to daughter equivalents (DE) l DE = k*Rel/(100 – Rel) l Calculate DE at each stage of evaluation l DEG = DEtotal - DEPA G. R. Wiggans
What are the relative weights given to SNP information for the. GPTAs of 1 st lactation cows, 1 st crop bulls, and 2 nd crop bulls? l Proportional to daughter equivalents (DE) l DE = k*Rel/(100 – Rel) l Calculate DE at each stage of evaluation l DEG = DEtotal - DEPA G. R. Wiggans
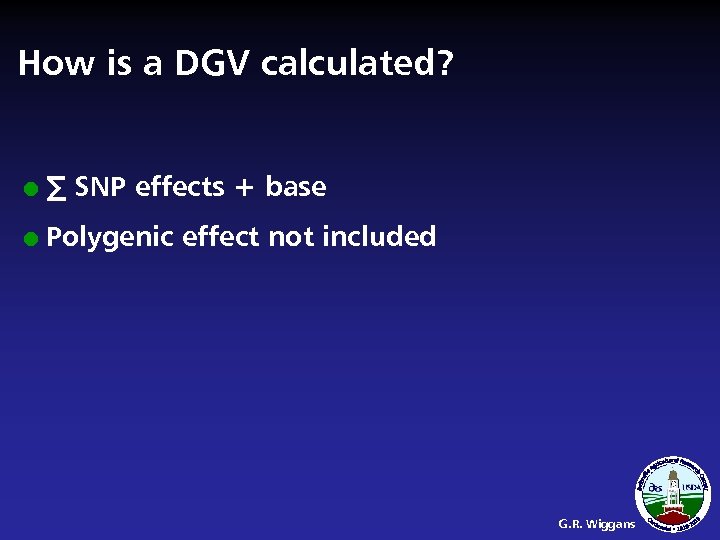 How is a DGV calculated? l ∑ SNP effects + base l Polygenic effect not included G. R. Wiggans
How is a DGV calculated? l ∑ SNP effects + base l Polygenic effect not included G. R. Wiggans
 Do SNP estimates change based on family? l No, SNP effect is change in PTA from having an A allele instead of a B allele (substitution effect) G. R. Wiggans
Do SNP estimates change based on family? l No, SNP effect is change in PTA from having an A allele instead of a B allele (substitution effect) G. R. Wiggans
 How can 99% Reliability bulls change between runs? l l l Traditional evaluations change High reliability evaluations force SNP effects to adjust to equal evaluation Possible because more SNP effects than predictor animals G. R. Wiggans
How can 99% Reliability bulls change between runs? l l l Traditional evaluations change High reliability evaluations force SNP effects to adjust to equal evaluation Possible because more SNP effects than predictor animals G. R. Wiggans


