21cf50cc0b9ea1a767023620a39ecd89.ppt
- Количество слайдов: 73
 Genetics for Epidemiologists National Human Genome Research Institute National Institutes of Health U. S. Department of Health and Human Services Lecture 2: Measurement of Genetic Exposures U. S. Department of Health and Human Services National Institutes of Health National Human Genome Research Institute Teri A. Manolio, M. D. , Ph. D. Director, Office of Population Genomics and Senior Advisor to the Director, NHGRI, for Population Genomics
Genetics for Epidemiologists National Human Genome Research Institute National Institutes of Health U. S. Department of Health and Human Services Lecture 2: Measurement of Genetic Exposures U. S. Department of Health and Human Services National Institutes of Health National Human Genome Research Institute Teri A. Manolio, M. D. , Ph. D. Director, Office of Population Genomics and Senior Advisor to the Director, NHGRI, for Population Genomics
 Topics to be Covered • Measuring genetic variation – Blood group markers – Restriction-fragment length polymorphisms – Variable number of tandem repeats (VNTRs, minisatellites and microsatellites) – Single nucleotide polymorphisms (SNPs) • Linkage disequilibrium (LD) • Familial resemblance and family history
Topics to be Covered • Measuring genetic variation – Blood group markers – Restriction-fragment length polymorphisms – Variable number of tandem repeats (VNTRs, minisatellites and microsatellites) – Single nucleotide polymorphisms (SNPs) • Linkage disequilibrium (LD) • Familial resemblance and family history
 Larson, G. The Complete Far Side. 2003.
Larson, G. The Complete Far Side. 2003.
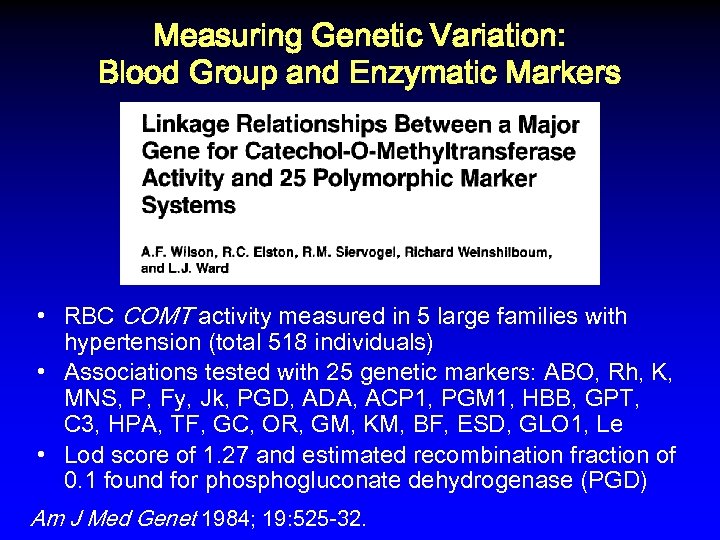 Measuring Genetic Variation: Blood Group and Enzymatic Markers • RBC COMT activity measured in 5 large families with hypertension (total 518 individuals) • Associations tested with 25 genetic markers: ABO, Rh, K, MNS, P, Fy, Jk, PGD, ADA, ACP 1, PGM 1, HBB, GPT, C 3, HPA, TF, GC, OR, GM, KM, BF, ESD, GLO 1, Le • Lod score of 1. 27 and estimated recombination fraction of 0. 1 found for phosphogluconate dehydrogenase (PGD) Am J Med Genet 1984; 19: 525 -32.
Measuring Genetic Variation: Blood Group and Enzymatic Markers • RBC COMT activity measured in 5 large families with hypertension (total 518 individuals) • Associations tested with 25 genetic markers: ABO, Rh, K, MNS, P, Fy, Jk, PGD, ADA, ACP 1, PGM 1, HBB, GPT, C 3, HPA, TF, GC, OR, GM, KM, BF, ESD, GLO 1, Le • Lod score of 1. 27 and estimated recombination fraction of 0. 1 found for phosphogluconate dehydrogenase (PGD) Am J Med Genet 1984; 19: 525 -32.
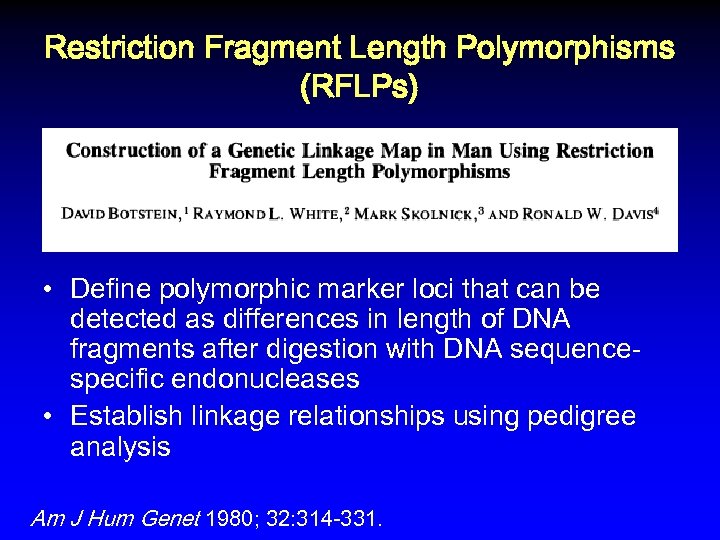 Restriction Fragment Length Polymorphisms (RFLPs) • Define polymorphic marker loci that can be detected as differences in length of DNA fragments after digestion with DNA sequencespecific endonucleases • Establish linkage relationships using pedigree analysis Am J Hum Genet 1980; 32: 314 -331.
Restriction Fragment Length Polymorphisms (RFLPs) • Define polymorphic marker loci that can be detected as differences in length of DNA fragments after digestion with DNA sequencespecific endonucleases • Establish linkage relationships using pedigree analysis Am J Hum Genet 1980; 32: 314 -331.
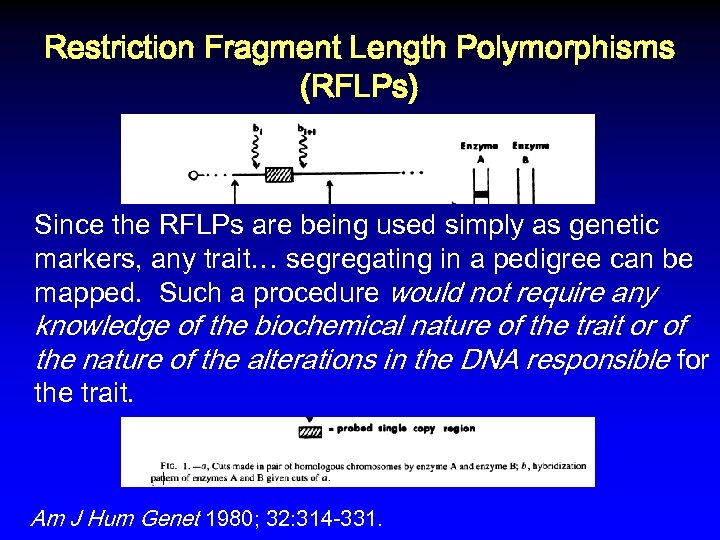 Restriction Fragment Length Polymorphisms (RFLPs) Since the RFLPs are being used simply as genetic markers, any trait… segregating in a pedigree can be mapped. Such a procedure would not require any knowledge of the biochemical nature of the trait or of the nature of the alterations in the DNA responsible for the trait. Am J Hum Genet 1980; 32: 314 -331.
Restriction Fragment Length Polymorphisms (RFLPs) Since the RFLPs are being used simply as genetic markers, any trait… segregating in a pedigree can be mapped. Such a procedure would not require any knowledge of the biochemical nature of the trait or of the nature of the alterations in the DNA responsible for the trait. Am J Hum Genet 1980; 32: 314 -331.
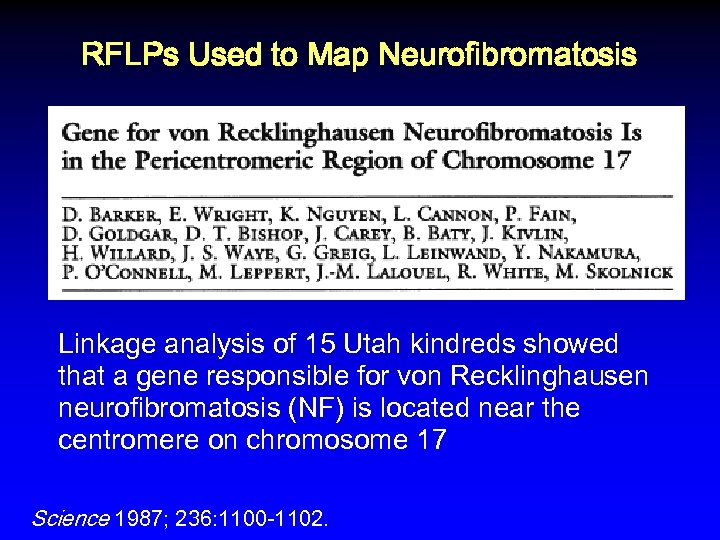 RFLPs Used to Map Neurofibromatosis Linkage analysis of 15 Utah kindreds showed that a gene responsible for von Recklinghausen neurofibromatosis (NF) is located near the centromere on chromosome 17 Science 1987; 236: 1100 -1102.
RFLPs Used to Map Neurofibromatosis Linkage analysis of 15 Utah kindreds showed that a gene responsible for von Recklinghausen neurofibromatosis (NF) is located near the centromere on chromosome 17 Science 1987; 236: 1100 -1102.
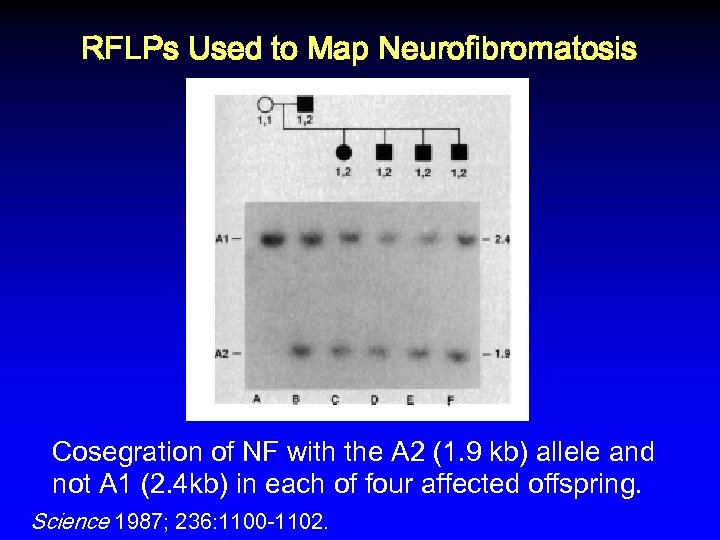 RFLPs Used to Map Neurofibromatosis Cosegration of NF with the A 2 (1. 9 kb) allele and not A 1 (2. 4 kb) in each of four affected offspring. Science 1987; 236: 1100 -1102.
RFLPs Used to Map Neurofibromatosis Cosegration of NF with the A 2 (1. 9 kb) allele and not A 1 (2. 4 kb) in each of four affected offspring. Science 1987; 236: 1100 -1102.
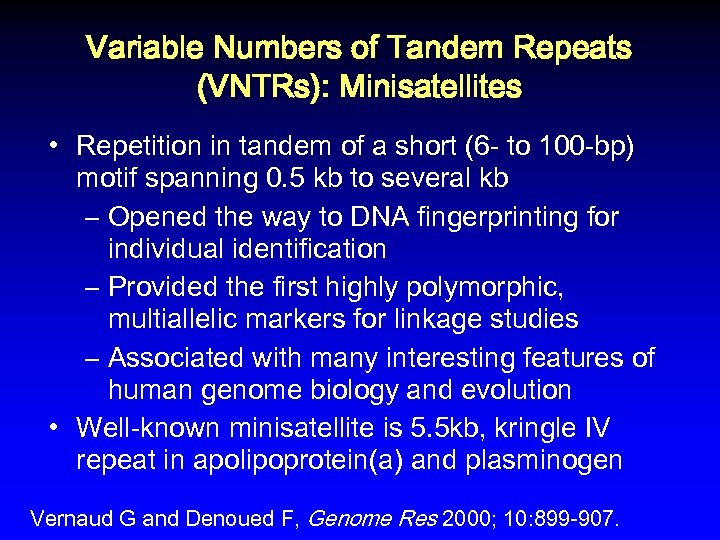 Variable Numbers of Tandem Repeats (VNTRs): Minisatellites • Repetition in tandem of a short (6 - to 100 -bp) motif spanning 0. 5 kb to several kb – Opened the way to DNA fingerprinting for individual identification – Provided the first highly polymorphic, multiallelic markers for linkage studies – Associated with many interesting features of human genome biology and evolution • Well-known minisatellite is 5. 5 kb, kringle IV repeat in apolipoprotein(a) and plasminogen Vernaud G and Denoued F, Genome Res 2000; 10: 899 -907.
Variable Numbers of Tandem Repeats (VNTRs): Minisatellites • Repetition in tandem of a short (6 - to 100 -bp) motif spanning 0. 5 kb to several kb – Opened the way to DNA fingerprinting for individual identification – Provided the first highly polymorphic, multiallelic markers for linkage studies – Associated with many interesting features of human genome biology and evolution • Well-known minisatellite is 5. 5 kb, kringle IV repeat in apolipoprotein(a) and plasminogen Vernaud G and Denoued F, Genome Res 2000; 10: 899 -907.
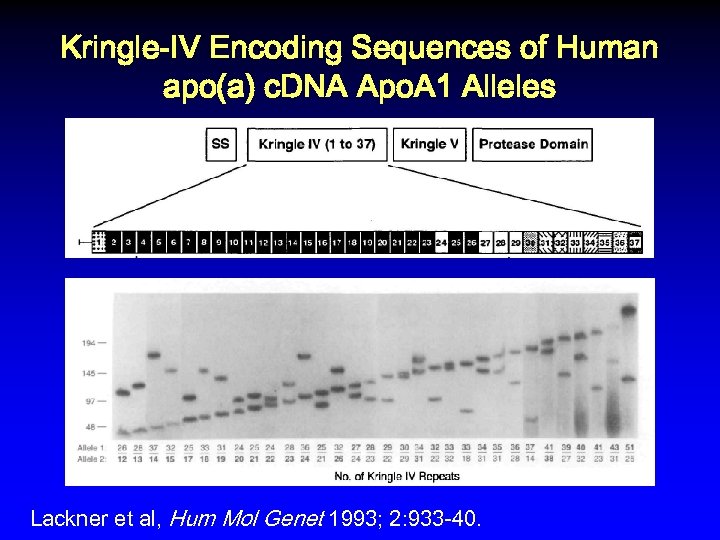 Kringle-IV Encoding Sequences of Human apo(a) c. DNA Apo. A 1 Alleles Lackner et al, Hum Mol Genet 1993; 2: 933 -40.
Kringle-IV Encoding Sequences of Human apo(a) c. DNA Apo. A 1 Alleles Lackner et al, Hum Mol Genet 1993; 2: 933 -40.
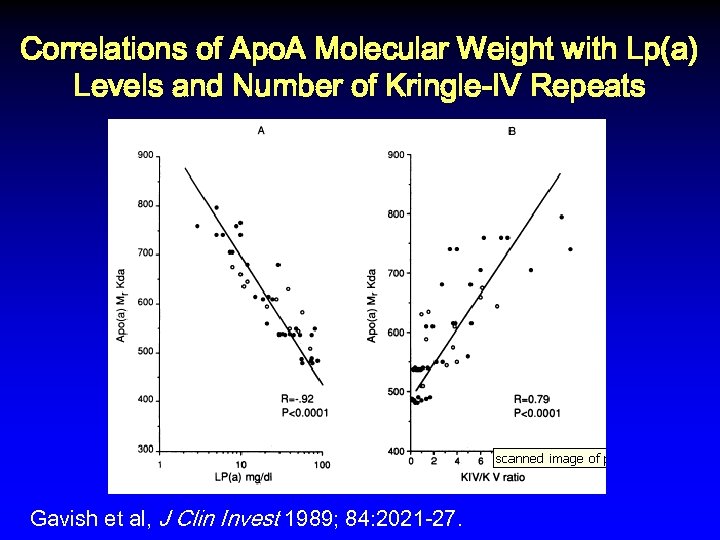 Correlations of Apo. A Molecular Weight with Lp(a) Levels and Number of Kringle-IV Repeats Gavish et al, J Clin Invest 1989; 84: 2021 -27.
Correlations of Apo. A Molecular Weight with Lp(a) Levels and Number of Kringle-IV Repeats Gavish et al, J Clin Invest 1989; 84: 2021 -27.
 Simple Sequence Repeats (also “VNTRs”): Microsatellites Repetition in tandem of a short (2 - to 6 -bp) motif from 5 -5, 000 times • Most are di-, tri-, and tetra-nucleotide repeats repeated 20 -50 times • Most are highly polymorphic making them enormously useful for mapping and linkage • Marshfield and similar maps placed ~400 microsatellites across genome, provided primers for analysis • Could be highly automated: NHLBI and CIDR large-scale genotyping services
Simple Sequence Repeats (also “VNTRs”): Microsatellites Repetition in tandem of a short (2 - to 6 -bp) motif from 5 -5, 000 times • Most are di-, tri-, and tetra-nucleotide repeats repeated 20 -50 times • Most are highly polymorphic making them enormously useful for mapping and linkage • Marshfield and similar maps placed ~400 microsatellites across genome, provided primers for analysis • Could be highly automated: NHLBI and CIDR large-scale genotyping services
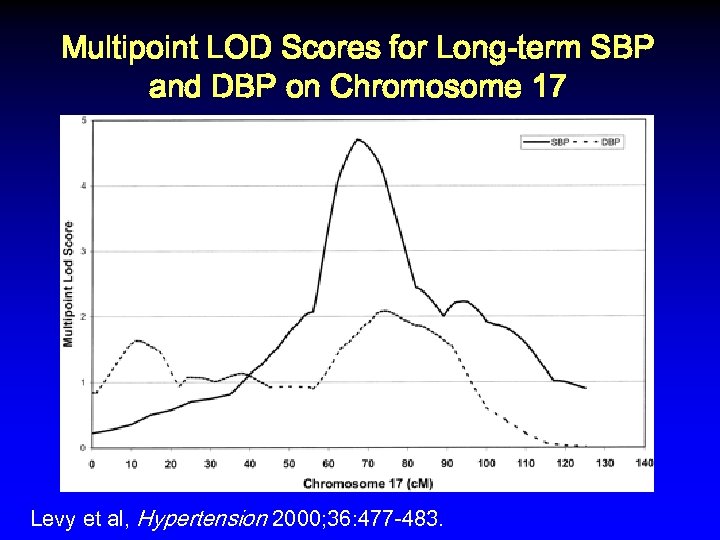 Multipoint LOD Scores for Long-term SBP and DBP on Chromosome 17 Levy et al, Hypertension 2000; 36: 477 -483.
Multipoint LOD Scores for Long-term SBP and DBP on Chromosome 17 Levy et al, Hypertension 2000; 36: 477 -483.
 Larson, G. The Complete Far Side. 2003.
Larson, G. The Complete Far Side. 2003.
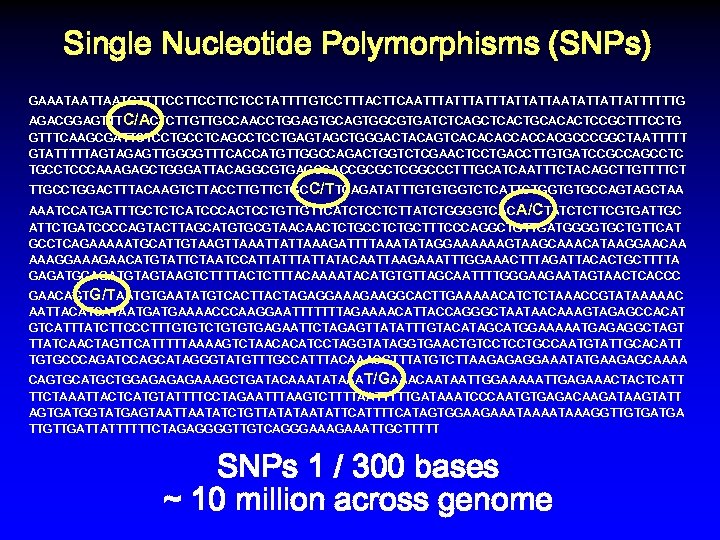 Single Nucleotide Polymorphisms (SNPs) GAAATAATGTTTTCCTTCTCCTATTTTGTCCTTTACTTCAATTTATTTATTATTAATATTATTATTTTTTG AGACGGAGTTTC/ACTCTTGTTGCCAACCTGGAGTGCAGTGGCGTGATCTCAGCTCACTGCACACTCCGCTTTCCTG GTTTCAAGCGATTCTCCTGCCTCAGCCTCCTGAGTAGCTGGGACTACAGTCACACACCACCACGCCCGGCTAATTTTT GTATTTTTAGTAGAGTTGGGGTTTCACCATGTTGGCCAGACTGGTCTCGAACTCCTGACCTTGTGATCCGCCAGCCTC TGCCTCCCAAAGAGCTGGGATTACAGGCGTGAGCCACCGCGCTCGGCCCTTTGCATCAATTTCTACAGCTTGTTTTCT TTGCCTGGACTTTACAAGTCTTACCTTGTTCTGC C/TTCAGATATTTGTGTGGTCTCATTCTGGTGTGCCAGTAGCTAA AAATCCATGATTTGCTCTCATCCCACTCCTGTTGTTCATCTCCTCTTATCTGGGGTCAC A/CTATCTCTTCGTGATTGC ATTCTGATCCCCAGTACTTAGCATGTGCGTAACAACTCTGCTTTCCCAGGCTGTTGATGGGGTGCTGTTCAT GCCTCAGAAAAATGCATTGTAAGTTAAATTATTAAAGATTTTAAATATAGGAAAAAAGTAAGCAAACATAAGGAACAA AAAGGAAAGAACATGTATTCTAATCCATTATTATACAATTAAGAAATTTGGAAACTTTAGATTACACTGCTTTTA GAGATGTAGTAAGTCTTTTACTCTTTACAAAATACATGTGTTAGCAATTTTGGGAAGAATAGTAACTCACCC GAACAGTG/TAATGTGAATATGTCACTTACTAGAGGAAAGAAGGCACTTGAAAAACATCTCTAAACCGTATAAAAAC AATTACATCATAATGATGAAAACCCAAGGAATTTTTTTAGAAAACATTACCAGGGCTAATAACAAAGTAGAGCCACAT GTCATTTATCTTCCCTTTGTGTCTGTGTGAGAATTCTAGAGTTATATTTGTACATAGCATGGAAAAATGAGAGGCTAGT TTATCAACTAGTTCATTTTTAAAAGTCTAACACATCCTAGGTATAGGTGAACTGTCCTCCTGCCAATGTATTGCACATT TGTGCCCAGATCCAGCATAGGGTATGTTTGCCATTTACAAACGTTTATGTCTTAAGAGAGGAAATATGAAGAGCAAAA CAGTGCATGCTGGAGAAAGCTGATACAAATATAAAT/GAAACAATAATTGGAAAAATTGAGAAACTACTCATT TTCTAAATTACTCATGTATTTTCCTAGAATTTAAGTCTTTTAATTTTTGATAAATCCCAATGTGAGACAAGATAAGTATT AGTGATGGTATGAGTAATATCTGTTATATAATATTCATTTTCATAGTGGAAGAAATAAAGGTTGTGATGA TTGTTGATTATTTTTTCTAGAGGGGTTGTCAGGGAAATTGCTTTTT SNPs 1 / 300 bases ~ 10 million across genome
Single Nucleotide Polymorphisms (SNPs) GAAATAATGTTTTCCTTCTCCTATTTTGTCCTTTACTTCAATTTATTTATTATTAATATTATTATTTTTTG AGACGGAGTTTC/ACTCTTGTTGCCAACCTGGAGTGCAGTGGCGTGATCTCAGCTCACTGCACACTCCGCTTTCCTG GTTTCAAGCGATTCTCCTGCCTCAGCCTCCTGAGTAGCTGGGACTACAGTCACACACCACCACGCCCGGCTAATTTTT GTATTTTTAGTAGAGTTGGGGTTTCACCATGTTGGCCAGACTGGTCTCGAACTCCTGACCTTGTGATCCGCCAGCCTC TGCCTCCCAAAGAGCTGGGATTACAGGCGTGAGCCACCGCGCTCGGCCCTTTGCATCAATTTCTACAGCTTGTTTTCT TTGCCTGGACTTTACAAGTCTTACCTTGTTCTGC C/TTCAGATATTTGTGTGGTCTCATTCTGGTGTGCCAGTAGCTAA AAATCCATGATTTGCTCTCATCCCACTCCTGTTGTTCATCTCCTCTTATCTGGGGTCAC A/CTATCTCTTCGTGATTGC ATTCTGATCCCCAGTACTTAGCATGTGCGTAACAACTCTGCTTTCCCAGGCTGTTGATGGGGTGCTGTTCAT GCCTCAGAAAAATGCATTGTAAGTTAAATTATTAAAGATTTTAAATATAGGAAAAAAGTAAGCAAACATAAGGAACAA AAAGGAAAGAACATGTATTCTAATCCATTATTATACAATTAAGAAATTTGGAAACTTTAGATTACACTGCTTTTA GAGATGTAGTAAGTCTTTTACTCTTTACAAAATACATGTGTTAGCAATTTTGGGAAGAATAGTAACTCACCC GAACAGTG/TAATGTGAATATGTCACTTACTAGAGGAAAGAAGGCACTTGAAAAACATCTCTAAACCGTATAAAAAC AATTACATCATAATGATGAAAACCCAAGGAATTTTTTTAGAAAACATTACCAGGGCTAATAACAAAGTAGAGCCACAT GTCATTTATCTTCCCTTTGTGTCTGTGTGAGAATTCTAGAGTTATATTTGTACATAGCATGGAAAAATGAGAGGCTAGT TTATCAACTAGTTCATTTTTAAAAGTCTAACACATCCTAGGTATAGGTGAACTGTCCTCCTGCCAATGTATTGCACATT TGTGCCCAGATCCAGCATAGGGTATGTTTGCCATTTACAAACGTTTATGTCTTAAGAGAGGAAATATGAAGAGCAAAA CAGTGCATGCTGGAGAAAGCTGATACAAATATAAAT/GAAACAATAATTGGAAAAATTGAGAAACTACTCATT TTCTAAATTACTCATGTATTTTCCTAGAATTTAAGTCTTTTAATTTTTGATAAATCCCAATGTGAGACAAGATAAGTATT AGTGATGGTATGAGTAATATCTGTTATATAATATTCATTTTCATAGTGGAAGAAATAAAGGTTGTGATGA TTGTTGATTATTTTTTCTAGAGGGGTTGTCAGGGAAATTGCTTTTT SNPs 1 / 300 bases ~ 10 million across genome
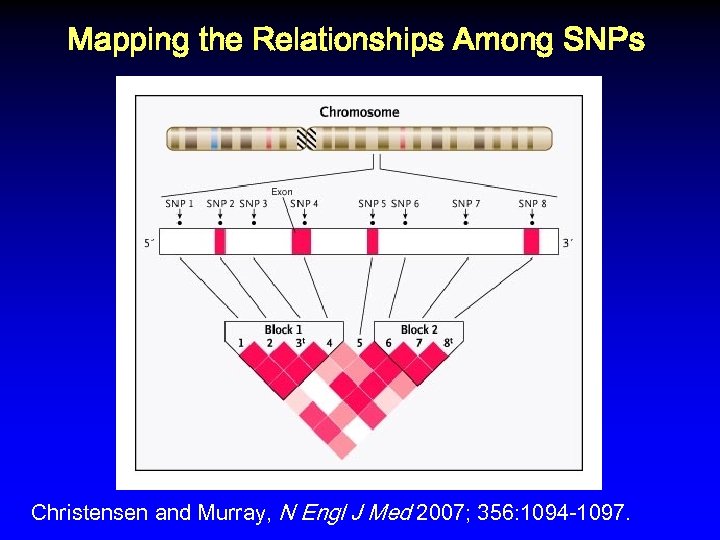 Mapping the Relationships Among SNPs Christensen and Murray, N Engl J Med 2007; 356: 1094 -1097.
Mapping the Relationships Among SNPs Christensen and Murray, N Engl J Med 2007; 356: 1094 -1097.
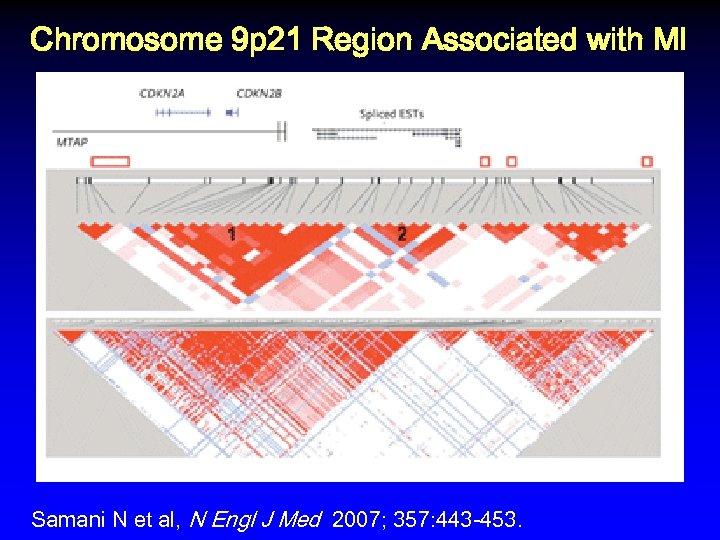 Chromosome 9 p 21 Region Associated with MI Samani N et al, N Engl J Med 2007; 357: 443 -453.
Chromosome 9 p 21 Region Associated with MI Samani N et al, N Engl J Med 2007; 357: 443 -453.
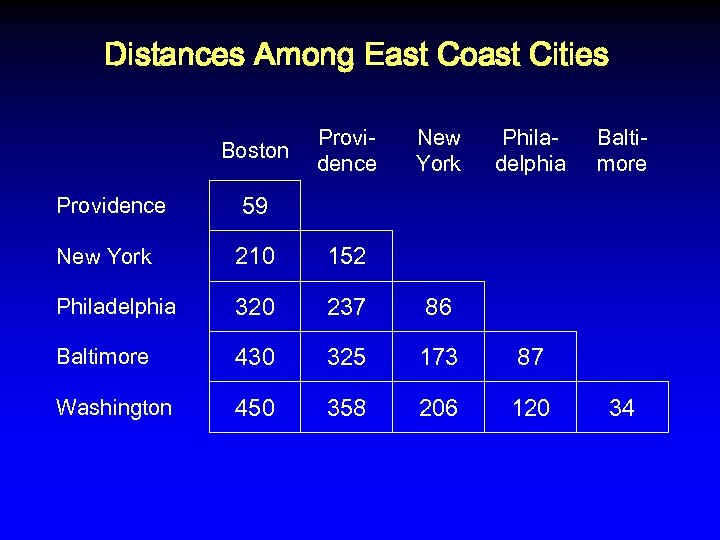 Distances Among East Coast Cities Boston Providence New York Philadelphia Baltimore 59 New York 210 152 Philadelphia 320 237 86 Baltimore 430 325 173 87 Washington 450 358 206 120 34
Distances Among East Coast Cities Boston Providence New York Philadelphia Baltimore 59 New York 210 152 Philadelphia 320 237 86 Baltimore 430 325 173 87 Washington 450 358 206 120 34
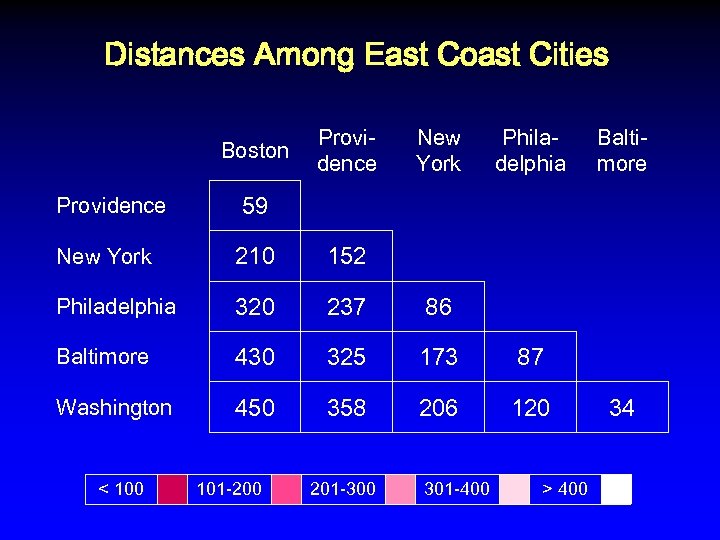 Distances Among East Coast Cities Boston Providence New York Philadelphia Baltimore 59 New York 210 152 Philadelphia 320 237 86 Baltimore 430 325 173 87 Washington 450 358 206 120 < 100 101 -200 201 -300 301 -400 > 400 34
Distances Among East Coast Cities Boston Providence New York Philadelphia Baltimore 59 New York 210 152 Philadelphia 320 237 86 Baltimore 430 325 173 87 Washington 450 358 206 120 < 100 101 -200 201 -300 301 -400 > 400 34
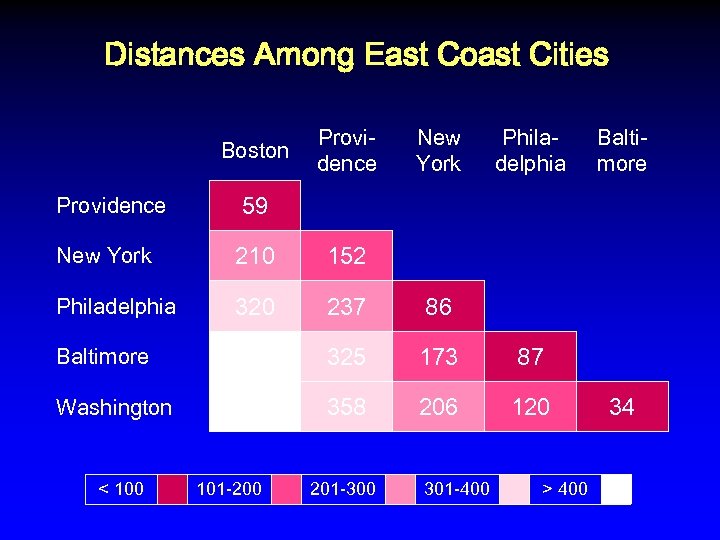 Distances Among East Coast Cities Boston Providence New York Philadelphia Baltimore 59 New York 210 152 Philadelphia 320 237 86 Baltimore 430 325 173 87 Washington 450 358 206 120 < 100 101 -200 201 -300 301 -400 > 400 34
Distances Among East Coast Cities Boston Providence New York Philadelphia Baltimore 59 New York 210 152 Philadelphia 320 237 86 Baltimore 430 325 173 87 Washington 450 358 206 120 < 100 101 -200 201 -300 301 -400 > 400 34
 Distances Among East Coast Cities n o ost B irov e P nc de w Ne rk Yo hila ia P ph l de alti e B r mo sh. Wa ton ing
Distances Among East Coast Cities n o ost B irov e P nc de w Ne rk Yo hila ia P ph l de alti e B r mo sh. Wa ton ing
 Distances Among East Coast Cities Boston Providence New York Philadelphia Baltimore Washington
Distances Among East Coast Cities Boston Providence New York Philadelphia Baltimore Washington
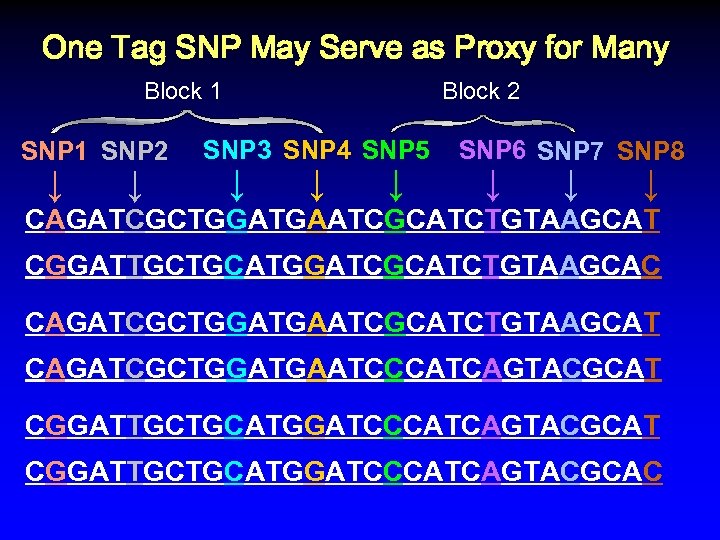 One Tag SNP May Serve as Proxy for Many Block 1 SNP 2 ↓ ↓ Block 2 SNP 3 SNP 4 SNP 5 ↓ ↓ ↓ SNP 6 SNP 7 SNP 8 ↓ ↓ ↓ CAGATCGCTGGATGAATCGCATCTGTAAGCAT CGGATTGCTGCATGGATCGCATCTGTAAGCAC CAGATCGCTGGATGAATCGCATCTGTAAGCAT CAGATCGCTGGATGAATCCCATCAGTACGCAT CGGATTGCTGCATGGATCCCATCAGTACGCAC
One Tag SNP May Serve as Proxy for Many Block 1 SNP 2 ↓ ↓ Block 2 SNP 3 SNP 4 SNP 5 ↓ ↓ ↓ SNP 6 SNP 7 SNP 8 ↓ ↓ ↓ CAGATCGCTGGATGAATCGCATCTGTAAGCAT CGGATTGCTGCATGGATCGCATCTGTAAGCAC CAGATCGCTGGATGAATCGCATCTGTAAGCAT CAGATCGCTGGATGAATCCCATCAGTACGCAT CGGATTGCTGCATGGATCCCATCAGTACGCAC
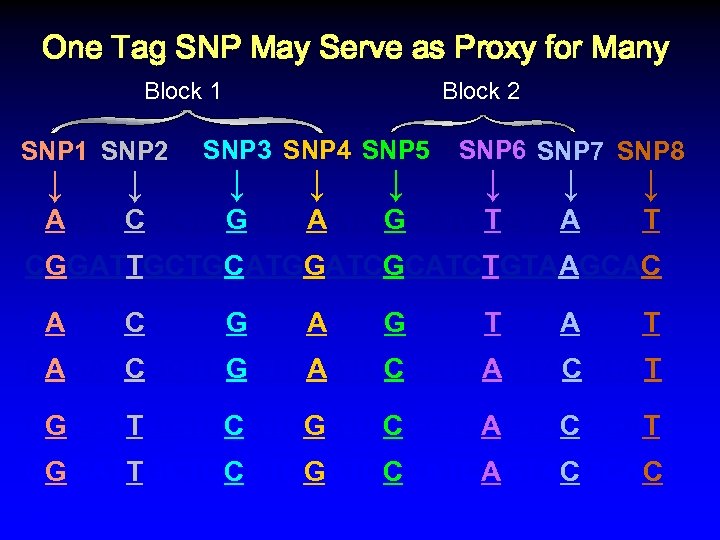 One Tag SNP May Serve as Proxy for Many Block 1 SNP 2 ↓ ↓ Block 2 SNP 3 SNP 4 SNP 5 ↓ ↓ ↓ SNP 6 SNP 7 SNP 8 ↓ ↓ ↓ CAGATCGCTGGATGAATCGCATCTGTAAGCAT CGGATTGCTGCATGGATCGCATCTGTAAGCAC CAGATCGCTGGATGAATCGCATCTGTAAGCAT CAGATCGCTGGATGAATCCCATCAGTACGCAT CGGATTGCTGCATGGATCCCATCAGTACGCAC %
One Tag SNP May Serve as Proxy for Many Block 1 SNP 2 ↓ ↓ Block 2 SNP 3 SNP 4 SNP 5 ↓ ↓ ↓ SNP 6 SNP 7 SNP 8 ↓ ↓ ↓ CAGATCGCTGGATGAATCGCATCTGTAAGCAT CGGATTGCTGCATGGATCGCATCTGTAAGCAC CAGATCGCTGGATGAATCGCATCTGTAAGCAT CAGATCGCTGGATGAATCCCATCAGTACGCAT CGGATTGCTGCATGGATCCCATCAGTACGCAC %
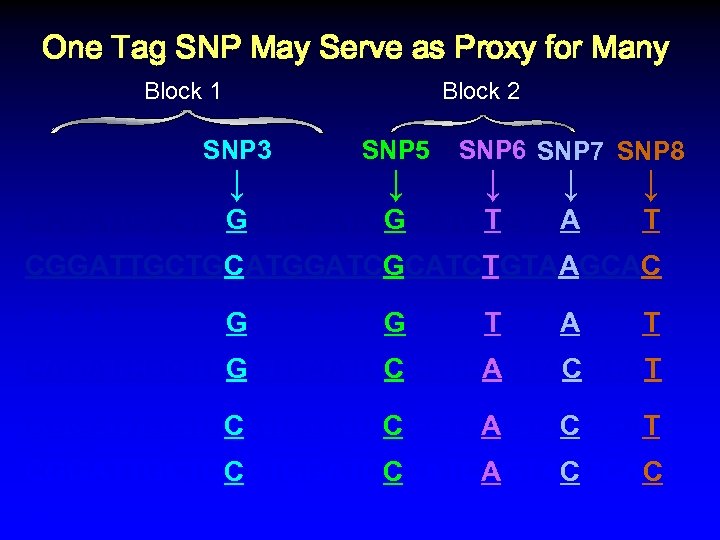 One Tag SNP May Serve as Proxy for Many Block 1 Block 2 SNP 3 ↓ SNP 5 ↓ SNP 6 SNP 7 SNP 8 ↓ ↓ ↓ CAGATCGCTGGATGAATCGCATCTGTAAGCAT CGGATTGCTGCATGGATCGCATCTGTAAGCAC CAGATCGCTGGATGAATCGCATCTGTAAGCAT CAGATCGCTGGATGAATCCCATCAGTACGCAT CGGATTGCTGCATGGATCCCATCAGTACGCAC %
One Tag SNP May Serve as Proxy for Many Block 1 Block 2 SNP 3 ↓ SNP 5 ↓ SNP 6 SNP 7 SNP 8 ↓ ↓ ↓ CAGATCGCTGGATGAATCGCATCTGTAAGCAT CGGATTGCTGCATGGATCGCATCTGTAAGCAC CAGATCGCTGGATGAATCGCATCTGTAAGCAT CAGATCGCTGGATGAATCCCATCAGTACGCAT CGGATTGCTGCATGGATCCCATCAGTACGCAC %
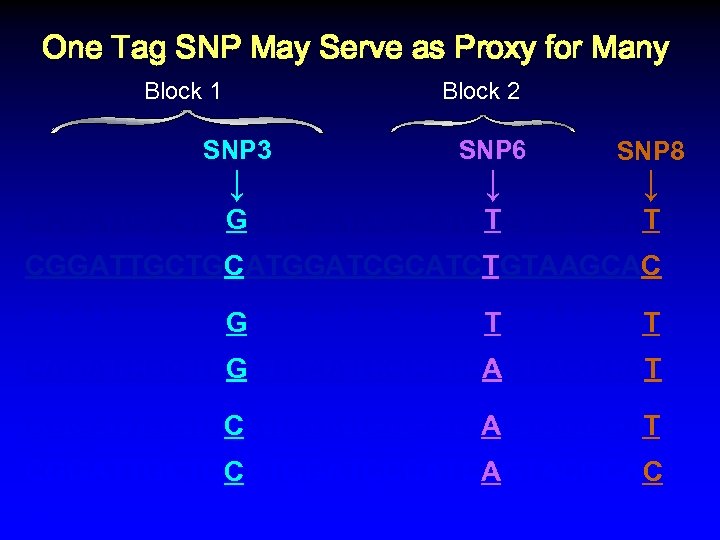 One Tag SNP May Serve as Proxy for Many Block 1 Block 2 SNP 3 ↓ SNP 6 ↓ SNP 8 ↓ CAGATCGCTGGATGAATCGCATCTGTAAGCAT CGGATTGCTGCATGGATCGCATCTGTAAGCAC CAGATCGCTGGATGAATCGCATCTGTAAGCAT CAGATCGCTGGATGAATCCCATCAGTACGCAT CGGATTGCTGCATGGATCCCATCAGTACGCAC %
One Tag SNP May Serve as Proxy for Many Block 1 Block 2 SNP 3 ↓ SNP 6 ↓ SNP 8 ↓ CAGATCGCTGGATGAATCGCATCTGTAAGCAT CGGATTGCTGCATGGATCGCATCTGTAAGCAC CAGATCGCTGGATGAATCGCATCTGTAAGCAT CAGATCGCTGGATGAATCCCATCAGTACGCAT CGGATTGCTGCATGGATCCCATCAGTACGCAC %
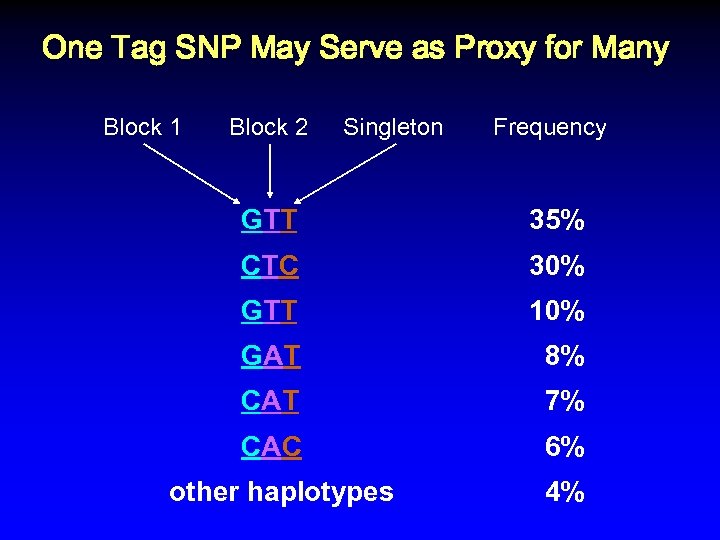 One Tag SNP May Serve as Proxy for Many Block 1 Block 2 GTT Singleton Frequency 35% CTC GTT 10% GAT 8% CAT 7% CAC 30% 6% other haplotypes 4%
One Tag SNP May Serve as Proxy for Many Block 1 Block 2 GTT Singleton Frequency 35% CTC GTT 10% GAT 8% CAT 7% CAC 30% 6% other haplotypes 4%
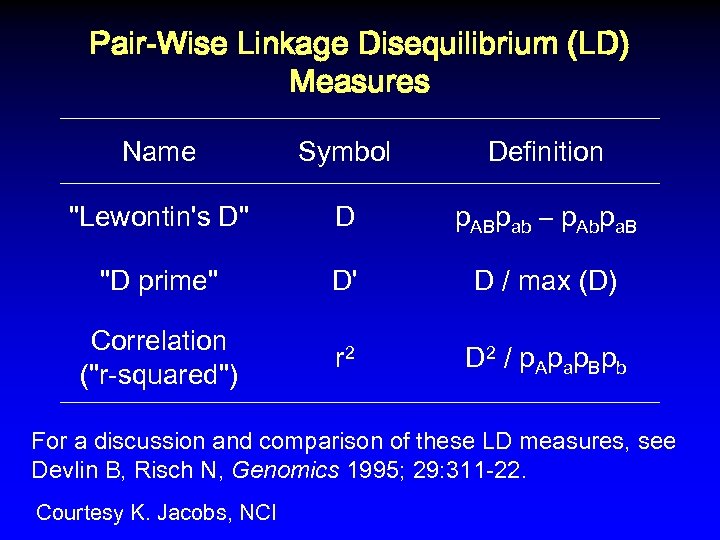 Pair-Wise Linkage Disequilibrium (LD) Measures Name Symbol Definition "Lewontin's D" D p. ABpab – p. Abpa. B "D prime" D' D / max (D) Correlation ("r-squared") r 2 D 2 / p A p ap B p b For a discussion and comparison of these LD measures, see Devlin B, Risch N, Genomics 1995; 29: 311 -22. Courtesy K. Jacobs, NCI
Pair-Wise Linkage Disequilibrium (LD) Measures Name Symbol Definition "Lewontin's D" D p. ABpab – p. Abpa. B "D prime" D' D / max (D) Correlation ("r-squared") r 2 D 2 / p A p ap B p b For a discussion and comparison of these LD measures, see Devlin B, Risch N, Genomics 1995; 29: 311 -22. Courtesy K. Jacobs, NCI
 Two Measures of LD: D' and r 2 • D' varies from 0 (complete equilibrium) to 1 (complete disequilibrium) • When D' = 0, typing one SNP provides no information on the other SNP • D' does not adequately account for allele frequencies; r 2 is correlation between SNPs, is preferred measure • When r 2 = 1, two SNPs are in perfect LD; allele frequencies are identical for both SNPs, and typing one SNP provides complete information on the other
Two Measures of LD: D' and r 2 • D' varies from 0 (complete equilibrium) to 1 (complete disequilibrium) • When D' = 0, typing one SNP provides no information on the other SNP • D' does not adequately account for allele frequencies; r 2 is correlation between SNPs, is preferred measure • When r 2 = 1, two SNPs are in perfect LD; allele frequencies are identical for both SNPs, and typing one SNP provides complete information on the other
 What can LD do for me? • Knowledge of patterns of LD can be quite useful in the design and analysis of genetic data • Design: – Estimation of theoretical power to detect associations – Evaluation of degree of completeness of sampling of genetic variants – Choice of most informative genetic variants to genotype • Sample size increases by ~1/r 2 to achieve same power to detect association with SNP 2 as SNP 1 Courtesy K. Jacobs, NCI
What can LD do for me? • Knowledge of patterns of LD can be quite useful in the design and analysis of genetic data • Design: – Estimation of theoretical power to detect associations – Evaluation of degree of completeness of sampling of genetic variants – Choice of most informative genetic variants to genotype • Sample size increases by ~1/r 2 to achieve same power to detect association with SNP 2 as SNP 1 Courtesy K. Jacobs, NCI
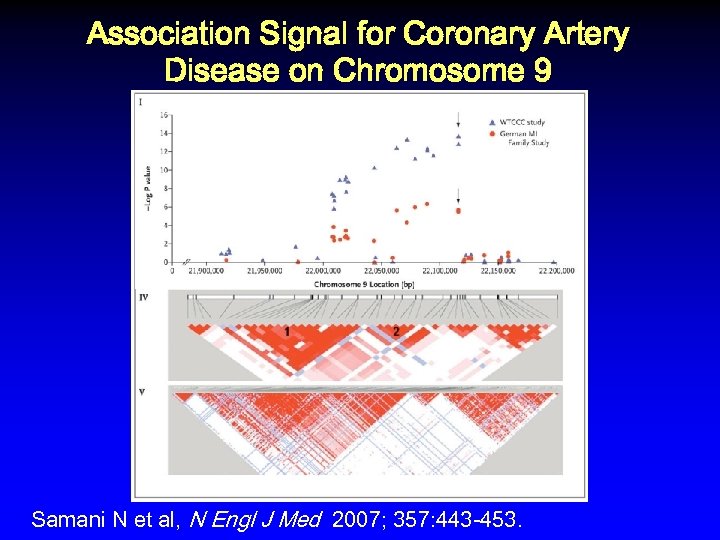 Association Signal for Coronary Artery Disease on Chromosome 9 Samani N et al, N Engl J Med 2007; 357: 443 -453.
Association Signal for Coronary Artery Disease on Chromosome 9 Samani N et al, N Engl J Med 2007; 357: 443 -453.
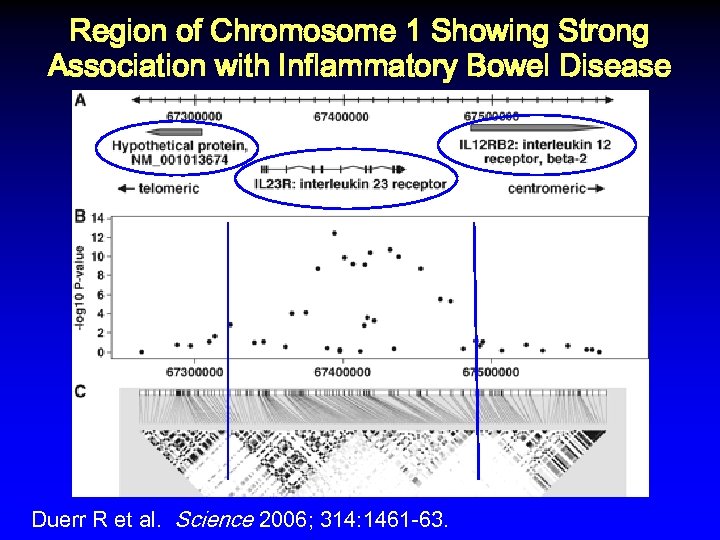 Region of Chromosome 1 Showing Strong Association with Inflammatory Bowel Disease Duerr R et al. Science 2006; 314: 1461 -63.
Region of Chromosome 1 Showing Strong Association with Inflammatory Bowel Disease Duerr R et al. Science 2006; 314: 1461 -63.
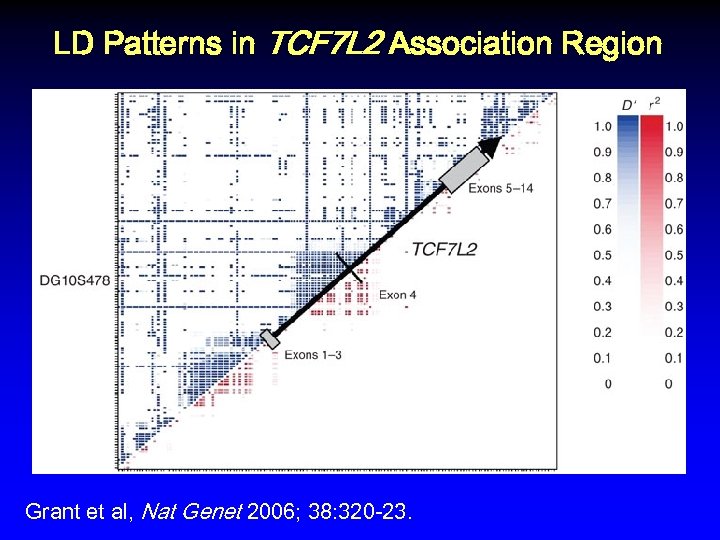 LD Patterns in TCF 7 L 2 Association Region Grant et al, Nat Genet 2006; 38: 320 -23.
LD Patterns in TCF 7 L 2 Association Region Grant et al, Nat Genet 2006; 38: 320 -23.
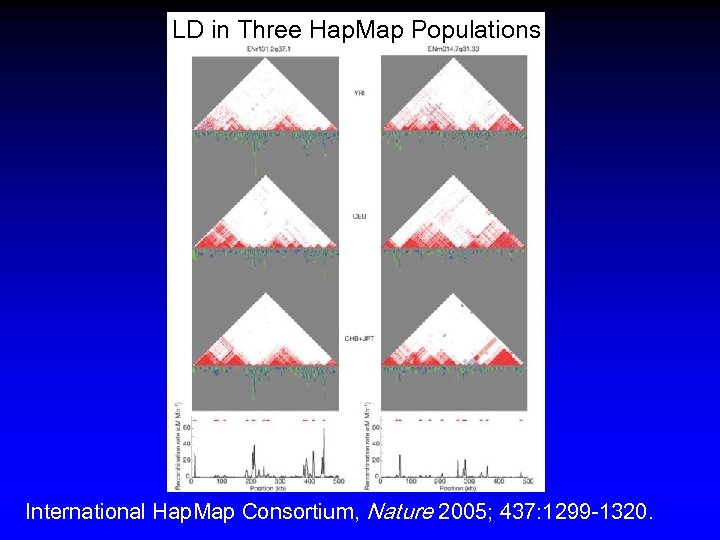 LD in Three Hap. Map Populations International Hap. Map Consortium, Nature 2005; 437: 1299 -1320.
LD in Three Hap. Map Populations International Hap. Map Consortium, Nature 2005; 437: 1299 -1320.
 A Hap. Map for More Efficient Association Studies: Goals • Use just the density of SNPs needed to find associations between SNPs and diseases • Do not miss chromosomal regions with disease association • Produce a tool to assist in finding genes affecting health and disease • Ancestral populations differ in their degree of LD; recent African ancestry populations are older and have shorter stretches of LD, need more SNPs for complete genome coverage
A Hap. Map for More Efficient Association Studies: Goals • Use just the density of SNPs needed to find associations between SNPs and diseases • Do not miss chromosomal regions with disease association • Produce a tool to assist in finding genes affecting health and disease • Ancestral populations differ in their degree of LD; recent African ancestry populations are older and have shorter stretches of LD, need more SNPs for complete genome coverage
 SNPs as Gateway to Genome-Wide Association (GWA) Studies • SNPs much more numerous than other markers and easier to assay • Genome-wide studies attempt to capture majority of genomic variation (10 M SNPs!) • Variation inherited in groups, or blocks, so not all 10 million points have to be tested • Blocks are shorter (so need to test more points) the less closely people are related • SNP technology allows studies in unrelated persons, assuming 5 kb – 10 kb lengths in common (300, 000 – 1, 000 markers)
SNPs as Gateway to Genome-Wide Association (GWA) Studies • SNPs much more numerous than other markers and easier to assay • Genome-wide studies attempt to capture majority of genomic variation (10 M SNPs!) • Variation inherited in groups, or blocks, so not all 10 million points have to be tested • Blocks are shorter (so need to test more points) the less closely people are related • SNP technology allows studies in unrelated persons, assuming 5 kb – 10 kb lengths in common (300, 000 – 1, 000 markers)
 www. hapmap. org International Hap. Map Consortium, Nature 2005; 437: 1299 -1320.
www. hapmap. org International Hap. Map Consortium, Nature 2005; 437: 1299 -1320.
 www. hapmap. org International Hap. Map Consortium, Nature 2007; 449: 851 -861.
www. hapmap. org International Hap. Map Consortium, Nature 2007; 449: 851 -861.
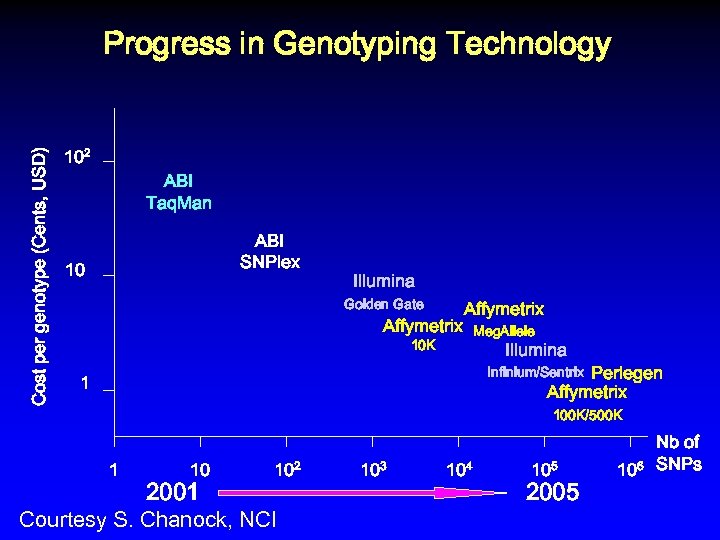 Cost per genotype (Cents, USD) Progress in Genotyping Technology 102 ABI Taq. Man ABI SNPlex 10 Illumina Golden Gate Affymetrix 10 K Meg. Allele Illumina Perlegen Affymetrix Infinium/Sentrix 1 100 K/500 K 1 10 2001 102 Courtesy S. Chanock, NCI 103 104 105 2005 106 Nb of SNPs
Cost per genotype (Cents, USD) Progress in Genotyping Technology 102 ABI Taq. Man ABI SNPlex 10 Illumina Golden Gate Affymetrix 10 K Meg. Allele Illumina Perlegen Affymetrix Infinium/Sentrix 1 100 K/500 K 1 10 2001 102 Courtesy S. Chanock, NCI 103 104 105 2005 106 Nb of SNPs
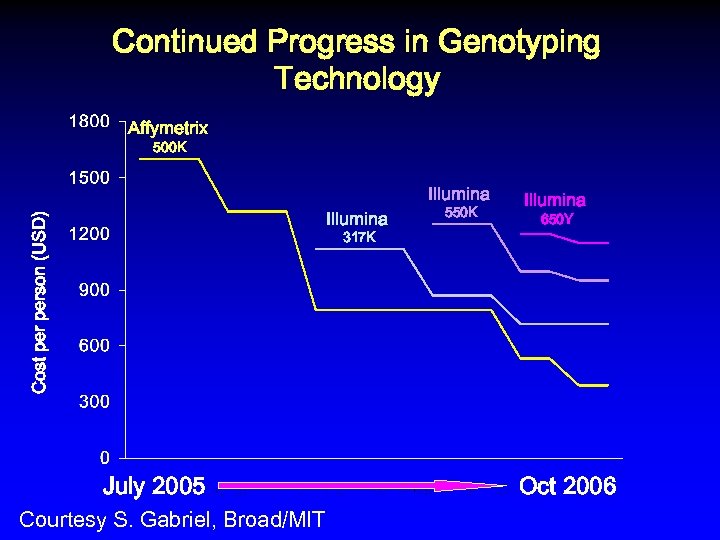 Continued Progress in Genotyping Technology Affymetrix 500 K Illumina Cost person (USD) Illumina 317 K July 2005 Courtesy S. Gabriel, Broad/MIT 550 K Illumina 650 Y Oct 2006
Continued Progress in Genotyping Technology Affymetrix 500 K Illumina Cost person (USD) Illumina 317 K July 2005 Courtesy S. Gabriel, Broad/MIT 550 K Illumina 650 Y Oct 2006
 Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs Cost/SNP Cost/Study
Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs Cost/SNP Cost/Study
 Cost of a Genome-Wide Association Study in 2, 000 People Year 2001 Number of SNPs Cost/SNP Cost/Study
Cost of a Genome-Wide Association Study in 2, 000 People Year 2001 Number of SNPs Cost/SNP Cost/Study
 Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs 2001 10, 000 Cost/SNP Cost/Study
Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs 2001 10, 000 Cost/SNP Cost/Study
 Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs Cost/SNP 2001 10, 000 $1. 00 Cost/Study
Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs Cost/SNP 2001 10, 000 $1. 00 Cost/Study
 Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs Cost/SNP Cost/Study 2001 10, 000 $1. 00 $20 billion
Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs Cost/SNP Cost/Study 2001 10, 000 $1. 00 $20 billion
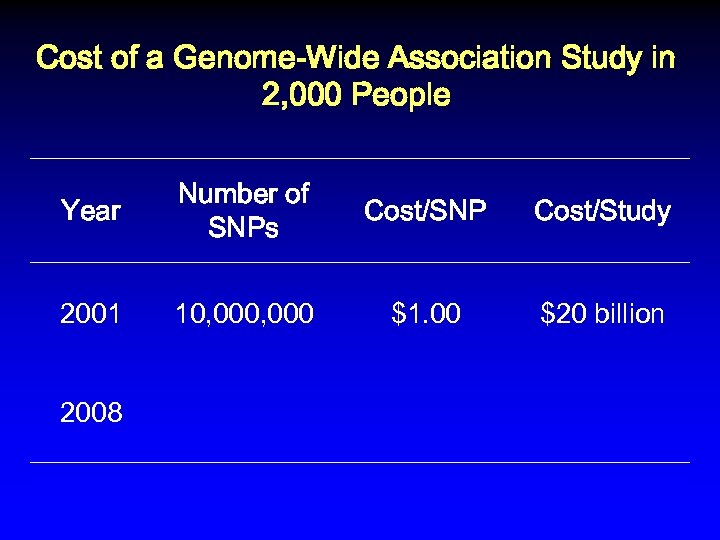 Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs Cost/SNP Cost/Study 2001 10, 000 $1. 00 $20 billion 2008
Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs Cost/SNP Cost/Study 2001 10, 000 $1. 00 $20 billion 2008
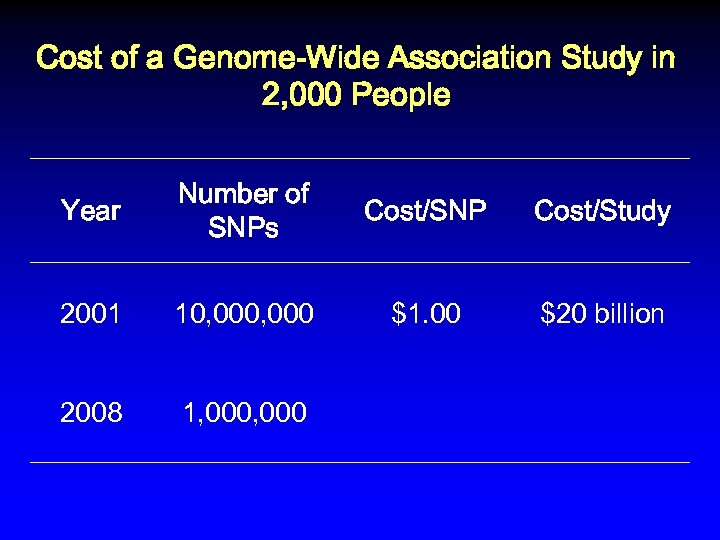 Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs Cost/SNP Cost/Study 2001 10, 000 $1. 00 $20 billion 2008 1, 000
Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs Cost/SNP Cost/Study 2001 10, 000 $1. 00 $20 billion 2008 1, 000
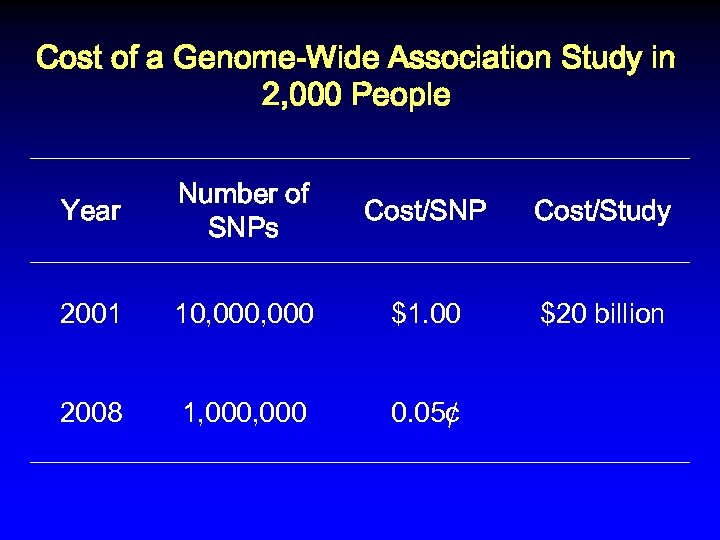 Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs Cost/SNP Cost/Study 2001 10, 000 $1. 00 $20 billion 2008 1, 000 0. 05¢
Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs Cost/SNP Cost/Study 2001 10, 000 $1. 00 $20 billion 2008 1, 000 0. 05¢
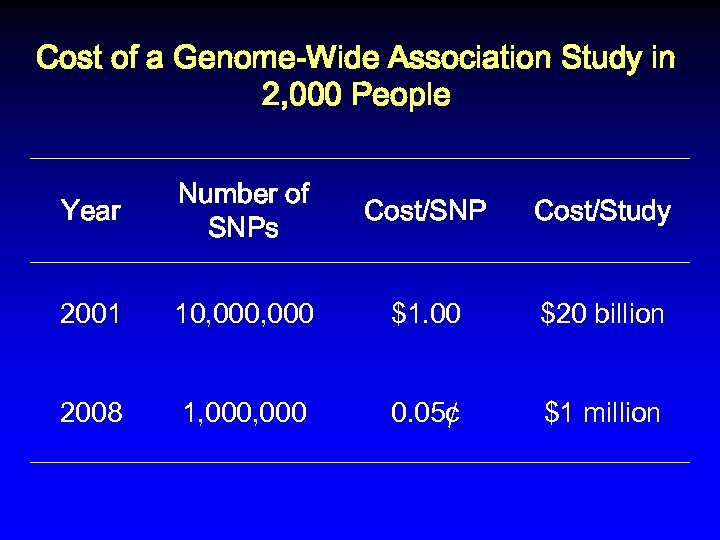 Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs Cost/SNP Cost/Study 2001 10, 000 $1. 00 $20 billion 2008 1, 000 0. 05¢ $1 million
Cost of a Genome-Wide Association Study in 2, 000 People Year Number of SNPs Cost/SNP Cost/Study 2001 10, 000 $1. 00 $20 billion 2008 1, 000 0. 05¢ $1 million
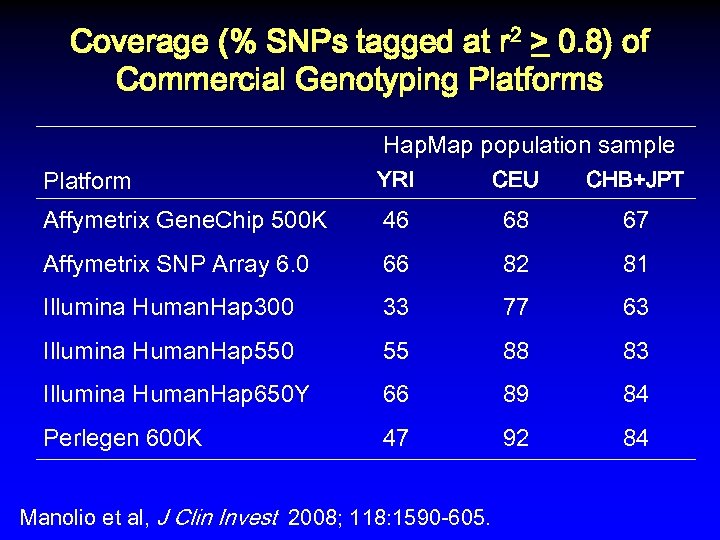 Coverage (% SNPs tagged at r 2 > 0. 8) of Commercial Genotyping Platforms Hap. Map population sample Platform YRI CEU CHB+JPT Affymetrix Gene. Chip 500 K 46 68 67 Affymetrix SNP Array 6. 0 66 82 81 Illumina Human. Hap 300 33 77 63 Illumina Human. Hap 550 55 88 83 Illumina Human. Hap 650 Y 66 89 84 Perlegen 600 K 47 92 84 Manolio et al, J Clin Invest 2008; 118: 1590 -605.
Coverage (% SNPs tagged at r 2 > 0. 8) of Commercial Genotyping Platforms Hap. Map population sample Platform YRI CEU CHB+JPT Affymetrix Gene. Chip 500 K 46 68 67 Affymetrix SNP Array 6. 0 66 82 81 Illumina Human. Hap 300 33 77 63 Illumina Human. Hap 550 55 88 83 Illumina Human. Hap 650 Y 66 89 84 Perlegen 600 K 47 92 84 Manolio et al, J Clin Invest 2008; 118: 1590 -605.
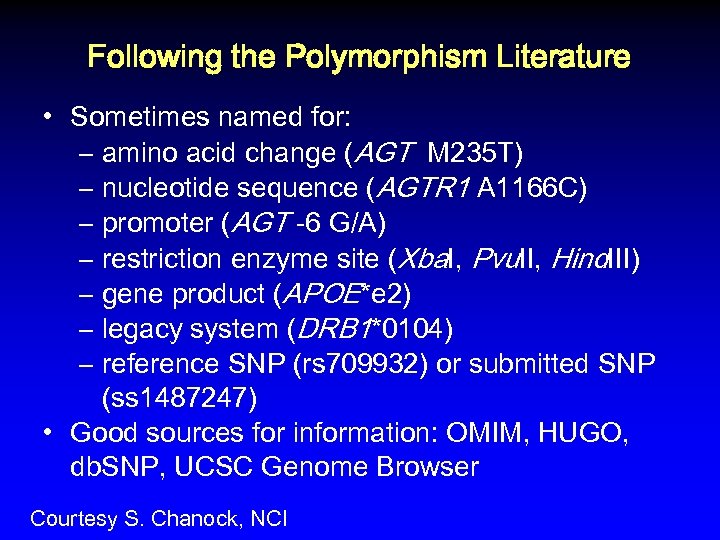 Following the Polymorphism Literature • Sometimes named for: – amino acid change (AGT M 235 T) – nucleotide sequence (AGTR 1 A 1166 C) – promoter (AGT -6 G/A) – restriction enzyme site (Xba. I, Pvu. II, Hind. III) – gene product (APOE*e 2) – legacy system (DRB 1*0104) – reference SNP (rs 709932) or submitted SNP (ss 1487247) • Good sources for information: OMIM, HUGO, db. SNP, UCSC Genome Browser Courtesy S. Chanock, NCI
Following the Polymorphism Literature • Sometimes named for: – amino acid change (AGT M 235 T) – nucleotide sequence (AGTR 1 A 1166 C) – promoter (AGT -6 G/A) – restriction enzyme site (Xba. I, Pvu. II, Hind. III) – gene product (APOE*e 2) – legacy system (DRB 1*0104) – reference SNP (rs 709932) or submitted SNP (ss 1487247) • Good sources for information: OMIM, HUGO, db. SNP, UCSC Genome Browser Courtesy S. Chanock, NCI
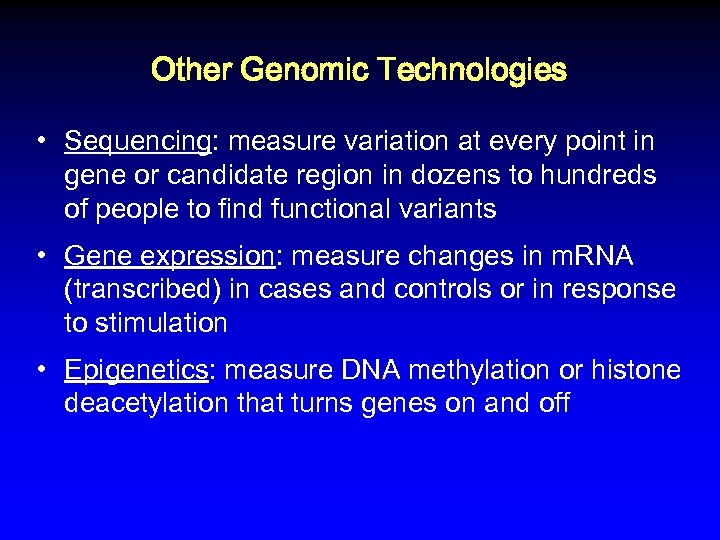 Other Genomic Technologies • Sequencing: measure variation at every point in gene or candidate region in dozens to hundreds of people to find functional variants • Gene expression: measure changes in m. RNA (transcribed) in cases and controls or in response to stimulation • Epigenetics: measure DNA methylation or histone deacetylation that turns genes on and off
Other Genomic Technologies • Sequencing: measure variation at every point in gene or candidate region in dozens to hundreds of people to find functional variants • Gene expression: measure changes in m. RNA (transcribed) in cases and controls or in response to stimulation • Epigenetics: measure DNA methylation or histone deacetylation that turns genes on and off
 Sidney Harris, http: //www. sciencecartoonsplus. com/gallery. htm.
Sidney Harris, http: //www. sciencecartoonsplus. com/gallery. htm.
 Summary Points: Genotyping Methods • Unbelievably rapid progress from small number of blood group markers to >10 M SNPs, CNVs, structural variants, sequence variants • Technology will continue to change and will be challenge to keep up with; difficult to know when ready to apply to population studies • SNPs are currently the dominant technology (more to come in Lecture 4) • Quality control is a major issue
Summary Points: Genotyping Methods • Unbelievably rapid progress from small number of blood group markers to >10 M SNPs, CNVs, structural variants, sequence variants • Technology will continue to change and will be challenge to keep up with; difficult to know when ready to apply to population studies • SNPs are currently the dominant technology (more to come in Lecture 4) • Quality control is a major issue
 Familial Resemblance? http: //en. wikipedia. org/wiki/Image: Kennedy_bros. jpg#file
Familial Resemblance? http: //en. wikipedia. org/wiki/Image: Kennedy_bros. jpg#file
 Evidence for Genetic Influence on Disease or Trait from Family Data • Familial resemblance: trait more similar among related than unrelated persons • Familial clustering: risk of disease in relative of case > risk in relative of non-case or of general population; (sibling relative risk, Risch's λS) • Distributions of continuous trait: mixtures of distributions or commingling analysis
Evidence for Genetic Influence on Disease or Trait from Family Data • Familial resemblance: trait more similar among related than unrelated persons • Familial clustering: risk of disease in relative of case > risk in relative of non-case or of general population; (sibling relative risk, Risch's λS) • Distributions of continuous trait: mixtures of distributions or commingling analysis
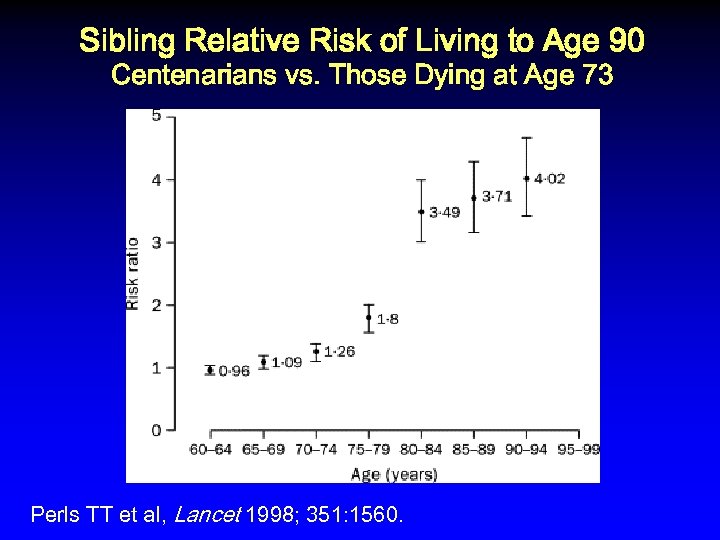 Sibling Relative Risk of Living to Age 90 Centenarians vs. Those Dying at Age 73 Perls TT et al, Lancet 1998; 351: 1560.
Sibling Relative Risk of Living to Age 90 Centenarians vs. Those Dying at Age 73 Perls TT et al, Lancet 1998; 351: 1560.
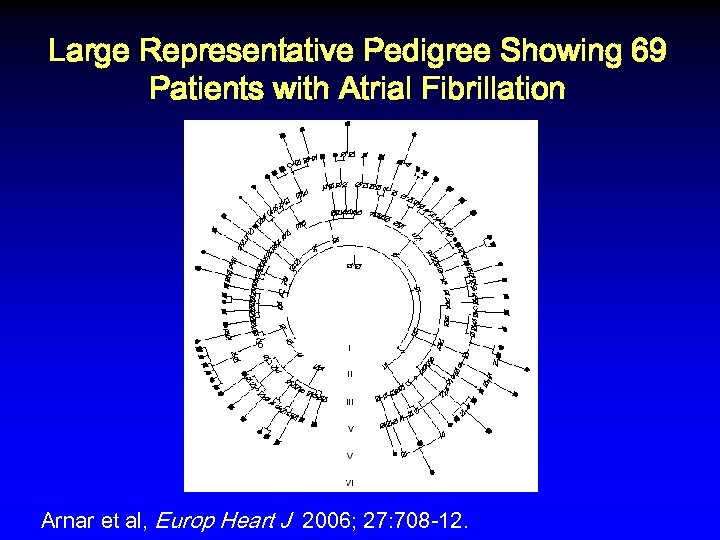 Large Representative Pedigree Showing 69 Patients with Atrial Fibrillation Arnar et al, Europ Heart J 2006; 27: 708 -12.
Large Representative Pedigree Showing 69 Patients with Atrial Fibrillation Arnar et al, Europ Heart J 2006; 27: 708 -12.
 Strength of Extensive Genealogies • Common diseases do not show Mendelian inheritance patterns • Affected siblings infrequent in common diseases, but many patients may have more distant relatives with same disease Degree of Relatives Risk Ratio [95% CI] P-Value 1 2 3 4 5 1. 77 [1. 67, 1. 88] 1. 36 [1. 27, 1. 44] 1. 18 [1. 14, 1. 23] 1. 10 [1. 06, 1. 13] 1. 05 [1. 02, 1. 07] < 0. 001 Arnar et al, Europ Heart J 2006; 27: 708 -12.
Strength of Extensive Genealogies • Common diseases do not show Mendelian inheritance patterns • Affected siblings infrequent in common diseases, but many patients may have more distant relatives with same disease Degree of Relatives Risk Ratio [95% CI] P-Value 1 2 3 4 5 1. 77 [1. 67, 1. 88] 1. 36 [1. 27, 1. 44] 1. 18 [1. 14, 1. 23] 1. 10 [1. 06, 1. 13] 1. 05 [1. 02, 1. 07] < 0. 001 Arnar et al, Europ Heart J 2006; 27: 708 -12.
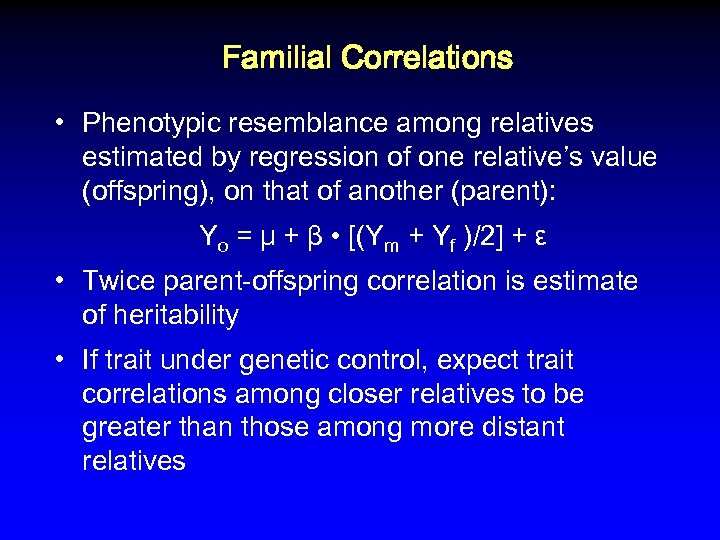 Familial Correlations • Phenotypic resemblance among relatives estimated by regression of one relative’s value (offspring), on that of another (parent): Yo = μ + β • [(Ym + Yf )/2] + ε • Twice parent-offspring correlation is estimate of heritability • If trait under genetic control, expect trait correlations among closer relatives to be greater than those among more distant relatives
Familial Correlations • Phenotypic resemblance among relatives estimated by regression of one relative’s value (offspring), on that of another (parent): Yo = μ + β • [(Ym + Yf )/2] + ε • Twice parent-offspring correlation is estimate of heritability • If trait under genetic control, expect trait correlations among closer relatives to be greater than those among more distant relatives
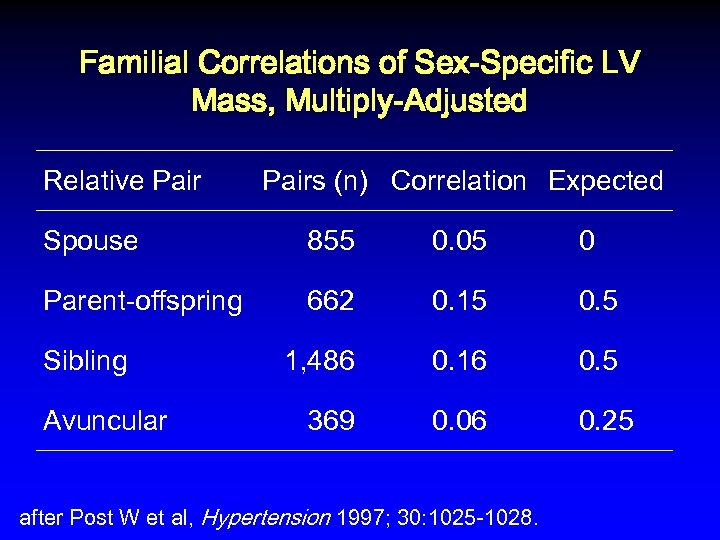 Familial Correlations of Sex-Specific LV Mass, Multiply-Adjusted Relative Pairs (n) Correlation Expected Spouse 855 0. 05 0 Parent-offspring 662 0. 15 0. 5 1, 486 0. 16 0. 5 369 0. 06 0. 25 Sibling Avuncular after Post W et al, Hypertension 1997; 30: 1025 -1028.
Familial Correlations of Sex-Specific LV Mass, Multiply-Adjusted Relative Pairs (n) Correlation Expected Spouse 855 0. 05 0 Parent-offspring 662 0. 15 0. 5 1, 486 0. 16 0. 5 369 0. 06 0. 25 Sibling Avuncular after Post W et al, Hypertension 1997; 30: 1025 -1028.
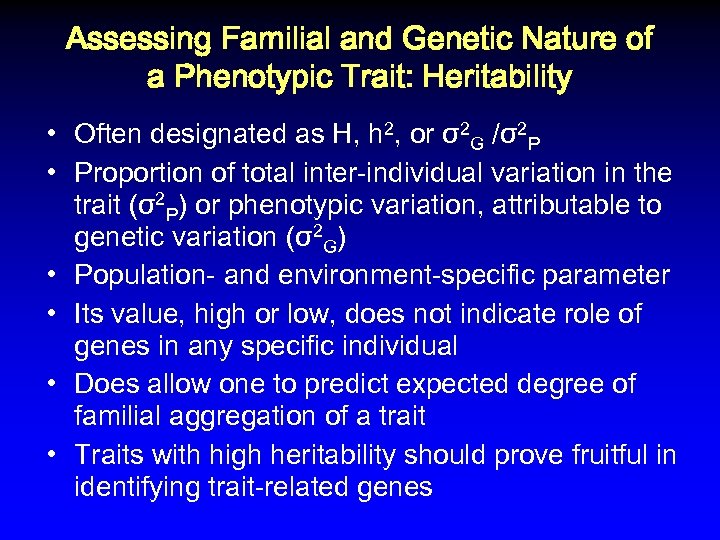 Assessing Familial and Genetic Nature of a Phenotypic Trait: Heritability • Often designated as H, h 2, or σ2 G /σ2 P • Proportion of total inter-individual variation in the trait (σ2 P) or phenotypic variation, attributable to genetic variation (σ2 G) • Population- and environment-specific parameter • Its value, high or low, does not indicate role of genes in any specific individual • Does allow one to predict expected degree of familial aggregation of a trait • Traits with high heritability should prove fruitful in identifying trait-related genes
Assessing Familial and Genetic Nature of a Phenotypic Trait: Heritability • Often designated as H, h 2, or σ2 G /σ2 P • Proportion of total inter-individual variation in the trait (σ2 P) or phenotypic variation, attributable to genetic variation (σ2 G) • Population- and environment-specific parameter • Its value, high or low, does not indicate role of genes in any specific individual • Does allow one to predict expected degree of familial aggregation of a trait • Traits with high heritability should prove fruitful in identifying trait-related genes
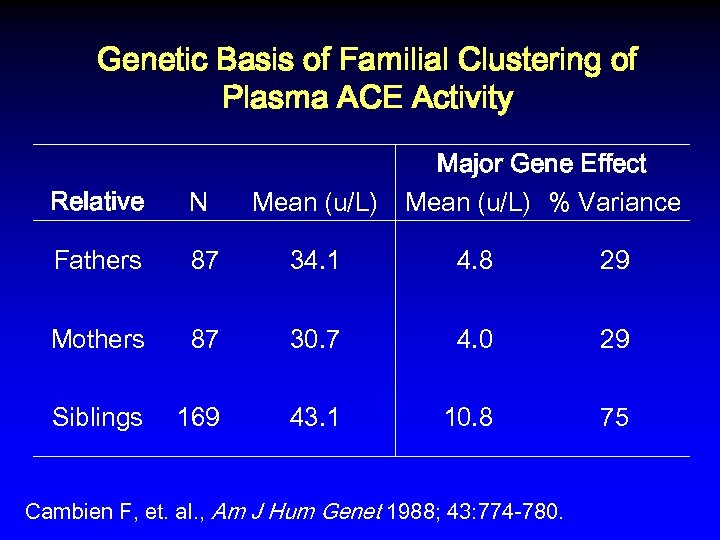 Genetic Basis of Familial Clustering of Plasma ACE Activity Major Gene Effect Mean (u/L) % Variance Relative N Mean (u/L) Fathers 87 34. 1 4. 8 29 Mothers 87 30. 7 4. 0 29 Siblings 169 43. 1 10. 8 75 Cambien F, et. al. , Am J Hum Genet 1988; 43: 774 -780.
Genetic Basis of Familial Clustering of Plasma ACE Activity Major Gene Effect Mean (u/L) % Variance Relative N Mean (u/L) Fathers 87 34. 1 4. 8 29 Mothers 87 30. 7 4. 0 29 Siblings 169 43. 1 10. 8 75 Cambien F, et. al. , Am J Hum Genet 1988; 43: 774 -780.
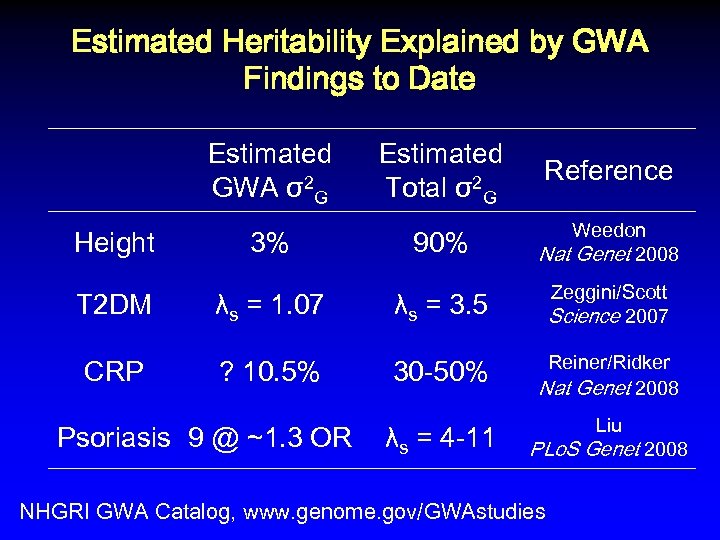 Estimated Heritability Explained by GWA Findings to Date Estimated GWA σ2 G Height 3% Estimated Total σ2 G Reference 90% Weedon Nat Genet 2008 T 2 DM λs = 1. 07 λs = 3. 5 Zeggini/Scott Science 2007 CRP ? 10. 5% 30 -50% Reiner/Ridker Nat Genet 2008 λs = 4 -11 Liu Psoriasis 9 @ ~1. 3 OR PLo. S Genet 2008 NHGRI GWA Catalog, www. genome. gov/GWAstudies
Estimated Heritability Explained by GWA Findings to Date Estimated GWA σ2 G Height 3% Estimated Total σ2 G Reference 90% Weedon Nat Genet 2008 T 2 DM λs = 1. 07 λs = 3. 5 Zeggini/Scott Science 2007 CRP ? 10. 5% 30 -50% Reiner/Ridker Nat Genet 2008 λs = 4 -11 Liu Psoriasis 9 @ ~1. 3 OR PLo. S Genet 2008 NHGRI GWA Catalog, www. genome. gov/GWAstudies
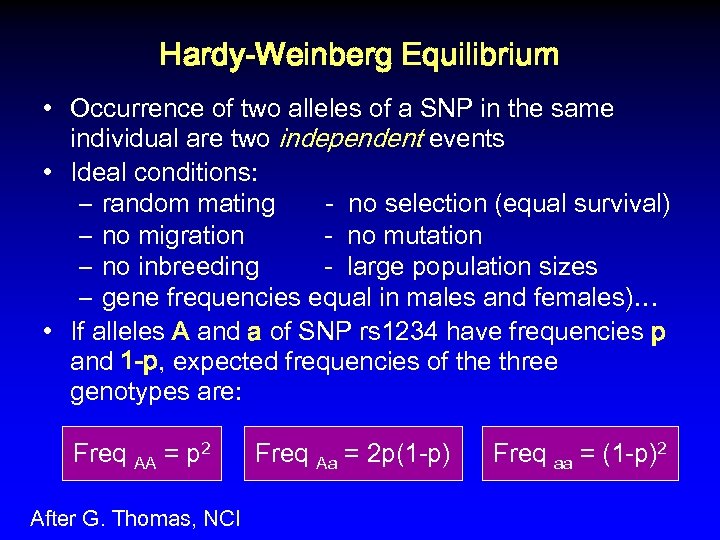 Hardy-Weinberg Equilibrium • Occurrence of two alleles of a SNP in the same individual are two independent events • Ideal conditions: – random mating - no selection (equal survival) – no migration - no mutation – no inbreeding - large population sizes – gene frequencies equal in males and females)… • If alleles A and a of SNP rs 1234 have frequencies p and 1 -p, expected frequencies of the three genotypes are: Freq AA = p 2 After G. Thomas, NCI Freq Aa = 2 p(1 -p) Freq aa = (1 -p)2
Hardy-Weinberg Equilibrium • Occurrence of two alleles of a SNP in the same individual are two independent events • Ideal conditions: – random mating - no selection (equal survival) – no migration - no mutation – no inbreeding - large population sizes – gene frequencies equal in males and females)… • If alleles A and a of SNP rs 1234 have frequencies p and 1 -p, expected frequencies of the three genotypes are: Freq AA = p 2 After G. Thomas, NCI Freq Aa = 2 p(1 -p) Freq aa = (1 -p)2
 Summary Points: Familial Clustering • Indicator of possible genetic influence • May over-estimate genetic component due to poor assessment and adjustment for shared environment • Methods include twin studies, parent-offspring correlation, “relative” relative risk, % variance explained • Current genes for complex disease explain only tiny fraction of total heritability
Summary Points: Familial Clustering • Indicator of possible genetic influence • May over-estimate genetic component due to poor assessment and adjustment for shared environment • Methods include twin studies, parent-offspring correlation, “relative” relative risk, % variance explained • Current genes for complex disease explain only tiny fraction of total heritability
 Larson, G. The Complete Far Side. 2003.
Larson, G. The Complete Far Side. 2003.
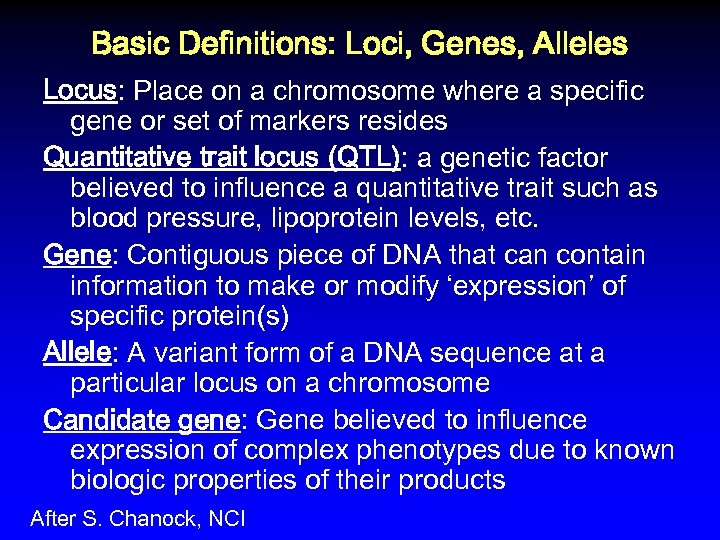 Basic Definitions: Loci, Genes, Alleles Locus: Place on a chromosome where a specific gene or set of markers resides Quantitative trait locus (QTL): a genetic factor believed to influence a quantitative trait such as blood pressure, lipoprotein levels, etc. Gene: Contiguous piece of DNA that can contain information to make or modify ‘expression’ of specific protein(s) Allele: A variant form of a DNA sequence at a particular locus on a chromosome Candidate gene: Gene believed to influence expression of complex phenotypes due to known biologic properties of their products After S. Chanock, NCI
Basic Definitions: Loci, Genes, Alleles Locus: Place on a chromosome where a specific gene or set of markers resides Quantitative trait locus (QTL): a genetic factor believed to influence a quantitative trait such as blood pressure, lipoprotein levels, etc. Gene: Contiguous piece of DNA that can contain information to make or modify ‘expression’ of specific protein(s) Allele: A variant form of a DNA sequence at a particular locus on a chromosome Candidate gene: Gene believed to influence expression of complex phenotypes due to known biologic properties of their products After S. Chanock, NCI
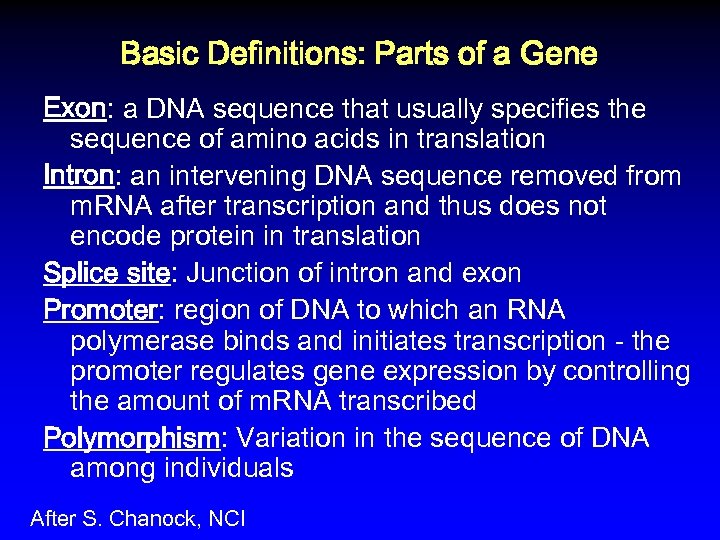 Basic Definitions: Parts of a Gene Exon: a DNA sequence that usually specifies the sequence of amino acids in translation Intron: an intervening DNA sequence removed from m. RNA after transcription and thus does not encode protein in translation Splice site: Junction of intron and exon Promoter: region of DNA to which an RNA polymerase binds and initiates transcription - the promoter regulates gene expression by controlling the amount of m. RNA transcribed Polymorphism: Variation in the sequence of DNA among individuals After S. Chanock, NCI
Basic Definitions: Parts of a Gene Exon: a DNA sequence that usually specifies the sequence of amino acids in translation Intron: an intervening DNA sequence removed from m. RNA after transcription and thus does not encode protein in translation Splice site: Junction of intron and exon Promoter: region of DNA to which an RNA polymerase binds and initiates transcription - the promoter regulates gene expression by controlling the amount of m. RNA transcribed Polymorphism: Variation in the sequence of DNA among individuals After S. Chanock, NCI
 SNPs and Function: We know so little… • Majority are “silent” – No known functional change • Some alter gene expression/regulation – Promoter/enhancer/silencer – m. RNA stability – Small RNAs • Some alter function of gene product – Change sequence of protein Courtesy S. Chanock, NCI
SNPs and Function: We know so little… • Majority are “silent” – No known functional change • Some alter gene expression/regulation – Promoter/enhancer/silencer – m. RNA stability – Small RNAs • Some alter function of gene product – Change sequence of protein Courtesy S. Chanock, NCI
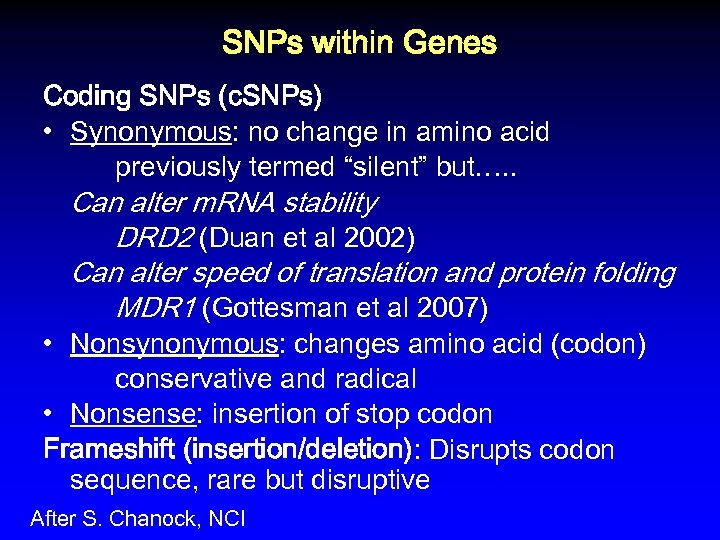 SNPs within Genes Coding SNPs (c. SNPs) • Synonymous: no change in amino acid previously termed “silent” but…. . Can alter m. RNA stability DRD 2 (Duan et al 2002) Can alter speed of translation and protein folding MDR 1 (Gottesman et al 2007) • Nonsynonymous: changes amino acid (codon) conservative and radical • Nonsense: insertion of stop codon Frameshift (insertion/deletion): Disrupts codon sequence, rare but disruptive After S. Chanock, NCI
SNPs within Genes Coding SNPs (c. SNPs) • Synonymous: no change in amino acid previously termed “silent” but…. . Can alter m. RNA stability DRD 2 (Duan et al 2002) Can alter speed of translation and protein folding MDR 1 (Gottesman et al 2007) • Nonsynonymous: changes amino acid (codon) conservative and radical • Nonsense: insertion of stop codon Frameshift (insertion/deletion): Disrupts codon sequence, rare but disruptive After S. Chanock, NCI
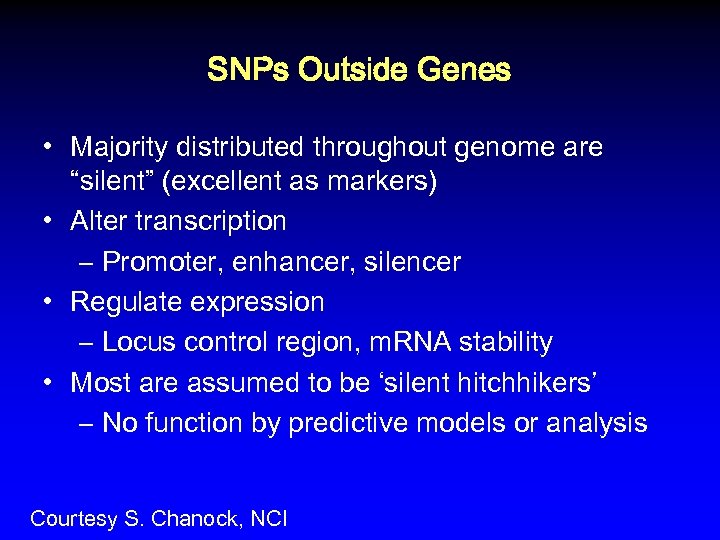 SNPs Outside Genes • Majority distributed throughout genome are “silent” (excellent as markers) • Alter transcription – Promoter, enhancer, silencer • Regulate expression – Locus control region, m. RNA stability • Most are assumed to be ‘silent hitchhikers’ – No function by predictive models or analysis Courtesy S. Chanock, NCI
SNPs Outside Genes • Majority distributed throughout genome are “silent” (excellent as markers) • Alter transcription – Promoter, enhancer, silencer • Regulate expression – Locus control region, m. RNA stability • Most are assumed to be ‘silent hitchhikers’ – No function by predictive models or analysis Courtesy S. Chanock, NCI
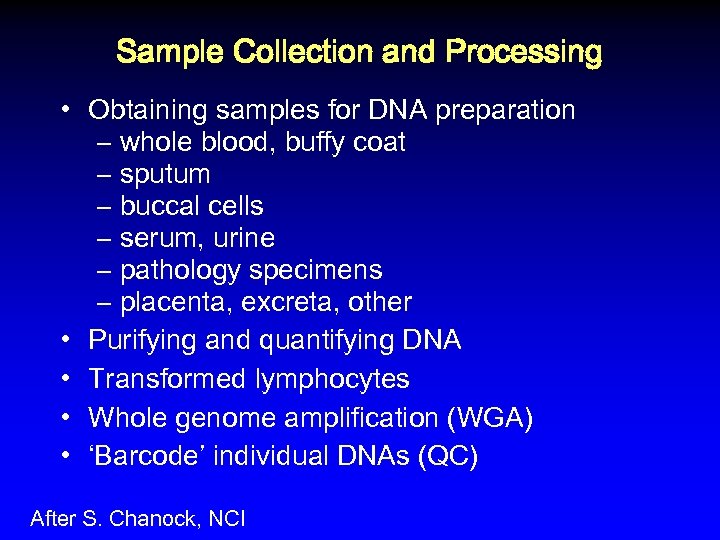 Sample Collection and Processing • Obtaining samples for DNA preparation – whole blood, buffy coat – sputum – buccal cells – serum, urine – pathology specimens – placenta, excreta, other • Purifying and quantifying DNA • Transformed lymphocytes • Whole genome amplification (WGA) • ‘Barcode’ individual DNAs (QC) After S. Chanock, NCI
Sample Collection and Processing • Obtaining samples for DNA preparation – whole blood, buffy coat – sputum – buccal cells – serum, urine – pathology specimens – placenta, excreta, other • Purifying and quantifying DNA • Transformed lymphocytes • Whole genome amplification (WGA) • ‘Barcode’ individual DNAs (QC) After S. Chanock, NCI


