980b4bd2e1b50c66088fa48ec0d6ec7d.ppt
- Количество слайдов: 17
 Genomic Arrays: Tools for cancer gene discovery Ian Roberts MRC Cancer Cell Unit Hutchison MRC Research Centre ir 210@cam. ac. uk
Genomic Arrays: Tools for cancer gene discovery Ian Roberts MRC Cancer Cell Unit Hutchison MRC Research Centre ir 210@cam. ac. uk
 What’s a genomic array? A platform of regularly spaced genomic sequences All known genes or a subset of genes of interest A tool for querying the genome about damage Genomic gains (oncogenes) Genomic losses (tumour suppressor genes) Applications Research disease gene discovery Clinical diagnostic tests 2/17
What’s a genomic array? A platform of regularly spaced genomic sequences All known genes or a subset of genes of interest A tool for querying the genome about damage Genomic gains (oncogenes) Genomic losses (tumour suppressor genes) Applications Research disease gene discovery Clinical diagnostic tests 2/17
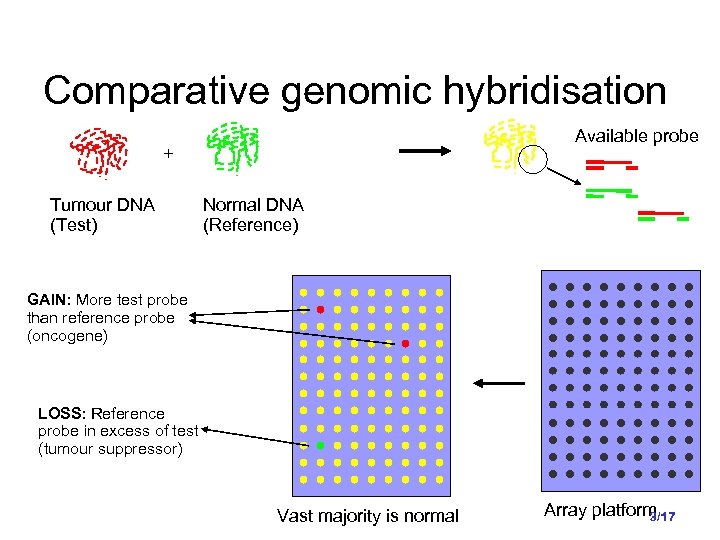 Comparative genomic hybridisation Available probe + Tumour DNA (Test) Normal DNA (Reference) GAIN: More test probe than reference probe (oncogene) LOSS: Reference probe in excess of test (tumour suppressor) Vast majority is normal Array platform 3/17
Comparative genomic hybridisation Available probe + Tumour DNA (Test) Normal DNA (Reference) GAIN: More test probe than reference probe (oncogene) LOSS: Reference probe in excess of test (tumour suppressor) Vast majority is normal Array platform 3/17
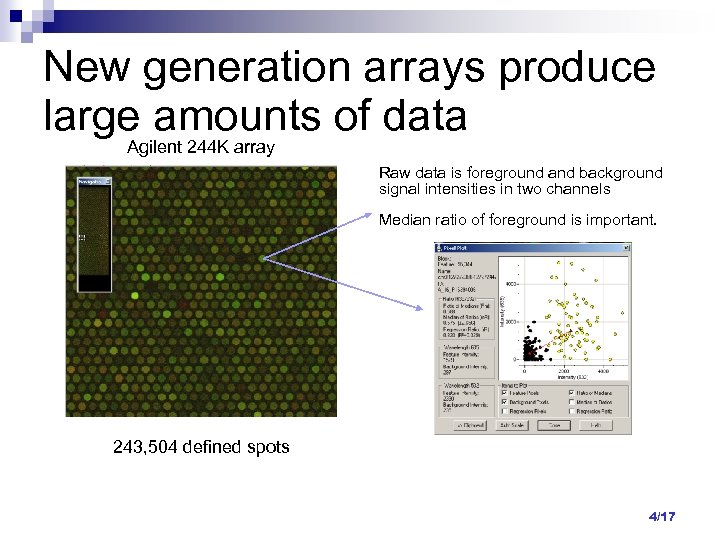 New generation arrays produce large amounts of data Agilent 244 K array Raw data is foreground and background signal intensities in two channels Median ratio of foreground is important. 243, 504 defined spots 4/17
New generation arrays produce large amounts of data Agilent 244 K array Raw data is foreground and background signal intensities in two channels Median ratio of foreground is important. 243, 504 defined spots 4/17
 a. CGH data analysis. . . using camgrid /17
a. CGH data analysis. . . using camgrid /17
 Genomic array analysis strategy using R 1. array data is processed by snap. CGH R package 2. Correct array data for background noise and mean distribution Order data by genomic location Apply an a. CGH segmentation algorithm Draw some plots Determine significant findings (in house R functions) Common and minimum genomic regions of gain and loss Summarise output R www. cran. r-project. org snap. CGH www. bioconductor. org parrot R on camgrid http: //www. bio. cam. ac. uk/local/condor-parrot. html 6/17
Genomic array analysis strategy using R 1. array data is processed by snap. CGH R package 2. Correct array data for background noise and mean distribution Order data by genomic location Apply an a. CGH segmentation algorithm Draw some plots Determine significant findings (in house R functions) Common and minimum genomic regions of gain and loss Summarise output R www. cran. r-project. org snap. CGH www. bioconductor. org parrot R on camgrid http: //www. bio. cam. ac. uk/local/condor-parrot. html 6/17
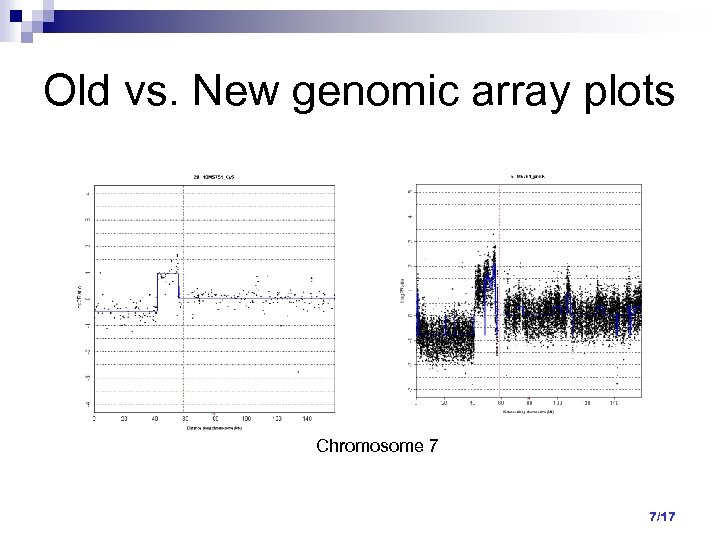 Old vs. New genomic array plots Chromosome 7 7/17
Old vs. New genomic array plots Chromosome 7 7/17
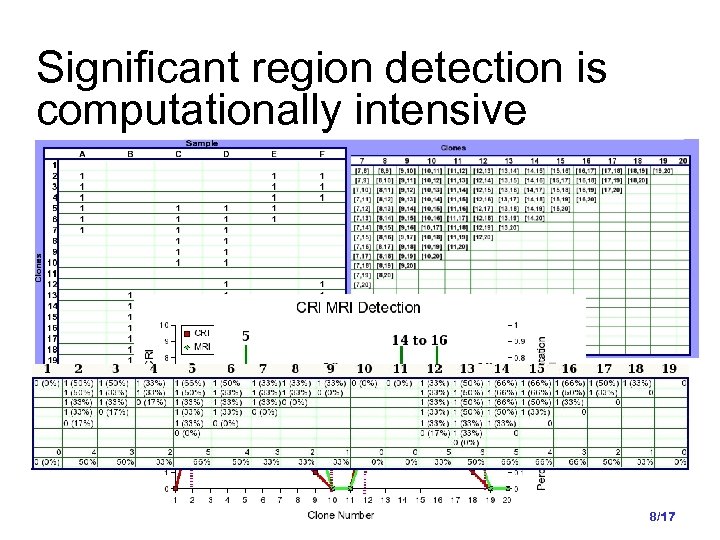 Significant region detection is computationally intensive 8/17
Significant region detection is computationally intensive 8/17
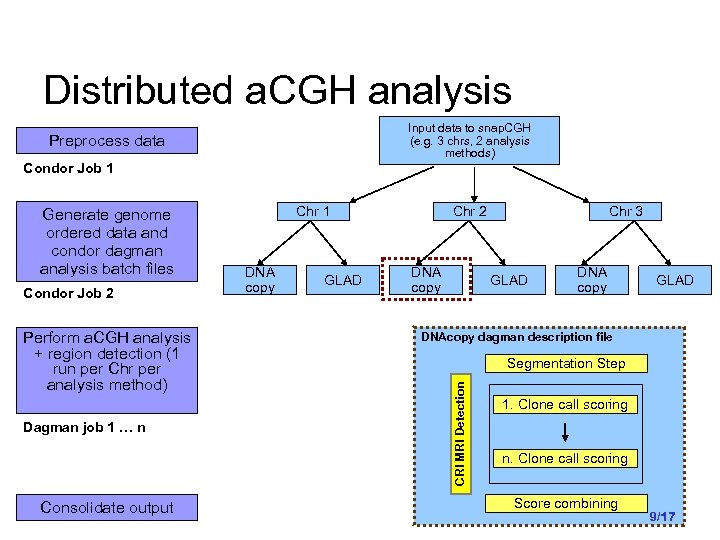 Distributed a. CGH analysis Input data to snap. CGH (e. g. 3 chrs, 2 analysis methods) Preprocess data Condor Job 1 Condor Job 2 Perform a. CGH analysis + region detection (1 run per Chr per analysis method) Dagman job 1 … n Consolidate output Chr 1 DNA copy GLAD Chr 2 DNA copy Chr 3 GLAD DNA copy GLAD DNAcopy dagman description file Segmentation Step CRI MRI Detection Generate genome ordered data and condor dagman analysis batch files 1. Clone call scoring n. Clone call scoring Score combining 9/17
Distributed a. CGH analysis Input data to snap. CGH (e. g. 3 chrs, 2 analysis methods) Preprocess data Condor Job 1 Condor Job 2 Perform a. CGH analysis + region detection (1 run per Chr per analysis method) Dagman job 1 … n Consolidate output Chr 1 DNA copy GLAD Chr 2 DNA copy Chr 3 GLAD DNA copy GLAD DNAcopy dagman description file Segmentation Step CRI MRI Detection Generate genome ordered data and condor dagman analysis batch files 1. Clone call scoring n. Clone call scoring Score combining 9/17
 Condor job scripting in BASH & R BASH function Responsible for producing required condor files for discrete jobs Default_submit has 2 positional parameters R script name $1 Data files $2 Initiates a. CGH analysis on grid. Condor dagman R function set R-scripter R-condor-submitter Writes the condor job submission file R-condor-executer Writes the appropriate R script for the current job Writes the condor job executable file R-job-descriptor Writes the condor dagman description file 10/17
Condor job scripting in BASH & R BASH function Responsible for producing required condor files for discrete jobs Default_submit has 2 positional parameters R script name $1 Data files $2 Initiates a. CGH analysis on grid. Condor dagman R function set R-scripter R-condor-submitter Writes the condor job submission file R-condor-executer Writes the appropriate R script for the current job Writes the condor job executable file R-job-descriptor Writes the condor dagman description file 10/17
 End user abstraction – start_a. CGH. sh a. CGH analysis undertaken by a single shell command Manages array data input Collects user specified parameters Chromosome range Segmentation algorithms Significance thresholds Links condor R job scripting 11/17
End user abstraction – start_a. CGH. sh a. CGH analysis undertaken by a single shell command Manages array data input Collects user specified parameters Chromosome range Segmentation algorithms Significance thresholds Links condor R job scripting 11/17
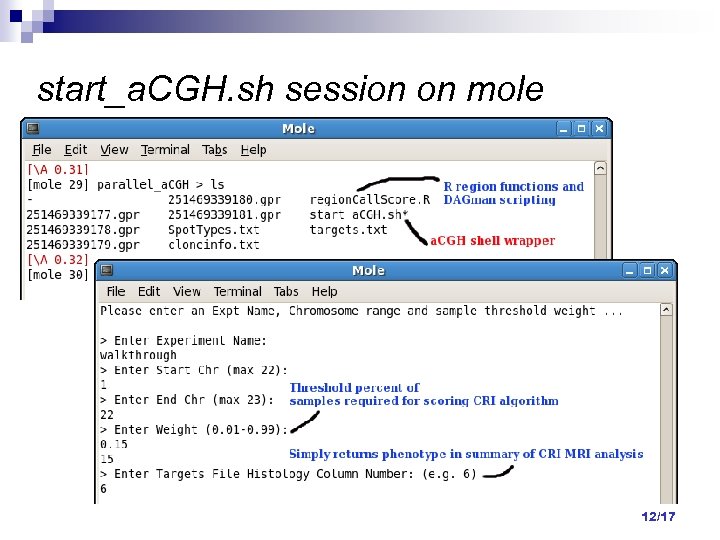 start_a. CGH. sh session on mole 12/17
start_a. CGH. sh session on mole 12/17
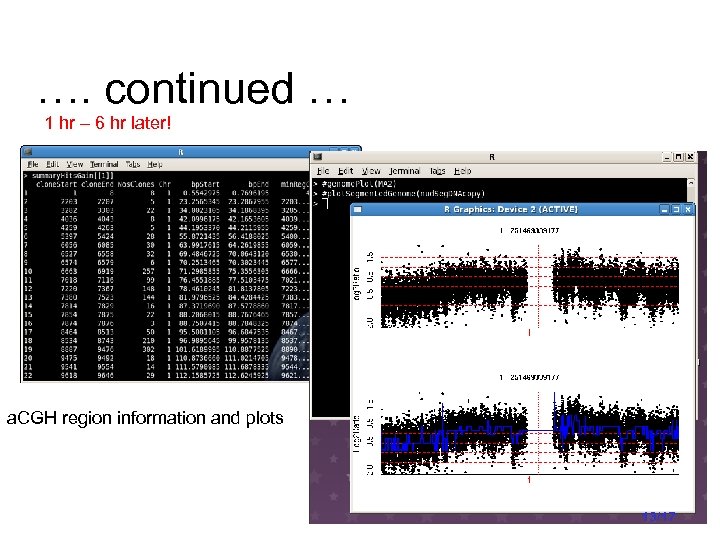 …. continued … 1 hr – 6 hr later! a. CGH region information and plots 13/17
…. continued … 1 hr – 6 hr later! a. CGH region information and plots 13/17
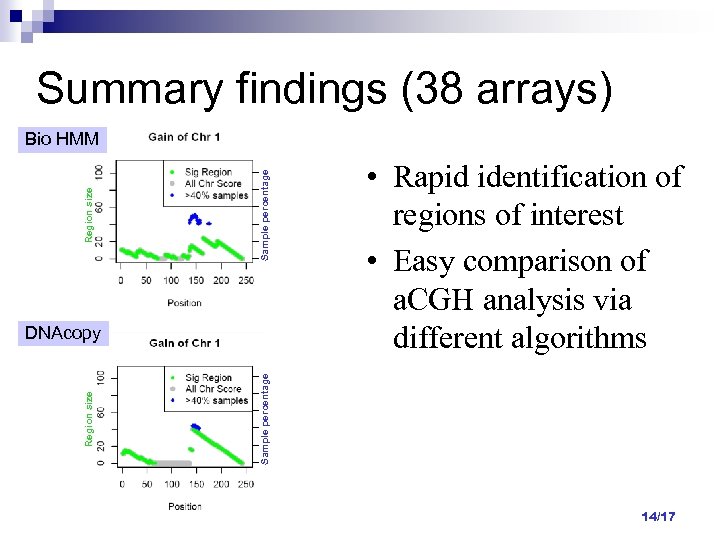 Summary findings (38 arrays) Sample percentage Region size Bio HMM Sample percentage Region size DNAcopy • Rapid identification of regions of interest • Easy comparison of a. CGH analysis via different algorithms 14/17
Summary findings (38 arrays) Sample percentage Region size Bio HMM Sample percentage Region size DNAcopy • Rapid identification of regions of interest • Easy comparison of a. CGH analysis via different algorithms 14/17
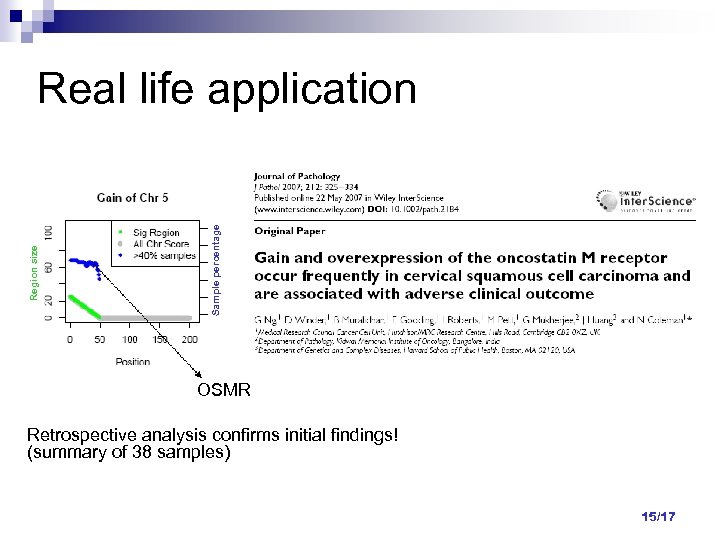 Sample percentage Region size Real life application OSMR Retrospective analysis confirms initial findings! (summary of 38 samples) 15/17
Sample percentage Region size Real life application OSMR Retrospective analysis confirms initial findings! (summary of 38 samples) 15/17
 Future development Tailor output for specific user requirements Produce overall summary plot Apply approach to expression arrays 16/17
Future development Tailor output for specific user requirements Produce overall summary plot Apply approach to expression arrays 16/17
 Grace Ng Steph Carter Konstantina Karagavriliidou Jenny Barna Mark Calleja Nick Coleman www. bio. cam. ac. uk/~ir 210 17/17
Grace Ng Steph Carter Konstantina Karagavriliidou Jenny Barna Mark Calleja Nick Coleman www. bio. cam. ac. uk/~ir 210 17/17


