d554d921db827cc9474b6b976a5aefc1.ppt
- Количество слайдов: 61
 GENETICS genetic mapping, classical approaches to study gene function
GENETICS genetic mapping, classical approaches to study gene function
 Basic aims: • uncovering gene function understand mechanisms of morphogenesis, development, metabolism, physiology etc. in connection with coordinated gene expression • breeding production of plants (organisms) with improved characteristics or their combination
Basic aims: • uncovering gene function understand mechanisms of morphogenesis, development, metabolism, physiology etc. in connection with coordinated gene expression • breeding production of plants (organisms) with improved characteristics or their combination
 Terminology Gene • segment of DNA (genetic information) that specifies a trait • basic unit of heridity in living organisms • Genotype + environment + ? = phenotype • Interactions between genes/proteins (epistasis – metabolic and signal pathways)
Terminology Gene • segment of DNA (genetic information) that specifies a trait • basic unit of heridity in living organisms • Genotype + environment + ? = phenotype • Interactions between genes/proteins (epistasis – metabolic and signal pathways)
 Allele – form of a gene • dominant vs. recesive (codominant, …) • genesis of new alleles by mutations Locus – location of a gene on a chromosome • Genetic linkage – inheriting of certain genes (their alleles) jointly, - consequence of their presence on the same chromosome (gene distance c. M = % of recombinant gametes) • Genetic (likage) maps x physical maps
Allele – form of a gene • dominant vs. recesive (codominant, …) • genesis of new alleles by mutations Locus – location of a gene on a chromosome • Genetic linkage – inheriting of certain genes (their alleles) jointly, - consequence of their presence on the same chromosome (gene distance c. M = % of recombinant gametes) • Genetic (likage) maps x physical maps
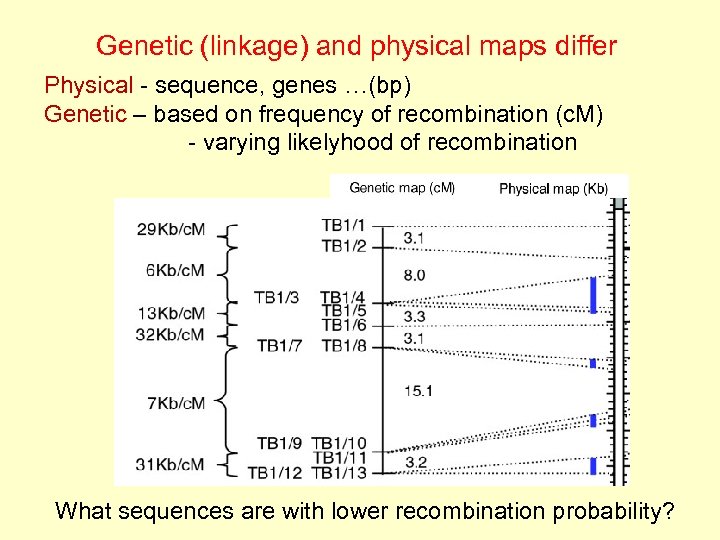 Genetic (linkage) and physical maps differ Physical - sequence, genes …(bp) Genetic – based on frequency of recombination (c. M) - varying likelyhood of recombination What sequences are with lower recombination probability?
Genetic (linkage) and physical maps differ Physical - sequence, genes …(bp) Genetic – based on frequency of recombination (c. M) - varying likelyhood of recombination What sequences are with lower recombination probability?
 Genetics classical (direct) x reverse Direct – from a trait (phenotype) to identification of corresponding gene Reverse – from a gene (with known sequence) to its function (phenotype) Both approaches are based on mutant analysis: Direct – looking for certain phenotype in mutagenized population Reverse – targeted mutagenesis/modification of selected gene
Genetics classical (direct) x reverse Direct – from a trait (phenotype) to identification of corresponding gene Reverse – from a gene (with known sequence) to its function (phenotype) Both approaches are based on mutant analysis: Direct – looking for certain phenotype in mutagenized population Reverse – targeted mutagenesis/modification of selected gene
 Mutagenesis • Classical: – chemical m. – EMS (ethane metyl sulfonate; point mutations) – physical m. – RTG, gama. . . (usually short deletions) – wide spektrum of affects (regulation, interaction) – even dominant mutations, resemble natural mutations, – BUT complicated identification of mutated gene
Mutagenesis • Classical: – chemical m. – EMS (ethane metyl sulfonate; point mutations) – physical m. – RTG, gama. . . (usually short deletions) – wide spektrum of affects (regulation, interaction) – even dominant mutations, resemble natural mutations, – BUT complicated identification of mutated gene
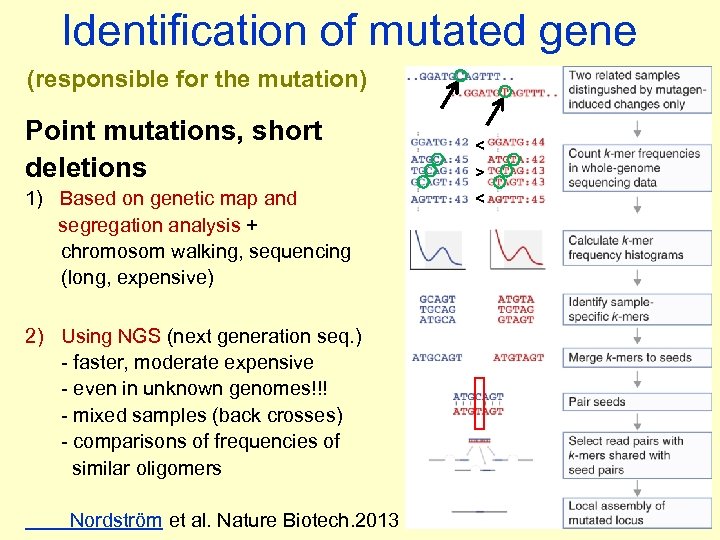 Identification of mutated gene (responsible for the mutation) Point mutations, short deletions 1) Based on genetic map and segregation analysis + chromosom walking, sequencing (long, expensive) 2) Using NGS (next generation seq. ) - faster, moderate expensive - even in unknown genomes!!! - mixed samples (back crosses) - comparisons of frequencies of similar oligomers Nordström et al. Nature Biotech. 2013 < > <
Identification of mutated gene (responsible for the mutation) Point mutations, short deletions 1) Based on genetic map and segregation analysis + chromosom walking, sequencing (long, expensive) 2) Using NGS (next generation seq. ) - faster, moderate expensive - even in unknown genomes!!! - mixed samples (back crosses) - comparisons of frequencies of similar oligomers Nordström et al. Nature Biotech. 2013 < > <
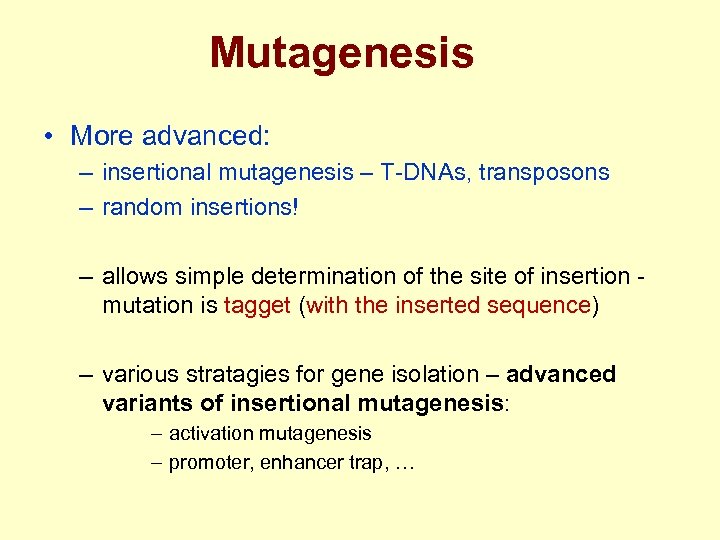 Mutagenesis • More advanced: – insertional mutagenesis – T-DNAs, transposons – random insertions! – allows simple determination of the site of insertion mutation is tagget (with the inserted sequence) – various stratagies for gene isolation – advanced variants of insertional mutagenesis: – activation mutagenesis – promoter, enhancer trap, …
Mutagenesis • More advanced: – insertional mutagenesis – T-DNAs, transposons – random insertions! – allows simple determination of the site of insertion mutation is tagget (with the inserted sequence) – various stratagies for gene isolation – advanced variants of insertional mutagenesis: – activation mutagenesis – promoter, enhancer trap, …
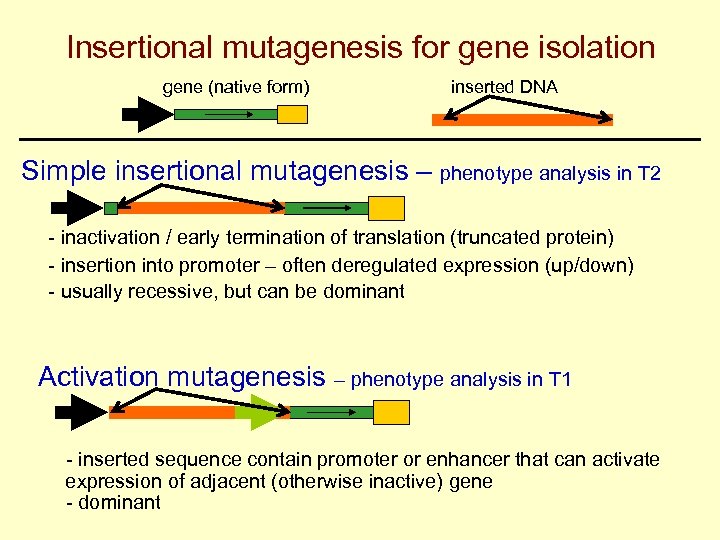 Insertional mutagenesis for gene isolation gene (native form) inserted DNA Simple insertional mutagenesis – phenotype analysis in T 2 - inactivation / early termination of translation (truncated protein) - insertion into promoter – often deregulated expression (up/down) - usually recessive, but can be dominant Activation mutagenesis – phenotype analysis in T 1 - inserted sequence contain promoter or enhancer that can activate expression of adjacent (otherwise inactive) gene - dominant
Insertional mutagenesis for gene isolation gene (native form) inserted DNA Simple insertional mutagenesis – phenotype analysis in T 2 - inactivation / early termination of translation (truncated protein) - insertion into promoter – often deregulated expression (up/down) - usually recessive, but can be dominant Activation mutagenesis – phenotype analysis in T 1 - inserted sequence contain promoter or enhancer that can activate expression of adjacent (otherwise inactive) gene - dominant
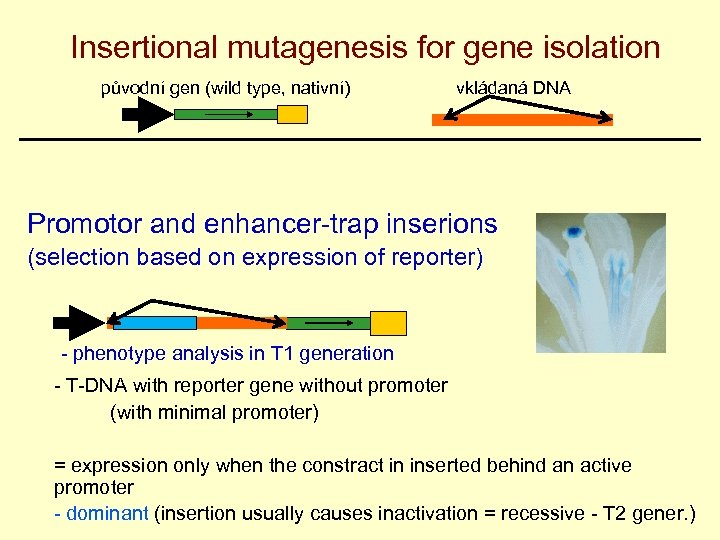 Insertional mutagenesis for gene isolation původní gen (wild type, nativní) vkládaná DNA Promotor and enhancer-trap inserions (selection based on expression of reporter) - phenotype analysis in T 1 generation - T-DNA with reporter gene without promoter (with minimal promoter) = expression only when the constract in inserted behind an active promoter - dominant (insertion usually causes inactivation = recessive - T 2 gener. )
Insertional mutagenesis for gene isolation původní gen (wild type, nativní) vkládaná DNA Promotor and enhancer-trap inserions (selection based on expression of reporter) - phenotype analysis in T 1 generation - T-DNA with reporter gene without promoter (with minimal promoter) = expression only when the constract in inserted behind an active promoter - dominant (insertion usually causes inactivation = recessive - T 2 gener. )
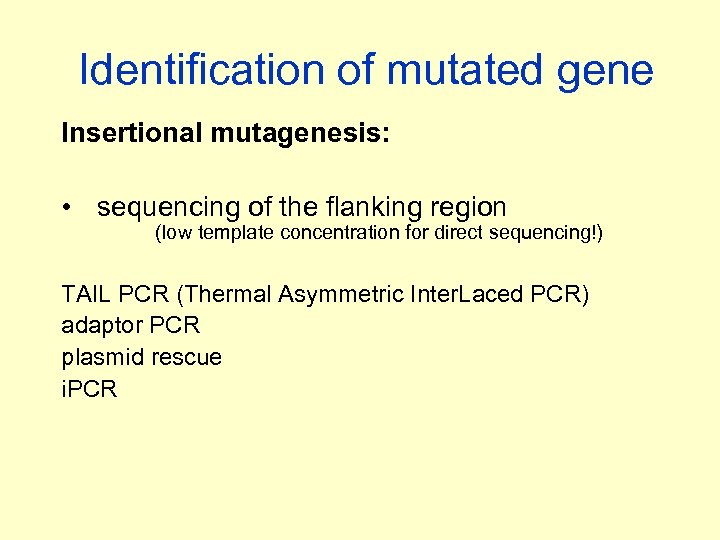 Identification of mutated gene Insertional mutagenesis: • sequencing of the flanking region (low template concentration for direct sequencing!) TAIL PCR (Thermal Asymmetric Inter. Laced PCR) adaptor PCR plasmid rescue i. PCR
Identification of mutated gene Insertional mutagenesis: • sequencing of the flanking region (low template concentration for direct sequencing!) TAIL PCR (Thermal Asymmetric Inter. Laced PCR) adaptor PCR plasmid rescue i. PCR
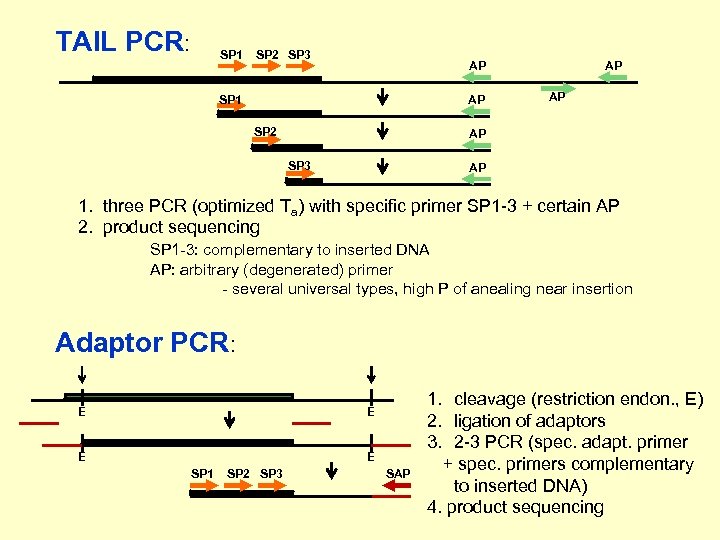 TAIL PCR: SP 1 SP 2 SP 3 AP SP 1 AP SP 2 AP AP AP SP 3 AP 1. three PCR (optimized Ta) with specific primer SP 1 -3 + certain AP 2. product sequencing SP 1 -3: complementary to inserted DNA AP: arbitrary (degenerated) primer - several universal types, high P of anealing near insertion Adaptor PCR: E E SP 1 SP 2 SP 3 SAP 1. cleavage (restriction endon. , E) 2. ligation of adaptors 3. 2 -3 PCR (spec. adapt. primer + spec. primers complementary to inserted DNA) 4. product sequencing
TAIL PCR: SP 1 SP 2 SP 3 AP SP 1 AP SP 2 AP AP AP SP 3 AP 1. three PCR (optimized Ta) with specific primer SP 1 -3 + certain AP 2. product sequencing SP 1 -3: complementary to inserted DNA AP: arbitrary (degenerated) primer - several universal types, high P of anealing near insertion Adaptor PCR: E E SP 1 SP 2 SP 3 SAP 1. cleavage (restriction endon. , E) 2. ligation of adaptors 3. 2 -3 PCR (spec. adapt. primer + spec. primers complementary to inserted DNA) 4. product sequencing
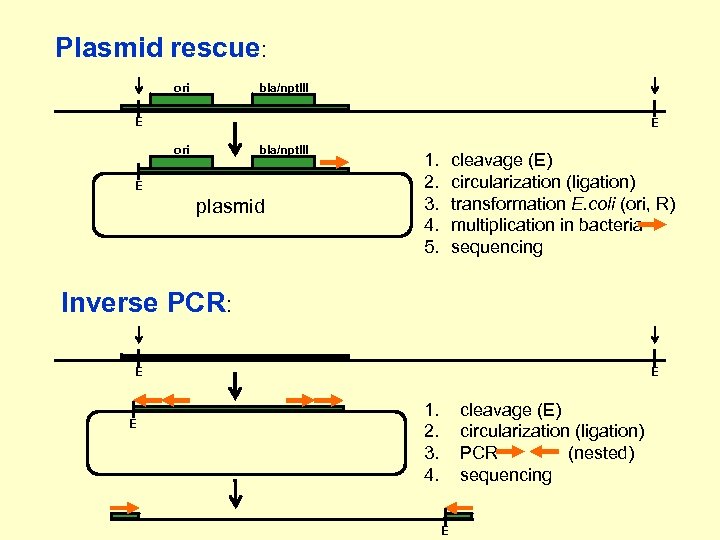 Plasmid rescue: ori bla/npt. III E E ori bla/npt. III E plasmid 1. 2. 3. 4. 5. cleavage (E) circularization (ligation) transformation E. coli (ori, R) multiplication in bacteria sequencing Inverse PCR: E E E 1. 2. 3. 4. cleavage (E) circularization (ligation) PCR (nested) sequencing E
Plasmid rescue: ori bla/npt. III E E ori bla/npt. III E plasmid 1. 2. 3. 4. 5. cleavage (E) circularization (ligation) transformation E. coli (ori, R) multiplication in bacteria sequencing Inverse PCR: E E E 1. 2. 3. 4. cleavage (E) circularization (ligation) PCR (nested) sequencing E
 Mutagenesis Most advanced genome editing = targeted mutagenesis within the genome: ZFN TALEN CRISPR/Cas 9 (oligonucleotide targeted mutagenesis) - mutation (usually inactivation) of selected gene
Mutagenesis Most advanced genome editing = targeted mutagenesis within the genome: ZFN TALEN CRISPR/Cas 9 (oligonucleotide targeted mutagenesis) - mutation (usually inactivation) of selected gene
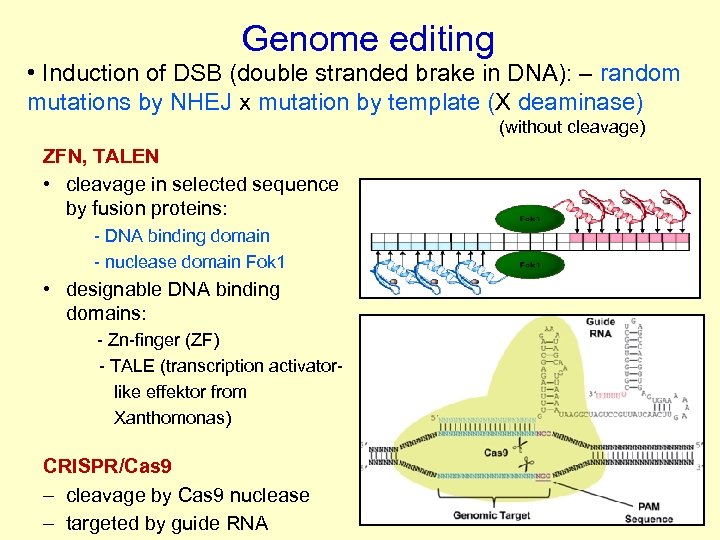 Genome editing • Induction of DSB (double stranded brake in DNA): – random mutations by NHEJ x mutation by template (X deaminase) (without cleavage) ZFN, TALEN • cleavage in selected sequence by fusion proteins: - DNA binding domain - nuclease domain Fok 1 • designable DNA binding domains: - Zn-finger (ZF) - TALE (transcription activatorlike effektor from Xanthomonas) CRISPR/Cas 9 – cleavage by Cas 9 nuclease – targeted by guide RNA
Genome editing • Induction of DSB (double stranded brake in DNA): – random mutations by NHEJ x mutation by template (X deaminase) (without cleavage) ZFN, TALEN • cleavage in selected sequence by fusion proteins: - DNA binding domain - nuclease domain Fok 1 • designable DNA binding domains: - Zn-finger (ZF) - TALE (transcription activatorlike effektor from Xanthomonas) CRISPR/Cas 9 – cleavage by Cas 9 nuclease – targeted by guide RNA
 Use of mutagenesis in genetics Direct: - classical mutagenesis - insertional mutagenesis (all types) Reverse: - directed mutagenesis - also random insertional mutagenesis - use of characterized mutant collections!!!
Use of mutagenesis in genetics Direct: - classical mutagenesis - insertional mutagenesis (all types) Reverse: - directed mutagenesis - also random insertional mutagenesis - use of characterized mutant collections!!!
 Reverse genetics – approaches - analyses of mutants with insertion in analyzed g. - modulation of gene expression - promoter activity analyses - analyses of cell localization (GFP fusion) - gene modification (within/outside genome) - TILLING, …. . - analysis of gene polymorphism in ecotypes (1000+1 genome project)
Reverse genetics – approaches - analyses of mutants with insertion in analyzed g. - modulation of gene expression - promoter activity analyses - analyses of cell localization (GFP fusion) - gene modification (within/outside genome) - TILLING, …. . - analysis of gene polymorphism in ecotypes (1000+1 genome project)
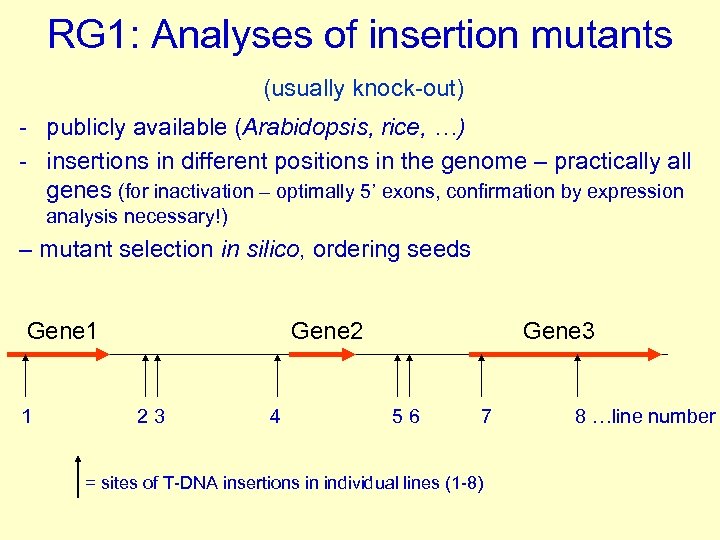 RG 1: Analyses of insertion mutants (usually knock-out) - publicly available (Arabidopsis, rice, …) - insertions in different positions in the genome – practically all genes (for inactivation – optimally 5’ exons, confirmation by expression analysis necessary!) – mutant selection in silico, ordering seeds Gene 1 1 Gene 2 23 4 Gene 3 56 7 = sites of T-DNA insertions in individual lines (1 -8) 8 …line number
RG 1: Analyses of insertion mutants (usually knock-out) - publicly available (Arabidopsis, rice, …) - insertions in different positions in the genome – practically all genes (for inactivation – optimally 5’ exons, confirmation by expression analysis necessary!) – mutant selection in silico, ordering seeds Gene 1 1 Gene 2 23 4 Gene 3 56 7 = sites of T-DNA insertions in individual lines (1 -8) 8 …line number
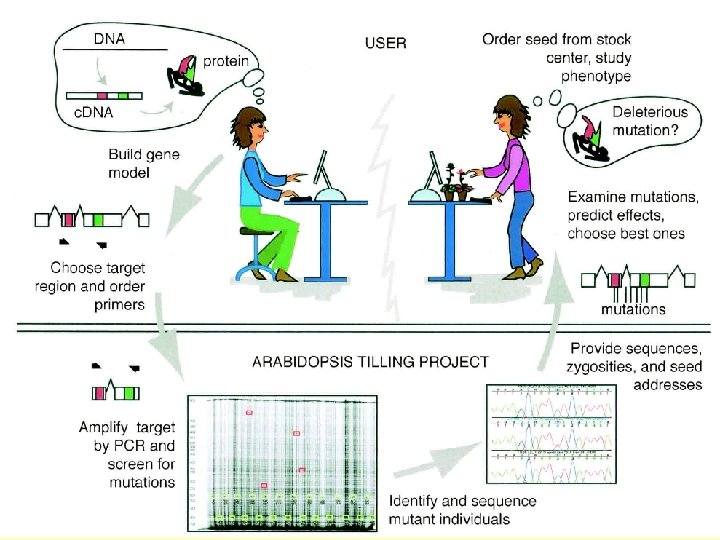
 WWW interphase http: //signal. salk. edu/cgi-bin/tdnaexpress
WWW interphase http: //signal. salk. edu/cgi-bin/tdnaexpress
 RG 2: Modulation of gene expression - increased protein level (overexpression) - introduction of a gene with a strong constitutive promoter (e. g. viral) - syn. gain-of-function mutations - decreased protein level by RNAi - silencing, knock-down - X knock–out, loss-of-function
RG 2: Modulation of gene expression - increased protein level (overexpression) - introduction of a gene with a strong constitutive promoter (e. g. viral) - syn. gain-of-function mutations - decreased protein level by RNAi - silencing, knock-down - X knock–out, loss-of-function
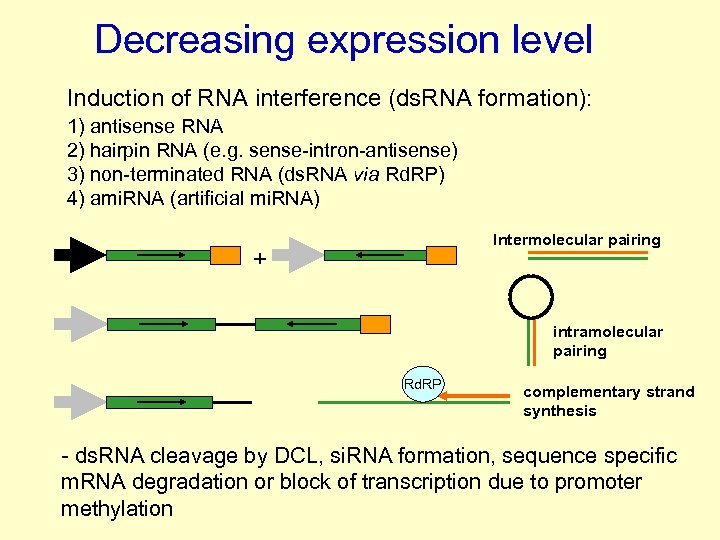 Decreasing expression level Induction of RNA interference (ds. RNA formation): 1) antisense RNA 2) hairpin RNA (e. g. sense-intron-antisense) 3) non-terminated RNA (ds. RNA via Rd. RP) 4) ami. RNA (artificial mi. RNA) Intermolecular pairing + intramolecular pairing Rd. RP complementary strand synthesis - ds. RNA cleavage by DCL, si. RNA formation, sequence specific m. RNA degradation or block of transcription due to promoter methylation
Decreasing expression level Induction of RNA interference (ds. RNA formation): 1) antisense RNA 2) hairpin RNA (e. g. sense-intron-antisense) 3) non-terminated RNA (ds. RNA via Rd. RP) 4) ami. RNA (artificial mi. RNA) Intermolecular pairing + intramolecular pairing Rd. RP complementary strand synthesis - ds. RNA cleavage by DCL, si. RNA formation, sequence specific m. RNA degradation or block of transcription due to promoter methylation
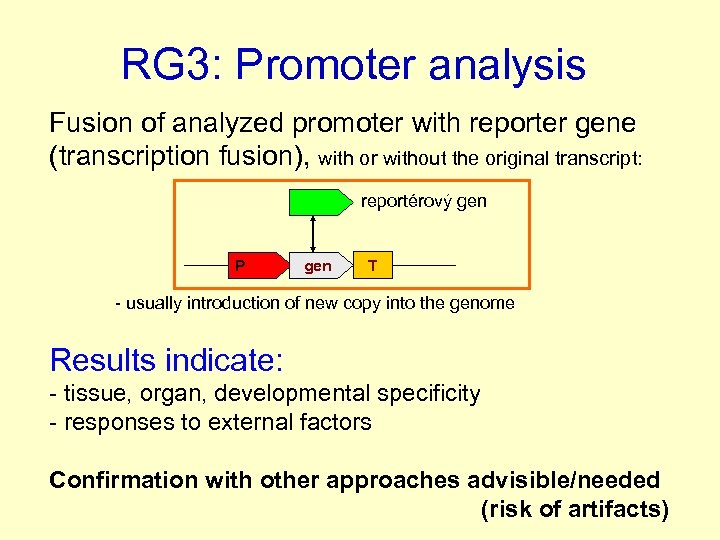 RG 3: Promoter analysis Fusion of analyzed promoter with reporter gene (transcription fusion), with or without the original transcript: reportérový gen P gen T - usually introduction of new copy into the genome Results indicate: - tissue, organ, developmental specificity - responses to external factors Confirmation with other approaches advisible/needed (risk of artifacts)
RG 3: Promoter analysis Fusion of analyzed promoter with reporter gene (transcription fusion), with or without the original transcript: reportérový gen P gen T - usually introduction of new copy into the genome Results indicate: - tissue, organ, developmental specificity - responses to external factors Confirmation with other approaches advisible/needed (risk of artifacts)
 Reporter genes • encode proteins that can be directly visualized or enzymes, whose activity can be visualized • quantitative or qualitative assessment • promoter activity analyses, subcellular localization of proteins, optimizing transformation procedure, …. Constrains: - background (autofluorescence, natural enzyme activity within the tissue) - protein stability (mask changes in promoter activity) - …. .
Reporter genes • encode proteins that can be directly visualized or enzymes, whose activity can be visualized • quantitative or qualitative assessment • promoter activity analyses, subcellular localization of proteins, optimizing transformation procedure, …. Constrains: - background (autofluorescence, natural enzyme activity within the tissue) - protein stability (mask changes in promoter activity) - …. .
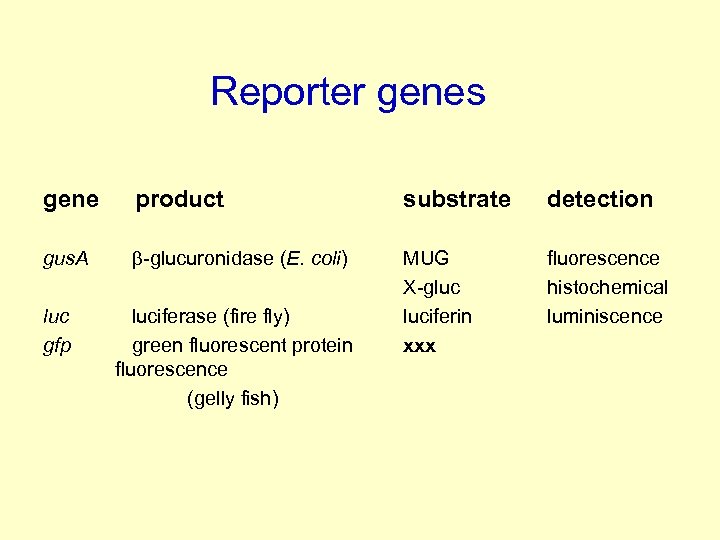 Reporter genes gene product substrate detection gus. A b-glucuronidase (E. coli) MUG X-gluc luciferin xxx fluorescence histochemical luminiscence luc gfp luciferase (fire fly) green fluorescent protein fluorescence (gelly fish)
Reporter genes gene product substrate detection gus. A b-glucuronidase (E. coli) MUG X-gluc luciferin xxx fluorescence histochemical luminiscence luc gfp luciferase (fire fly) green fluorescent protein fluorescence (gelly fish)
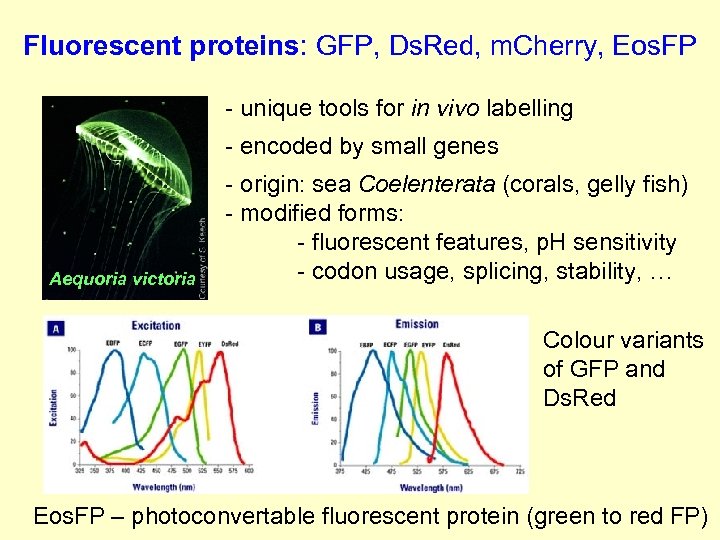 Fluorescent proteins: GFP, Ds. Red, m. Cherry, Eos. FP - unique tools for in vivo labelling - encoded by small genes Aequoria victoria - origin: sea Coelenterata (corals, gelly fish) - modified forms: - fluorescent features, p. H sensitivity - codon usage, splicing, stability, … Colour variants of GFP and Ds. Red Eos. FP – photoconvertable fluorescent protein (green to red FP)
Fluorescent proteins: GFP, Ds. Red, m. Cherry, Eos. FP - unique tools for in vivo labelling - encoded by small genes Aequoria victoria - origin: sea Coelenterata (corals, gelly fish) - modified forms: - fluorescent features, p. H sensitivity - codon usage, splicing, stability, … Colour variants of GFP and Ds. Red Eos. FP – photoconvertable fluorescent protein (green to red FP)
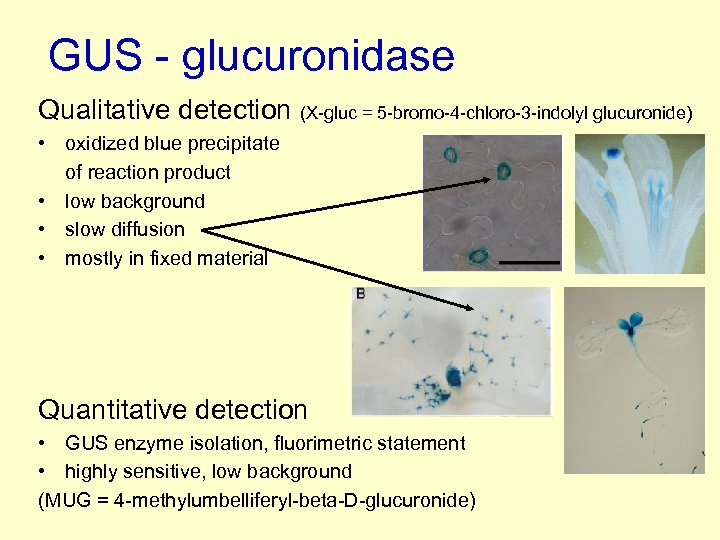 GUS - glucuronidase Qualitative detection (X-gluc = 5 -bromo-4 -chloro-3 -indolyl glucuronide) • oxidized blue precipitate of reaction product • low background • slow diffusion • mostly in fixed material Quantitative detection • GUS enzyme isolation, fluorimetric statement • highly sensitive, low background (MUG = 4 -methylumbelliferyl-beta-D-glucuronide)
GUS - glucuronidase Qualitative detection (X-gluc = 5 -bromo-4 -chloro-3 -indolyl glucuronide) • oxidized blue precipitate of reaction product • low background • slow diffusion • mostly in fixed material Quantitative detection • GUS enzyme isolation, fluorimetric statement • highly sensitive, low background (MUG = 4 -methylumbelliferyl-beta-D-glucuronide)
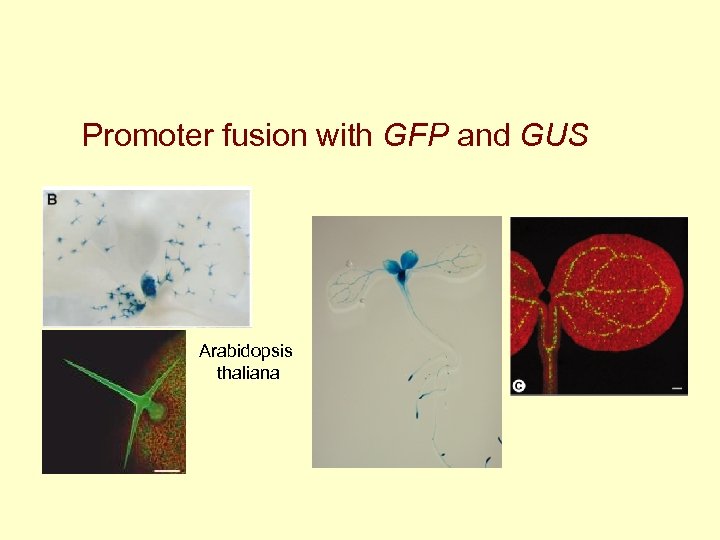 Promoter fusion with GFP and GUS Arabidopsis thaliana
Promoter fusion with GFP and GUS Arabidopsis thaliana
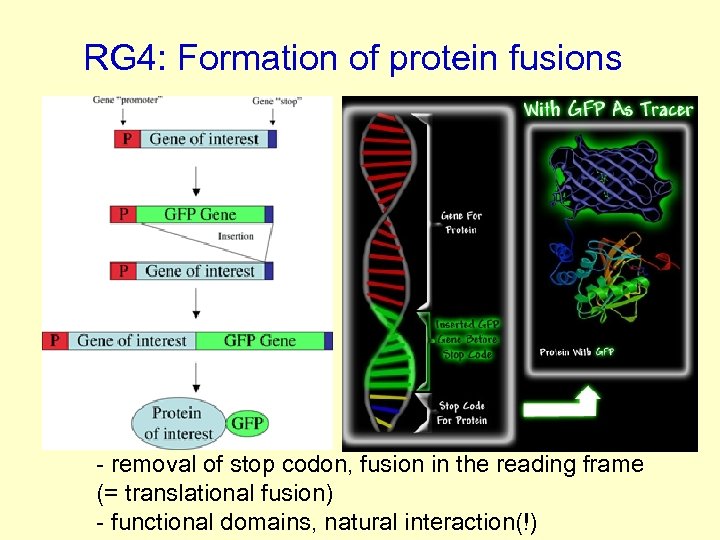 RG 4: Formation of protein fusions - removal of stop codon, fusion in the reading frame (= translational fusion) - functional domains, natural interaction(!)
RG 4: Formation of protein fusions - removal of stop codon, fusion in the reading frame (= translational fusion) - functional domains, natural interaction(!)
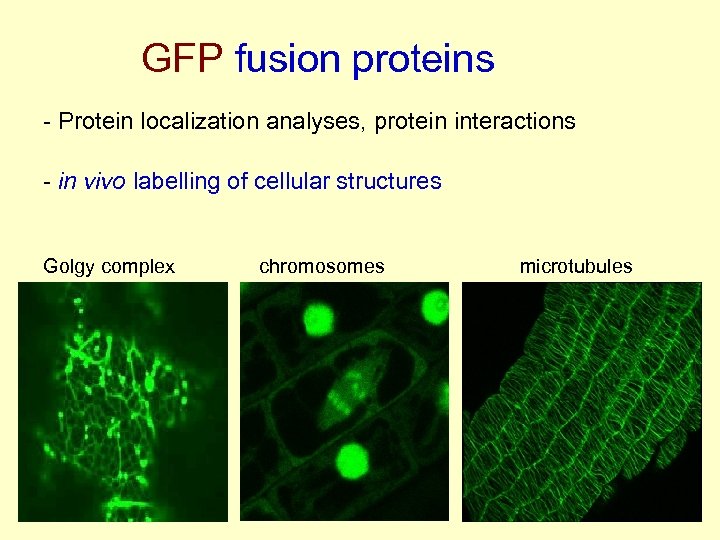 GFP fusion proteins - Protein localization analyses, protein interactions - in vivo labelling of cellular structures Golgy complex chromosomes microtubules
GFP fusion proteins - Protein localization analyses, protein interactions - in vivo labelling of cellular structures Golgy complex chromosomes microtubules
 RG 5: Modification of the gene (outside genome) - introduction of modified gene copy to knock-out mutant • gene isolation (including native promoter) • directed mutagenesis ex vivo by PCR (primers) • selection of homozygous knock-out mutant in the gene (heterozygous mutant in case of lethal mutations) • introduction of modified gene (analysis of numerous lines to avoid position effect artifacts) RG 6: Gene editing in the genome (see earlier) (ZFN, TALEN, CRISPR/Cas 9 + oligonukleotidy)
RG 5: Modification of the gene (outside genome) - introduction of modified gene copy to knock-out mutant • gene isolation (including native promoter) • directed mutagenesis ex vivo by PCR (primers) • selection of homozygous knock-out mutant in the gene (heterozygous mutant in case of lethal mutations) • introduction of modified gene (analysis of numerous lines to avoid position effect artifacts) RG 6: Gene editing in the genome (see earlier) (ZFN, TALEN, CRISPR/Cas 9 + oligonukleotidy)
 RG 7: TILLING isolation of mutants with point mutations in certain gene Targeting induced local lesions in genomes • chemical mutagenesis (EMS) – seed and DNA stocks • PCR- and heteroduplex analysis-based screen for mutants in certain gene • Point mutations! (changed regulation, interactions, …)
RG 7: TILLING isolation of mutants with point mutations in certain gene Targeting induced local lesions in genomes • chemical mutagenesis (EMS) – seed and DNA stocks • PCR- and heteroduplex analysis-based screen for mutants in certain gene • Point mutations! (changed regulation, interactions, …)
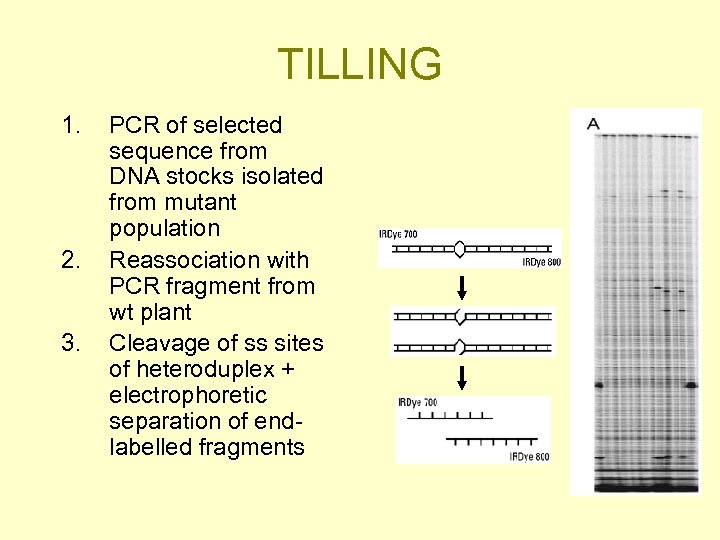 TILLING 1. 2. 3. PCR of selected sequence from DNA stocks isolated from mutant population Reassociation with PCR fragment from wt plant Cleavage of ss sites of heteroduplex + electrophoretic separation of endlabelled fragments
TILLING 1. 2. 3. PCR of selected sequence from DNA stocks isolated from mutant population Reassociation with PCR fragment from wt plant Cleavage of ss sites of heteroduplex + electrophoretic separation of endlabelled fragments
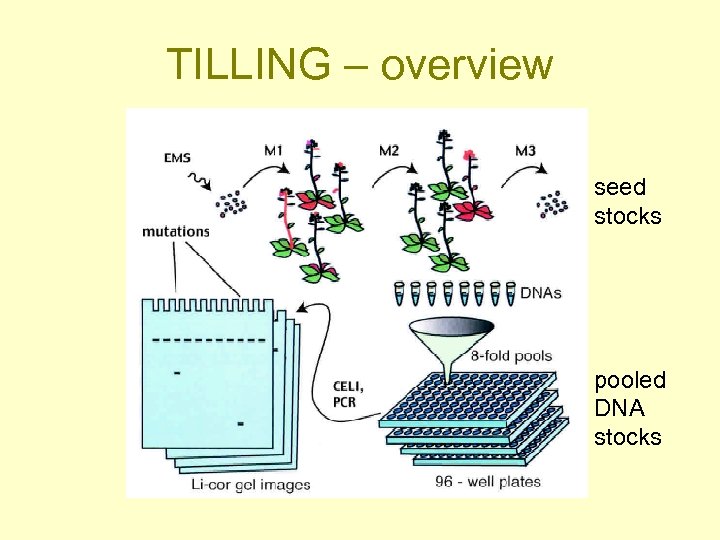 TILLING – overview seed stocks pooled DNA stocks
TILLING – overview seed stocks pooled DNA stocks
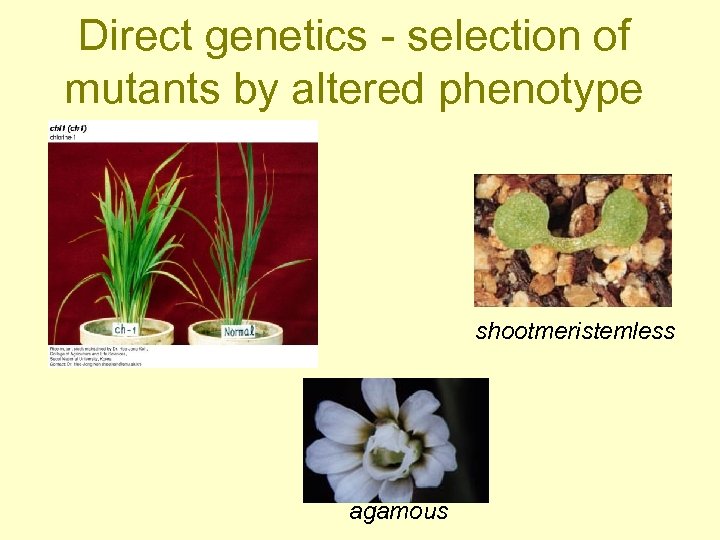 Direct genetics - selection of mutants by altered phenotype shootmeristemless agamous
Direct genetics - selection of mutants by altered phenotype shootmeristemless agamous
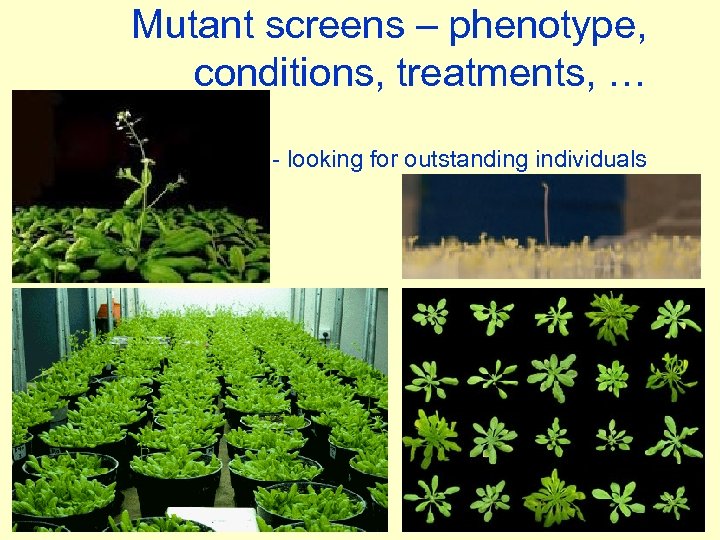 Mutant screens – phenotype, conditions, treatments, … - looking for outstanding individuals
Mutant screens – phenotype, conditions, treatments, … - looking for outstanding individuals
 Direct genetics – identification of mutated gene - the same phenotypic change can result from different mutations „there are numerous ways how to build up house incorrectly“ - allelic mutations – mutation in the same gene (x different g. ) How to distinguish (recesive mutation)? Crossing of homozygous mutants F 1 - wt phenotype = different genes (complementation, both heterozygous) - mutant phenotype = allelic (same genes were mutated)
Direct genetics – identification of mutated gene - the same phenotypic change can result from different mutations „there are numerous ways how to build up house incorrectly“ - allelic mutations – mutation in the same gene (x different g. ) How to distinguish (recesive mutation)? Crossing of homozygous mutants F 1 - wt phenotype = different genes (complementation, both heterozygous) - mutant phenotype = allelic (same genes were mutated)
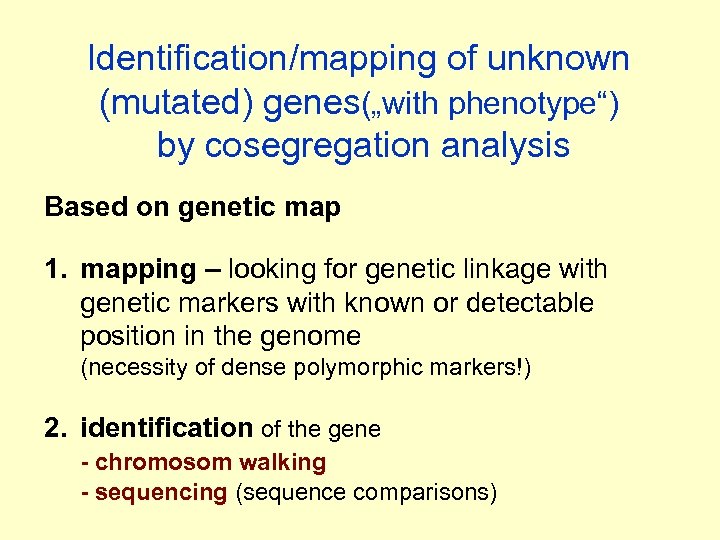 Identification/mapping of unknown (mutated) genes(„with phenotype“) by cosegregation analysis Based on genetic map 1. mapping – looking for genetic linkage with genetic markers with known or detectable position in the genome (necessity of dense polymorphic markers!) 2. identification of the gene - chromosom walking - sequencing (sequence comparisons)
Identification/mapping of unknown (mutated) genes(„with phenotype“) by cosegregation analysis Based on genetic map 1. mapping – looking for genetic linkage with genetic markers with known or detectable position in the genome (necessity of dense polymorphic markers!) 2. identification of the gene - chromosom walking - sequencing (sequence comparisons)
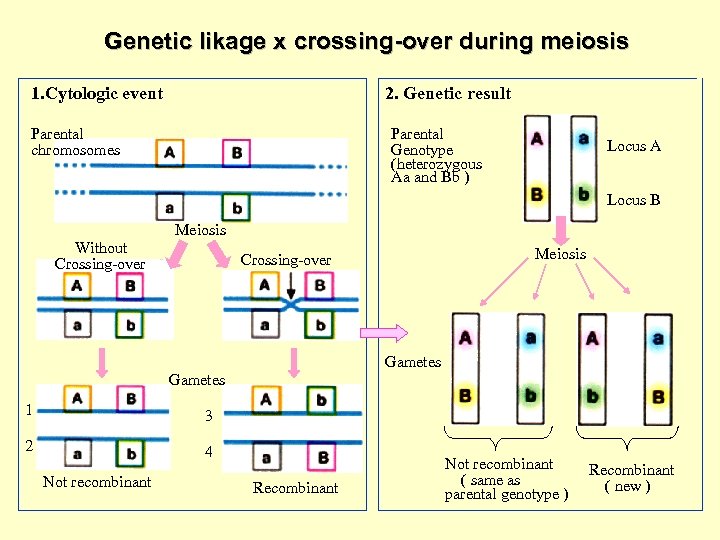 Genetic likage x crossing-over during meiosis 1. Cytologic event 2. Genetic result Parental chromosomes Parental Genotype (heterozygous Aa and Bb ) Locus A Locus B Meiosis Without Crossing-over Meiosis Crossing-over Gametes 1 3 2 4 Not recombinant Recombinant Not recombinant ( same as parental genotype ) Recombinant ( new )
Genetic likage x crossing-over during meiosis 1. Cytologic event 2. Genetic result Parental chromosomes Parental Genotype (heterozygous Aa and Bb ) Locus A Locus B Meiosis Without Crossing-over Meiosis Crossing-over Gametes 1 3 2 4 Not recombinant Recombinant Not recombinant ( same as parental genotype ) Recombinant ( new )
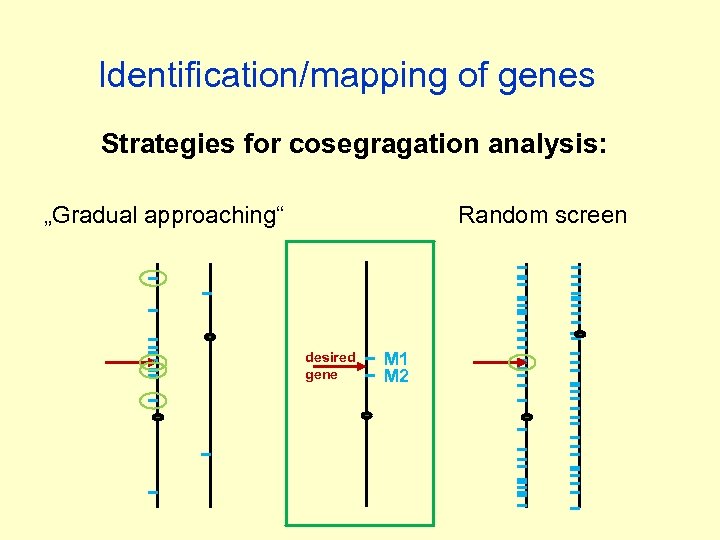 Identification/mapping of genes Strategies for cosegragation analysis: „Gradual approaching“ Random screen desired gene M 1 M 2
Identification/mapping of genes Strategies for cosegragation analysis: „Gradual approaching“ Random screen desired gene M 1 M 2
 Types of genetic markers = trait with known or identifiable position in genetic map with polymorphism between parental genotypes (e. g. different ecotypes) • Morphological (limited number) • Molecular
Types of genetic markers = trait with known or identifiable position in genetic map with polymorphism between parental genotypes (e. g. different ecotypes) • Morphological (limited number) • Molecular
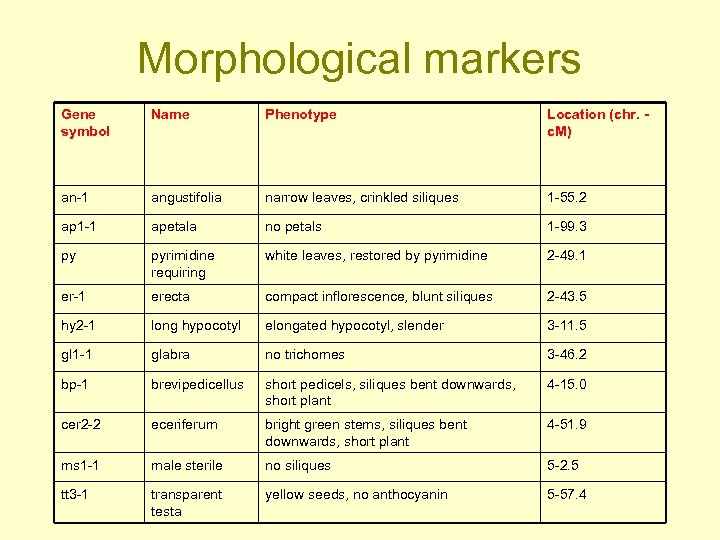 Morphological markers Gene symbol Name Phenotype Location (chr. c. M) an-1 angustifolia narrow leaves, crinkled siliques 1 -55. 2 ap 1 -1 apetala no petals 1 -99. 3 py pyrimidine requiring white leaves, restored by pyrimidine 2 -49. 1 er-1 erecta compact inflorescence, blunt siliques 2 -43. 5 hy 2 -1 long hypocotyl elongated hypocotyl, slender 3 -11. 5 gl 1 -1 glabra no trichomes 3 -46. 2 bp-1 brevipedicellus short pedicels, siliques bent downwards, short plant 4 -15. 0 cer 2 -2 eceriferum bright green stems, siliques bent downwards, short plant 4 -51. 9 ms 1 -1 male sterile no siliques 5 -2. 5 tt 3 -1 transparent testa yellow seeds, no anthocyanin 5 -57. 4
Morphological markers Gene symbol Name Phenotype Location (chr. c. M) an-1 angustifolia narrow leaves, crinkled siliques 1 -55. 2 ap 1 -1 apetala no petals 1 -99. 3 py pyrimidine requiring white leaves, restored by pyrimidine 2 -49. 1 er-1 erecta compact inflorescence, blunt siliques 2 -43. 5 hy 2 -1 long hypocotyl elongated hypocotyl, slender 3 -11. 5 gl 1 -1 glabra no trichomes 3 -46. 2 bp-1 brevipedicellus short pedicels, siliques bent downwards, short plant 4 -15. 0 cer 2 -2 eceriferum bright green stems, siliques bent downwards, short plant 4 -51. 9 ms 1 -1 male sterile no siliques 5 -2. 5 tt 3 -1 transparent testa yellow seeds, no anthocyanin 5 -57. 4
 Types of genetic markers = trait with known or identifiable position in genetic map with polymorphism between parental genotypes (e. g. different ecotypes) • Morphological (limited number) • Molecular – isozymes – DNA markers – differences in DNA sequence known or detectable position in the genome!
Types of genetic markers = trait with known or identifiable position in genetic map with polymorphism between parental genotypes (e. g. different ecotypes) • Morphological (limited number) • Molecular – isozymes – DNA markers – differences in DNA sequence known or detectable position in the genome!
 DNA molecular markers (= usually an electrophoretic band) • RFLP (Restriction fragment length polymorfism) + Southern • RAPD (Random amplified polymorphism detection) • AFLP (Amplified fragment length polymorphism)
DNA molecular markers (= usually an electrophoretic band) • RFLP (Restriction fragment length polymorfism) + Southern • RAPD (Random amplified polymorphism detection) • AFLP (Amplified fragment length polymorphism)
 Examples of DNA molecular markers Known position in the genome • RFLP (Restriction fragment length polymorfism) + Southern hybridization • SSR (Simple sequence repeats) Unknown sequence and position - randomly amplified DNA segments - sequence and position determined subsequently (only for those of interest - in genetic linkage with a trait) • RAPD (Random amplified polymorphism detection) • AFLP (Amplified fragment length polymorphism)
Examples of DNA molecular markers Known position in the genome • RFLP (Restriction fragment length polymorfism) + Southern hybridization • SSR (Simple sequence repeats) Unknown sequence and position - randomly amplified DNA segments - sequence and position determined subsequently (only for those of interest - in genetic linkage with a trait) • RAPD (Random amplified polymorphism detection) • AFLP (Amplified fragment length polymorphism)
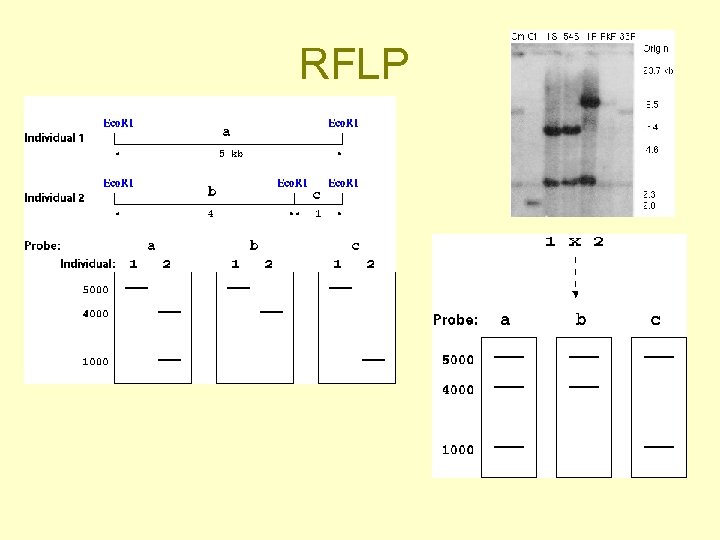 RFLP
RFLP
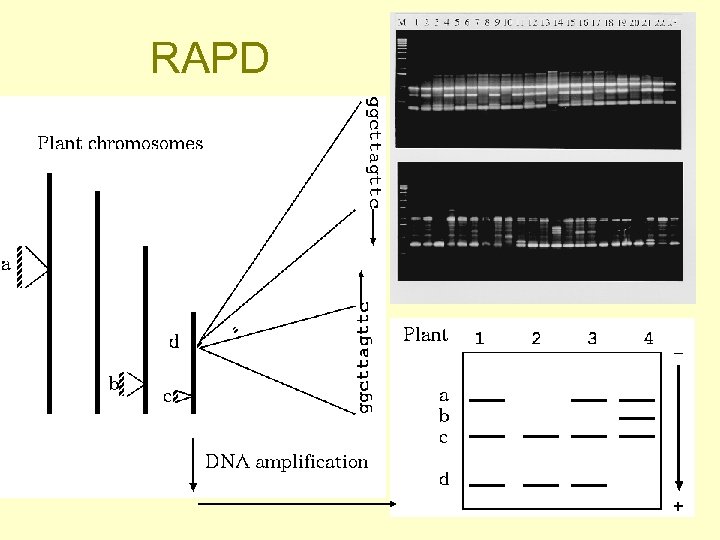 RAPD
RAPD
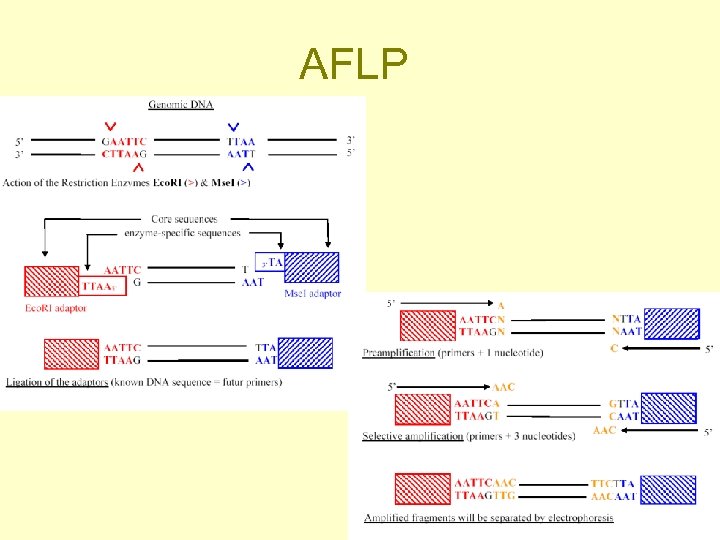 AFLP
AFLP
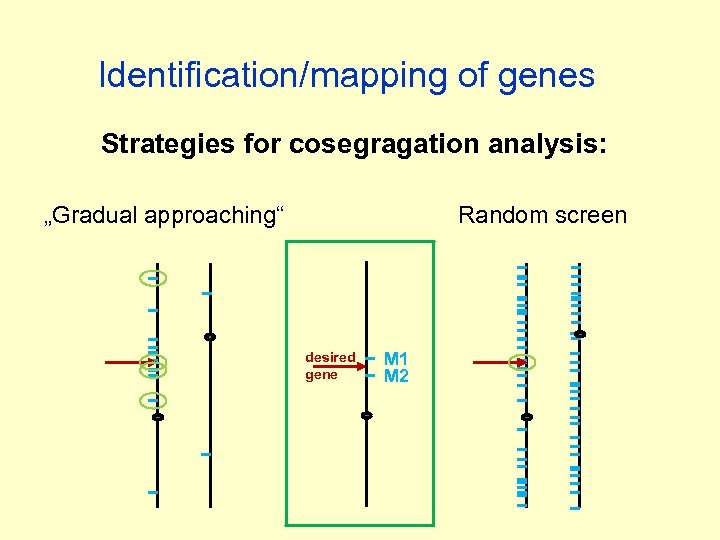 Identification/mapping of genes Strategies for cosegragation analysis: „Gradual approaching“ Random screen desired gene M 1 M 2
Identification/mapping of genes Strategies for cosegragation analysis: „Gradual approaching“ Random screen desired gene M 1 M 2
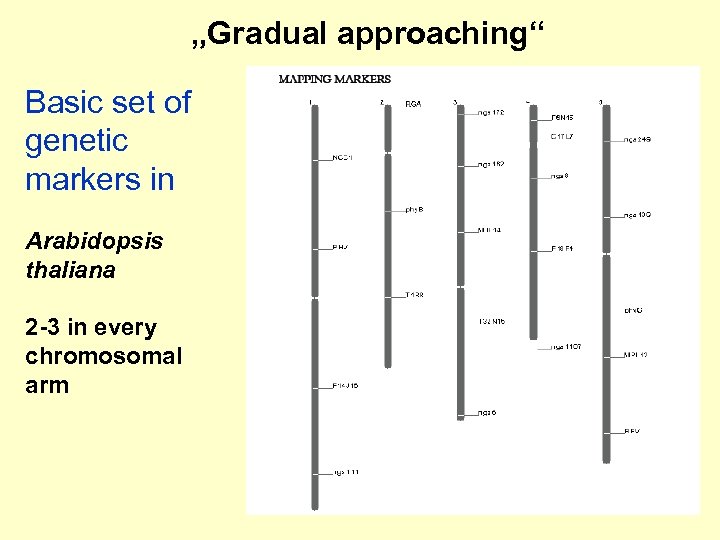 „Gradual approaching“ Basic set of genetic markers in Arabidopsis thaliana 2 -3 in every chromosomal arm
„Gradual approaching“ Basic set of genetic markers in Arabidopsis thaliana 2 -3 in every chromosomal arm
 Molecular markers in Arabidopsis
Molecular markers in Arabidopsis
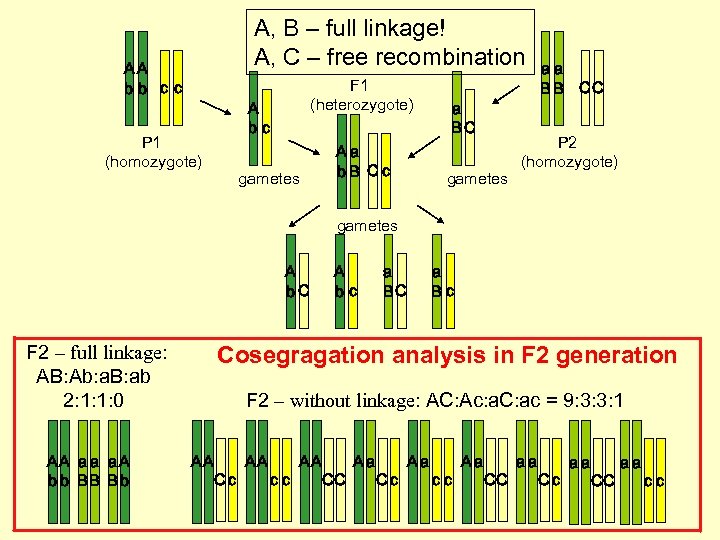 A, B – full linkage! A, C – free recombination AA bb cc F 1 (heterozygote) A bc P 1 (homozygote) gametes Aa b. B Cc a BC gametes aa B B CC P 2 (homozygote) gametes A b. C F 2 – full linkage: AB: Ab: a. B: ab 2: 1: 1: 0 AA a a a A b b BB B b A bc a BC a Bc Cosegragation analysis in F 2 generation F 2 – without linkage: AC: Ac: a. C: ac = 9: 3: 3: 1 AA Cc AA cc AA CC Aa Cc Aa cc Aa CC aa Cc aa CC aa cc
A, B – full linkage! A, C – free recombination AA bb cc F 1 (heterozygote) A bc P 1 (homozygote) gametes Aa b. B Cc a BC gametes aa B B CC P 2 (homozygote) gametes A b. C F 2 – full linkage: AB: Ab: a. B: ab 2: 1: 1: 0 AA a a a A b b BB B b A bc a BC a Bc Cosegragation analysis in F 2 generation F 2 – without linkage: AC: Ac: a. C: ac = 9: 3: 3: 1 AA Cc AA cc AA CC Aa Cc Aa cc Aa CC aa Cc aa CC aa cc
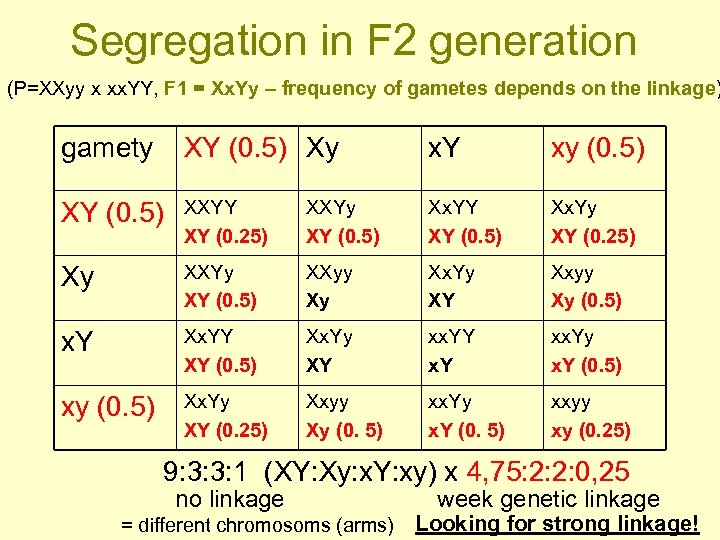 Segregation in F 2 generation (P=XXyy x xx. YY, F 1 = Xx. Yy – frequency of gametes depends on the linkage) gamety XY (0. 5) Xy x. Y xy (0. 5) XY (0. 5) XXYY XY (0. 25) XXYy XY (0. 5) Xx. YY XY (0. 5) Xx. Yy XY (0. 25) Xy XXYy XY (0. 5) XXyy Xy Xx. Yy XY Xxyy Xy (0. 5) x. Y Xx. YY XY (0. 5) Xx. Yy XY xx. YY x. Y xx. Yy x. Y (0. 5) xy (0. 5) Xx. Yy XY (0. 25) Xxyy Xy (0. 5) xx. Yy x. Y (0. 5) xxyy xy (0. 25) 9: 3: 3: 1 (XY: Xy: x. Y: xy) x 4, 75: 2: 2: 0, 25 no linkage = different chromosoms (arms) week genetic linkage Looking for strong linkage!
Segregation in F 2 generation (P=XXyy x xx. YY, F 1 = Xx. Yy – frequency of gametes depends on the linkage) gamety XY (0. 5) Xy x. Y xy (0. 5) XY (0. 5) XXYY XY (0. 25) XXYy XY (0. 5) Xx. YY XY (0. 5) Xx. Yy XY (0. 25) Xy XXYy XY (0. 5) XXyy Xy Xx. Yy XY Xxyy Xy (0. 5) x. Y Xx. YY XY (0. 5) Xx. Yy XY xx. YY x. Y xx. Yy x. Y (0. 5) xy (0. 5) Xx. Yy XY (0. 25) Xxyy Xy (0. 5) xx. Yy x. Y (0. 5) xxyy xy (0. 25) 9: 3: 3: 1 (XY: Xy: x. Y: xy) x 4, 75: 2: 2: 0, 25 no linkage = different chromosoms (arms) week genetic linkage Looking for strong linkage!
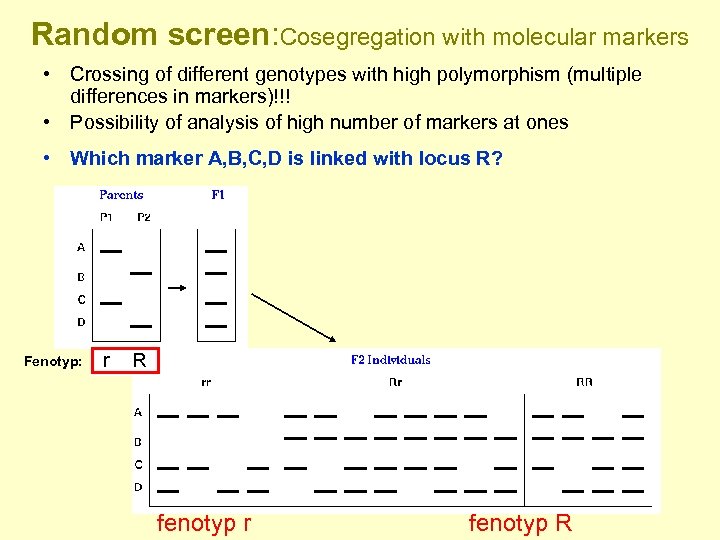 Random screen: Cosegregation with molecular markers • Crossing of different genotypes with high polymorphism (multiple differences in markers)!!! • Possibility of analysis of high number of markers at ones • Which marker A, B, C, D is linked with locus R? Fenotyp: r R fenotyp r fenotyp R
Random screen: Cosegregation with molecular markers • Crossing of different genotypes with high polymorphism (multiple differences in markers)!!! • Possibility of analysis of high number of markers at ones • Which marker A, B, C, D is linked with locus R? Fenotyp: r R fenotyp r fenotyp R
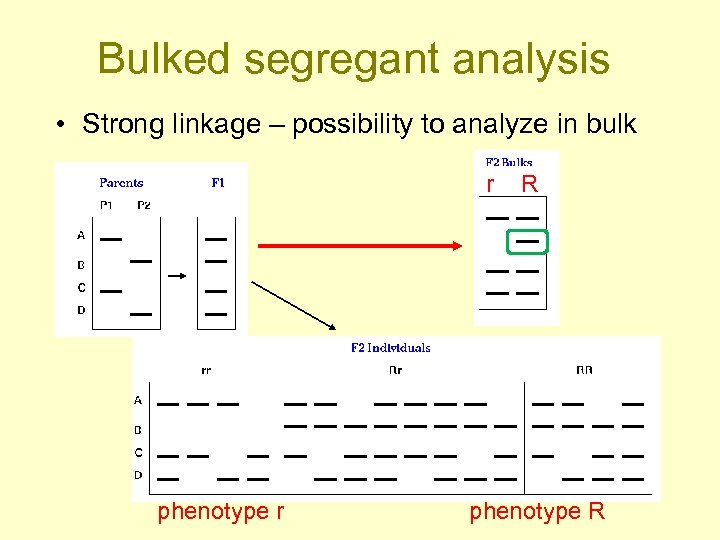 Bulked segregant analysis • Strong linkage – possibility to analyze in bulk r phenotype r R phenotype R
Bulked segregant analysis • Strong linkage – possibility to analyze in bulk r phenotype r R phenotype R
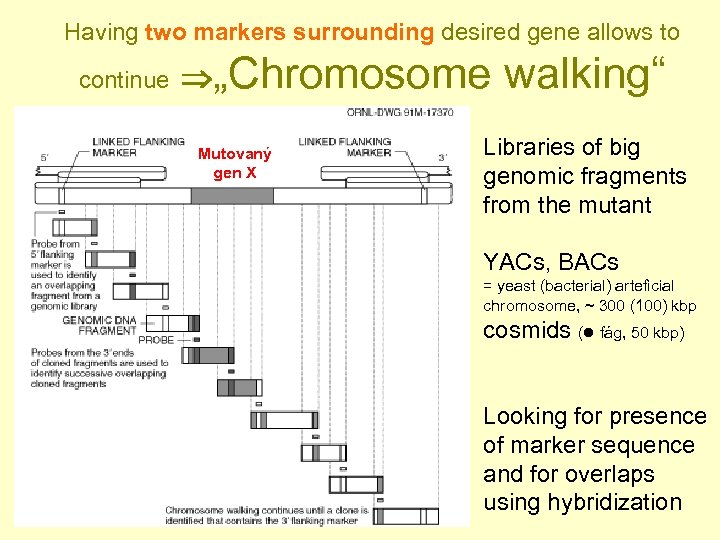 Having two markers surrounding desired gene allows to continue „Chromosome Mutovaný gen X walking“ Libraries of big genomic fragments from the mutant YACs, BACs = yeast (bacterial) arteficial chromosome, ~ 300 (100) kbp cosmids ( fág, 50 kbp) Looking for presence of marker sequence and for overlaps using hybridization
Having two markers surrounding desired gene allows to continue „Chromosome Mutovaný gen X walking“ Libraries of big genomic fragments from the mutant YACs, BACs = yeast (bacterial) arteficial chromosome, ~ 300 (100) kbp cosmids ( fág, 50 kbp) Looking for presence of marker sequence and for overlaps using hybridization
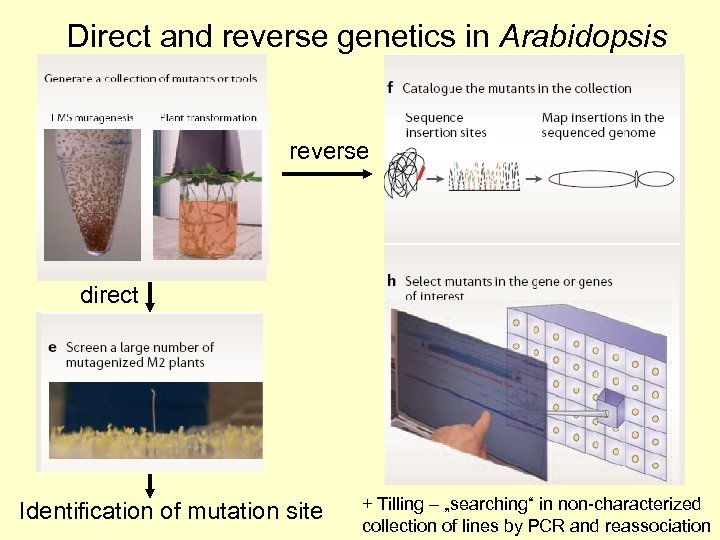 Direct and reverse genetics in Arabidopsis reverse direct Identification of mutation site + Tilling – „searching“ in non-characterized collection of lines by PCR and reassociation
Direct and reverse genetics in Arabidopsis reverse direct Identification of mutation site + Tilling – „searching“ in non-characterized collection of lines by PCR and reassociation
 Marker assisted selection (MAS) Molecular marker in strong genetic linkage with certain trait can be used for screening of hybrids instead of the phenotypic characterization Advantages: • Not influenced by environmental conditions • Screens of seedlings • Often simple and cheaper • Possibility to distinguish between homo- and heterozygots (using certain markers)
Marker assisted selection (MAS) Molecular marker in strong genetic linkage with certain trait can be used for screening of hybrids instead of the phenotypic characterization Advantages: • Not influenced by environmental conditions • Screens of seedlings • Often simple and cheaper • Possibility to distinguish between homo- and heterozygots (using certain markers)
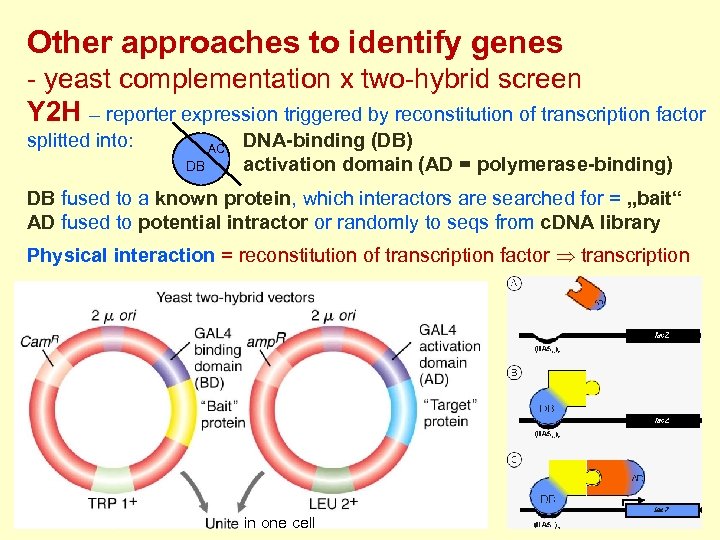 Other approaches to identify genes - yeast complementation x two-hybrid screen Y 2 H – reporter expression triggered by reconstitution of transcription factor splitted into: AC DB DNA-binding (DB) activation domain (AD = polymerase-binding) DB fused to a known protein, which interactors are searched for = „bait“ AD fused to potential intractor or randomly to seqs from c. DNA library Physical interaction = reconstitution of transcription factor transcription in one cell
Other approaches to identify genes - yeast complementation x two-hybrid screen Y 2 H – reporter expression triggered by reconstitution of transcription factor splitted into: AC DB DNA-binding (DB) activation domain (AD = polymerase-binding) DB fused to a known protein, which interactors are searched for = „bait“ AD fused to potential intractor or randomly to seqs from c. DNA library Physical interaction = reconstitution of transcription factor transcription in one cell
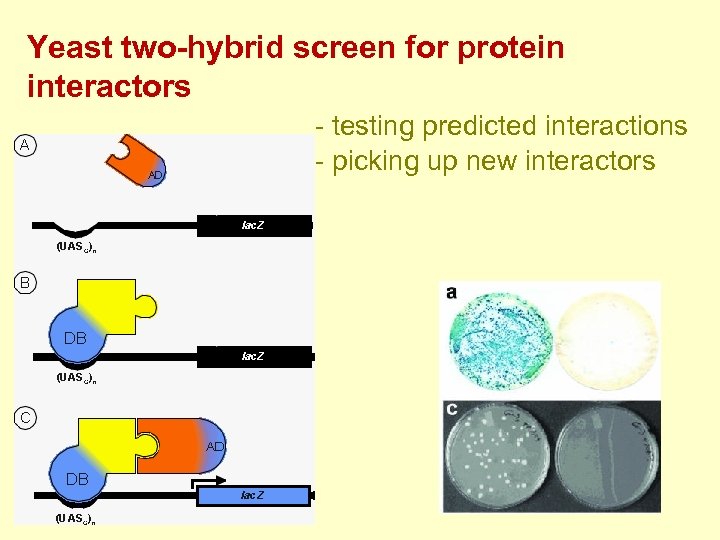 Yeast two-hybrid screen for protein interactors - testing predicted interactions - picking up new interactors
Yeast two-hybrid screen for protein interactors - testing predicted interactions - picking up new interactors


