a45f218ab156f72074e6dc90acf281cb.ppt
- Количество слайдов: 76
 GENOME ANNOTATION AND FUNCTIONAL GENOMICS The protein sequence perspective
GENOME ANNOTATION AND FUNCTIONAL GENOMICS The protein sequence perspective
 GENOME ANNOTATION • Two main levels: – STRUCTURAL ANNOTATION – Finding genes and other biologically relevant sites thus building up a model of genome as objects with specific locations – FUNCTIONAL ANNOTATION – Objects are used in database searches (and expts) aim is attributing biologically relevant information to whole sequence and individual objects
GENOME ANNOTATION • Two main levels: – STRUCTURAL ANNOTATION – Finding genes and other biologically relevant sites thus building up a model of genome as objects with specific locations – FUNCTIONAL ANNOTATION – Objects are used in database searches (and expts) aim is attributing biologically relevant information to whole sequence and individual objects
 WHY PROTEIN RATHER THAN DNA? • • Larger alphabet -more sensitive comparisons Protein sequences lower signal to noise ratio Less redundancy and no frameshifts Each aa has different properties like size, charge etc Closer to biological function 3 D structure of similar proteins may be known Evolutionary relationships more evident Availability of good, well annotated protein sequence and pattern databases
WHY PROTEIN RATHER THAN DNA? • • Larger alphabet -more sensitive comparisons Protein sequences lower signal to noise ratio Less redundancy and no frameshifts Each aa has different properties like size, charge etc Closer to biological function 3 D structure of similar proteins may be known Evolutionary relationships more evident Availability of good, well annotated protein sequence and pattern databases
 Large-scale genome analysis projects • Rate-limiting step is annotation • Whole genome availability provides context information • Main goal is to bridge gap between genotype and phenotype
Large-scale genome analysis projects • Rate-limiting step is annotation • Whole genome availability provides context information • Main goal is to bridge gap between genotype and phenotype
 Definitions of Annotation • Addition of as much reliable and up-to-date information as possible to describe a sequence • Identification, structural description, characterisation of putative protein products and other features in primary genomic sequence • Information attached to genomic coordinates with start and end point, can occur at different levels • Interpreting raw sequence data into useful biological information
Definitions of Annotation • Addition of as much reliable and up-to-date information as possible to describe a sequence • Identification, structural description, characterisation of putative protein products and other features in primary genomic sequence • Information attached to genomic coordinates with start and end point, can occur at different levels • Interpreting raw sequence data into useful biological information
 ANNOTATION/FUNCTION CAN BE MAPPED TO DIFFERENT LEVELS: ORGANISM -phenotypic function (morphology, physiology, behavior, environmental response), context NB CELLULAR -metabolic pathway, signal cascades, cellular localization. Context dependent MOLECULAR -binding sites, catalytic activity, PTM, 3 D structure DOMAIN SINGLE RESIDUE
ANNOTATION/FUNCTION CAN BE MAPPED TO DIFFERENT LEVELS: ORGANISM -phenotypic function (morphology, physiology, behavior, environmental response), context NB CELLULAR -metabolic pathway, signal cascades, cellular localization. Context dependent MOLECULAR -binding sites, catalytic activity, PTM, 3 D structure DOMAIN SINGLE RESIDUE
 Annotation is the description of: • • Function(s) of the protein Post-translational modification(s) Domains and sites Secondary structure Quaternary structure Similarities to other proteins Disease(s) associated with deficiencie(s) in the protein • Sequence conflicts, variants, etc.
Annotation is the description of: • • Function(s) of the protein Post-translational modification(s) Domains and sites Secondary structure Quaternary structure Similarities to other proteins Disease(s) associated with deficiencie(s) in the protein • Sequence conflicts, variants, etc.
 Additional information for proteins • • FUNCTION CATALYTIC ACTIVITY COFACTOR INDUCTION ENZYME REGULATION PATHWAY SUBUNIT DOMAIN • • • SPLICE PRODUCTS POLYMORPHISM DISEASE TISSUE SPECIFICITY DEVELOPMENTAL STAGE • SUBCELLULAR LOCATION • TRANSMEMBRANE
Additional information for proteins • • FUNCTION CATALYTIC ACTIVITY COFACTOR INDUCTION ENZYME REGULATION PATHWAY SUBUNIT DOMAIN • • • SPLICE PRODUCTS POLYMORPHISM DISEASE TISSUE SPECIFICITY DEVELOPMENTAL STAGE • SUBCELLULAR LOCATION • TRANSMEMBRANE
 Amino-acid sites are: • • • Post-translational modification of a residue Covalent binding of a lipidic moiety Disulfide bond Thiolester bond Thioether bond Active site Glycosylation site Binding site for a metal ion Binding site for any chemical group (co-enzyme, prosthetic group, etc. )
Amino-acid sites are: • • • Post-translational modification of a residue Covalent binding of a lipidic moiety Disulfide bond Thiolester bond Thioether bond Active site Glycosylation site Binding site for a metal ion Binding site for any chemical group (co-enzyme, prosthetic group, etc. )
 Annotation sources: • Publications that report experimental data • Review articles on specific protein families or groups of proteins • Protein sequence analysis • External experts on the organism • Comparison with other, related sequenced organisms
Annotation sources: • Publications that report experimental data • Review articles on specific protein families or groups of proteins • Protein sequence analysis • External experts on the organism • Comparison with other, related sequenced organisms
 Approaches to functional annotation: · Automatic annotation (sequence homology, rules, transfer info from protein databases) · Automatic classification (pattern databases, sequence clustering, protein structure) · Automatic characterisation (functional databases) · Context information (comparative genome analysis, metabolic pathway databases) · Experimental results (2 D gels, microarrays) · Full manual annotation (SWISS-PROT style)
Approaches to functional annotation: · Automatic annotation (sequence homology, rules, transfer info from protein databases) · Automatic classification (pattern databases, sequence clustering, protein structure) · Automatic characterisation (functional databases) · Context information (comparative genome analysis, metabolic pathway databases) · Experimental results (2 D gels, microarrays) · Full manual annotation (SWISS-PROT style)
 PROTEIN SEQUENCE ANALYSIS FROM HOMOLOGY • Protein sequence can come from gene predictions, literature or peptide sequencing • Simplest case- match for whole sequence in database - determination of structure and function • In between- partial matches across sequence to diverse or hypothetical proteins • Difficult case- no match, have to derive information from amino acid properties, pattern searches etc
PROTEIN SEQUENCE ANALYSIS FROM HOMOLOGY • Protein sequence can come from gene predictions, literature or peptide sequencing • Simplest case- match for whole sequence in database - determination of structure and function • In between- partial matches across sequence to diverse or hypothetical proteins • Difficult case- no match, have to derive information from amino acid properties, pattern searches etc
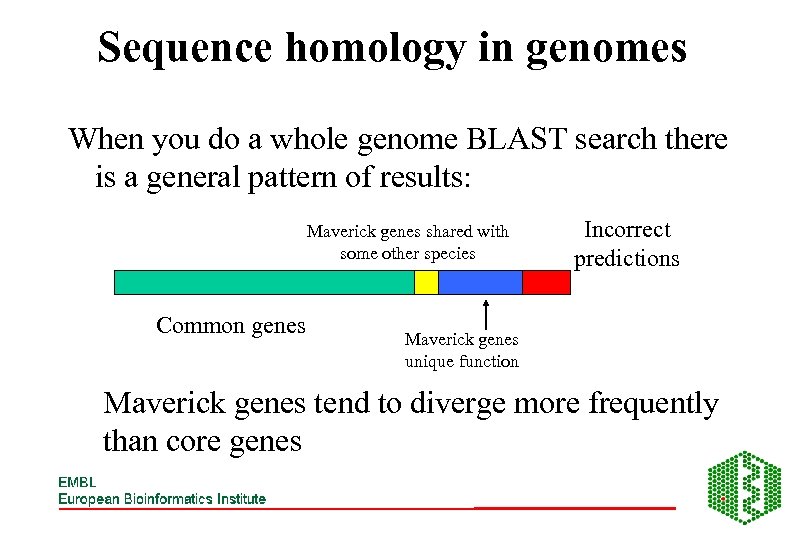 Sequence homology in genomes When you do a whole genome BLAST search there is a general pattern of results: Maverick genes shared with some other species Common genes Incorrect predictions Maverick genes unique function Maverick genes tend to diverge more frequently than core genes
Sequence homology in genomes When you do a whole genome BLAST search there is a general pattern of results: Maverick genes shared with some other species Common genes Incorrect predictions Maverick genes unique function Maverick genes tend to diverge more frequently than core genes
 From sequence to function
From sequence to function
 Predicting function from sequence similarity • Orthologs- arose from speciation, same gene in different organisms -can have <30% homology • Paralogs- from duplication within a genome, second copy may have new or changed function (difficult to distinguish between otho- and paralogues unless whole genome is available) • Equivalog- proteins with equivalent functions • Analog- proteins catalyzing same reaction but not structurally related • Some enzymes may have sequence similarity simply because common catalytic site, substrate, pathway.
Predicting function from sequence similarity • Orthologs- arose from speciation, same gene in different organisms -can have <30% homology • Paralogs- from duplication within a genome, second copy may have new or changed function (difficult to distinguish between otho- and paralogues unless whole genome is available) • Equivalog- proteins with equivalent functions • Analog- proteins catalyzing same reaction but not structurally related • Some enzymes may have sequence similarity simply because common catalytic site, substrate, pathway.
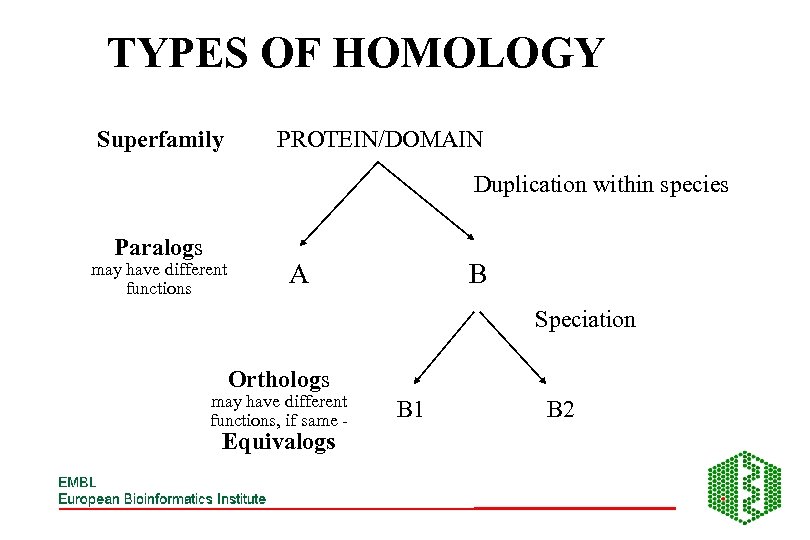 TYPES OF HOMOLOGY Superfamily PROTEIN/DOMAIN Duplication within species Paralogs may have different functions A B Speciation Orthologs may have different functions, if same - Equivalogs B 1 B 2
TYPES OF HOMOLOGY Superfamily PROTEIN/DOMAIN Duplication within species Paralogs may have different functions A B Speciation Orthologs may have different functions, if same - Equivalogs B 1 B 2
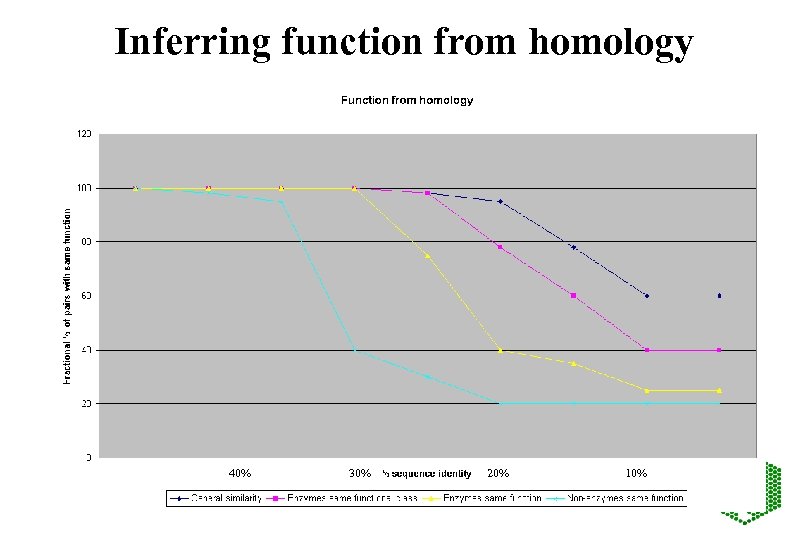 Inferring function from homology 40% 30% 20% 10%
Inferring function from homology 40% 30% 20% 10%
 Using homology information for automatic annotation- automatic annotation of Tr. EMBL as an example
Using homology information for automatic annotation- automatic annotation of Tr. EMBL as an example
 Requirements for automatic annotation • Well-annotated reference database (eg SWISS-PROT or PIR) • Highly reliable diagnostic protein family signature database with the means to assign proteins to groups (eg CDD, Inter. Pro) • A Rule. Base to store and manage the annotation rules, their sources and their usage
Requirements for automatic annotation • Well-annotated reference database (eg SWISS-PROT or PIR) • Highly reliable diagnostic protein family signature database with the means to assign proteins to groups (eg CDD, Inter. Pro) • A Rule. Base to store and manage the annotation rules, their sources and their usage
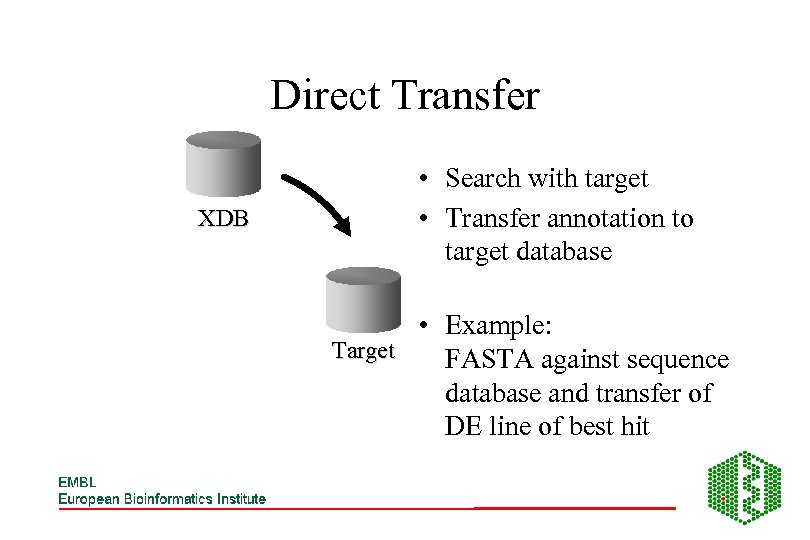 Direct Transfer XDB • Search with target • Transfer annotation to target database • Example: Target FASTA against sequence database and transfer of DE line of best hit
Direct Transfer XDB • Search with target • Transfer annotation to target database • Example: Target FASTA against sequence database and transfer of DE line of best hit
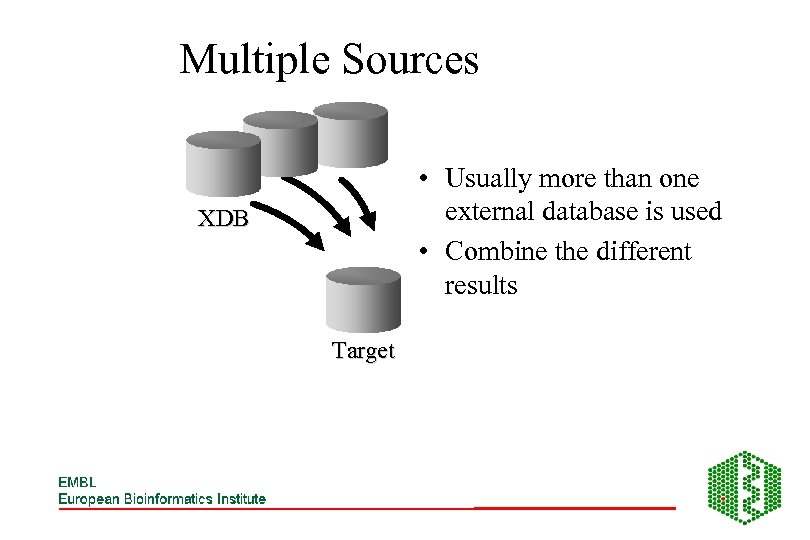 Multiple Sources • Usually more than one external database is used • Combine the different results XDB Target
Multiple Sources • Usually more than one external database is used • Combine the different results XDB Target
 CONFLICTS • • Contradiction Inconsistencies Synonyms Redundancy
CONFLICTS • • Contradiction Inconsistencies Synonyms Redundancy
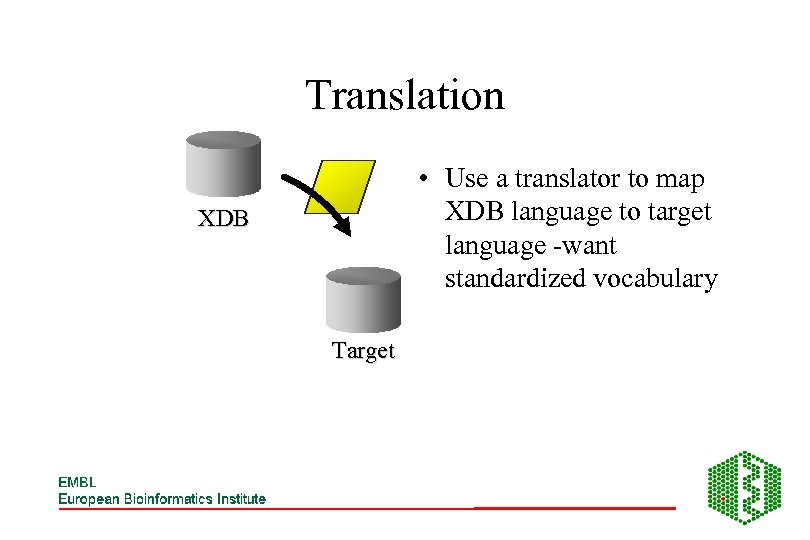 Translation • Use a translator to map XDB language to target language -want standardized vocabulary XDB Target
Translation • Use a translator to map XDB language to target language -want standardized vocabulary XDB Target
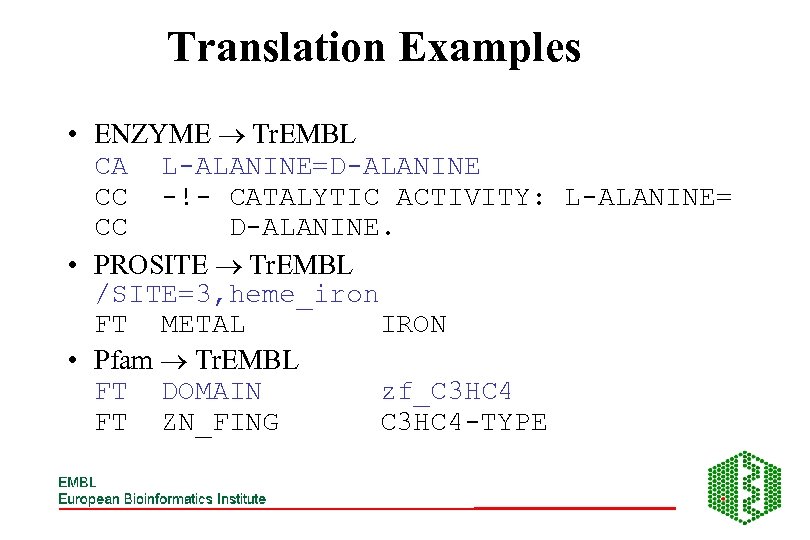 Translation Examples • ENZYME Tr. EMBL CA L-ALANINE=D-ALANINE CC -!- CATALYTIC ACTIVITY: L-ALANINE= CC D-ALANINE. • PROSITE Tr. EMBL /SITE=3, heme_iron FT METAL IRON • Pfam Tr. EMBL FT DOMAIN zf_C 3 HC 4 FT ZN_FING C 3 HC 4 -TYPE
Translation Examples • ENZYME Tr. EMBL CA L-ALANINE=D-ALANINE CC -!- CATALYTIC ACTIVITY: L-ALANINE= CC D-ALANINE. • PROSITE Tr. EMBL /SITE=3, heme_iron FT METAL IRON • Pfam Tr. EMBL FT DOMAIN zf_C 3 HC 4 FT ZN_FING C 3 HC 4 -TYPE
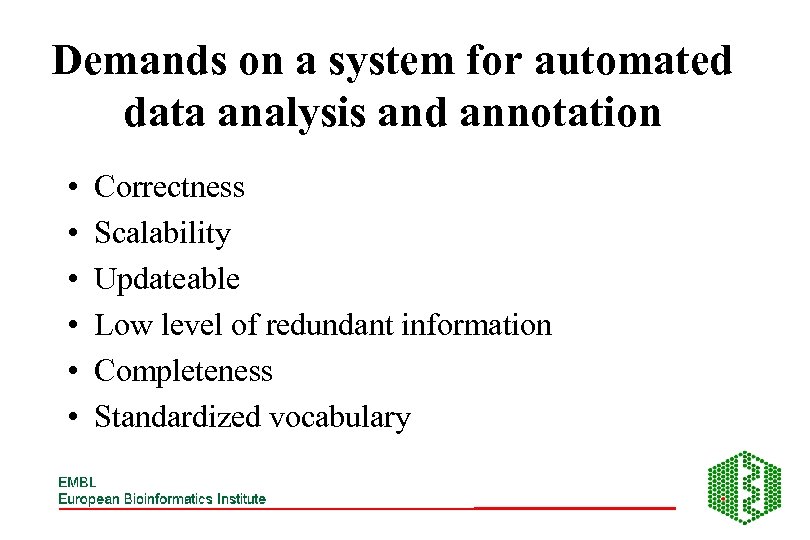 Demands on a system for automated data analysis and annotation • • • Correctness Scalability Updateable Low level of redundant information Completeness Standardized vocabulary
Demands on a system for automated data analysis and annotation • • • Correctness Scalability Updateable Low level of redundant information Completeness Standardized vocabulary
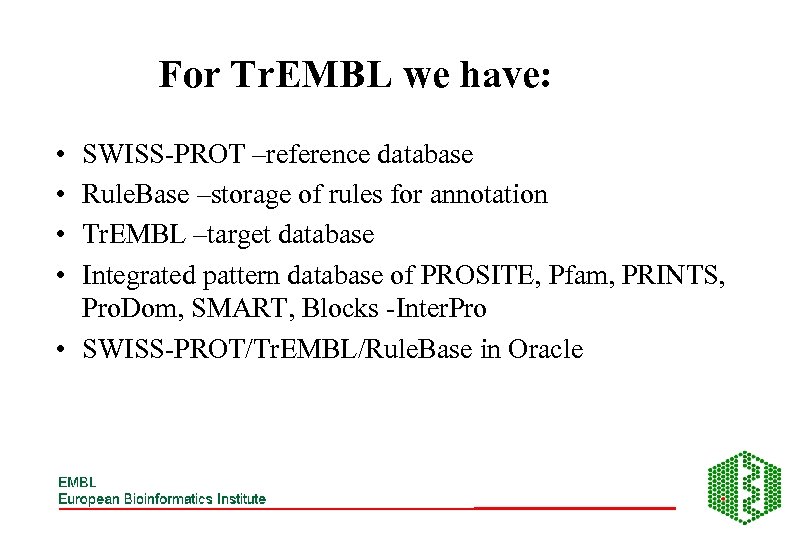 For Tr. EMBL we have: • • SWISS-PROT –reference database Rule. Base –storage of rules for annotation Tr. EMBL –target database Integrated pattern database of PROSITE, Pfam, PRINTS, Pro. Dom, SMART, Blocks -Inter. Pro • SWISS-PROT/Tr. EMBL/Rule. Base in Oracle
For Tr. EMBL we have: • • SWISS-PROT –reference database Rule. Base –storage of rules for annotation Tr. EMBL –target database Integrated pattern database of PROSITE, Pfam, PRINTS, Pro. Dom, SMART, Blocks -Inter. Pro • SWISS-PROT/Tr. EMBL/Rule. Base in Oracle
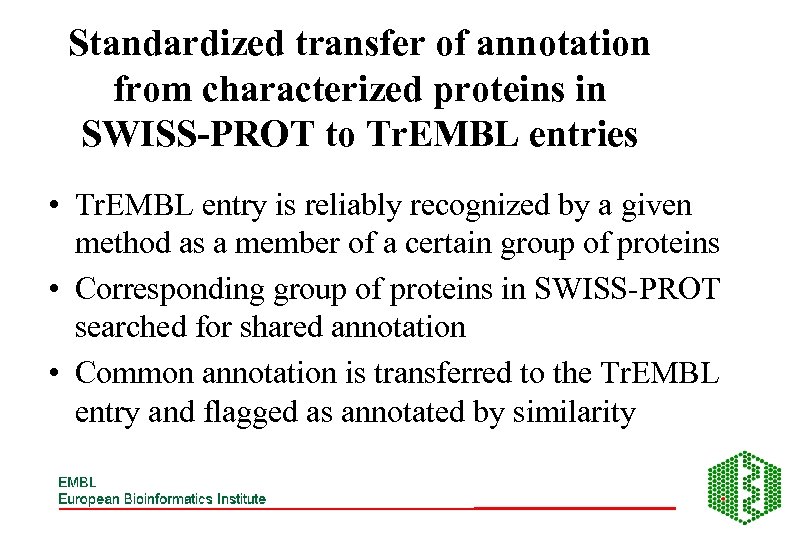 Standardized transfer of annotation from characterized proteins in SWISS-PROT to Tr. EMBL entries • Tr. EMBL entry is reliably recognized by a given method as a member of a certain group of proteins • Corresponding group of proteins in SWISS-PROT searched for shared annotation • Common annotation is transferred to the Tr. EMBL entry and flagged as annotated by similarity
Standardized transfer of annotation from characterized proteins in SWISS-PROT to Tr. EMBL entries • Tr. EMBL entry is reliably recognized by a given method as a member of a certain group of proteins • Corresponding group of proteins in SWISS-PROT searched for shared annotation • Common annotation is transferred to the Tr. EMBL entry and flagged as annotated by similarity
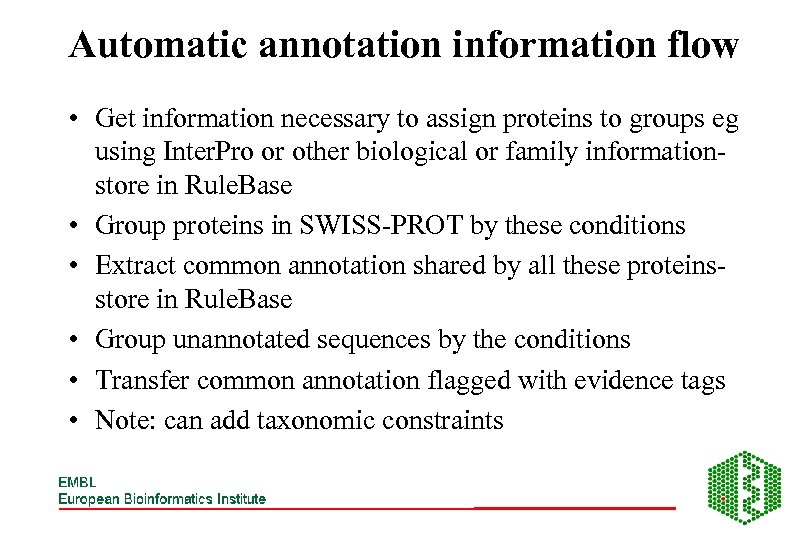 Automatic annotation information flow • Get information necessary to assign proteins to groups eg using Inter. Pro or other biological or family informationstore in Rule. Base • Group proteins in SWISS-PROT by these conditions • Extract common annotation shared by all these proteinsstore in Rule. Base • Group unannotated sequences by the conditions • Transfer common annotation flagged with evidence tags • Note: can add taxonomic constraints
Automatic annotation information flow • Get information necessary to assign proteins to groups eg using Inter. Pro or other biological or family informationstore in Rule. Base • Group proteins in SWISS-PROT by these conditions • Extract common annotation shared by all these proteinsstore in Rule. Base • Group unannotated sequences by the conditions • Transfer common annotation flagged with evidence tags • Note: can add taxonomic constraints
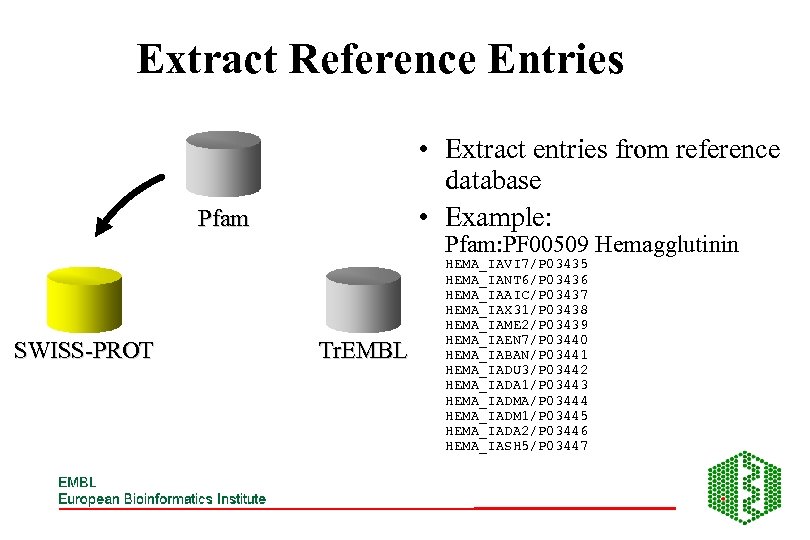 Extract Reference Entries • Extract entries from reference database • Example: Pfam SWISS-PROT Pfam: PF 00509 Hemagglutinin Tr. EMBL HEMA_IAVI 7/P 03435 HEMA_IANT 6/P 03436 HEMA_IAAIC/P 03437 HEMA_IAX 31/P 03438 HEMA_IAME 2/P 03439 HEMA_IAEN 7/P 03440 HEMA_IABAN/P 03441 HEMA_IADU 3/P 03442 HEMA_IADA 1/P 03443 HEMA_IADMA/P 03444 HEMA_IADM 1/P 03445 HEMA_IADA 2/P 03446 HEMA_IASH 5/P 03447
Extract Reference Entries • Extract entries from reference database • Example: Pfam SWISS-PROT Pfam: PF 00509 Hemagglutinin Tr. EMBL HEMA_IAVI 7/P 03435 HEMA_IANT 6/P 03436 HEMA_IAAIC/P 03437 HEMA_IAX 31/P 03438 HEMA_IAME 2/P 03439 HEMA_IAEN 7/P 03440 HEMA_IABAN/P 03441 HEMA_IADU 3/P 03442 HEMA_IADA 1/P 03443 HEMA_IADMA/P 03444 HEMA_IADM 1/P 03445 HEMA_IADA 2/P 03446 HEMA_IASH 5/P 03447
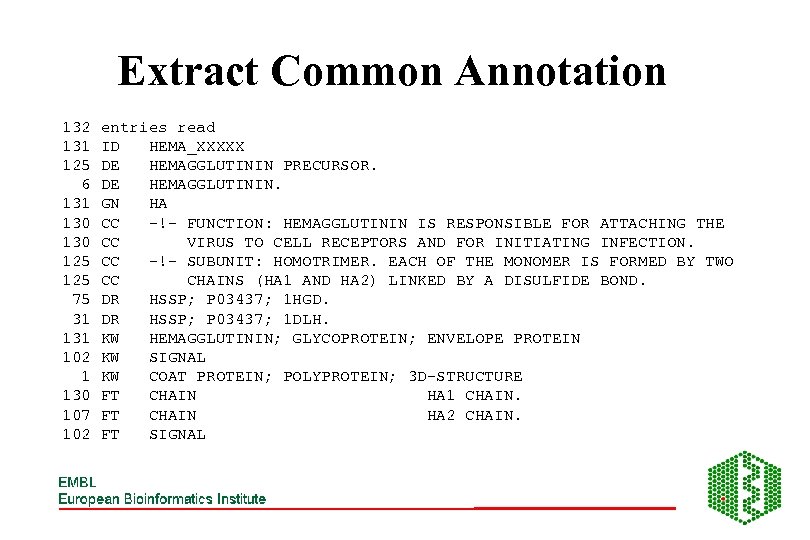 Extract Common Annotation 132 131 125 6 131 130 125 75 31 102 1 130 107 102 entries read ID HEMA_XXXXX DE HEMAGGLUTININ PRECURSOR. DE HEMAGGLUTININ. GN HA CC -!- FUNCTION: HEMAGGLUTININ IS RESPONSIBLE FOR ATTACHING THE CC VIRUS TO CELL RECEPTORS AND FOR INITIATING INFECTION. CC -!- SUBUNIT: HOMOTRIMER. EACH OF THE MONOMER IS FORMED BY TWO CC CHAINS (HA 1 AND HA 2) LINKED BY A DISULFIDE BOND. DR HSSP; P 03437; 1 HGD. DR HSSP; P 03437; 1 DLH. KW HEMAGGLUTININ; GLYCOPROTEIN; ENVELOPE PROTEIN KW SIGNAL KW COAT PROTEIN; POLYPROTEIN; 3 D-STRUCTURE FT CHAIN HA 1 CHAIN. FT CHAIN HA 2 CHAIN. FT SIGNAL
Extract Common Annotation 132 131 125 6 131 130 125 75 31 102 1 130 107 102 entries read ID HEMA_XXXXX DE HEMAGGLUTININ PRECURSOR. DE HEMAGGLUTININ. GN HA CC -!- FUNCTION: HEMAGGLUTININ IS RESPONSIBLE FOR ATTACHING THE CC VIRUS TO CELL RECEPTORS AND FOR INITIATING INFECTION. CC -!- SUBUNIT: HOMOTRIMER. EACH OF THE MONOMER IS FORMED BY TWO CC CHAINS (HA 1 AND HA 2) LINKED BY A DISULFIDE BOND. DR HSSP; P 03437; 1 HGD. DR HSSP; P 03437; 1 DLH. KW HEMAGGLUTININ; GLYCOPROTEIN; ENVELOPE PROTEIN KW SIGNAL KW COAT PROTEIN; POLYPROTEIN; 3 D-STRUCTURE FT CHAIN HA 1 CHAIN. FT CHAIN HA 2 CHAIN. FT SIGNAL
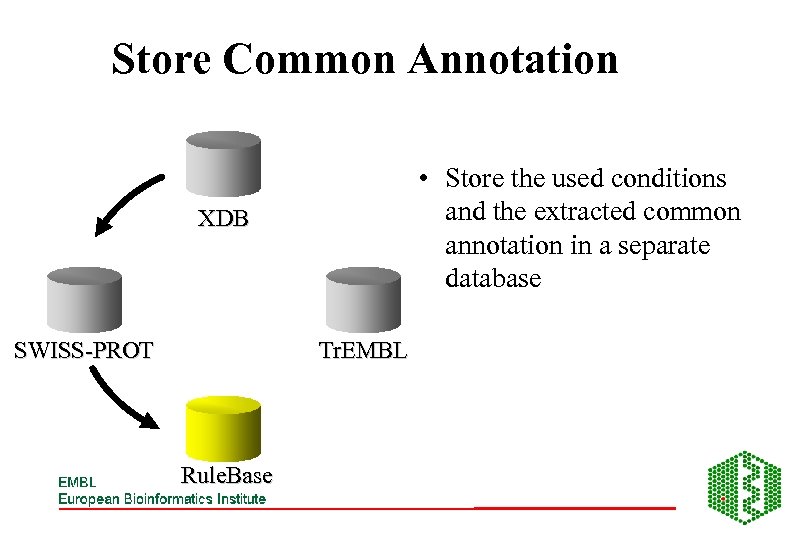 Store Common Annotation • Store the used conditions and the extracted common annotation in a separate database XDB SWISS-PROT Tr. EMBL Rule. Base
Store Common Annotation • Store the used conditions and the extracted common annotation in a separate database XDB SWISS-PROT Tr. EMBL Rule. Base
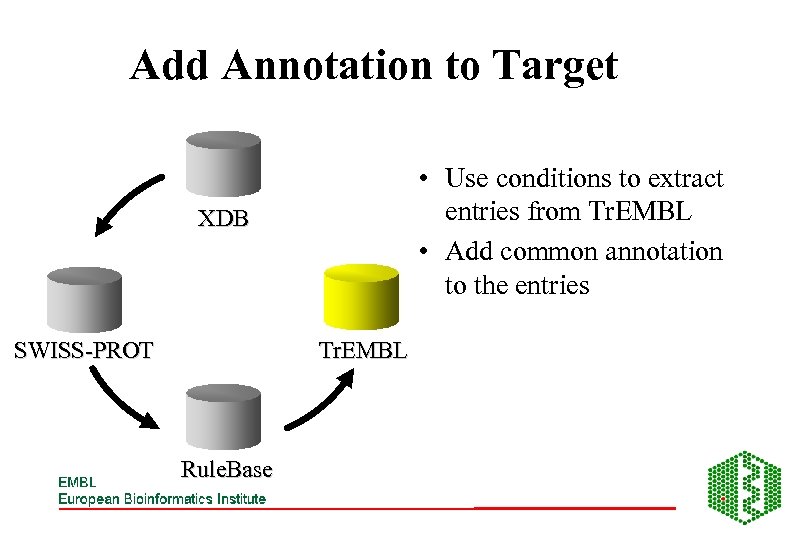 Add Annotation to Target • Use conditions to extract entries from Tr. EMBL • Add common annotation to the entries XDB SWISS-PROT Tr. EMBL Rule. Base
Add Annotation to Target • Use conditions to extract entries from Tr. EMBL • Add common annotation to the entries XDB SWISS-PROT Tr. EMBL Rule. Base
 RULES • Rules describe: – the content of the annotation to be transferred (ACTIONS), – the CONDITIONS which the target Tr. EMBL entry must fulfill in order to allow transfer of the annotation. • Rules uniquely describe or delineate a set of SWISSPROT entries. – The common annotation in these entries is transferred to Tr. EMBL.
RULES • Rules describe: – the content of the annotation to be transferred (ACTIONS), – the CONDITIONS which the target Tr. EMBL entry must fulfill in order to allow transfer of the annotation. • Rules uniquely describe or delineate a set of SWISSPROT entries. – The common annotation in these entries is transferred to Tr. EMBL.
 // #RULE RU 000482 #DATE 2001 -01 -11 #USER OPS$WFL #PACK PROSITE ? PSAC PS 00449 ACTIONS ? EMOT PS 00449 }CONDITIONS !ECNO 3. 6. 1. 34 !SPDE ATP synthase A chain !CCFU KEY COMPONENT OF THE PROTON CHANNEL; IT MAY PLAY A DIRECT ROLE IN THE TRANSLOCATION OF PROTONS ACROSS THE MEMBRANE (BY SIMILARITY) !CCSU F-TYPE ATPASES HAVE 2 COMPONENTS, CF(1) - THE CATALYTIC CORE - AND CF(0) - THE MEMBRANE PROTON CHANNEL. CF(1) HAS FIVE SUBUNITS: ALPHA(3), BETA(3), GAMM A(1), DELTA(1), EPSILON(1). CF(0) HAS THREE MAIN SUBUNITS: A, B AND C (BY SIMILARITY) !CCLO INTEGRAL MEMBRANE PROTEIN (By Similarity) !CCSI TO THE ATPASE A CHAIN FAMILY !SPKW CF(0) !SPKW Hydrogen ion transport !SPKW Transmembrane //
// #RULE RU 000482 #DATE 2001 -01 -11 #USER OPS$WFL #PACK PROSITE ? PSAC PS 00449 ACTIONS ? EMOT PS 00449 }CONDITIONS !ECNO 3. 6. 1. 34 !SPDE ATP synthase A chain !CCFU KEY COMPONENT OF THE PROTON CHANNEL; IT MAY PLAY A DIRECT ROLE IN THE TRANSLOCATION OF PROTONS ACROSS THE MEMBRANE (BY SIMILARITY) !CCSU F-TYPE ATPASES HAVE 2 COMPONENTS, CF(1) - THE CATALYTIC CORE - AND CF(0) - THE MEMBRANE PROTON CHANNEL. CF(1) HAS FIVE SUBUNITS: ALPHA(3), BETA(3), GAMM A(1), DELTA(1), EPSILON(1). CF(0) HAS THREE MAIN SUBUNITS: A, B AND C (BY SIMILARITY) !CCLO INTEGRAL MEMBRANE PROTEIN (By Similarity) !CCSI TO THE ATPASE A CHAIN FAMILY !SPKW CF(0) !SPKW Hydrogen ion transport !SPKW Transmembrane //
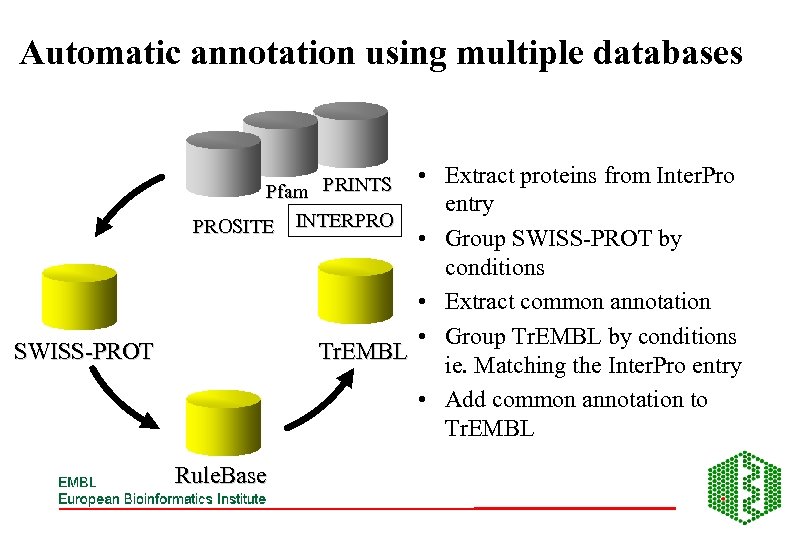 Automatic annotation using multiple databases • Extract proteins from Inter. Pro entry • Group SWISS-PROT by conditions • Extract common annotation • Group Tr. EMBL by conditions Tr. EMBL ie. Matching the Inter. Pro entry • Add common annotation to Tr. EMBL Pfam PRINTS PROSITE INTERPRO SWISS-PROT Rule. Base
Automatic annotation using multiple databases • Extract proteins from Inter. Pro entry • Group SWISS-PROT by conditions • Extract common annotation • Group Tr. EMBL by conditions Tr. EMBL ie. Matching the Inter. Pro entry • Add common annotation to Tr. EMBL Pfam PRINTS PROSITE INTERPRO SWISS-PROT Rule. Base
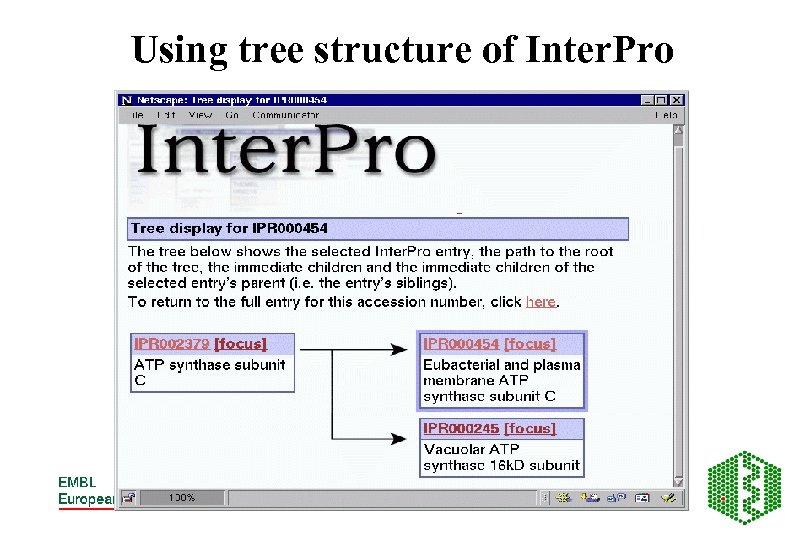 Using tree structure of Inter. Pro
Using tree structure of Inter. Pro
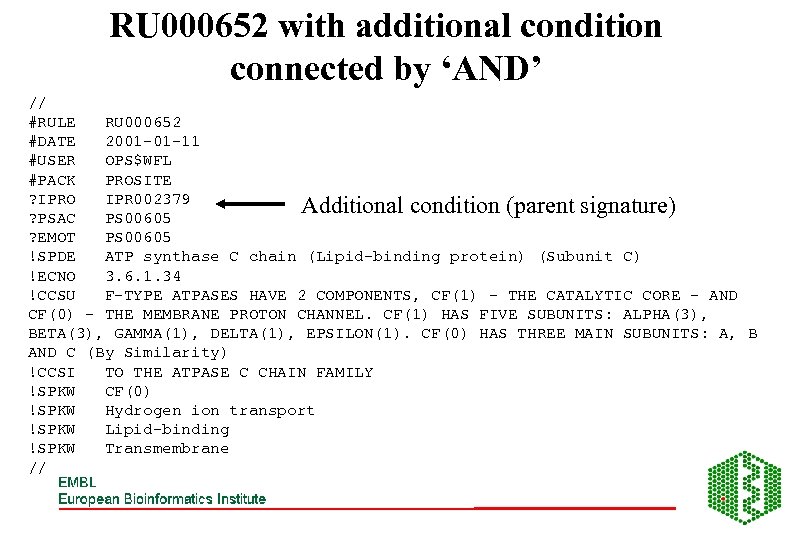 RU 000652 with additional condition connected by ‘AND’ // #RULE RU 000652 #DATE 2001 -01 -11 #USER OPS$WFL #PACK PROSITE ? IPRO IPR 002379 Additional condition (parent signature) ? PSAC PS 00605 ? EMOT PS 00605 !SPDE ATP synthase C chain (Lipid-binding protein) (Subunit C) !ECNO 3. 6. 1. 34 !CCSU F-TYPE ATPASES HAVE 2 COMPONENTS, CF(1) - THE CATALYTIC CORE - AND CF(0) - THE MEMBRANE PROTON CHANNEL. CF(1) HAS FIVE SUBUNITS: ALPHA(3), BETA(3), GAMMA(1), DELTA(1), EPSILON(1). CF(0) HAS THREE MAIN SUBUNITS: A, B AND C (By Similarity) !CCSI TO THE ATPASE C CHAIN FAMILY !SPKW CF(0) !SPKW Hydrogen ion transport !SPKW Lipid-binding !SPKW Transmembrane //
RU 000652 with additional condition connected by ‘AND’ // #RULE RU 000652 #DATE 2001 -01 -11 #USER OPS$WFL #PACK PROSITE ? IPRO IPR 002379 Additional condition (parent signature) ? PSAC PS 00605 ? EMOT PS 00605 !SPDE ATP synthase C chain (Lipid-binding protein) (Subunit C) !ECNO 3. 6. 1. 34 !CCSU F-TYPE ATPASES HAVE 2 COMPONENTS, CF(1) - THE CATALYTIC CORE - AND CF(0) - THE MEMBRANE PROTON CHANNEL. CF(1) HAS FIVE SUBUNITS: ALPHA(3), BETA(3), GAMMA(1), DELTA(1), EPSILON(1). CF(0) HAS THREE MAIN SUBUNITS: A, B AND C (By Similarity) !CCSI TO THE ATPASE C CHAIN FAMILY !SPKW CF(0) !SPKW Hydrogen ion transport !SPKW Lipid-binding !SPKW Transmembrane //
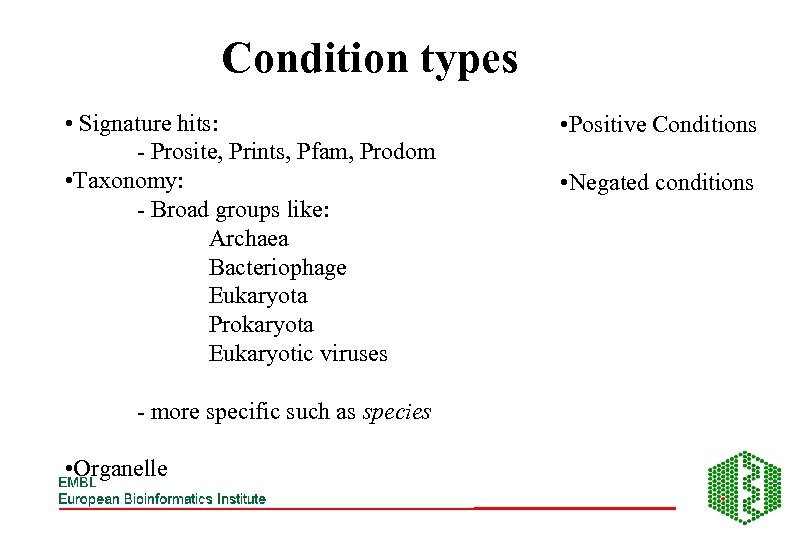 Condition types • Signature hits: - Prosite, Prints, Pfam, Prodom • Taxonomy: - Broad groups like: Archaea Bacteriophage Eukaryota Prokaryota Eukaryotic viruses - more specific such as species • Organelle • Positive Conditions • Negated conditions
Condition types • Signature hits: - Prosite, Prints, Pfam, Prodom • Taxonomy: - Broad groups like: Archaea Bacteriophage Eukaryota Prokaryota Eukaryotic viruses - more specific such as species • Organelle • Positive Conditions • Negated conditions
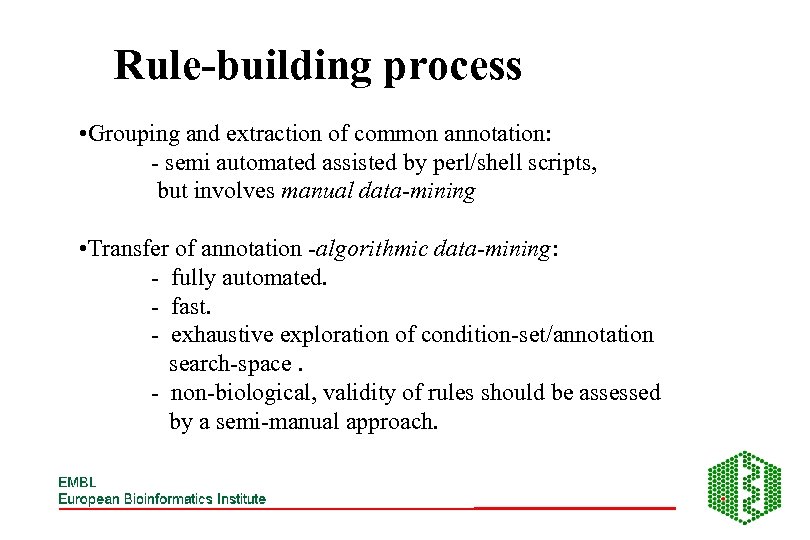 Rule-building process • Grouping and extraction of common annotation: - semi automated assisted by perl/shell scripts, but involves manual data-mining • Transfer of annotation -algorithmic data-mining: - fully automated. - fast. - exhaustive exploration of condition-set/annotation search-space. - non-biological, validity of rules should be assessed by a semi-manual approach.
Rule-building process • Grouping and extraction of common annotation: - semi automated assisted by perl/shell scripts, but involves manual data-mining • Transfer of annotation -algorithmic data-mining: - fully automated. - fast. - exhaustive exploration of condition-set/annotation search-space. - non-biological, validity of rules should be assessed by a semi-manual approach.
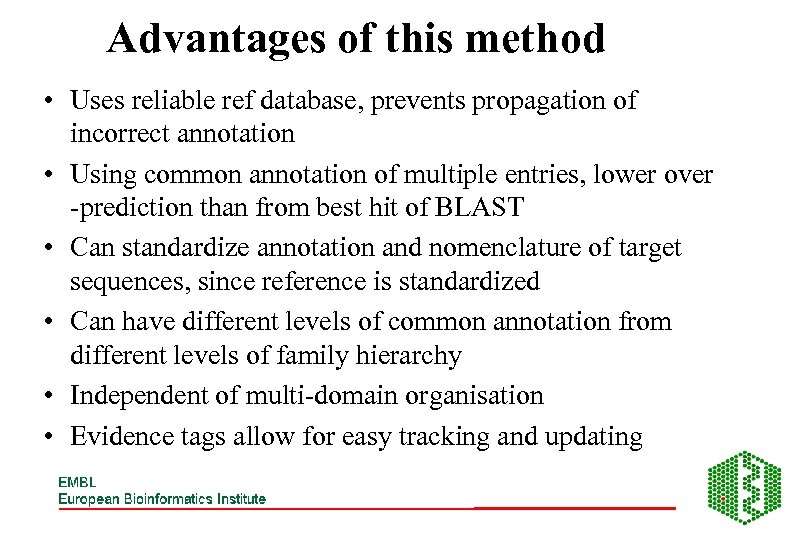 Advantages of this method • Uses reliable ref database, prevents propagation of incorrect annotation • Using common annotation of multiple entries, lower over -prediction than from best hit of BLAST • Can standardize annotation and nomenclature of target sequences, since reference is standardized • Can have different levels of common annotation from different levels of family hierarchy • Independent of multi-domain organisation • Evidence tags allow for easy tracking and updating
Advantages of this method • Uses reliable ref database, prevents propagation of incorrect annotation • Using common annotation of multiple entries, lower over -prediction than from best hit of BLAST • Can standardize annotation and nomenclature of target sequences, since reference is standardized • Can have different levels of common annotation from different levels of family hierarchy • Independent of multi-domain organisation • Evidence tags allow for easy tracking and updating
 Pitfalls of automatic functional analysis • Multifunctional proteins- genome projects often assign single function, info is lost in homology search • No coverage of position-specific annotation eg active sites • Relies on coverage by reference databases including pattern daabases (60 -65%) • Hypothetical proteins (40% ORFs unknown), and poorly or even wrongly annotated proteins It is important to have evidence for all annotation added
Pitfalls of automatic functional analysis • Multifunctional proteins- genome projects often assign single function, info is lost in homology search • No coverage of position-specific annotation eg active sites • Relies on coverage by reference databases including pattern daabases (60 -65%) • Hypothetical proteins (40% ORFs unknown), and poorly or even wrongly annotated proteins It is important to have evidence for all annotation added
 Evidence tags • All annotation of proteins should have evidence or status • Necessary to trace level of confidence for information so that second user can see what is automatic and what is manual • Example –evidence tags to be introduced for SPTR
Evidence tags • All annotation of proteins should have evidence or status • Necessary to trace level of confidence for information so that second user can see what is automatic and what is manual • Example –evidence tags to be introduced for SPTR
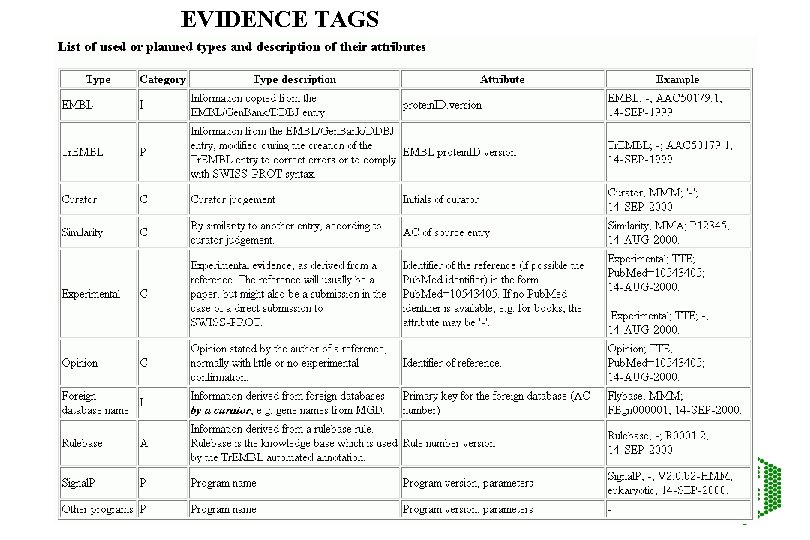 EVIDENCE TAGS
EVIDENCE TAGS

 Predicting function from non-homology • Look at position of genes relative to others, compare with other organisms- use reverse approach, finding proteins for functions • Can still build up rules from annotated sequences using information you have on other features like fold, physical properties etc. • Use physical properties and known attributes
Predicting function from non-homology • Look at position of genes relative to others, compare with other organisms- use reverse approach, finding proteins for functions • Can still build up rules from annotated sequences using information you have on other features like fold, physical properties etc. • Use physical properties and known attributes
 Protein functions from regions • Active sites- short, highly conserved regions • Loops- charged residues and variable sequence • Interior of protein- conservation of charged amino acids
Protein functions from regions • Active sites- short, highly conserved regions • Loops- charged residues and variable sequence • Interior of protein- conservation of charged amino acids
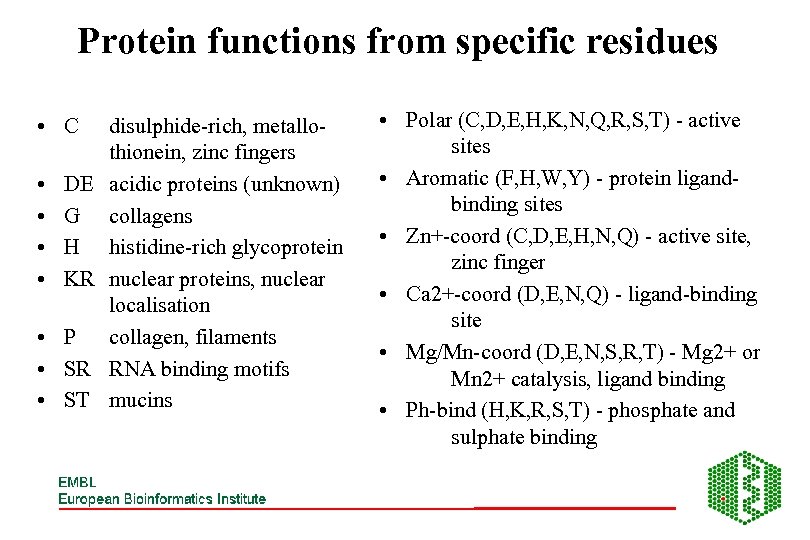 Protein functions from specific residues • C • • DE G H KR • P • SR • ST disulphide-rich, metallothionein, zinc fingers acidic proteins (unknown) collagens histidine-rich glycoprotein nuclear proteins, nuclear localisation collagen, filaments RNA binding motifs mucins • Polar (C, D, E, H, K, N, Q, R, S, T) - active sites • Aromatic (F, H, W, Y) - protein ligandbinding sites • Zn+-coord (C, D, E, H, N, Q) - active site, zinc finger • Ca 2+-coord (D, E, N, Q) - ligand-binding site • Mg/Mn-coord (D, E, N, S, R, T) - Mg 2+ or Mn 2+ catalysis, ligand binding • Ph-bind (H, K, R, S, T) - phosphate and sulphate binding
Protein functions from specific residues • C • • DE G H KR • P • SR • ST disulphide-rich, metallothionein, zinc fingers acidic proteins (unknown) collagens histidine-rich glycoprotein nuclear proteins, nuclear localisation collagen, filaments RNA binding motifs mucins • Polar (C, D, E, H, K, N, Q, R, S, T) - active sites • Aromatic (F, H, W, Y) - protein ligandbinding sites • Zn+-coord (C, D, E, H, N, Q) - active site, zinc finger • Ca 2+-coord (D, E, N, Q) - ligand-binding site • Mg/Mn-coord (D, E, N, S, R, T) - Mg 2+ or Mn 2+ catalysis, ligand binding • Ph-bind (H, K, R, S, T) - phosphate and sulphate binding
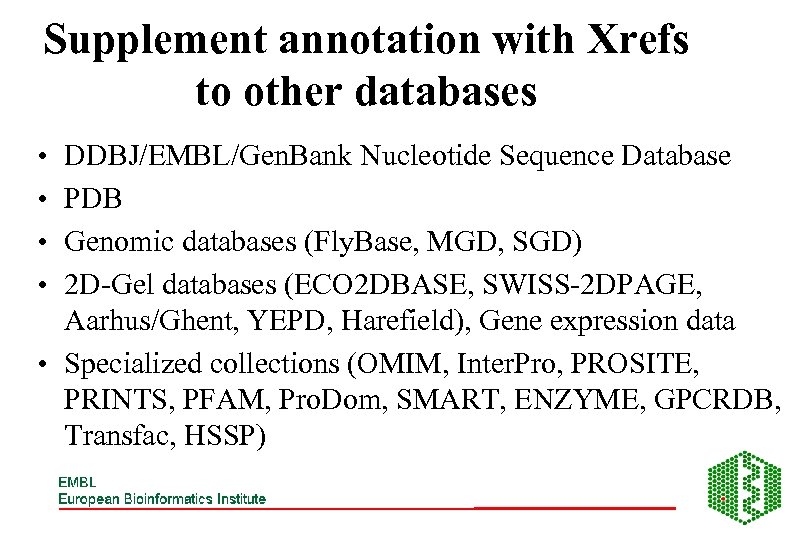 Supplement annotation with Xrefs to other databases • • DDBJ/EMBL/Gen. Bank Nucleotide Sequence Database PDB Genomic databases (Fly. Base, MGD, SGD) 2 D-Gel databases (ECO 2 DBASE, SWISS-2 DPAGE, Aarhus/Ghent, YEPD, Harefield), Gene expression data • Specialized collections (OMIM, Inter. Pro, PROSITE, PRINTS, PFAM, Pro. Dom, SMART, ENZYME, GPCRDB, Transfac, HSSP)
Supplement annotation with Xrefs to other databases • • DDBJ/EMBL/Gen. Bank Nucleotide Sequence Database PDB Genomic databases (Fly. Base, MGD, SGD) 2 D-Gel databases (ECO 2 DBASE, SWISS-2 DPAGE, Aarhus/Ghent, YEPD, Harefield), Gene expression data • Specialized collections (OMIM, Inter. Pro, PROSITE, PRINTS, PFAM, Pro. Dom, SMART, ENZYME, GPCRDB, Transfac, HSSP)
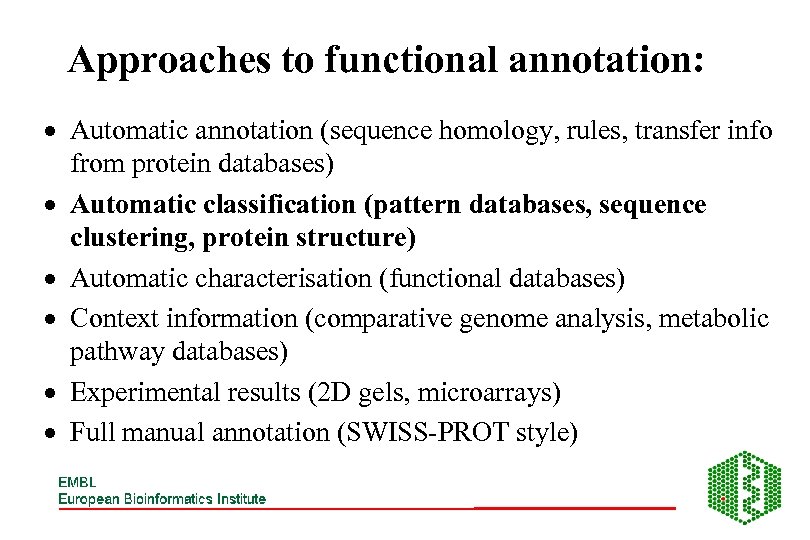 Approaches to functional annotation: · Automatic annotation (sequence homology, rules, transfer info from protein databases) · Automatic classification (pattern databases, sequence clustering, protein structure) · Automatic characterisation (functional databases) · Context information (comparative genome analysis, metabolic pathway databases) · Experimental results (2 D gels, microarrays) · Full manual annotation (SWISS-PROT style)
Approaches to functional annotation: · Automatic annotation (sequence homology, rules, transfer info from protein databases) · Automatic classification (pattern databases, sequence clustering, protein structure) · Automatic characterisation (functional databases) · Context information (comparative genome analysis, metabolic pathway databases) · Experimental results (2 D gels, microarrays) · Full manual annotation (SWISS-PROT style)
 AUTOMATIC CLASSIFICATION Annotation using Clustering methods eg Clu. STR (EBI), and pattern searches (Inter. Pro etc)classification of proteins into different families Clusters of human sequences:
AUTOMATIC CLASSIFICATION Annotation using Clustering methods eg Clu. STR (EBI), and pattern searches (Inter. Pro etc)classification of proteins into different families Clusters of human sequences:
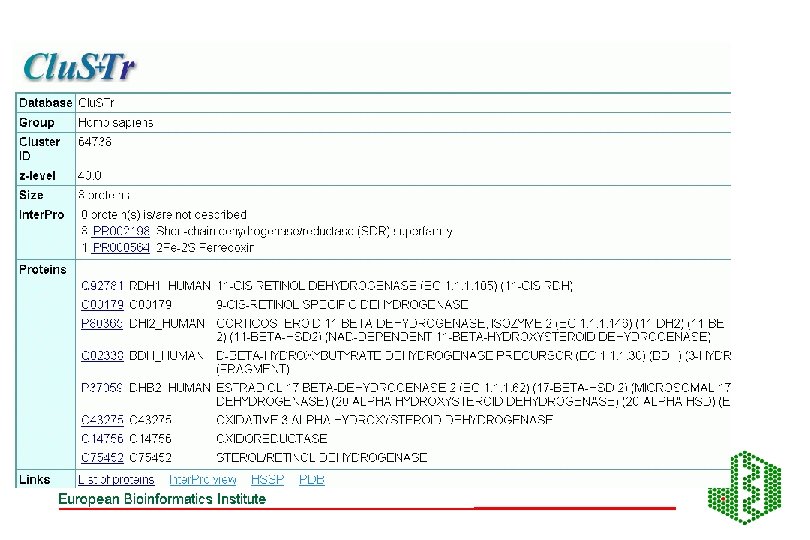
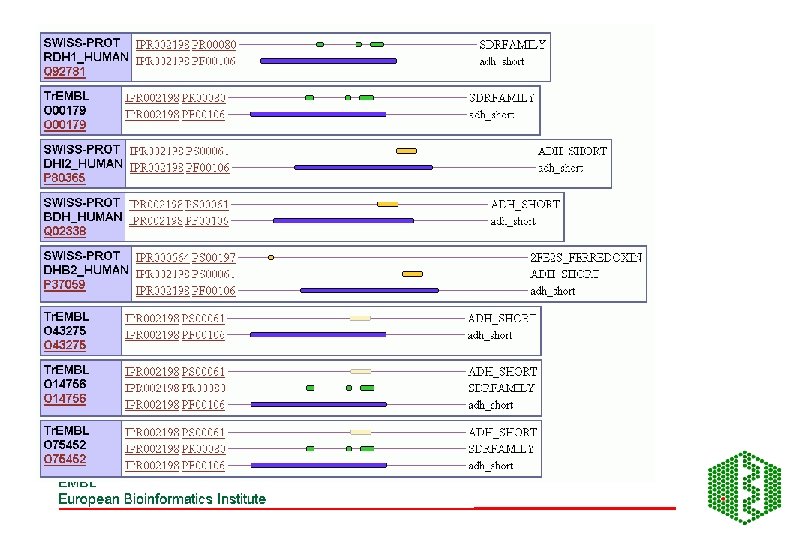
 Using Clustering for annotation • Find a good clustering database • Link clusters to functional information eg Inter. Pro, PDB etc • For unknown sequences see where they cluster, may be able to infer function
Using Clustering for annotation • Find a good clustering database • Link clusters to functional information eg Inter. Pro, PDB etc • For unknown sequences see where they cluster, may be able to infer function
 Approaches to functional annotation: · Automatic annotation (sequence homology, rules, transfer info from protein databases) · Automatic classification (pattern databases, clustering, structure) · Automatic characterisation (functional databases) · Context information (comparative genome analysis, metabolic pathway databases) · Experimental results (2 D gels, microarrays) · Full manual annotation (SWISS-PROT style)
Approaches to functional annotation: · Automatic annotation (sequence homology, rules, transfer info from protein databases) · Automatic classification (pattern databases, clustering, structure) · Automatic characterisation (functional databases) · Context information (comparative genome analysis, metabolic pathway databases) · Experimental results (2 D gels, microarrays) · Full manual annotation (SWISS-PROT style)
 Automatic characterization- Functional annotation schemes • • First attempt –Riley classification of E. coli Genome sequencing projects driving force Need standardised system and vocabulary Functional schemes normally hierarchies of different levels of generalisation
Automatic characterization- Functional annotation schemes • • First attempt –Riley classification of E. coli Genome sequencing projects driving force Need standardised system and vocabulary Functional schemes normally hierarchies of different levels of generalisation
 Databases for Functional Information • KEGG -Kyoto encyclopedia of genes and genomes – (http: //www. genome. ad. jp/kegg/) – Links genome information (GENES database) to high order functional information stored in PATHWAY database. – Also has LIGAND database for chemical compounds, molecules and reactions. • PEDANT -Protein Extraction, Description and Analysis Tool – (http: //pedant. gsf. de/) – Annotation for complete and incomplete genomes eg. List of ORFs, EC numbers, functional categories, list seqs with homologs, gene clusters, domain hits, TM, structure links, search facility for sequences etc • WIT –What is there – ( http: //www. cme. msu. edu/WIT) – Database of metabolic pathways, can text search for ORFs, pathways, enzymes
Databases for Functional Information • KEGG -Kyoto encyclopedia of genes and genomes – (http: //www. genome. ad. jp/kegg/) – Links genome information (GENES database) to high order functional information stored in PATHWAY database. – Also has LIGAND database for chemical compounds, molecules and reactions. • PEDANT -Protein Extraction, Description and Analysis Tool – (http: //pedant. gsf. de/) – Annotation for complete and incomplete genomes eg. List of ORFs, EC numbers, functional categories, list seqs with homologs, gene clusters, domain hits, TM, structure links, search facility for sequences etc • WIT –What is there – ( http: //www. cme. msu. edu/WIT) – Database of metabolic pathways, can text search for ORFs, pathways, enzymes
 Databases for Functional Information (2) • COG -Clusters of Orthologous Groups – – (http: //www. ncbi. nlm. nih. gov/COG) Phylogenetic classification of proteins encoded in complete genomes. Contains 2791 COGs including 30 genomes. COGs thought to contain orthologous proteins, classified into broad functional categories (transciption, replication, cell division). – COGNITOR assigns proteins to COGs based on best-hit, divides multi-domain proteins – Can compare results with complete genomes, look for missing functions • GO –Gene Ontology – (http: //www. geneontology. org) – Standard vocabulary first used for mouse, fly and yeast – Three ontologies: molecular function, biological process and cellular component
Databases for Functional Information (2) • COG -Clusters of Orthologous Groups – – (http: //www. ncbi. nlm. nih. gov/COG) Phylogenetic classification of proteins encoded in complete genomes. Contains 2791 COGs including 30 genomes. COGs thought to contain orthologous proteins, classified into broad functional categories (transciption, replication, cell division). – COGNITOR assigns proteins to COGs based on best-hit, divides multi-domain proteins – Can compare results with complete genomes, look for missing functions • GO –Gene Ontology – (http: //www. geneontology. org) – Standard vocabulary first used for mouse, fly and yeast – Three ontologies: molecular function, biological process and cellular component
 Databases for Functional Information (3) • MIPS: MYGD Fun. Cat –Functional catalogue (yeast) http: //www. mips. biochem. mpg. de/proj/yeast • Eco. Cyc -Encyclopedia of E. coli Genes and Metabolism http: //ecocyc. doubletwist. com/ecocyc. html • Enzyme database http: //wwwexpasy. ch/sprot/enzyme. html • TIGR –Gene identification list http: //www. tigr. org/tdb/mdb. html - All schemes have different depths, breadths and resolutions - Schemes need to be applicable to all organisms, standardized for comparisons and permit multiple assignments
Databases for Functional Information (3) • MIPS: MYGD Fun. Cat –Functional catalogue (yeast) http: //www. mips. biochem. mpg. de/proj/yeast • Eco. Cyc -Encyclopedia of E. coli Genes and Metabolism http: //ecocyc. doubletwist. com/ecocyc. html • Enzyme database http: //wwwexpasy. ch/sprot/enzyme. html • TIGR –Gene identification list http: //www. tigr. org/tdb/mdb. html - All schemes have different depths, breadths and resolutions - Schemes need to be applicable to all organisms, standardized for comparisons and permit multiple assignments
 Assignment of function • Use a combination of databases, especially those with standardised functional information • Search function databases with sequences to find matches -assign function eg PENDANT, PIR superfamilies, COGs, GO (via Inter. Pro or other mappings)
Assignment of function • Use a combination of databases, especially those with standardised functional information • Search function databases with sequences to find matches -assign function eg PENDANT, PIR superfamilies, COGs, GO (via Inter. Pro or other mappings)
 FUNCTIONAL CLASSIFICATION USING INTERPRO • Inter. Pro classification with 3 -4 letter codes • Mapping of Inter. Pro entries to GO • For a whole genome, can count number of proteins hitting Inter. Pro (50 -70%) with particular functions • Can represent this in charts and use the data in genome comparisons
FUNCTIONAL CLASSIFICATION USING INTERPRO • Inter. Pro classification with 3 -4 letter codes • Mapping of Inter. Pro entries to GO • For a whole genome, can count number of proteins hitting Inter. Pro (50 -70%) with particular functions • Can represent this in charts and use the data in genome comparisons
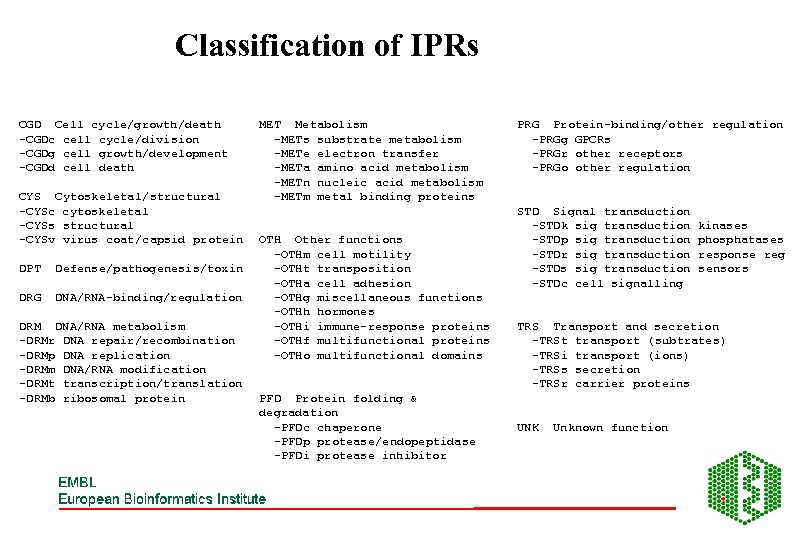 Classification of IPRs CGD Cell cycle/growth/death -CGDc cell cycle/division -CGDg cell growth/development -CGDd cell death CYS Cytoskeletal/structural -CYSc cytoskeletal -CYSs structural -CYSv virus coat/capsid protein DPT Defense/pathogenesis/toxin DRG DNA/RNA-binding/regulation DRM DNA/RNA metabolism -DRMr DNA repair/recombination -DRMp DNA replication -DRMm DNA/RNA modification -DRMt transcription/translation -DRMb ribosomal protein MET Metabolism -METs substrate metabolism -METe electron transfer -METa amino acid metabolism -METn nucleic acid metabolism -METm metal binding proteins OTH Other functions -OTHm cell motility -OTHt transposition -OTHa cell adhesion -OTHg miscellaneous functions -OTHh hormones -OTHi immune-response proteins -OTHf multifunctional proteins -OTHo multifunctional domains PFD Protein folding & degradation -PFDc chaperone -PFDp protease/endopeptidase -PFDi protease inhibitor PRG Protein-binding/other regulation -PRGg GPCRs -PRGr other receptors -PRGo other regulation STD Signal transduction -STDk sig transduction -STDp sig transduction -STDr sig transduction -STDs sig transduction -STDc cell signalling kinases phosphatases response reg sensors TRS Transport and secretion -TRSt transport (subtrates) -TRSi transport (ions) -TRSs secretion -TRSr carrier proteins UNK Unknown function
Classification of IPRs CGD Cell cycle/growth/death -CGDc cell cycle/division -CGDg cell growth/development -CGDd cell death CYS Cytoskeletal/structural -CYSc cytoskeletal -CYSs structural -CYSv virus coat/capsid protein DPT Defense/pathogenesis/toxin DRG DNA/RNA-binding/regulation DRM DNA/RNA metabolism -DRMr DNA repair/recombination -DRMp DNA replication -DRMm DNA/RNA modification -DRMt transcription/translation -DRMb ribosomal protein MET Metabolism -METs substrate metabolism -METe electron transfer -METa amino acid metabolism -METn nucleic acid metabolism -METm metal binding proteins OTH Other functions -OTHm cell motility -OTHt transposition -OTHa cell adhesion -OTHg miscellaneous functions -OTHh hormones -OTHi immune-response proteins -OTHf multifunctional proteins -OTHo multifunctional domains PFD Protein folding & degradation -PFDc chaperone -PFDp protease/endopeptidase -PFDi protease inhibitor PRG Protein-binding/other regulation -PRGg GPCRs -PRGr other receptors -PRGo other regulation STD Signal transduction -STDk sig transduction -STDp sig transduction -STDr sig transduction -STDs sig transduction -STDc cell signalling kinases phosphatases response reg sensors TRS Transport and secretion -TRSt transport (subtrates) -TRSi transport (ions) -TRSs secretion -TRSr carrier proteins UNK Unknown function
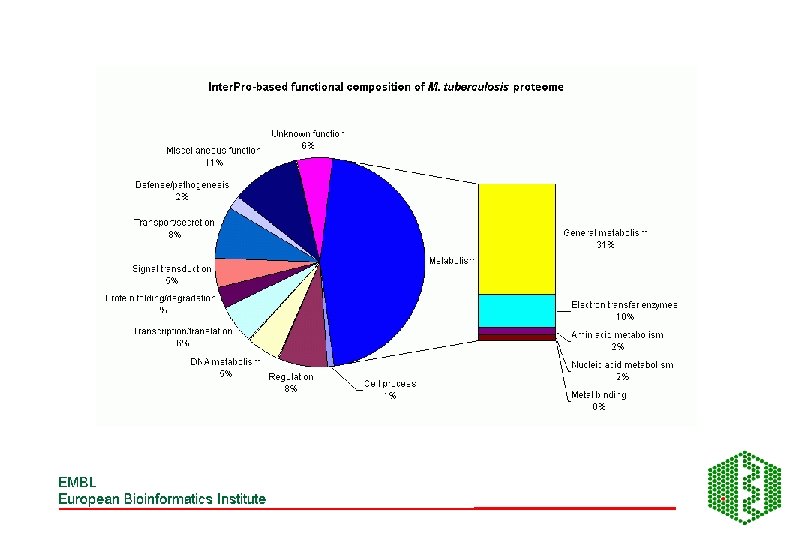
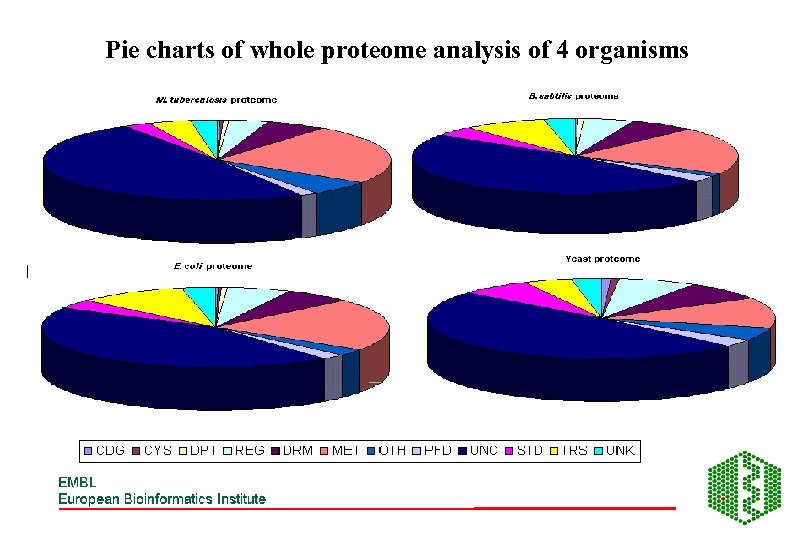 Pie charts of whole proteome analysis of 4 organisms
Pie charts of whole proteome analysis of 4 organisms
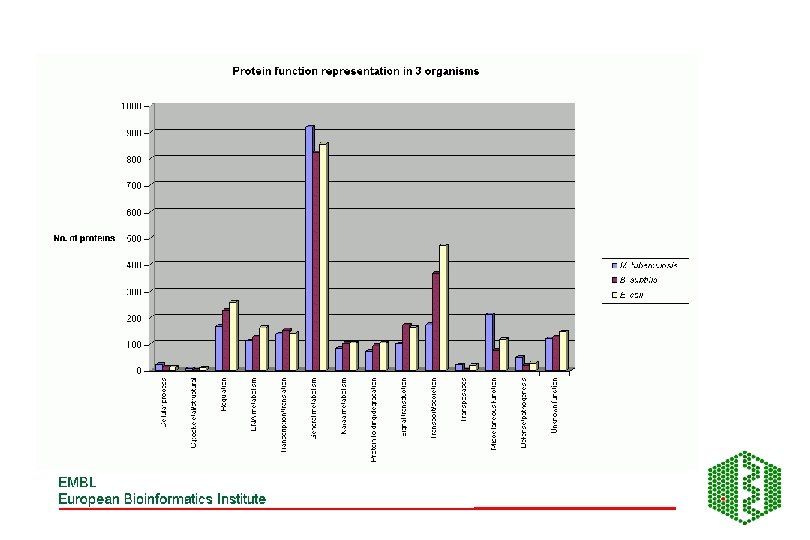
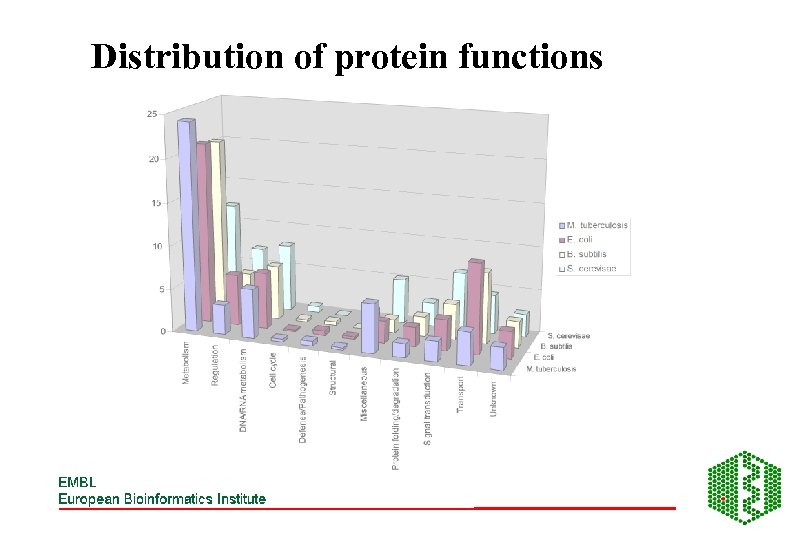 Distribution of protein functions
Distribution of protein functions
 Approaches to functional annotation: · Automatic annotation (sequence homology, rules, transfer info from protein databases) · Automatic classification (pattern databases, sequence clustering, protein structure) · Automatic characterisation (functional databases) · Context information (comparative genome analysis, metabolic pathway databases) · Experimental results (2 D gels, microarrays) · Full manual annotation (SWISS-PROT style)
Approaches to functional annotation: · Automatic annotation (sequence homology, rules, transfer info from protein databases) · Automatic classification (pattern databases, sequence clustering, protein structure) · Automatic characterisation (functional databases) · Context information (comparative genome analysis, metabolic pathway databases) · Experimental results (2 D gels, microarrays) · Full manual annotation (SWISS-PROT style)
 GENOME ANNOTATION TOOLS • Oakridge Genome Annotation Channel (http: //compbio. ornl. gov/channel/) • ENSEMBL (http: //ensembl. ebi. ac. uk) • Artemis (http: //www. sanger. ac. uk/Software/Artemis) Sequence viewer and annotation tool • Gene. Quiz (http//www. sander. ebi. ac. uk/genequiz/) System for automated annotation of sequences, web access required • Genome Annotation Assessment Project (GASP 1) (http: //www. fruitfly. org/GASP 1)
GENOME ANNOTATION TOOLS • Oakridge Genome Annotation Channel (http: //compbio. ornl. gov/channel/) • ENSEMBL (http: //ensembl. ebi. ac. uk) • Artemis (http: //www. sanger. ac. uk/Software/Artemis) Sequence viewer and annotation tool • Gene. Quiz (http//www. sander. ebi. ac. uk/genequiz/) System for automated annotation of sequences, web access required • Genome Annotation Assessment Project (GASP 1) (http: //www. fruitfly. org/GASP 1)
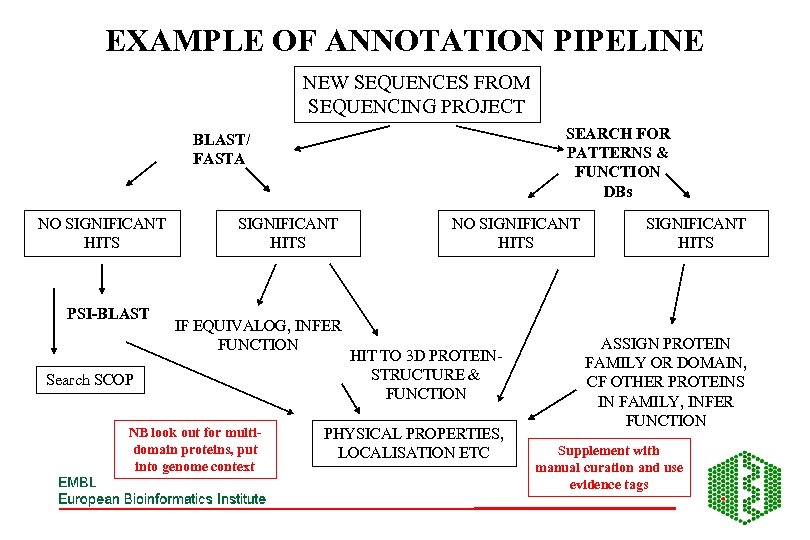 EXAMPLE OF ANNOTATION PIPELINE NEW SEQUENCES FROM SEQUENCING PROJECT SEARCH FOR PATTERNS & FUNCTION DBs BLAST/ FASTA NO SIGNIFICANT HITS PSI-BLAST SIGNIFICANT HITS IF EQUIVALOG, INFER FUNCTION Search SCOP NB look out for multidomain proteins, put into genome context NO SIGNIFICANT HITS HIT TO 3 D PROTEINSTRUCTURE & FUNCTION PHYSICAL PROPERTIES, LOCALISATION ETC SIGNIFICANT HITS ASSIGN PROTEIN FAMILY OR DOMAIN, CF OTHER PROTEINS IN FAMILY, INFER FUNCTION Supplement with manual curation and use evidence tags
EXAMPLE OF ANNOTATION PIPELINE NEW SEQUENCES FROM SEQUENCING PROJECT SEARCH FOR PATTERNS & FUNCTION DBs BLAST/ FASTA NO SIGNIFICANT HITS PSI-BLAST SIGNIFICANT HITS IF EQUIVALOG, INFER FUNCTION Search SCOP NB look out for multidomain proteins, put into genome context NO SIGNIFICANT HITS HIT TO 3 D PROTEINSTRUCTURE & FUNCTION PHYSICAL PROPERTIES, LOCALISATION ETC SIGNIFICANT HITS ASSIGN PROTEIN FAMILY OR DOMAIN, CF OTHER PROTEINS IN FAMILY, INFER FUNCTION Supplement with manual curation and use evidence tags
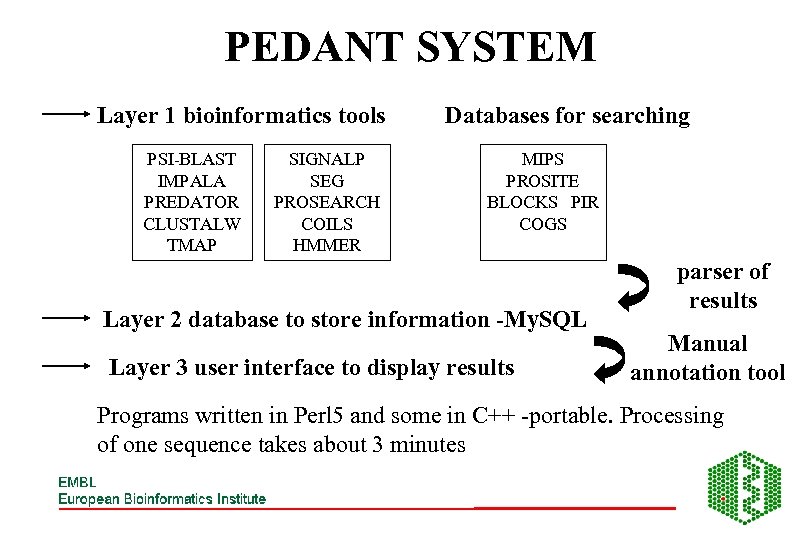 PEDANT SYSTEM Layer 1 bioinformatics tools PSI-BLAST IMPALA PREDATOR CLUSTALW TMAP SIGNALP SEG PROSEARCH COILS HMMER Databases for searching MIPS PROSITE BLOCKS PIR COGS Layer 2 database to store information -My. SQL Layer 3 user interface to display results parser of results Manual annotation tool Programs written in Perl 5 and some in C++ -portable. Processing of one sequence takes about 3 minutes
PEDANT SYSTEM Layer 1 bioinformatics tools PSI-BLAST IMPALA PREDATOR CLUSTALW TMAP SIGNALP SEG PROSEARCH COILS HMMER Databases for searching MIPS PROSITE BLOCKS PIR COGS Layer 2 database to store information -My. SQL Layer 3 user interface to display results parser of results Manual annotation tool Programs written in Perl 5 and some in C++ -portable. Processing of one sequence takes about 3 minutes
 Summary of protein sequence annotation • • • Mask compositionally-biased and coiled-coil regions Identify transmembrane regions, signal peptides, GPI anchors Predict secondary structure Look for known domains from protein pattern databases Search sequence database for similar sequences If no or few results search with subsequences, do iterative searches • Functional annotation: consider function of each domain present, annotation from database homologs, function from hits with 3 D structure
Summary of protein sequence annotation • • • Mask compositionally-biased and coiled-coil regions Identify transmembrane regions, signal peptides, GPI anchors Predict secondary structure Look for known domains from protein pattern databases Search sequence database for similar sequences If no or few results search with subsequences, do iterative searches • Functional annotation: consider function of each domain present, annotation from database homologs, function from hits with 3 D structure
 Limits of protein sequence annotation (1) • Predicting function from sequence requires another sequence to be mapped to a function –many hypothetical proteins in database and UPFs • If sequence homologues are found, may not be functional homologs -qualitative rather than quantitative process - orthologs may have different functions -enzyme homologs may be inactive -equivalent functions may use different genes, not ortholog • Analogy can often infer molecular function, but not necessarily cellular function
Limits of protein sequence annotation (1) • Predicting function from sequence requires another sequence to be mapped to a function –many hypothetical proteins in database and UPFs • If sequence homologues are found, may not be functional homologs -qualitative rather than quantitative process - orthologs may have different functions -enzyme homologs may be inactive -equivalent functions may use different genes, not ortholog • Analogy can often infer molecular function, but not necessarily cellular function
 Limits of protein sequence annotation (2) • Databases are biased in sequence and aa composition and search is dependent on size • If no homology found- limited amount of information can be inferred • Incorrect annotation can be propagated when similarity is over part on sequence not used in annotation • No answers to tissue-specificity, binding of ligands, relationship between genotype and phenotype
Limits of protein sequence annotation (2) • Databases are biased in sequence and aa composition and search is dependent on size • If no homology found- limited amount of information can be inferred • Incorrect annotation can be propagated when similarity is over part on sequence not used in annotation • No answers to tissue-specificity, binding of ligands, relationship between genotype and phenotype
 Limits of protein sequence annotation (3) • Need additional information from experiments, eg can predict glycosylation sites, but not kind of sugar attached • Problem with multidomain proteins (Do you assign orthology on basis of domains or domain composition of whole protein? ) -check also known domain architectures and their taxonomic limitations
Limits of protein sequence annotation (3) • Need additional information from experiments, eg can predict glycosylation sites, but not kind of sugar attached • Problem with multidomain proteins (Do you assign orthology on basis of domains or domain composition of whole protein? ) -check also known domain architectures and their taxonomic limitations
 Using different approaches to functional annotation: Status for SPTR • Automatic annotation (Rule. Base): 20% of all protein sequences and 20% of all new sequences • Automatic classification (Inter. Pro, Clu. STr, Structure): 60% of all protein sequences and 60% of all new sequences • Automatic characterisation (GO): 40% of all protein sequences and 40% of all new sequences • Full annotation (SWISS-PROT style): 20% of all protein sequences and 5% of all new sequences
Using different approaches to functional annotation: Status for SPTR • Automatic annotation (Rule. Base): 20% of all protein sequences and 20% of all new sequences • Automatic classification (Inter. Pro, Clu. STr, Structure): 60% of all protein sequences and 60% of all new sequences • Automatic characterisation (GO): 40% of all protein sequences and 40% of all new sequences • Full annotation (SWISS-PROT style): 20% of all protein sequences and 5% of all new sequences
 Using different approaches to functional annotation: Future for SPTR • Automatic annotation (Rule. Base): 50% of all protein sequences in 2004 • Automatic classification (Inter. Pro, Clu. STr, Structure): 90% of all protein sequences in 2004 • Automatic characterisation (GO): 70% of all protein sequences in 2004 • Full annotation (SWISS-PROT style): 10% of all protein sequences in 2004
Using different approaches to functional annotation: Future for SPTR • Automatic annotation (Rule. Base): 50% of all protein sequences in 2004 • Automatic classification (Inter. Pro, Clu. STr, Structure): 90% of all protein sequences in 2004 • Automatic characterisation (GO): 70% of all protein sequences in 2004 • Full annotation (SWISS-PROT style): 10% of all protein sequences in 2004
 IMPORTANT TO NOTE: • DON’T COMPLETELY TRUST COMPUTER RESULTS • CHECK LITERATURE • CONFIRM WITH WETLAB WORK- mutational analysis gives valuable info about function • COMPROMISE BETWEEN OVER AND UNDERPREDICTIONS -overpredictions can be checked by curators, easier to delete than find missing info.
IMPORTANT TO NOTE: • DON’T COMPLETELY TRUST COMPUTER RESULTS • CHECK LITERATURE • CONFIRM WITH WETLAB WORK- mutational analysis gives valuable info about function • COMPROMISE BETWEEN OVER AND UNDERPREDICTIONS -overpredictions can be checked by curators, easier to delete than find missing info.


