12b37ecef6293b4d8d25076afd74efb3.ppt
- Количество слайдов: 36

Genome Assembly and Finishing Alla Lapidus, Ph. D. Associate Professor Fox Chase Cancer Center
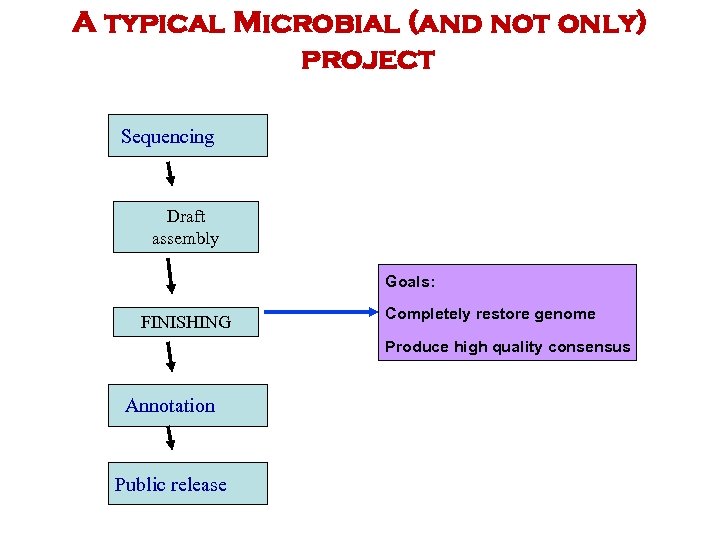
A typical Microbial (and not only) project Sequencing Draft assembly Goals: FINISHING Completely restore genome Produce high quality consensus Annotation Public release
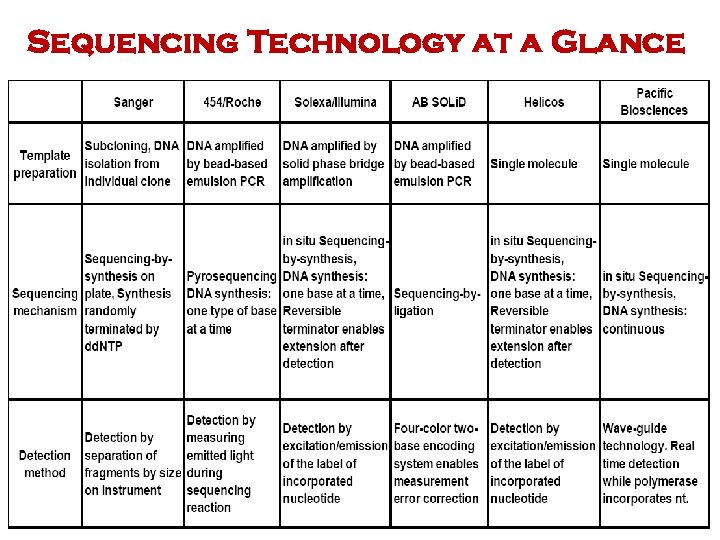
Sequencing Technology at a Glance

Evolution of Microbial Drafts Sanger only – 4 x of 3 kb plasmids + 4 x of 8 kb plasmids + 1 x of fosmids – ~ $50 k for 5 MB genome draft Hybrid Sanger/pyrosequence/Illumina – 4 x 8 kb Sanger + 15 x coverage 454 shotgun + 20 x Illumina (quality improvement) – ~ $35 k for 5 MB genome draft 454 + Solexa - 20 x coverage 454 standard + 4 x coverage 454 paired end (PE) + 50 x coverage Illumina shotgun (quality improvement; gaps) - ~ $10 k per 5 MB genome Solexa only - low cost; too fragmented; good assembler is needed! Solexa +Pac. Bio - low cost; better sachffolding
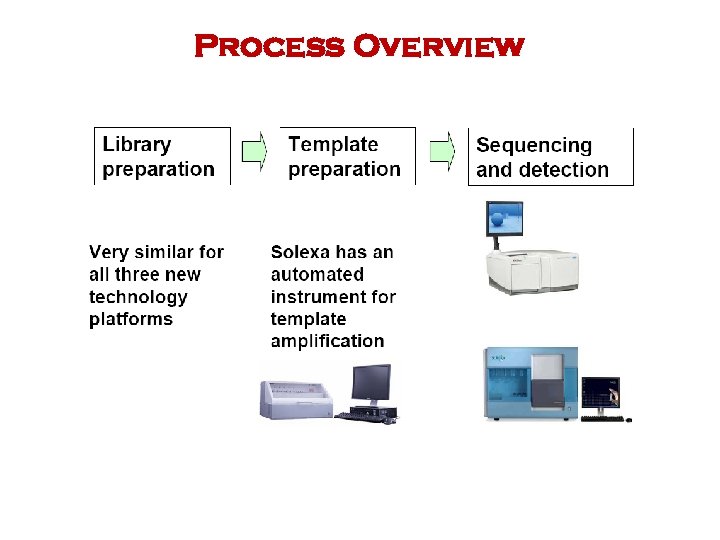
Process Overview
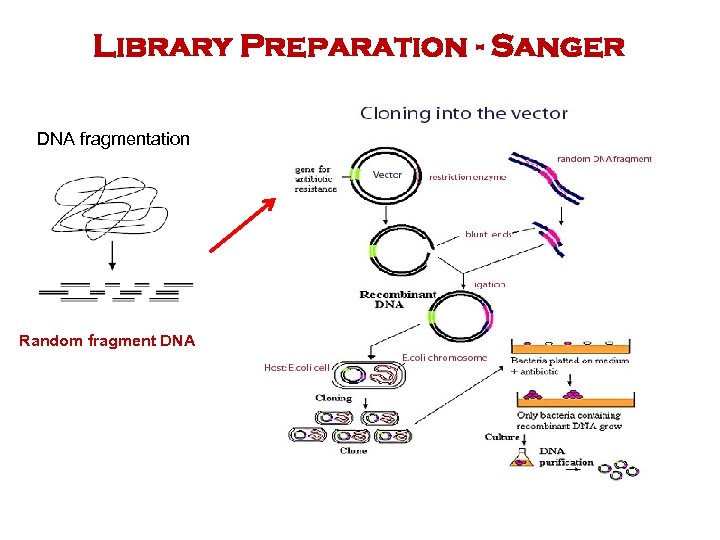
Library Preparation - Sanger DNA fragmentation Random fragment DNA
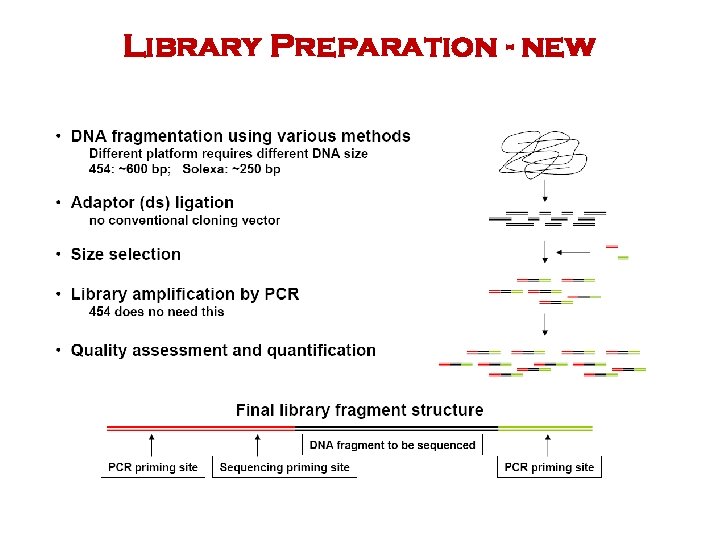
Library Preparation - new
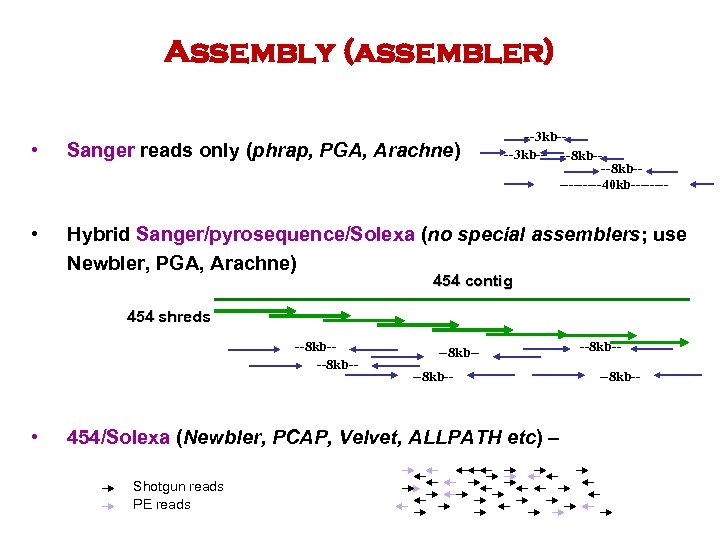
Assembly (assembler) --3 kb-- --8 kb------40 kb---- • Sanger reads only (phrap, PGA, Arachne) • Hybrid Sanger/pyrosequence/Solexa (no special assemblers; use Newbler, PGA, Arachne) 454 contig 454 shreds --8 kb-- • --8 kb-- 454/Solexa (Newbler, PCAP, Velvet, ALLPATH etc) – Shotgun reads PE reads --8 kb--
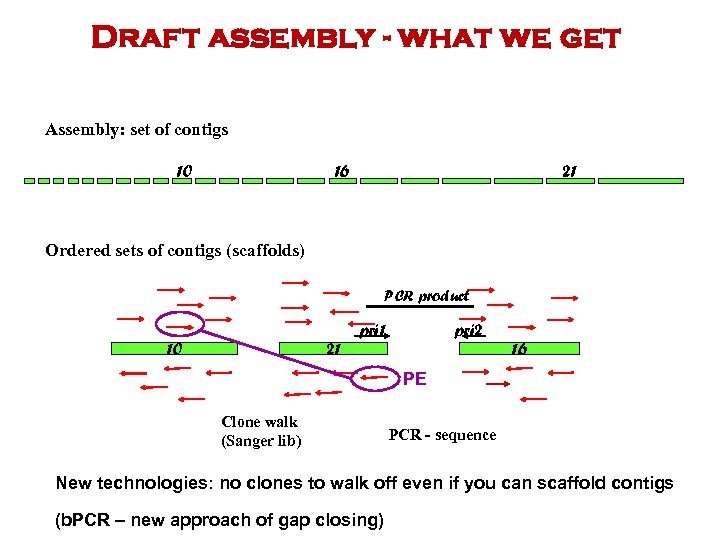
Draft assembly - what we get Assembly: set of contigs 10 16 21 Ordered sets of contigs (scaffolds) PCR product 10 21 pri 2 16 PE Clone walk (Sanger lib) PCR - sequence New technologies: no clones to walk off even if you can scaffold contigs (b. PCR – new approach of gap closing)
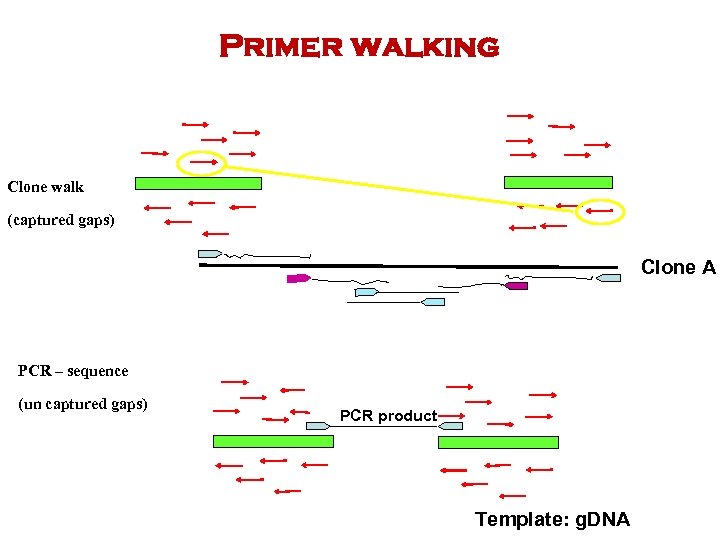
Primer walking Clone walk (captured gaps) Clone A PCR – sequence (un captured gaps) PCR product Template: g. DNA

Why do we have gaps What are gaps ? - Genome areas not covered by random shotgun • Sequencing coverage may not span all regions of the genome, thus producing gaps in the assembly – colony picking • Assembly results of the shotgun reads may produce misassembled regions due to repetitive sequences (new and old tech) • A biased base content (this can result in failure to be cloned, poor stability in the chosen host-vector system, or inability of the polymerase to reliably copy the sequence): ~ AT-rich DNA clones poorly in bacteria (cloning bias; promoters like structures {Sanger} )=> uncaptured gaps ~GC rich DNA is difficult to PCR and to sequence and often requires the use of special chemistry => captured gaps ~ high AT and GC content caused by problematic PCR (new tech)

Assembling repeats Actual genome
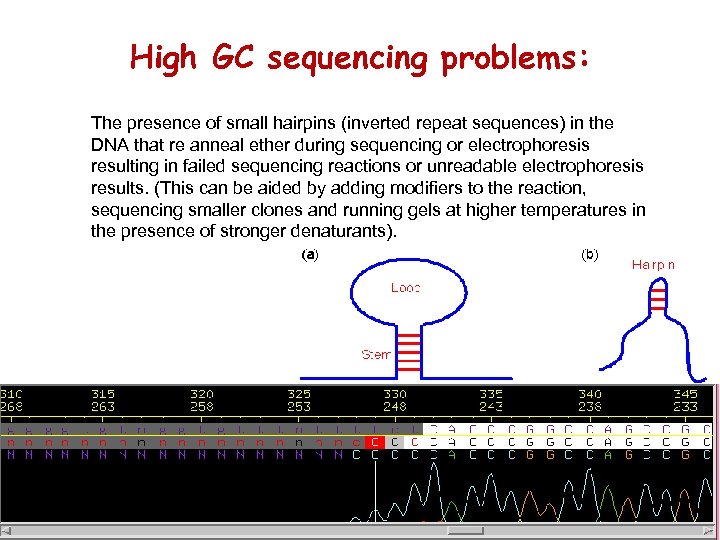
High GC sequencing problems: The presence of small hairpins (inverted repeat sequences) in the DNA that re anneal ether during sequencing or electrophoresis resulting in failed sequencing reactions or unreadable electrophoresis results. (This can be aided by adding modifiers to the reaction, sequencing smaller clones and running gels at higher temperatures in the presence of stronger denaturants).
Why more than one platform? • 454 - high quality reliable skeletons of genomes (454 std + 454 PE): correctly assembled contigs; problems with repeats (unassembled or assembled in contigs outside of main scaffolds); homopolymer related frame shifts • Illumina data is used to help improve the overall consensus quality, correct frameshifts and to close secondary structure related gaps; not ready for de-novo assembly of complex genomes (too many gaps!) • Sanger – finishing reads; fosmids – larger repeats and templates for primer walk – less cost effective but very useful in many cases
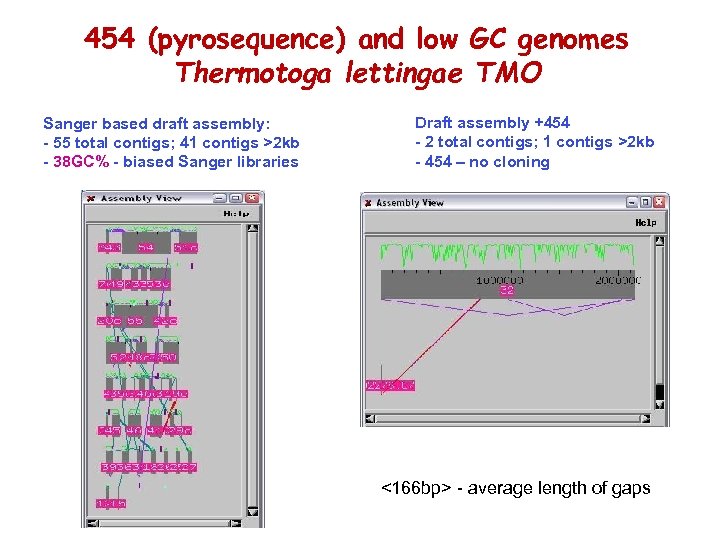
454 (pyrosequence) and low GC genomes Thermotoga lettingae TMO Sanger based draft assembly: - 55 total contigs; 41 contigs >2 kb - 38 GC% - biased Sanger libraries Draft assembly +454 - 2 total contigs; 1 contigs >2 kb - 454 – no cloning <166 bp> - average length of gaps
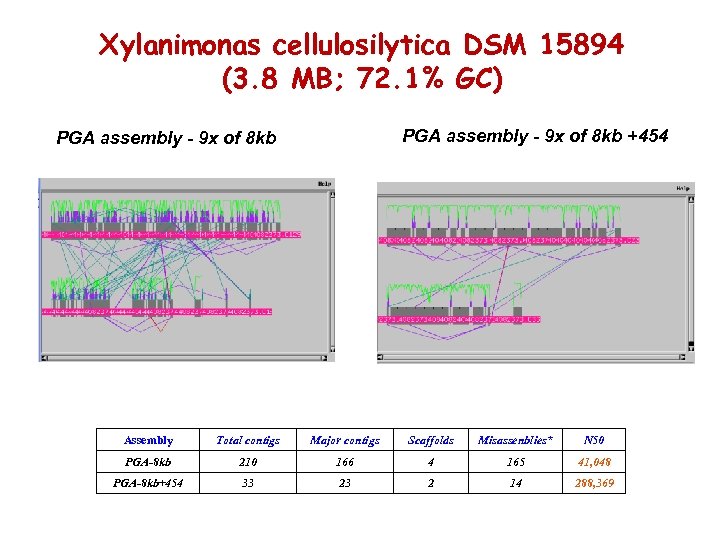
Xylanimonas cellulosilytica DSM 15894 454 and High GC projects (3. 8 MB; 72. 1% GC) PGA assembly - 9 x of 8 kb +454 PGA assembly - 9 x of 8 kb Assembly Total contigs Major contigs Scaffolds Misassenblies* N 50 PGA-8 kb 210 166 4 165 41, 048 PGA-8 kb+454 33 23 2 14 288, 369
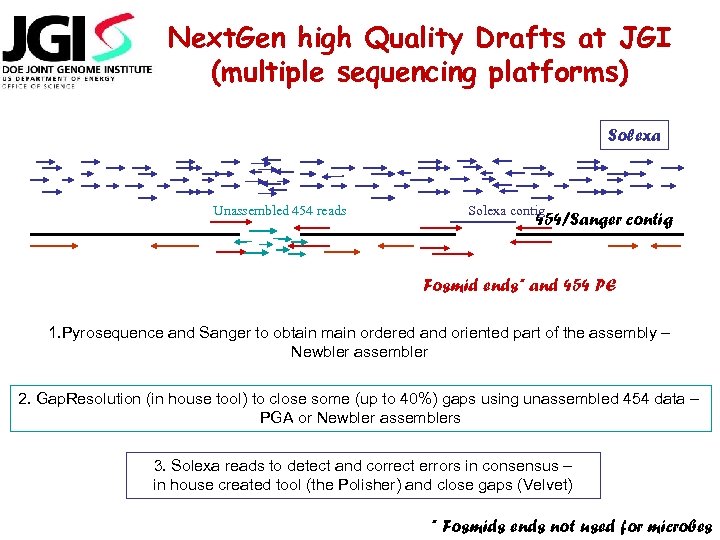
Next. Gen high Quality Drafts at JGI (multiple sequencing platforms) Solexa Unassembled 454 reads Solexa contig 454/Sanger contig Fosmid ends* and 454 PE 1. Pyrosequence and Sanger to obtain main ordered and oriented part of the assembly – Newbler assembler 2. Gap. Resolution (in house tool) to close some (up to 40%) gaps using unassembled 454 data – PGA or Newbler assemblers 3. Solexa reads to detect and correct errors in consensus – in house created tool (the Polisher) and close gaps (Velvet) * Fosmids ends not used for microbes
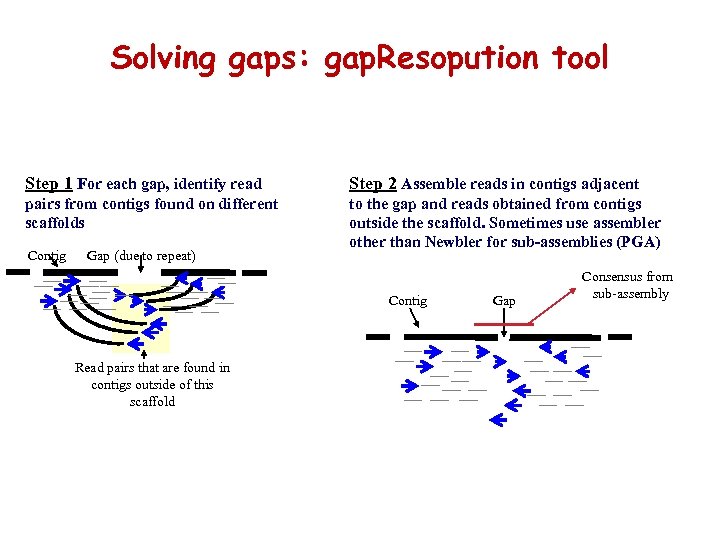
Solving gaps: gap. Resopution tool Step 1 For each gap, identify read Step 2 Assemble reads in contigs adjacent pairs from contigs found on different scaffolds to the gap and reads obtained from contigs outside the scaffold. Sometimes use assembler other than Newbler for sub-assemblies (PGA) Contig Gap (due to repeat) Contig Read pairs that are found in contigs outside of this scaffold Gap Consensus from sub-assembly
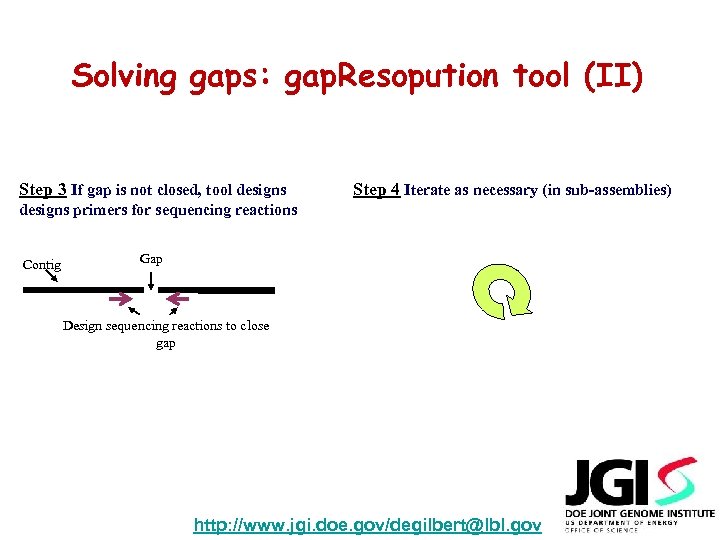
Solving gaps: gap. Resopution tool (II) Step 3 If gap is not closed, tool designs Step 4 Iterate as necessary (in sub-assemblies) designs primers for sequencing reactions Contig Gap Design sequencing reactions to close gap http: //www. jgi. doe. gov/degilbert@lbl. gov
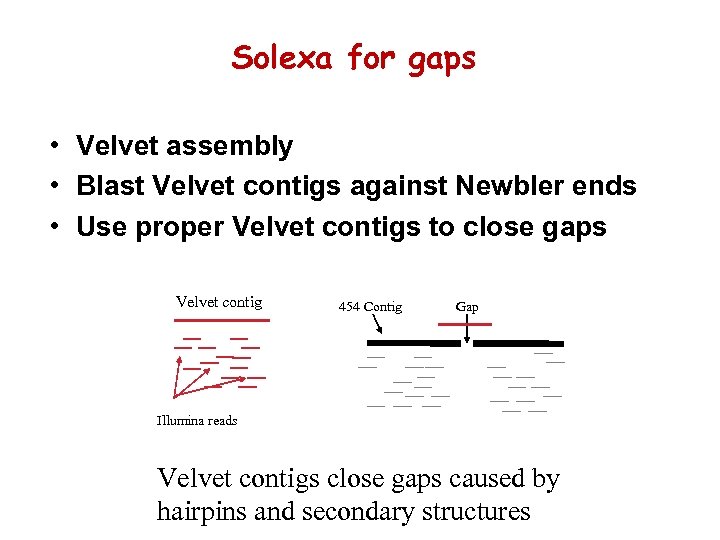
Solexa for gaps • Velvet assembly • Blast Velvet contigs against Newbler ends • Use proper Velvet contigs to close gaps Velvet contig 454 Contig Gap Illumina reads Velvet contigs close gaps caused by hairpins and secondary structures
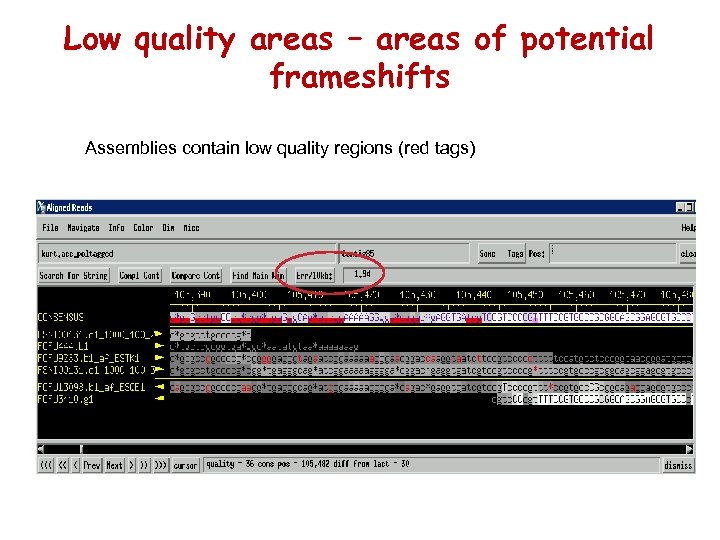
Low quality areas – areas of potential frameshifts Assemblies contain low quality regions (red tags)
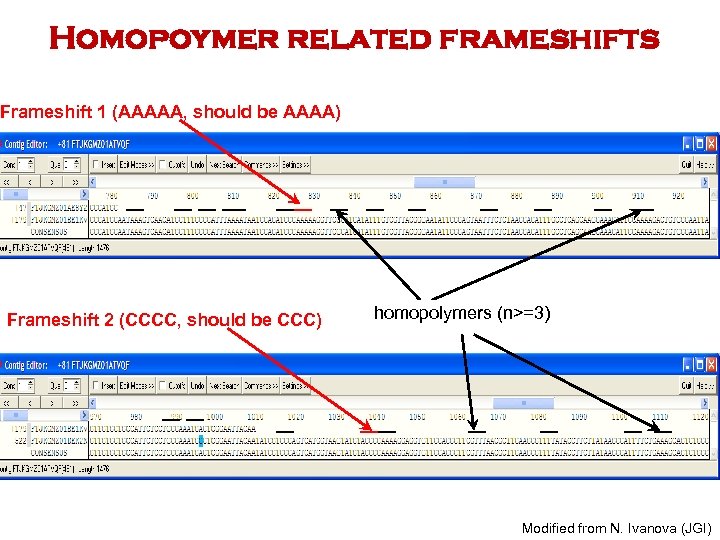
Homopoymer related frameshifts Frameshift 1 (AAAAA, should be AAAA) Frameshift 2 (CCCC, should be CCC) homopolymers (n>=3) Modified from N. Ivanova (JGI)
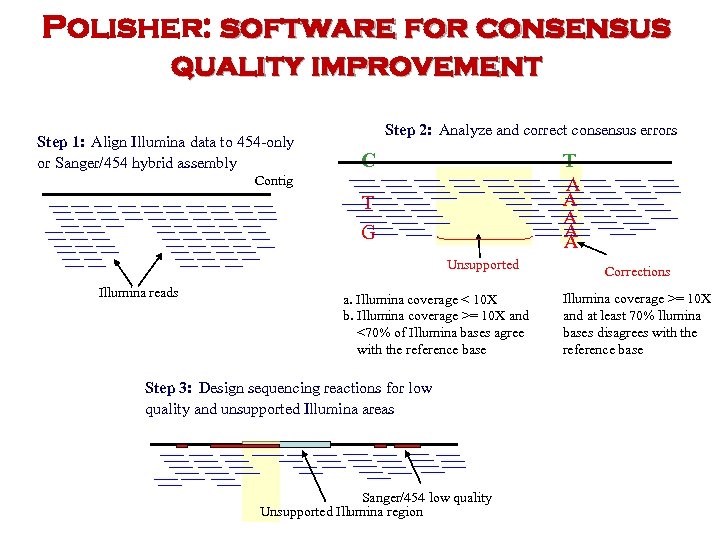
Polisher: software for consensus quality improvement Step 1: Align Illumina data to 454 -only or Sanger/454 hybrid assembly Step 2: Analyze and correct consensus errors C T A A A Contig T G Unsupported Illumina reads a. Illumina coverage < 10 X b. Illumina coverage >= 10 X and <70% of Illumina bases agree with the reference base Step 3: Design sequencing reactions for low quality and unsupported Illumina areas Sanger/454 low quality Unsupported Illumina region Corrections Illumina coverage >= 10 X and at least 70% llumina bases disagrees with the reference base

Errors corrected by Solexa Frame shift detected (454 contig) Finished consensus 454 contig Sanger reads CCTCTTTGATGGAAATGATA**TCTTCGAGCATCGCCTC**GGGTTTTCCATACAGAGAACCTTTGATGATGAACCGGTTGAAGATCTGCGGGTCAAA CCTCTTTGATGGAAATAATA**TATTCGAGCATC TTAGTGGAAATGATA**TCTTCGAGCATCGCCTC CGAGCNTCGCCTC**GGGCTTTCCCT CGAGCATCGCCTC**GGGTTCTCCATACACAGA GCATCGCCTC**GGGTTTTCAATACAGAGAACCT CAGCGCCTC**GGGTTTTCCATACAGAGAACCTT ATCGCCTC**GGGTTTTCCAGAGAACCTTT GGTTC**GGGTTTTCCATACAGAGAACCTTTGAT GTTTTCCATACAGAGAACATTTGATGATGAAC GTTGTCCATACAGAGAACTTTTGATGATGAAC TATANCATACAGAGAACCTTTGATGATGAACC ATTTCCAGAGAACCNTTGATGATGAACC CAAACAGAGAACCTTTGAGGATGAACCGGTTG ACAGGGAACCTTAGATGATGAACCGGTTGAAG ACAGAGAACCTTAGATGATGAACCGGTTGAAG ACCGTTGATGATGAACCGGTTGAAGATCTGCG GATGGTGAACGGGTTGAAGATCTGCGGGTCAA GGTTTGAAGATCTGCGGGTCAAACCAGTCCTC GGTGGAAGATCTGCGGGTAAAACCAGTCCTCT GGT. GNAGAGCTGCGGGTCAAACCAGTCCTCTG TGAAGATCTGCGGTTCAAACCAGTCCTCTCCC GATCGGCGTGTCAAACCAGTCCTCTGCCTCGT TCTGCGGGTCAAACCAGTACTCTGCCTCGTTC

So, what is Finishing? The process of taking a rough draft assembly composed of shotgun sequencing reads, identifying and resolving miss assemblies, sequence gaps and regions of low quality to produce a highly accurate finished DNA sequence. Final quality: Final error rate should be less than 1 per 50 Kb. No gaps, no misassembled areas, no characters other than ACGT
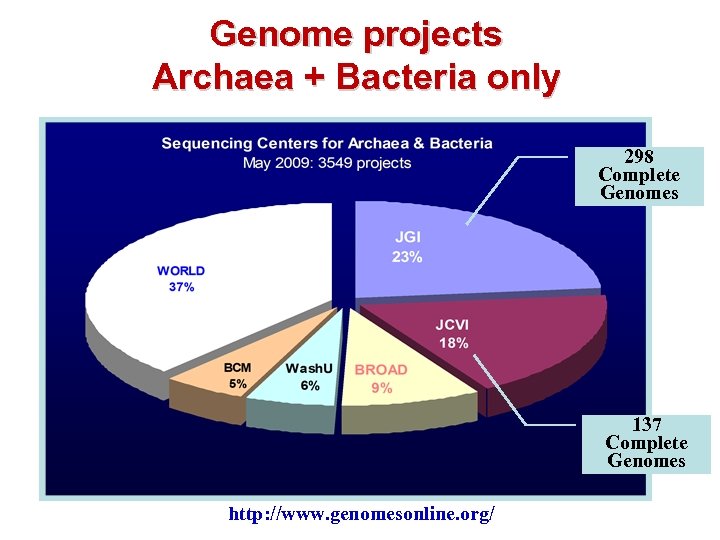
Genome projects Archaea + Bacteria only 298 Complete Genomes 137 Complete Genomes http: //www. genomesonline. org/
Metagenomic assembly and Finishing The whole-genome shotgun sequencing approach was used for a number of microbial community projects, however useful quality control and assembly of these data require reassessing methods developed to handle relatively uniform sequences derived from isolate microbes. • Typically size of metagenomic sequencing project is very large • Different organisms have different coverage. Non-uniform sequence coverage results in significant under- and over-representation of certain community members • Low coverage for the majority of organisms in highly complex communities leads to poor (if any) assemblies • Chimerical contigs produced by co-assembly of sequencing reads originating from different species. • Genome rearrangements and the presence of mobile genetic elements (phages, transposons) in closely related organisms further complicate assembly. • No assemblers developed for metagenomic data sets
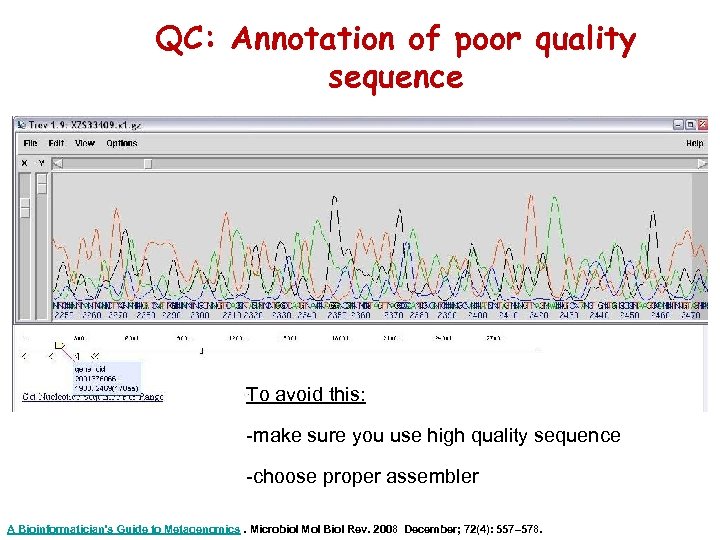
QC: Annotation of poor quality sequence To avoid this: -make sure you use high quality sequence -choose proper assembler A Bioinformatician's Guide to Metagenomics. Microbiol Mol Biol Rev. 2008 December; 72(4): 557– 578.
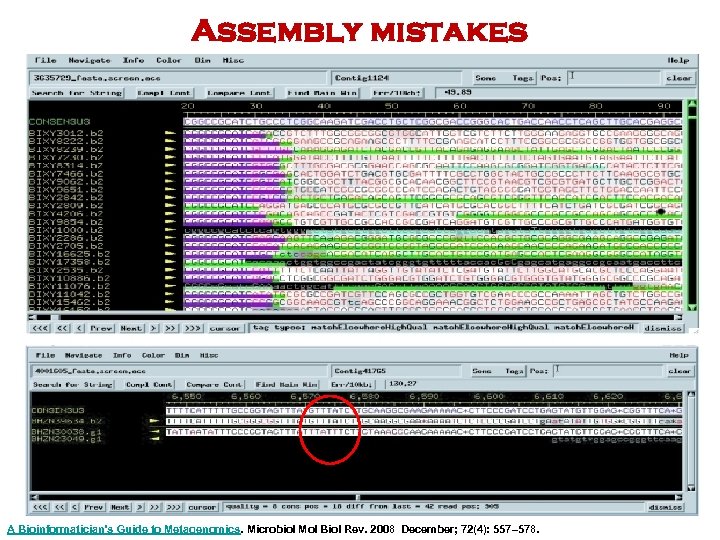
Assembly mistakes A Bioinformatician's Guide to Metagenomics. Microbiol Mol Biol Rev. 2008 December; 72(4): 557– 578.
Recommendations for metagenomic assembly - Use Trimmer (Lucy etc) to treat reads PRIOR to assembly - None of the existing assemblers designed for metagenomic data but assemblers like PGA work better with paired reads information and produce better assemblies. - We currently test Newbler assembler for second generation sequencing: 454 only and 454/Solexa co-assembly
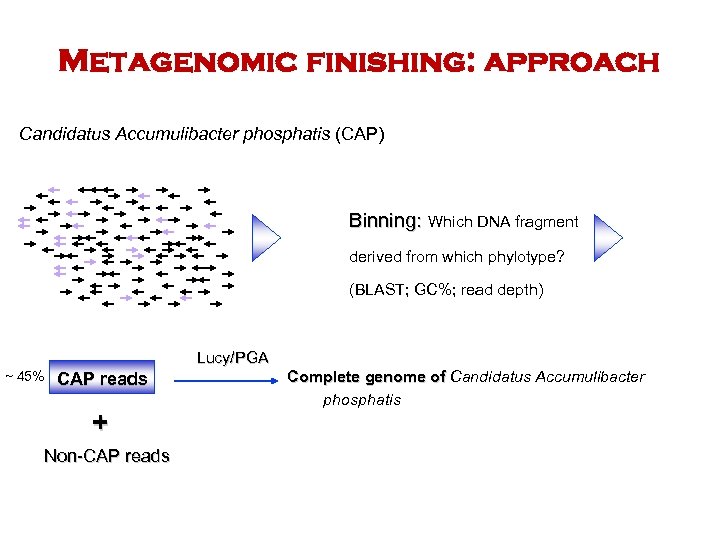
Metagenomic finishing: approach Candidatus Accumulibacter phosphatis (CAP) Binning: Which DNA fragment derived from which phylotype? (BLAST; GC%; read depth) Lucy/PGA ~ 45% CAP reads + Non-CAP reads Complete genome of Candidatus Accumulibacter phosphatis
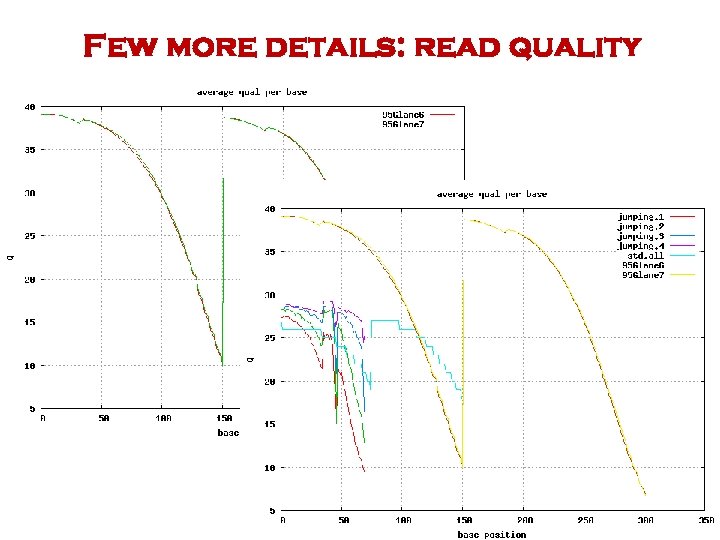
Few more details: read quality
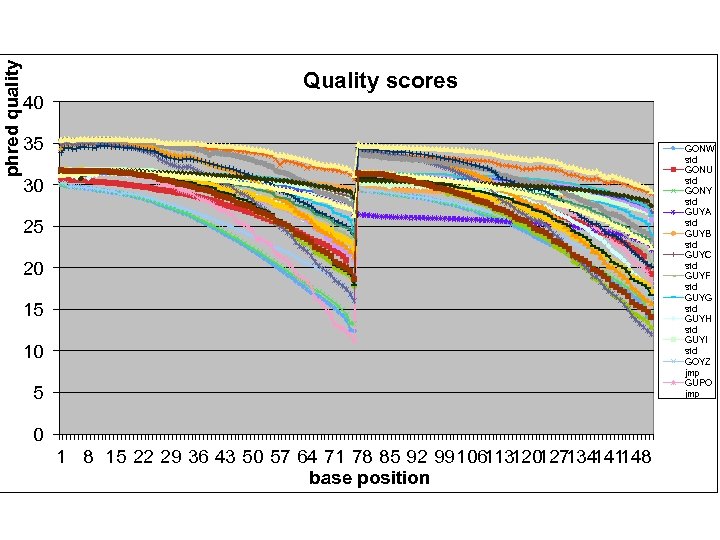
phred quality 40 Quality scores 35 GONW std GONU std GONY std GUYA std GUYB std GUYC std GUYF std GUYG std GUYH std GUYI std GOYZ jmp GUPO jmp 30 25 20 15 10 5 0 1 8 15 22 29 36 43 50 57 64 71 78 85 92 99106 120 134 148 113 127 141 base position
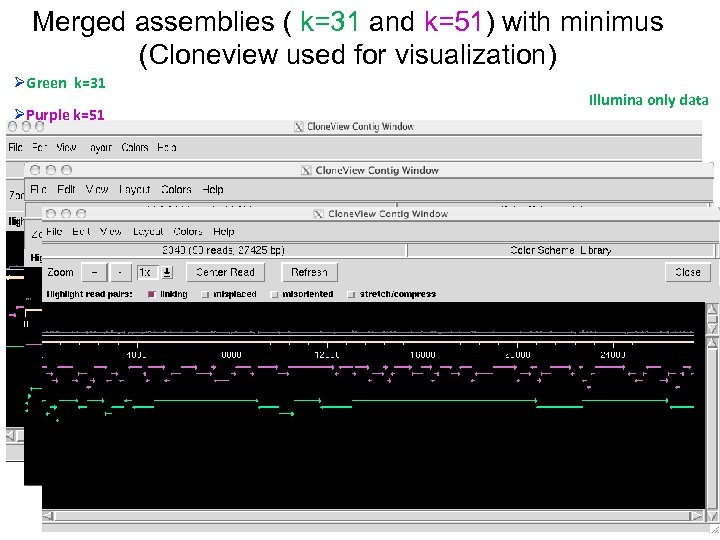
Merged assemblies ( k=31 and k=51) with minimus (Cloneview used for visualization) ØGreen k=31 ØPurple k=51 Illumina only data
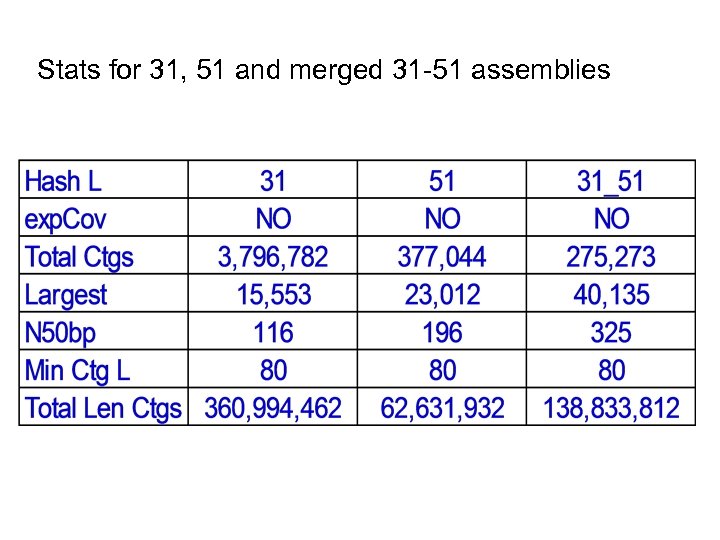
Stats for 31, 51 and merged 31 -51 assemblies

Thank you!
12b37ecef6293b4d8d25076afd74efb3.ppt