4dd684b15e99e72874976215409c2a1f.ppt
- Количество слайдов: 49

Genome Annotation Daniel Lawson Vector. Base @ EBI August 2008 Bioinformatics tools for Comparative Genomics of Vectors 1
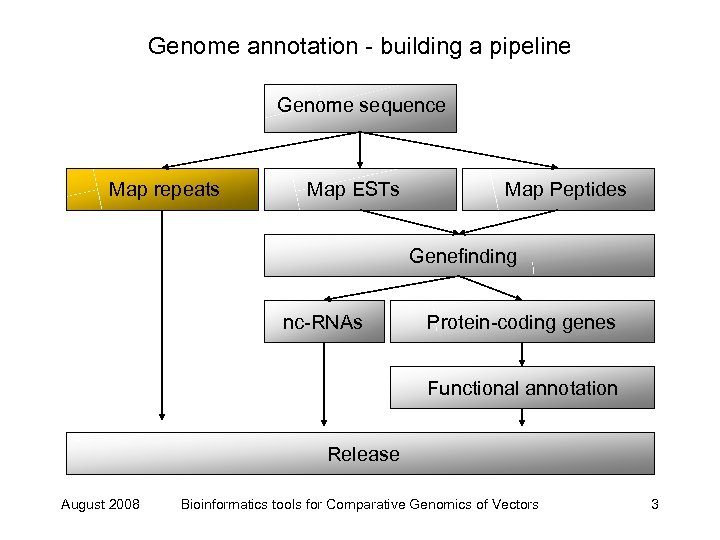
Genome annotation - building a pipeline Genome sequence Map repeats Map ESTs Map Peptides Genefinding nc-RNAs Protein-coding genes Functional annotation Release August 2008 Bioinformatics tools for Comparative Genomics of Vectors 3
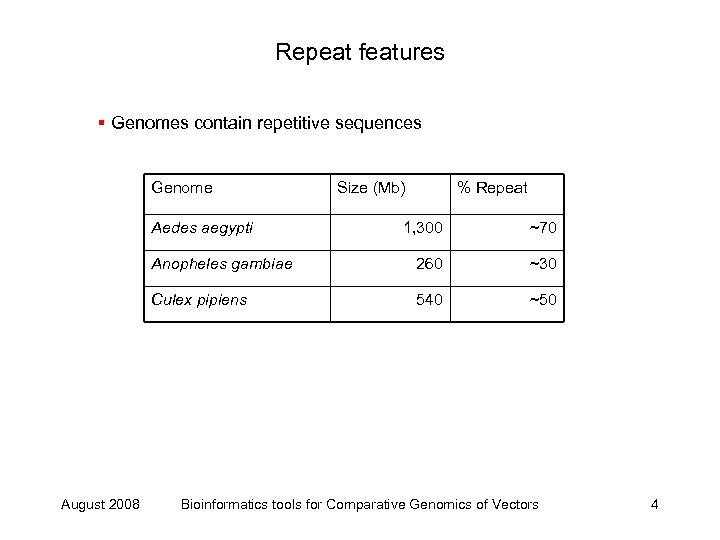
Repeat features § Genomes contain repetitive sequences Genome Aedes aegypti Size (Mb) % Repeat ~70 Anopheles gambiae 260 ~30 Culex pipiens August 2008 1, 300 540 ~50 Bioinformatics tools for Comparative Genomics of Vectors 4
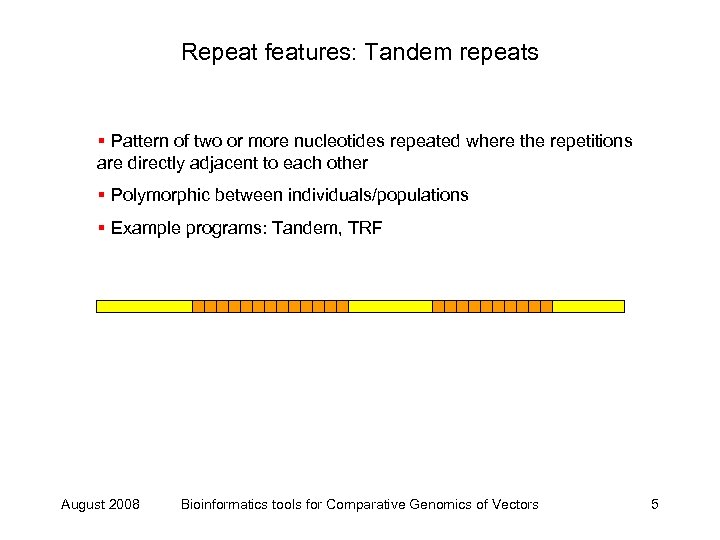
Repeat features: Tandem repeats § Pattern of two or more nucleotides repeated where the repetitions are directly adjacent to each other § Polymorphic between individuals/populations § Example programs: Tandem, TRF August 2008 Bioinformatics tools for Comparative Genomics of Vectors 5
Repeat features: Interspersed elements § Transposable elements (TEs) § Transposons, Retrotransposons etc § Entire research field in itself § Example programs: Repeatscout, RECON August 2008 Bioinformatics tools for Comparative Genomics of Vectors 6
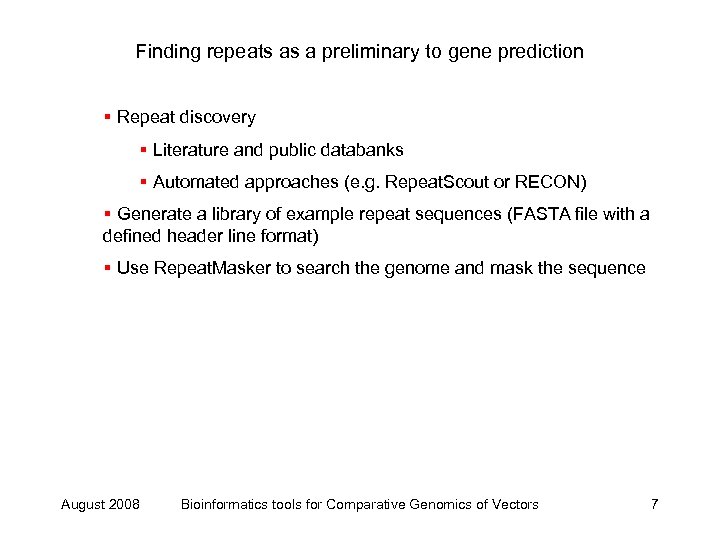
Finding repeats as a preliminary to gene prediction § Repeat discovery § Literature and public databanks § Automated approaches (e. g. Repeat. Scout or RECON) § Generate a library of example repeat sequences (FASTA file with a defined header line format) § Use Repeat. Masker to search the genome and mask the sequence August 2008 Bioinformatics tools for Comparative Genomics of Vectors 7

Masked sequence § § Repeatmasked sequence is an artificial construction where those regions which are thought to be repetitive are marked with X’s Widely used to reduce the overhead of subsequent computational analyses and to reduce the impact of TE’s in the final annotation set >my sequence (repeatmasked) atgagcttcgatagcgatcagctagcgatcaggctactattggct tctctagactcgtctatctctattagctatcatctcgatagcgatcag ctagcgatcaggctactattggcttcgatagcgatcagctagcgatcaggct actattggctgatcttaggtcttctgatcttct atgagcttcgatagcgatcagctagcgatcaggctactattxxxxxxxxxxxxxxxxatctcgatagcg atcagctagcgatcaggctactattxxxxxxxxxx tagcgatcaggctactattggcttcgatagcgatcagctagcgat caggctxxxxxxxxxxtcttctgatcttct August 2008 Bioinformatics tools for Comparative Genomics of Vectors 8

Masked sequence - Hard or Soft? § Sometimes we want to mark up repetitive sequence but not to exclude it from downstream analyses. This is achieved using a format known as soft-masked >my sequence (softmasked) ATGAGCTTCGATAGCGCATCAGCTAGCGAT CAGGCTACTATTGGCTTCTCTAGACTCGTCT ATCTCTATTAGTATCATCTCGATAGCGATCA GCTAGCGATCAGGCTACTATTGGCTTCGAT AGCGATCAGCTAGCGATCAGGCTACTATTG GCTTCGATAGCGATCAGCTAGCGATCAGGC TACTATTGGCTGATCTTAGGTCTTCTGATCT ATGAGCTTCGATAGCGCATCAGCTAGCGAT CAGGCTACTATTggcttctctagactcgtctatctctattag tatc. ATCTCGATAGCGATCAGCTAGCGATCAG GCTACTATTggcttcgatagcgatcagc. TAGCGATC AGGCTACTATTggcttcgatagcgatcagc. TAGCGA TCAGGCTACTATTGGCTGATCTTAGGTCTTC TGATCTTCT August 2008 Bioinformatics tools for Comparative Genomics of Vectors 9
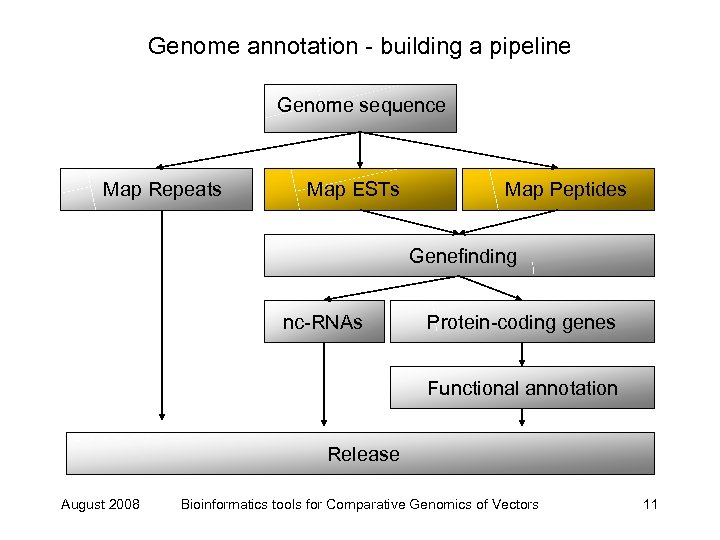
Genome annotation - building a pipeline Genome sequence Map Repeats Map ESTs Map Peptides Genefinding nc-RNAs Protein-coding genes Functional annotation Release August 2008 Bioinformatics tools for Comparative Genomics of Vectors 11
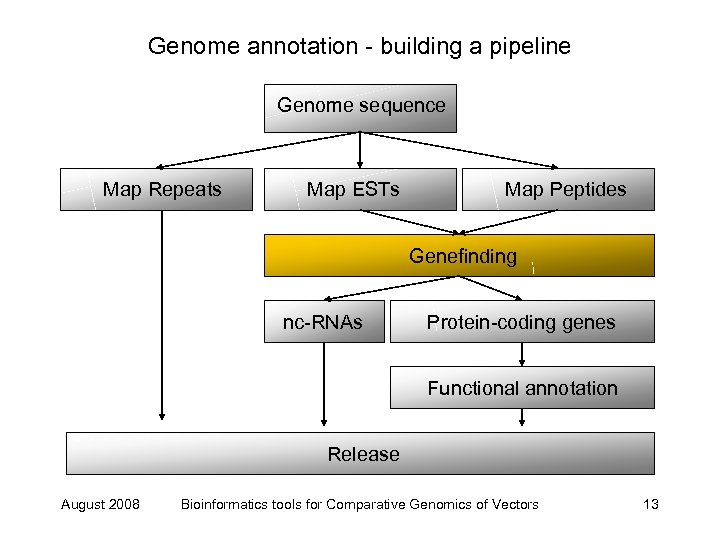
Genome annotation - building a pipeline Genome sequence Map Repeats Map ESTs Map Peptides Genefinding nc-RNAs Protein-coding genes Functional annotation Release August 2008 Bioinformatics tools for Comparative Genomics of Vectors 13
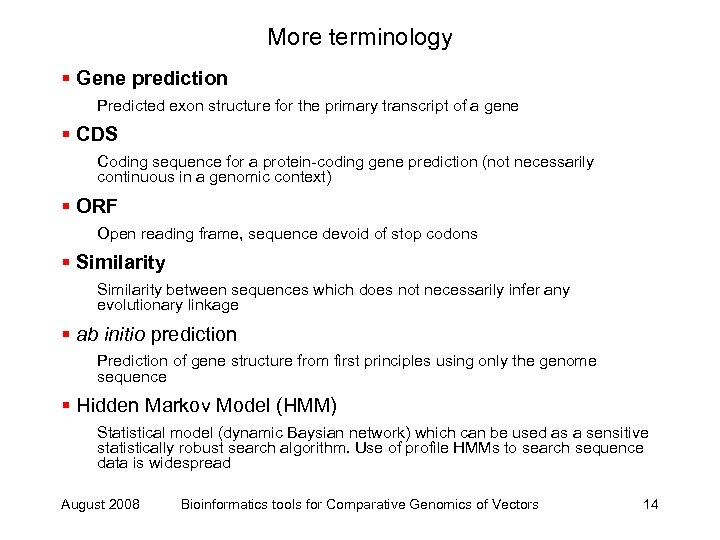
More terminology § Gene prediction Predicted exon structure for the primary transcript of a gene § CDS Coding sequence for a protein-coding gene prediction (not necessarily continuous in a genomic context) § ORF Open reading frame, sequence devoid of stop codons § Similarity between sequences which does not necessarily infer any evolutionary linkage § ab initio prediction Prediction of gene structure from first principles using only the genome sequence § Hidden Markov Model (HMM) Statistical model (dynamic Baysian network) which can be used as a sensitive statistically robust search algorithm. Use of profile HMMs to search sequence data is widespread August 2008 Bioinformatics tools for Comparative Genomics of Vectors 14
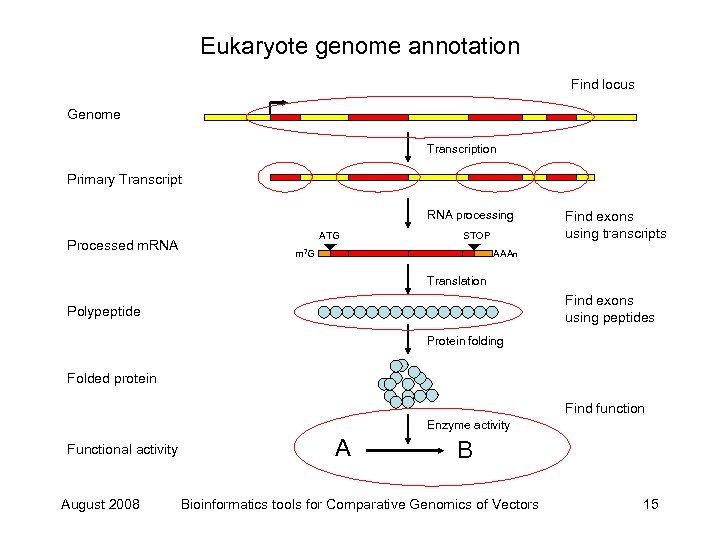
Eukaryote genome annotation Find locus Genome Transcription Primary Transcript RNA processing Processed m. RNA ATG STOP m 7 G Find exons using transcripts AAAn Translation Find exons using peptides Polypeptide Protein folding Folded protein Find function Enzyme activity Functional activity August 2008 A B Bioinformatics tools for Comparative Genomics of Vectors 15
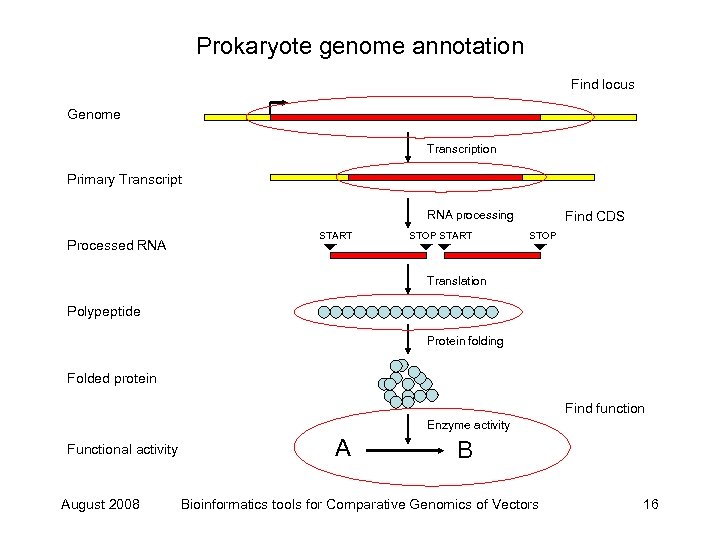
Prokaryote genome annotation Find locus Genome Transcription Primary Transcript RNA processing Processed RNA START STOP START Find CDS STOP Translation Polypeptide Protein folding Folded protein Find function Enzyme activity Functional activity August 2008 A B Bioinformatics tools for Comparative Genomics of Vectors 16
Genefinding ab initio August 2008 similarity Bioinformatics tools for Comparative Genomics of Vectors 17
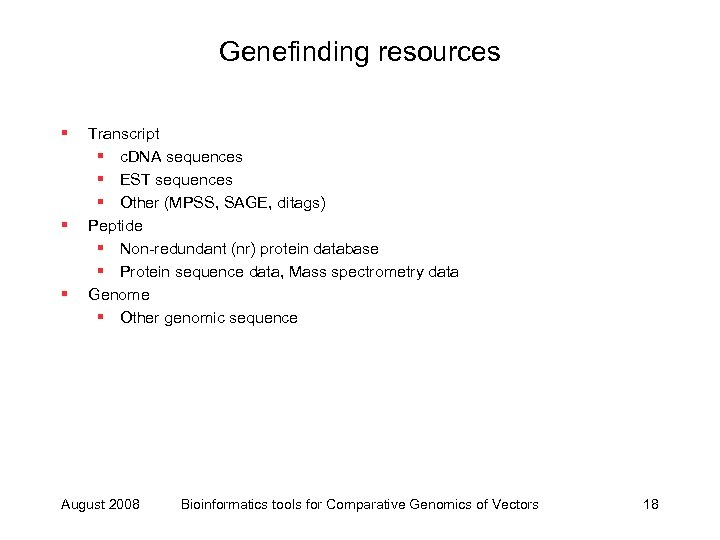
Genefinding resources § § § Transcript § c. DNA sequences § EST sequences § Other (MPSS, SAGE, ditags) Peptide § Non-redundant (nr) protein database § Protein sequence data, Mass spectrometry data Genome § Other genomic sequence August 2008 Bioinformatics tools for Comparative Genomics of Vectors 18
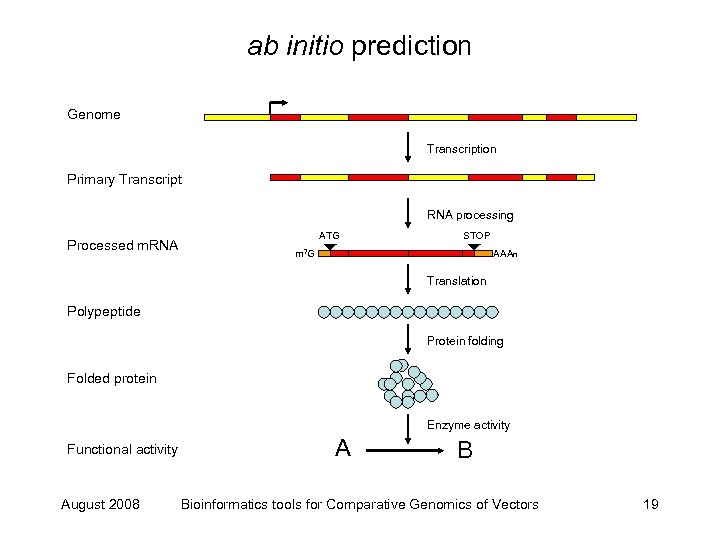
ab initio prediction Genome Transcription Primary Transcript RNA processing Processed m. RNA ATG STOP m 7 G AAAn Translation Polypeptide Protein folding Folded protein Enzyme activity Functional activity August 2008 A B Bioinformatics tools for Comparative Genomics of Vectors 19
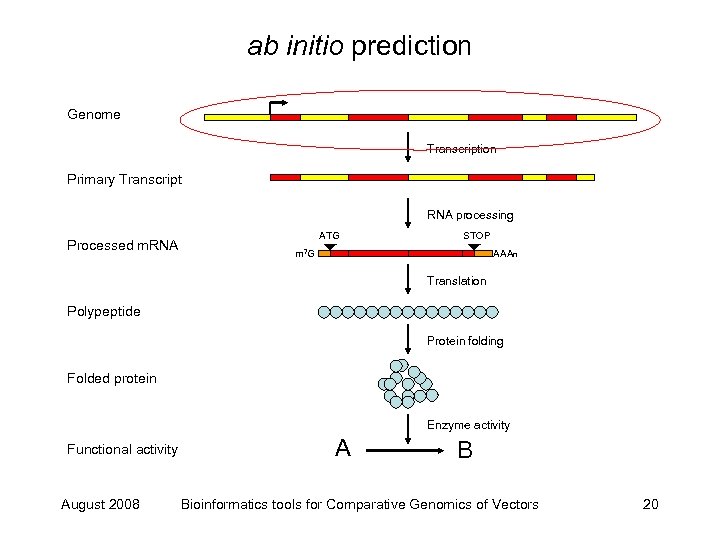
ab initio prediction Genome Transcription Primary Transcript RNA processing Processed m. RNA ATG STOP m 7 G AAAn Translation Polypeptide Protein folding Folded protein Enzyme activity Functional activity August 2008 A B Bioinformatics tools for Comparative Genomics of Vectors 20
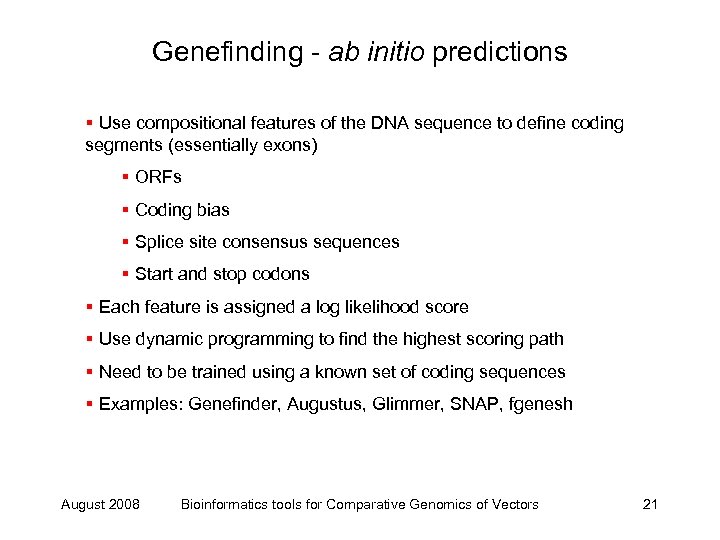
Genefinding - ab initio predictions § Use compositional features of the DNA sequence to define coding segments (essentially exons) § ORFs § Coding bias § Splice site consensus sequences § Start and stop codons § Each feature is assigned a log likelihood score § Use dynamic programming to find the highest scoring path § Need to be trained using a known set of coding sequences § Examples: Genefinder, Augustus, Glimmer, SNAP, fgenesh August 2008 Bioinformatics tools for Comparative Genomics of Vectors 21
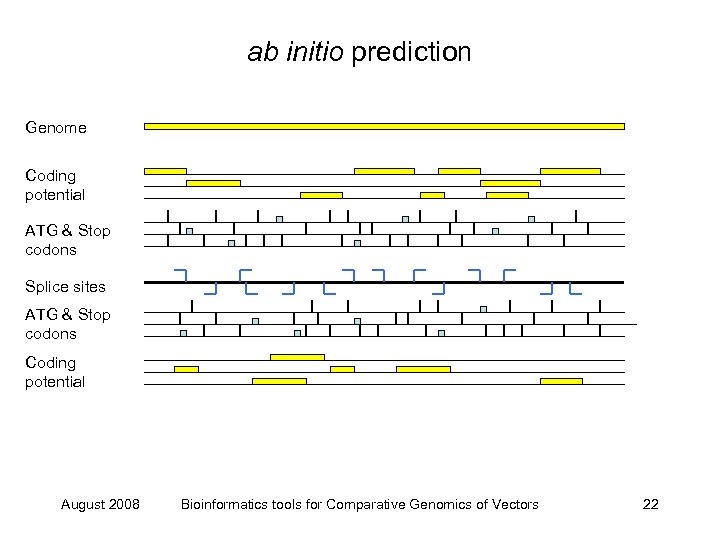
ab initio prediction Genome Coding potential ATG & Stop codons Splice sites ATG & Stop codons Coding potential August 2008 Bioinformatics tools for Comparative Genomics of Vectors 22
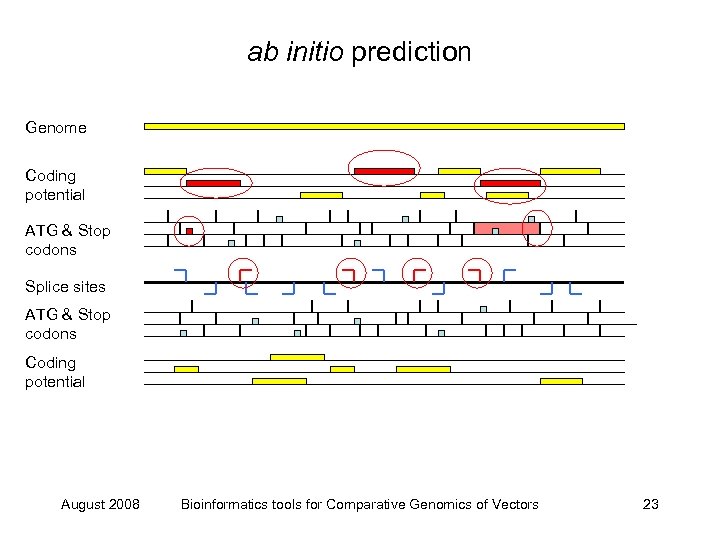
ab initio prediction Genome Coding potential ATG & Stop codons Splice sites ATG & Stop codons Coding potential August 2008 Bioinformatics tools for Comparative Genomics of Vectors 23
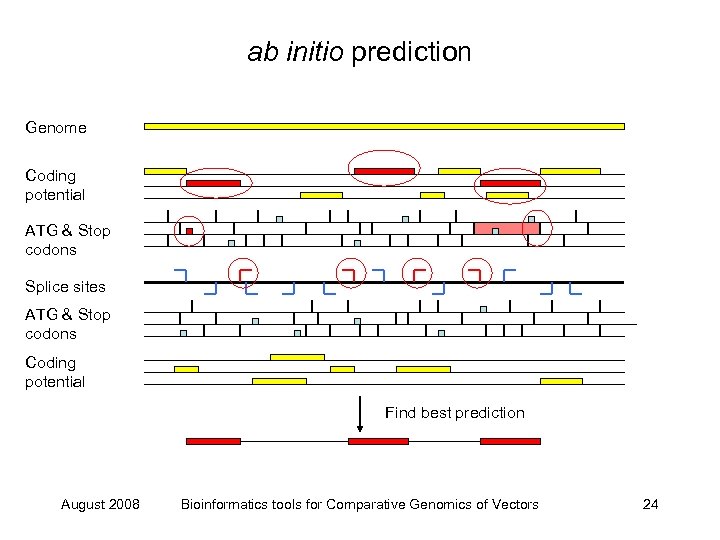
ab initio prediction Genome Coding potential ATG & Stop codons Splice sites ATG & Stop codons Coding potential Find best prediction August 2008 Bioinformatics tools for Comparative Genomics of Vectors 24
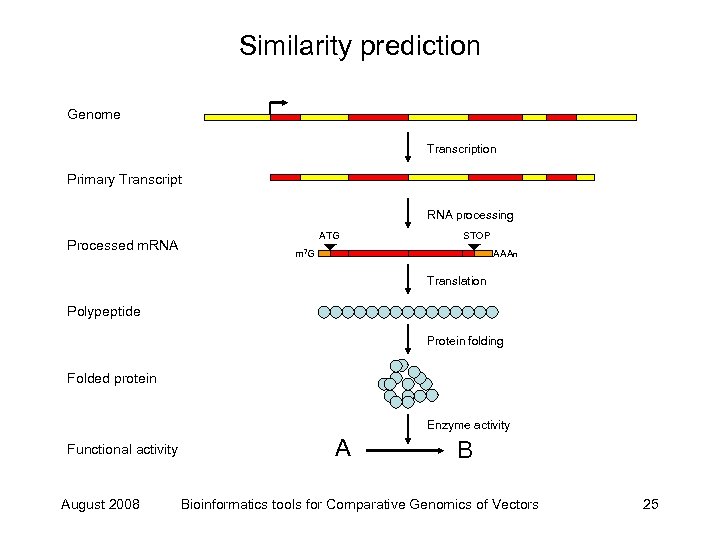
Similarity prediction Genome Transcription Primary Transcript RNA processing Processed m. RNA ATG STOP m 7 G AAAn Translation Polypeptide Protein folding Folded protein Enzyme activity Functional activity August 2008 A B Bioinformatics tools for Comparative Genomics of Vectors 25
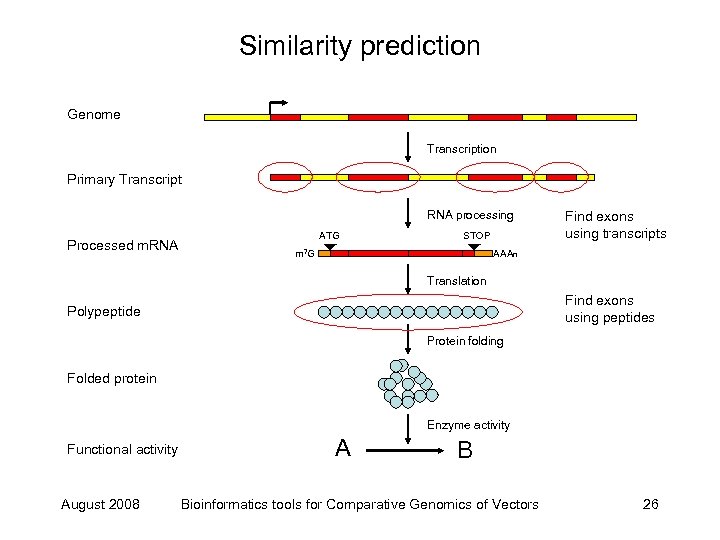
Similarity prediction Genome Transcription Primary Transcript RNA processing Processed m. RNA ATG STOP m 7 G Find exons using transcripts AAAn Translation Find exons using peptides Polypeptide Protein folding Folded protein Enzyme activity Functional activity August 2008 A B Bioinformatics tools for Comparative Genomics of Vectors 26
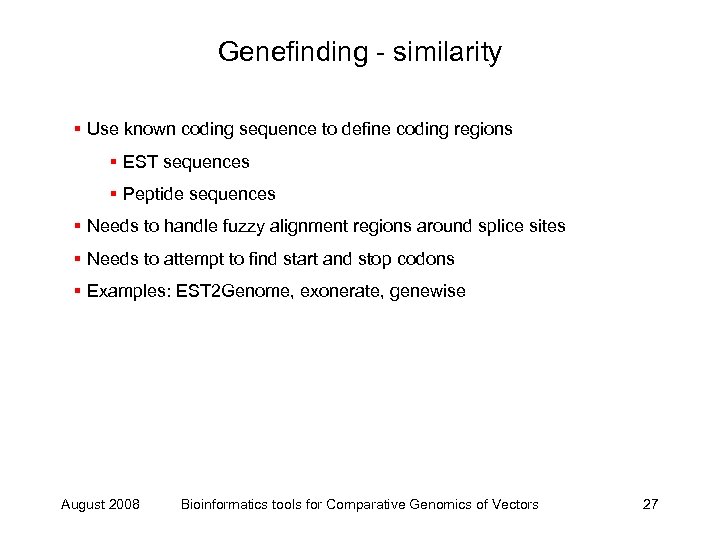
Genefinding - similarity § Use known coding sequence to define coding regions § EST sequences § Peptide sequences § Needs to handle fuzzy alignment regions around splice sites § Needs to attempt to find start and stop codons § Examples: EST 2 Genome, exonerate, genewise August 2008 Bioinformatics tools for Comparative Genomics of Vectors 27
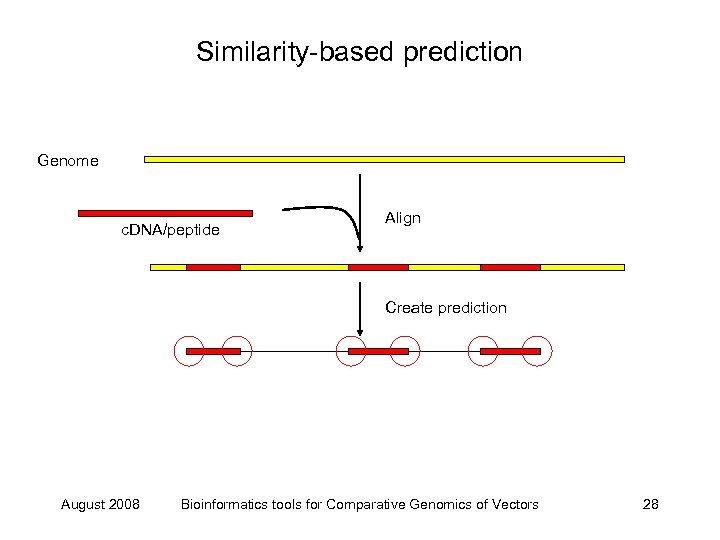
Similarity-based prediction Genome c. DNA/peptide Align Create prediction August 2008 Bioinformatics tools for Comparative Genomics of Vectors 28

Genefinding - comparative § Use 2 or more genomic sequences to predict genes based on conservation of exon sequences § Examples: Twinscan and SLAM August 2008 Bioinformatics tools for Comparative Genomics of Vectors 29
Genefinding - manual § § Manual annotation is time consuming Annotators use specialized utilities to view genomic regions with tiers/columns of data from which they construct a gene prediction Most decisions are subjective and tedious to document Avoids the systematic problems of ab initio predictors and automated annotation pipeline August 2008 Bioinformatics tools for Comparative Genomics of Vectors 30
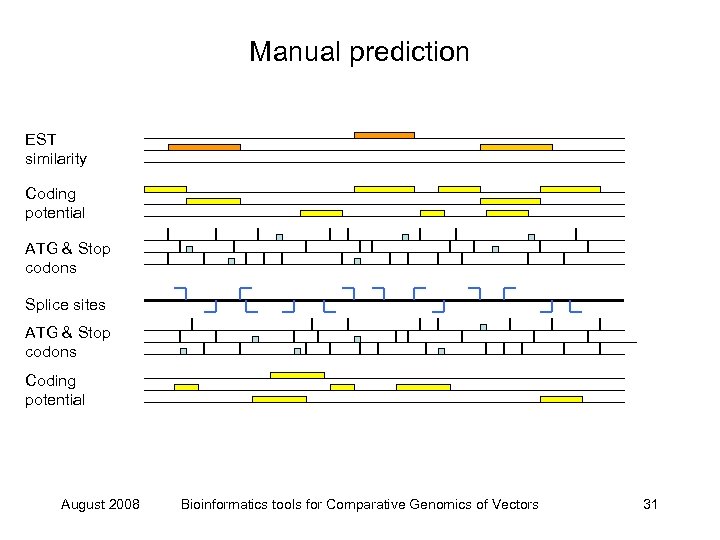
Manual prediction EST similarity Coding potential ATG & Stop codons Splice sites ATG & Stop codons Coding potential August 2008 Bioinformatics tools for Comparative Genomics of Vectors 31
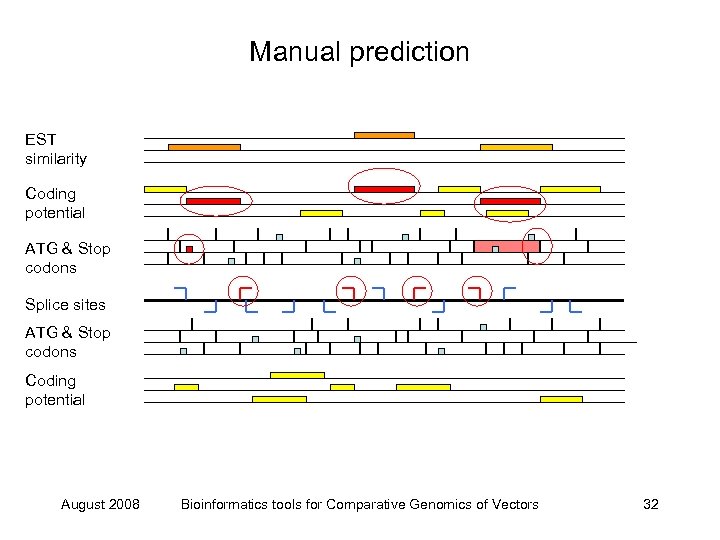
Manual prediction EST similarity Coding potential ATG & Stop codons Splice sites ATG & Stop codons Coding potential August 2008 Bioinformatics tools for Comparative Genomics of Vectors 32
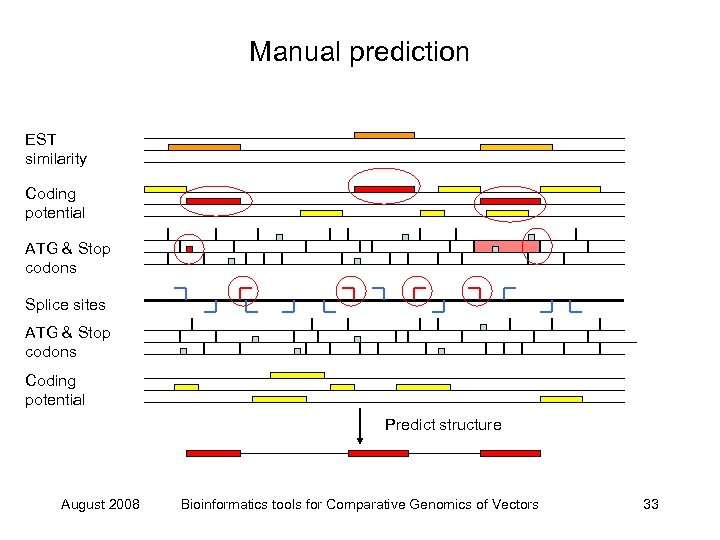
Manual prediction EST similarity Coding potential ATG & Stop codons Splice sites ATG & Stop codons Coding potential Predict structure August 2008 Bioinformatics tools for Comparative Genomics of Vectors 33
Genefinding - non-coding RNA genes § Non-coding RNA genes can be predicted using knowledge of their structure or by similarity with known examples § t. RNAscan - uses an HMM and co-variance model for prediction of t. RNA genes § Rfam - a suite of HMM’s trained against a large number of different RNA genes August 2008 Bioinformatics tools for Comparative Genomics of Vectors 34
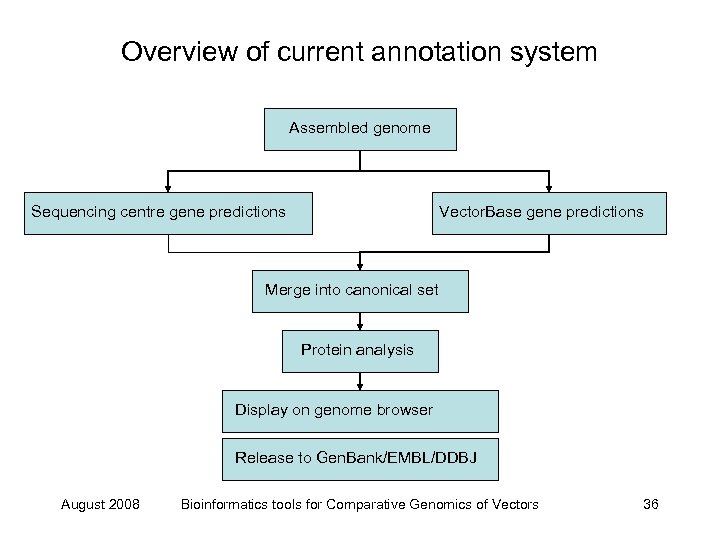
Overview of current annotation system Assembled genome Sequencing centre gene predictions Vector. Base gene predictions Merge into canonical set Protein analysis Display on genome browser Release to Gen. Bank/EMBL/DDBJ August 2008 Bioinformatics tools for Comparative Genomics of Vectors 36
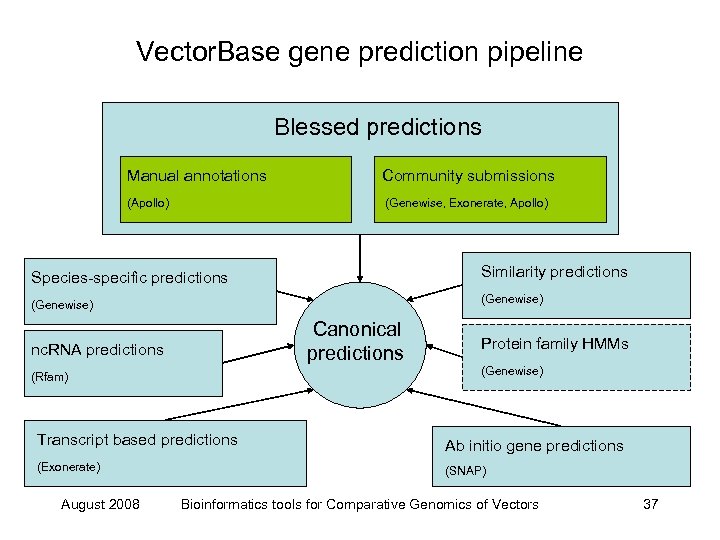
Vector. Base gene prediction pipeline Blessed predictions Manual annotations Community submissions (Apollo) (Genewise, Exonerate, Apollo) Species-specific predictions Similarity predictions (Genewise) Canonical predictions nc. RNA predictions (Rfam) Protein family HMMs (Genewise) Transcript based predictions Ab initio gene predictions (Exonerate) (SNAP) August 2008 Bioinformatics tools for Comparative Genomics of Vectors 37
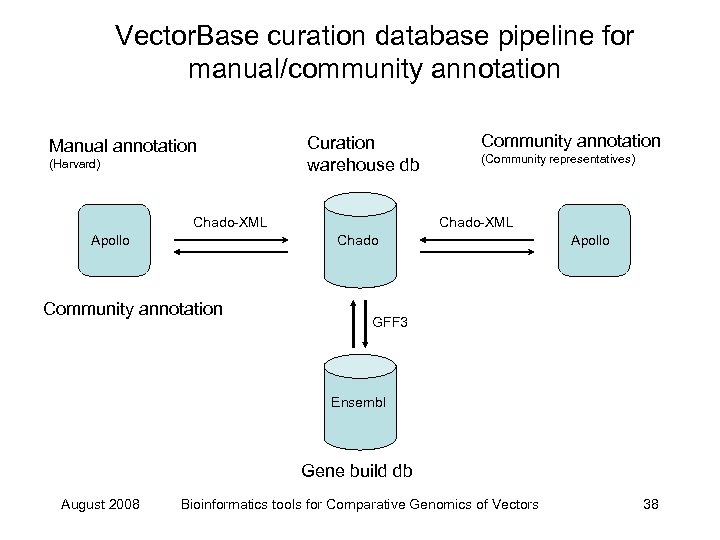
Vector. Base curation database pipeline for manual/community annotation Manual annotation (Harvard) Curation warehouse db Chado-XML Apollo Community annotation (Community representatives) Chado-XML Chado Community annotation Apollo GFF 3 Ensembl Gene build db August 2008 Bioinformatics tools for Comparative Genomics of Vectors 38

Genefinding - Review § § Gene prediction relies heavily on similarity data EST/c. DNA sequences are vital for genefinding § Training for ab initio approaches § Similarity builds § Validating predictions Protein data is the predominant supporting evidence for prediction in most vector genomes § Need to be wary of predicting from predictions Genefinding is still something of a dark art § Efforts to standardize and document supporting evidence for prediction and modifications are ongoing August 2008 Bioinformatics tools for Comparative Genomics of Vectors 39
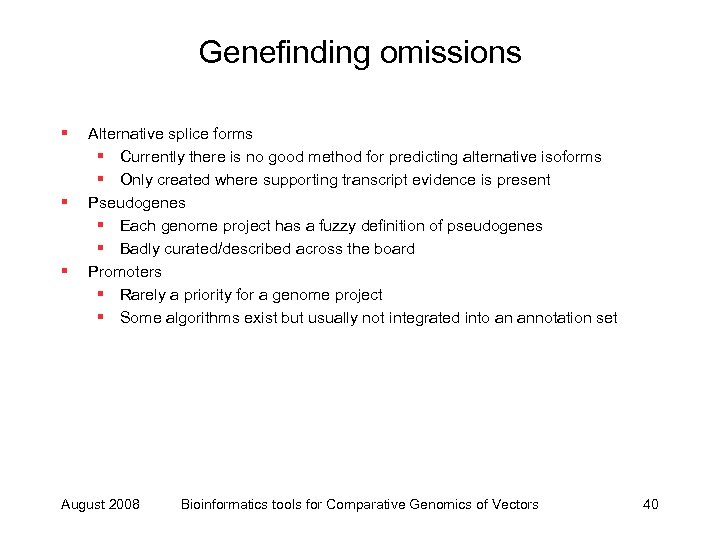
Genefinding omissions § § § Alternative splice forms § Currently there is no good method for predicting alternative isoforms § Only created where supporting transcript evidence is present Pseudogenes § Each genome project has a fuzzy definition of pseudogenes § Badly curated/described across the board Promoters § Rarely a priority for a genome project § Some algorithms exist but usually not integrated into an annotation set August 2008 Bioinformatics tools for Comparative Genomics of Vectors 40

Functional annotation August 2008 Bioinformatics tools for Comparative Genomics of Vectors 41

Functional annotation § § Utilise known structure/function information to infer facts related to the predicted protein sequence Provide users with results from a number of standard algorithms/searches Provide users with cross-references (dbxrefs) to other resources Assign a simple one line description for each gene product § § This will never be comprehensive This will always be somewhat general August 2008 Bioinformatics tools for Comparative Genomics of Vectors 42
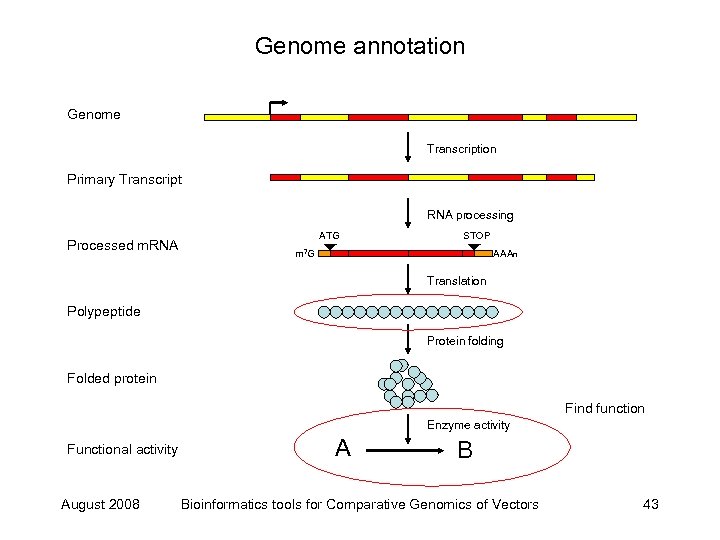
Genome annotation Genome Transcription Primary Transcript RNA processing Processed m. RNA ATG STOP m 7 G AAAn Translation Polypeptide Protein folding Folded protein Find function Enzyme activity Functional activity August 2008 A B Bioinformatics tools for Comparative Genomics of Vectors 43
Functional annotation - protein similarities § § § Predicted proteins are searched against the non-redundant protein database to look for similarities Visually assess the top 5 -10 hits to identify whether these have been assigned a function It is important to check how the function of the top hits has been assigned in order not to transfer erroneous annotations August 2008 Bioinformatics tools for Comparative Genomics of Vectors 44
Functional annotation - Protein domains § § § Protein domains have a number of definitions based on their size, folding and function/evolution. Domains are a part of protein structure description Domains with a similar structure are likely to be related evolutionarily and have a similar function We can use this to infer function (& structure) for an unknown protein be comparison to known proteins The tool of choice here is a Hidden Markov Model (HMM) August 2008 Bioinformatics tools for Comparative Genomics of Vectors 45
Protein Domain databases § Inter. Pro § § § August 2008 Uni. Prot - protein database Prosite - database of regular expressions Pfam - profile HMMs PRINTS - conserved protein signatures Prodom - collection of multiple sequence alignments SMART - HMMs TIGRfams - HMMs PIRSF Superfamily Gene 3 D Panther - HMMs Bioinformatics tools for Comparative Genomics of Vectors 46
Functional annotation - Other features § Other features which can be determined § Signal peptides § Transmembrane domains § Low complexity regions § Various binding sites, glycosylation sites etc. See http: //expasy. org/tools/ for a good list of possible prediction algorithms August 2008 Bioinformatics tools for Comparative Genomics of Vectors 47
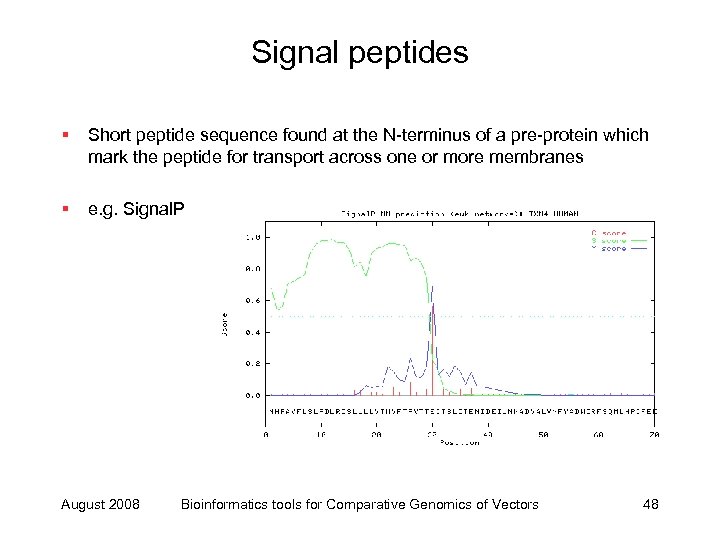
Signal peptides § Short peptide sequence found at the N-terminus of a pre-protein which mark the peptide for transport across one or more membranes § e. g. Signal. P August 2008 Bioinformatics tools for Comparative Genomics of Vectors 48
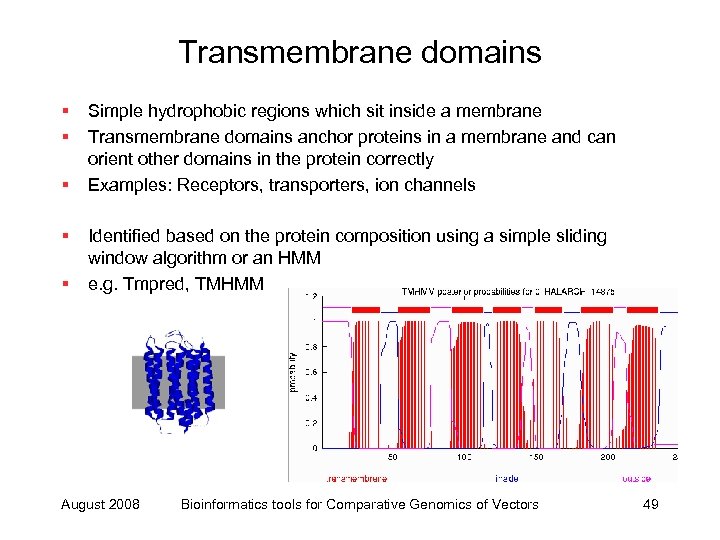
Transmembrane domains § § § Simple hydrophobic regions which sit inside a membrane Transmembrane domains anchor proteins in a membrane and can orient other domains in the protein correctly Examples: Receptors, transporters, ion channels Identified based on the protein composition using a simple sliding window algorithm or an HMM e. g. Tmpred, TMHMM August 2008 Bioinformatics tools for Comparative Genomics of Vectors 49

Ontologies § Use of ontologies to annotate gene products § Gene Ontology (GO) § Cellular component § Molecular function § Biological process § Sequence Ontology (SO) August 2008 Bioinformatics tools for Comparative Genomics of Vectors 50
Other data to look at § § § Enzyme classification (EC) numbers Phenotype information § Alleles § Gene knockouts § RNAi knockdowns Expression data § EST libraries (source of RNA material) § Microarrays § SAGE tags August 2008 Bioinformatics tools for Comparative Genomics of Vectors 51

Functional assignment § § The assignment of a function to a gene product can be made by a human curator by assessing all of the data (similarities, protein domains, signal peptide etc) This is a labour intensive process and like gene prediction is subjective § There automated approaches (based on family and domain databases such as Panther or Inter. Pro) but these are under-developed § Large number of predictions from a genome project remain ‘hypothetical protein’ or ‘conserved hypothetical protein’. August 2008 Bioinformatics tools for Comparative Genomics of Vectors 52

Caveats to genome annotation § § § Annotation accuracy is only as good as the available supporting data at the time of annotation Gene predictions will change over time as new data becomes available (ESTs, related genomes) Functional assignments will change over time as new data becomes available (characterisation of hypothetical proteins) § § Gene predictions are ‘best guess’ Functional annotations are not definitive and only a guide § If you want the annotation to improve you should get involved with whoever is (or has) sequenced your genome of interest. § For vectors you can mail info@vectorbase. org with suggestions and corrections. August 2008 Bioinformatics tools for Comparative Genomics of Vectors 53
4dd684b15e99e72874976215409c2a1f.ppt