15fcc9e61feff327c961f906dc33d704.ppt
- Количество слайдов: 47

Genome Browser Background and Strategy 12 April 2010 I. Background 1. 2. Project definition Survey of genome browsers II. Strategy Alejandro Caro, Chandni Desai, Neha Gupta, Jay Humphrey, Chengwei Luo, Sandeep Namburi, Aarthi Talla
I. Background Project definition

Project definition We have been directed: • to build a genome browser featuring strains M 13519 and M 16917. • to build on the work of previous groups • to make a significant contribution beyond what has already been achieved.
Project definition-Introduction • • A genome browser is a website that provides access to an organism-specific genomic database. There are different visualization programs and databases for genome browser, but many of them provide similar tools for the users, such as: – Interactive graphical map of the genome – regions of interest and features – search tools for relevant tags or sequence homology (BLAST)
Project definition General Description GENERAL Genome data in our genome browser will include : 1. The assembled genome sequence from N. meningiditis M 16917 and M 13519. 2. Features (genes, s. RNAs, ) identified by homology (BLAST) and ab -initio gene predictors. 3. Functional annotation of predicted genes, operon prediction and metabolic pathway reconstruction 4. Ontological linkages between features.
Project definition Specific Description SPECIFIC Our genome browser will explore the findings of the functional annotation and comparative genomic group related to: 1. Phylogenetic relation of the strains • Whole genome tree • ANI • Intra-contig local syntheny 2. Genomic basis of pathogenicity • Capsule genes (capsule polymerase) • Opa, Opc, (recognition of different host receptors ) 3. Genomic basis for non- groupability (M 16917) • (polyaglutinator) but serogroup B 4. Genomic basis for discrepancy on test results (M 13519): • Serogroup C SGS-PCR (serogroup-specific PCR assays) • but serogroup W 135 agglutination test.

I. Background 2. Survey of genome browsers
Scale of a genome browser • One model genome, or a few representatives of one species • Flybase, Wormbase • Several genomes of a certain type • Geno. List (microbial genomes) • Psuedomonas (7 species, 18 strains) • Hundreds or thousands – Separated into organism families • Geno. Scope (the platform shared by Ne. Me. Sys) • UCSC
genolist home
genolist search
genolist results
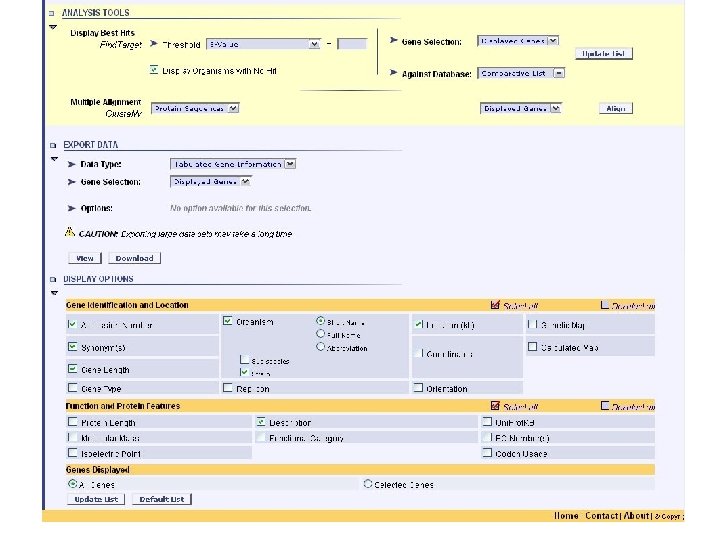
genolist analysis tools
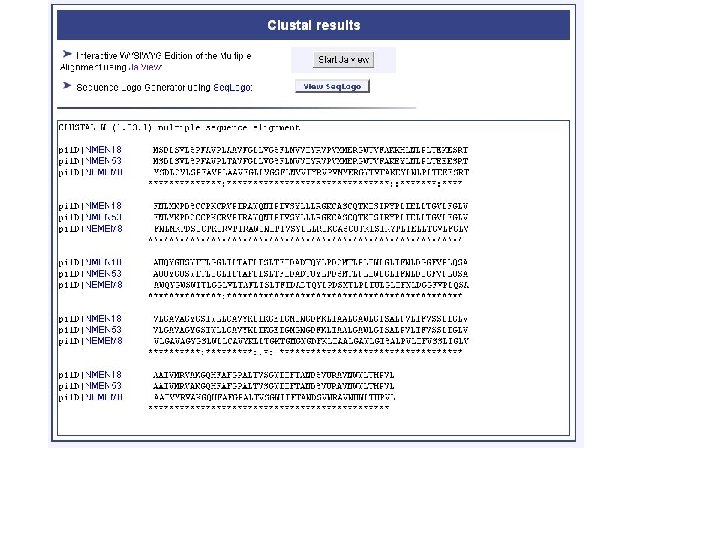
genolist alignment

genolist seqlogo
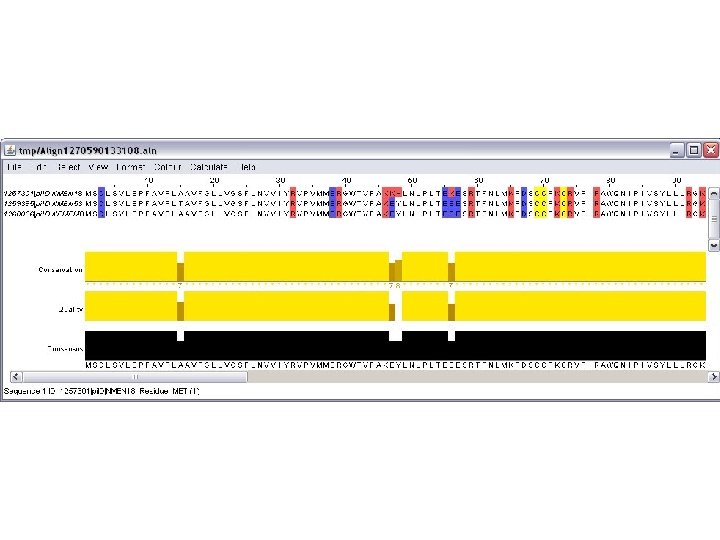
genolist jalview
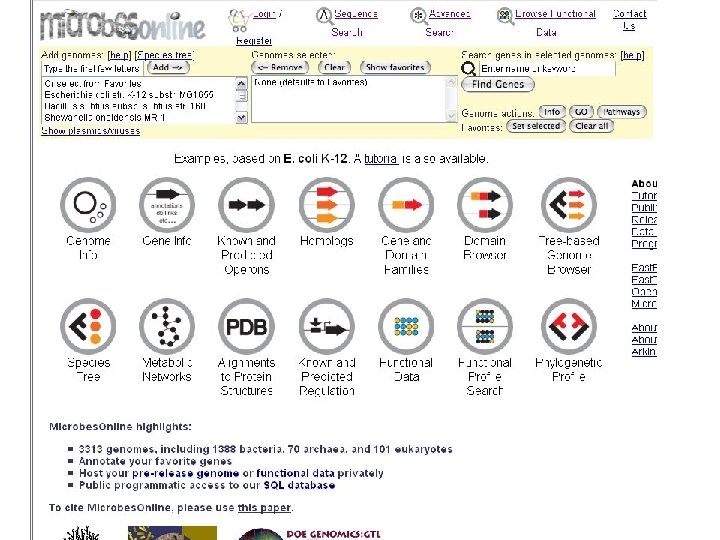
microbes home
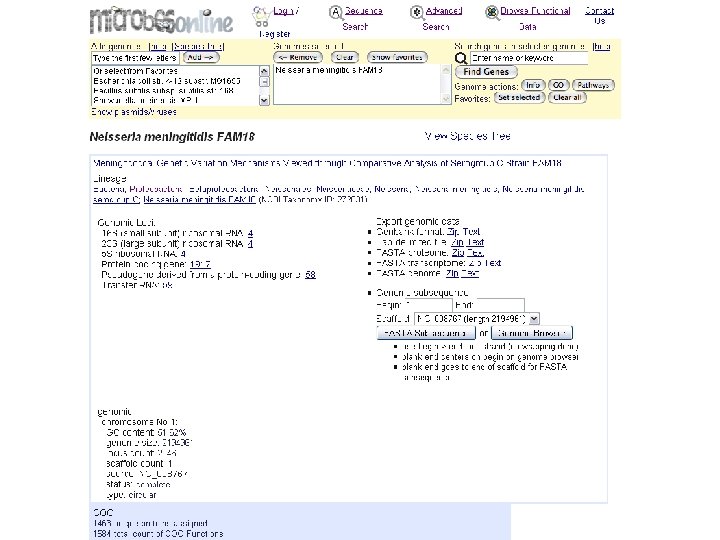
microbes genome info top
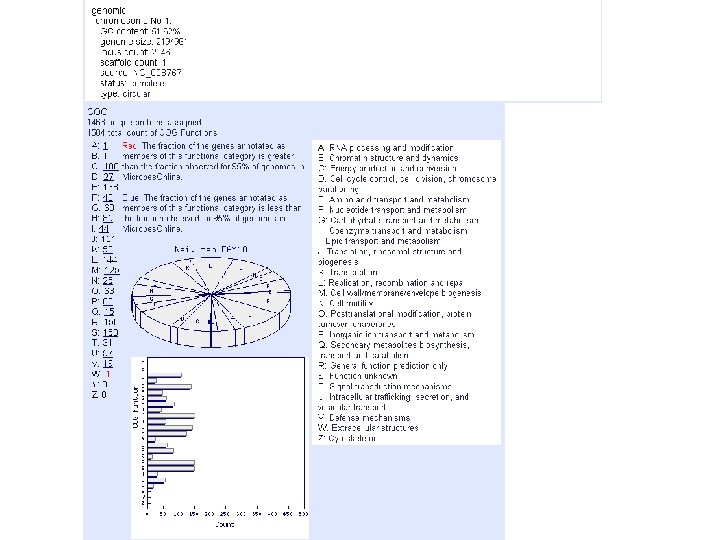
microbes genome info foot
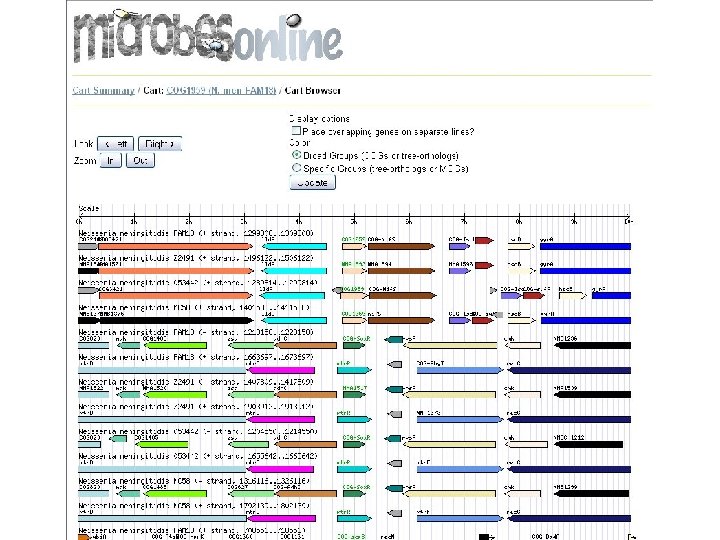
microbes browser
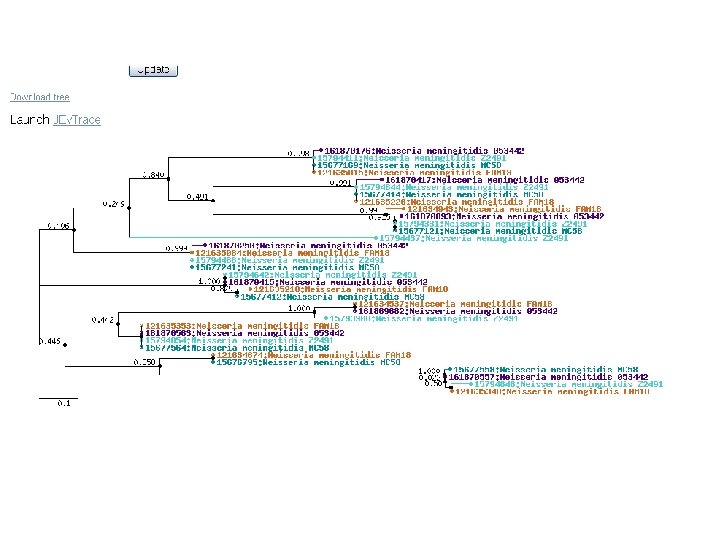
microbes phylo tree
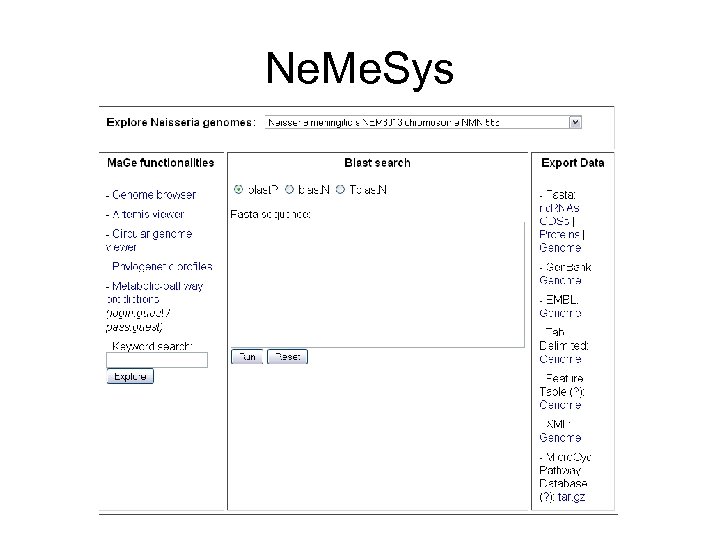
Ne. Me. Sys
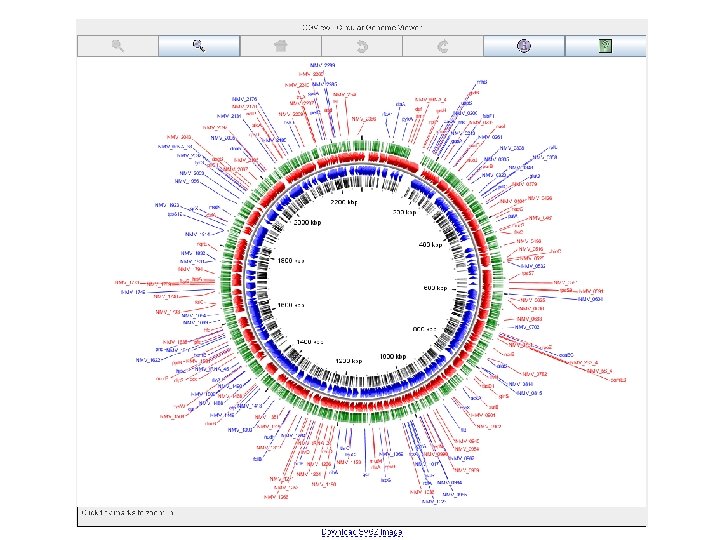
nemesys circular genome viewer
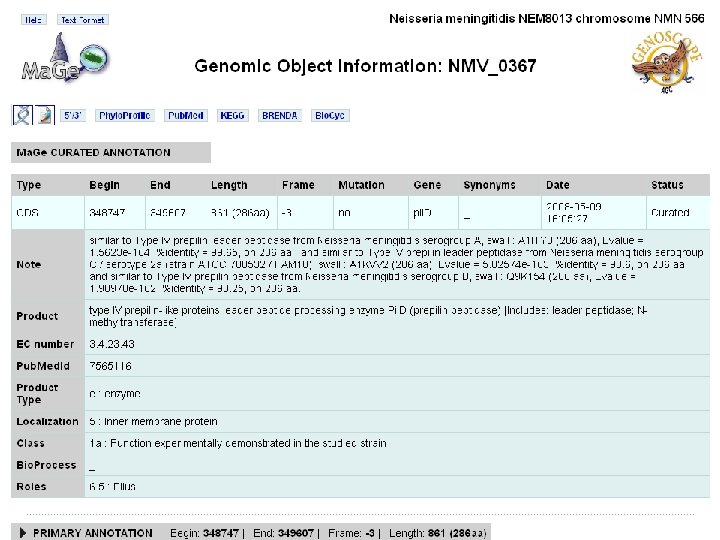
nemesys details
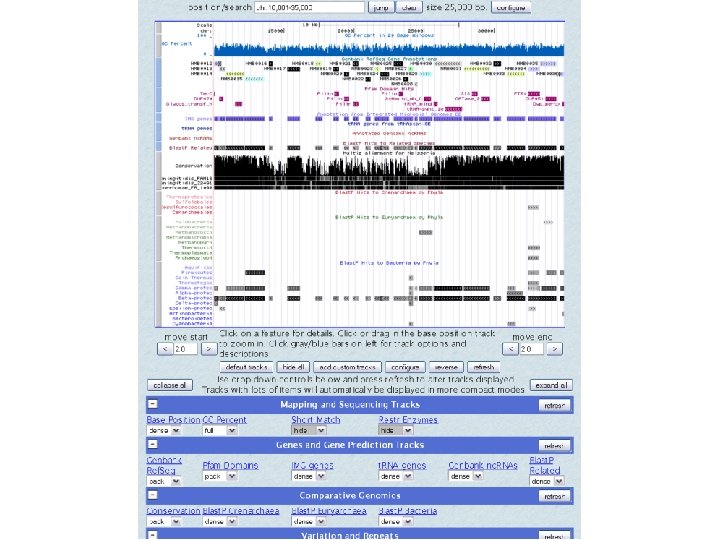
UCSC browser MC 58
Psuedomonas aeruginosa search
Psuedo tools
Psuedo tools
Psuedo synteny
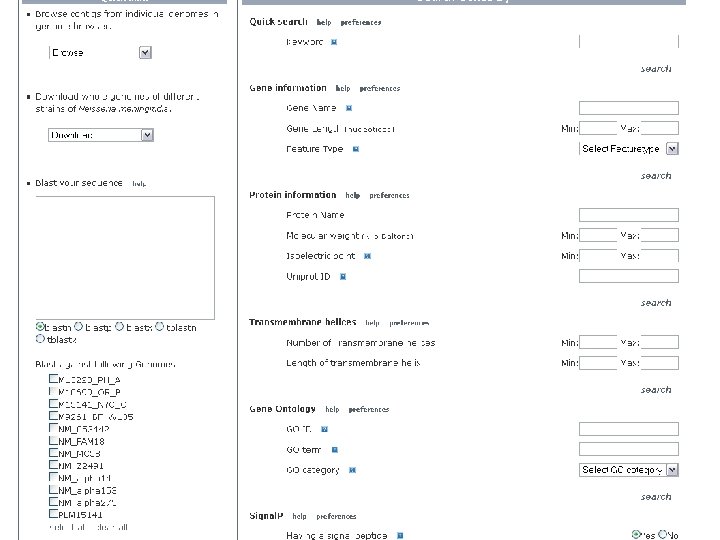
NBase
Database models • How are the databases for these web applications implemented? – Flat files - plain text tables • examples include GBrowse, CGView – RDBMS • May be standard (like Chado, an extensive model organism db schema) • May be created in-house, which is probably the case with many of these multiple genome online databases

NBase has a hybrid schema • The next few slides show it • Bio: : DB: : GFF which is the GBrowse default
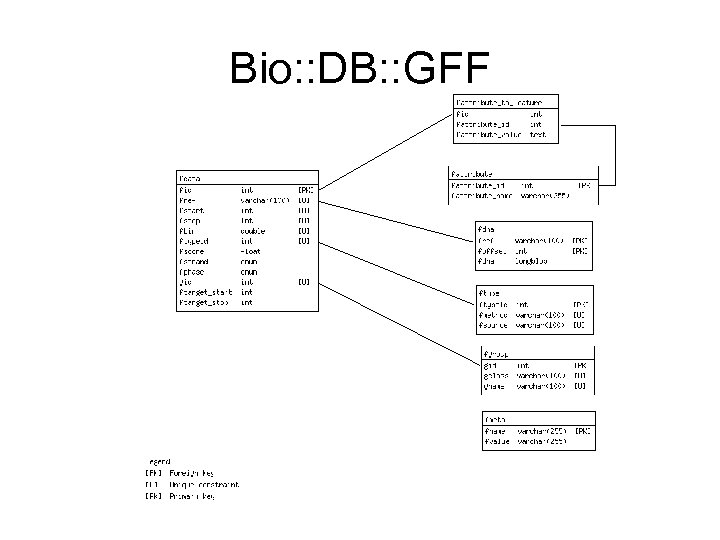
Bio: : DB: : GFF
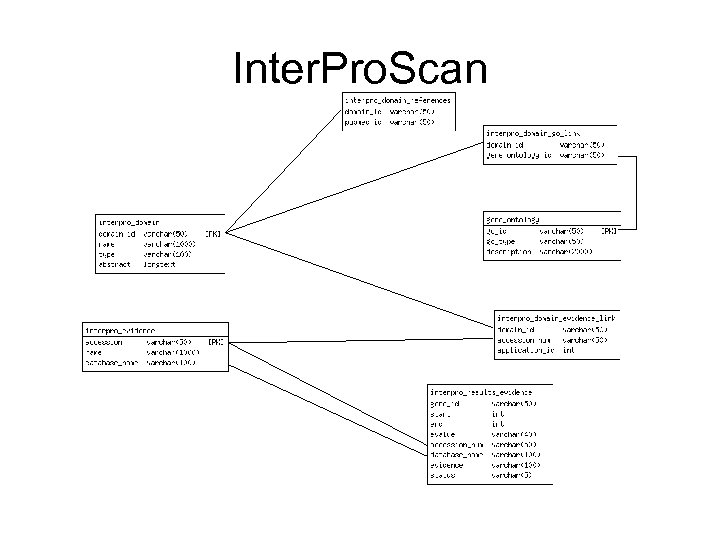
Inter. Pro. Scan
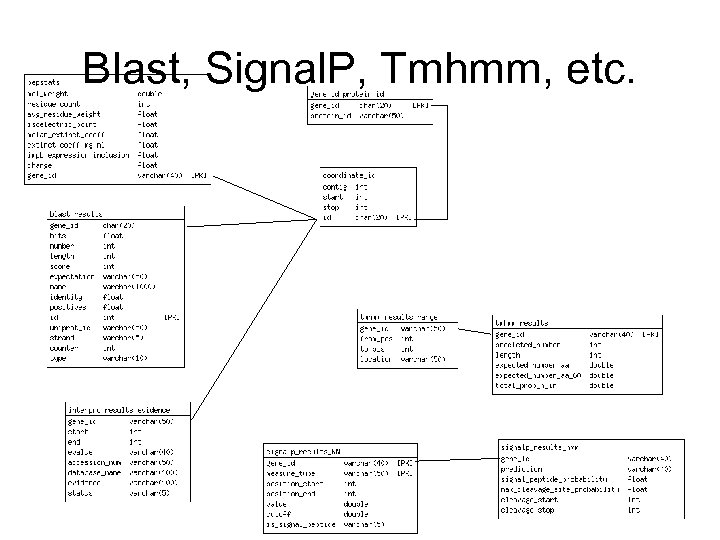
Blast, Signal. P, Tmhmm, etc.

Chado
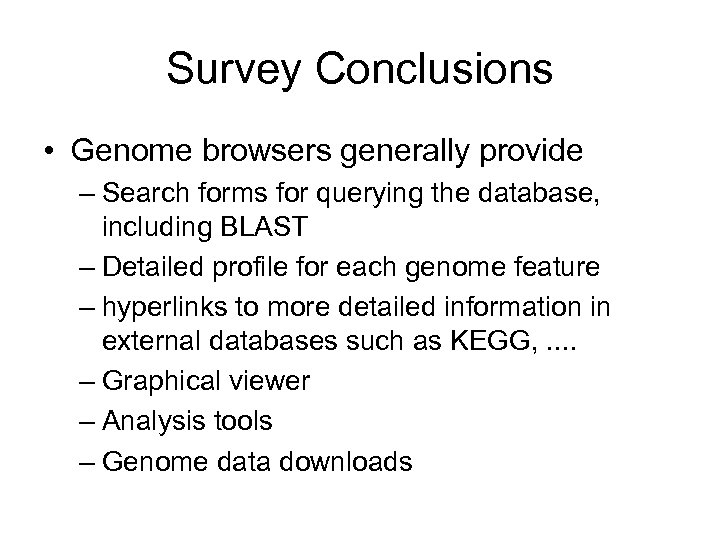
Survey Conclusions • Genome browsers generally provide – Search forms for querying the database, including BLAST – Detailed profile for each genome feature – hyperlinks to more detailed information in external databases such as KEGG, . . – Graphical viewer – Analysis tools – Genome data downloads
Survey Conclusions – Backend implementation, the database design of these genome browsers, is kind of a mystery, we don't have access to their schemas unless they are using a standard one like Chado, GFF, Bio. SQL – They might be using a home-built or modified schema customized for their purposes

III. Strategy 1. Data Collation 2. Browser Design

DATA COLLATION 1) Assembly, Prediction, Annotation and Comparative Genomics provide us with data which are compiled into a searchable database. 2)Along with that we will also compile this data in special formats used by other tools like BLAST and the SYNTENY VIEWER.

BASIC BROWSER DESIGN While using our website, the user may follow certain paths in their workflow, starting with the home page. • From Home access is provided to a genome viewer (to a selected contig in a selected genome) • Search forms for querying the database – From search results, access to Feature Details • A BLAST search – From BLAST results, access to Feature Details • An MLST typing tool • Comparative genomics data ( Synteny, Phylogeny, Genome characteristics profiles)

• From the viewer : In the viewer, you see a positional overview of selected feature types. • Feature Details: a page where all data in the database on a feature is shown – From Details, link back to the viewer – From Details, links to external databases (KEGG, Pubmed) – From Details, link to a BLAST search (homology search)
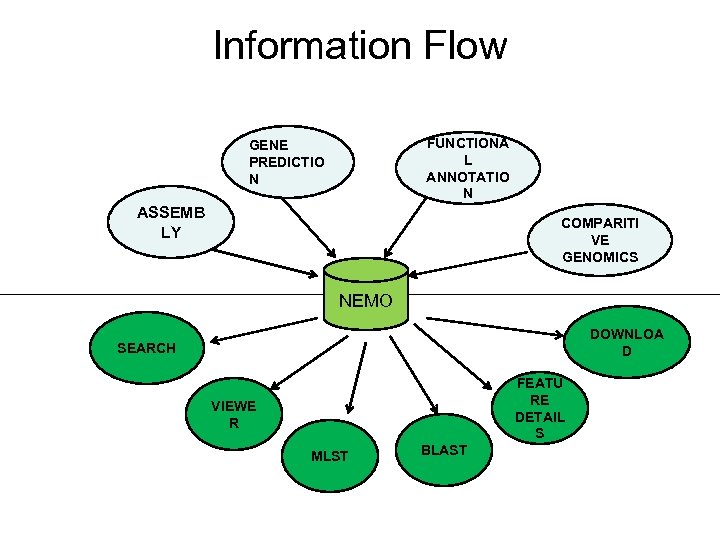
Information Flow FUNCTIONA L ANNOTATIO N GENE PREDICTIO N ASSEMB LY COMPARITI VE GENOMICS NEMO DOWNLOA D SEARCH FEATU RE DETAIL S VIEWE R MLST BLAST
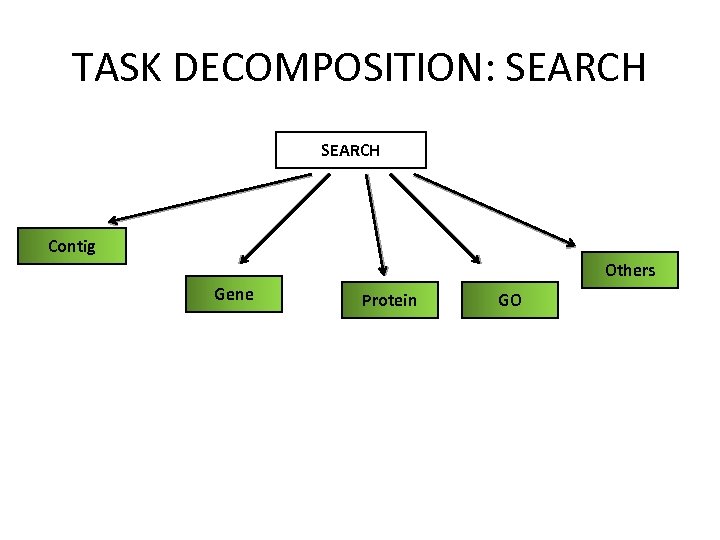
TASK DECOMPOSITION: SEARCH Contig Others Gene Protein GO
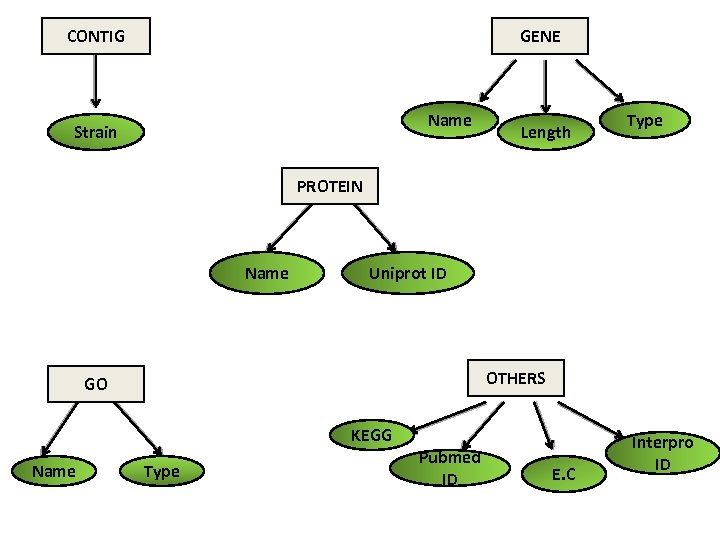
CONTIG GENE Name Strain Length Type PROTEIN Name Uniprot ID OTHERS GO KEGG Name Type Pubmed ID E. C Interpro ID
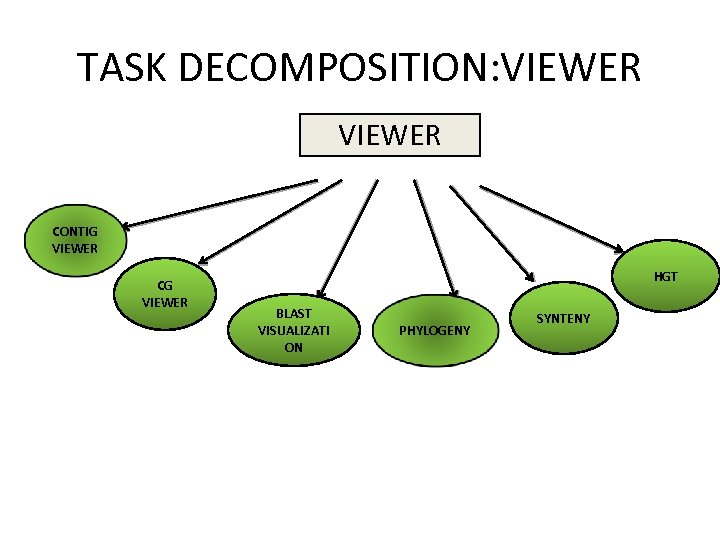
TASK DECOMPOSITION: VIEWER CONTIG VIEWER CG VIEWER HGT BLAST VISUALIZATI ON PHYLOGENY SYNTENY
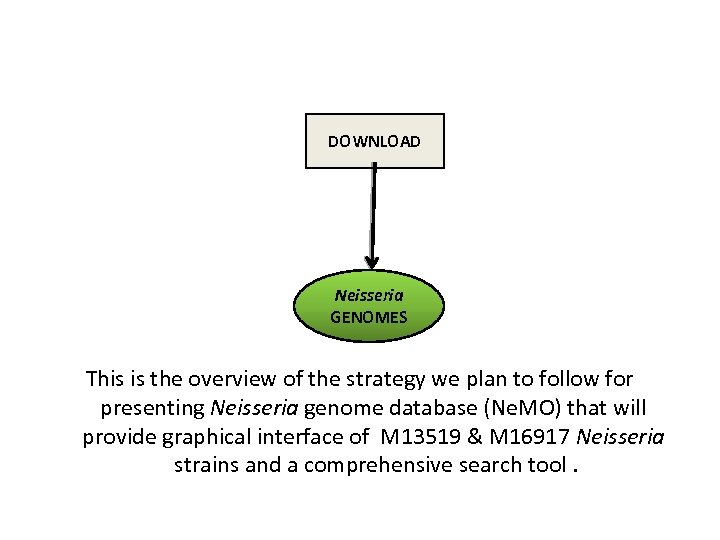
DOWNLOAD Neisseria GENOMES This is the overview of the strategy we plan to follow for presenting Neisseria genome database (Ne. MO) that will provide graphical interface of M 13519 & M 16917 Neisseria strains and a comprehensive search tool.

Questions / Feedback Next: 14 April 2010 • Preliminary Results – Data Collation – Implementation of browser applications • Lab Exercise – Data Collation and provisioning GBrowse with FAM 18 (Tentative)
15fcc9e61feff327c961f906dc33d704.ppt