b9c5341ca3e465fd4d0629e4534e6f5a.ppt
- Количество слайдов: 112

Genetic Association Studies and GWAS October 16, 2015

Topics • Study Design • Potential Threats to validity: - sample recruitment - genotyping error -errors in data analysis - replication - population structure
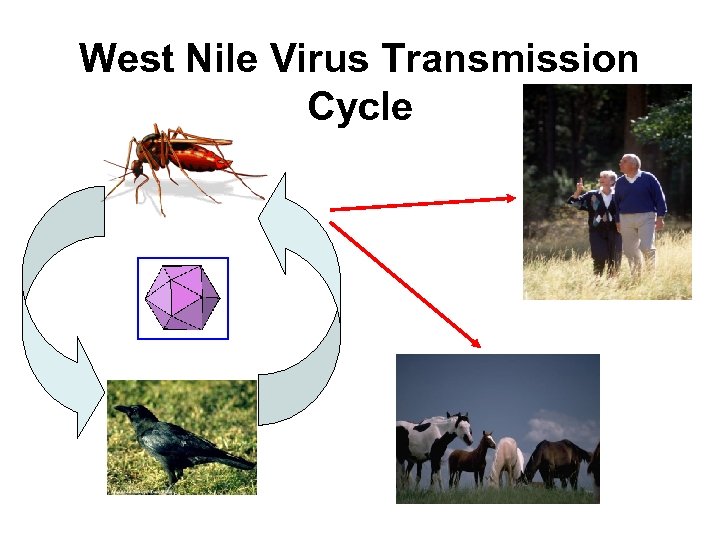
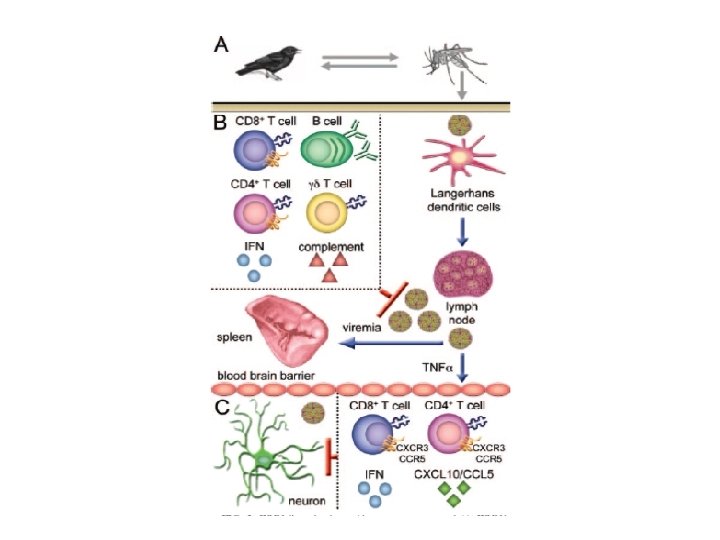
West Nile Virus Transmission Cycle

West Nile Outbreaks • • • Israel - 1951 -1954, 1957 France - 1962 South Africa - 1974 Romania – 1996 Italy 1998 Russia - 1999
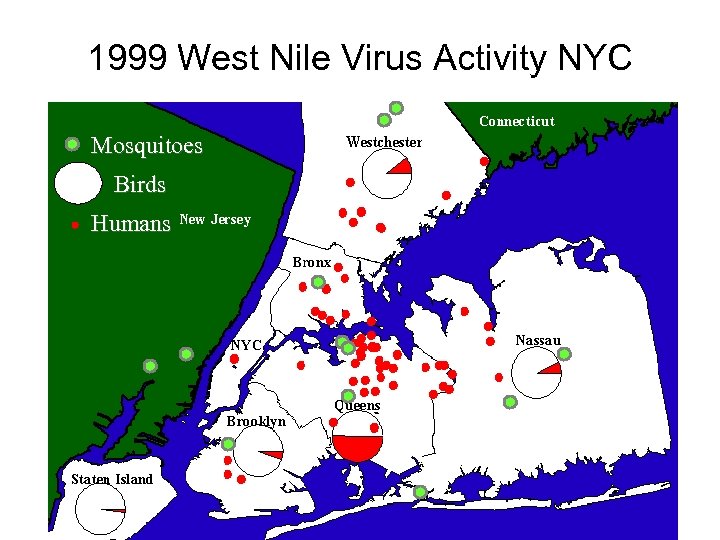
1999 West Nile Virus Activity NYC Mosquitoes Birds Humans

Clinical Syndromes • 80% asymptomatic • 20% “West Nile Fever” • 1 in 20 of symptomatic patients develop neuroinvasive disease - Meningitis - Encephalitis - Acute Flaccid Paralysis • Apart from increased age, risk factors ill defined

Hypothesis • WNV neuroinvasive disease is a consequence of genetic factors that result in increased WNV replication and subsequent pathology

Q. Why detect Genes associated with Disease ?

• Diagnosis • Prognosis • Therapeutics • Basic Mechanisms of disease
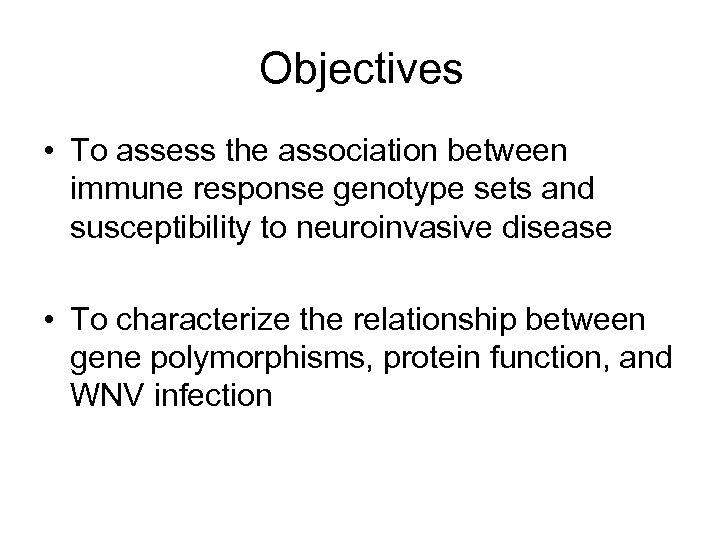
Objectives • To assess the association between immune response genotype sets and susceptibility to neuroinvasive disease • To characterize the relationship between gene polymorphisms, protein function, and WNV infection

Q. What sort of evidence do you look for to see if the question is worthwhile?
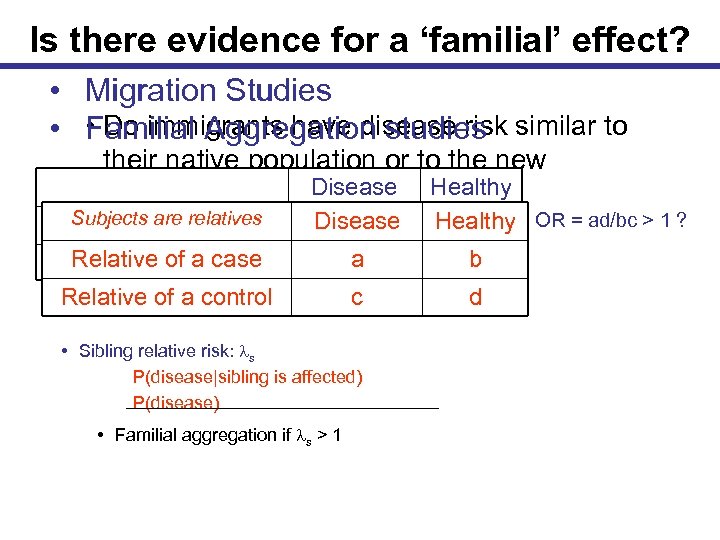
Is there evidence for a ‘familial’ effect? • Migration Studies • Do immigrants have disease risk • Familial Aggregation studies similar to their native population or to the new Disease Healthy population? Subjects are relatives FH+ Relative of a case FHRelative of a control Disease a a c c • Sibling relative risk: ls P(disease|sibling is affected) P(disease) • Familial aggregation if ls > 1 Healthy OR = ad/bc > 1 ? b b d d
Is there evidence for a ‘genetic’ effect? • Familial correlations in phenotype? • Heritability can be thought of as the similarity between related individuals that is due to shared genes. • If trait is heritable, individuals who share genes should have higher correlation between trait values than individuals who do not share genes – Parent & offspring trait values should be correlated – Identical twins should be more correlated than siblings – Sibling values should be more correlated than cousins
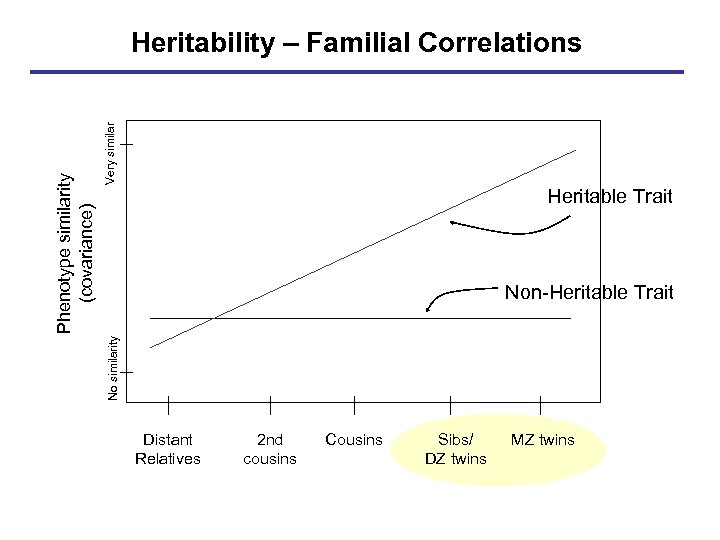
Very similar Heritable Trait Non-Heritable Trait No similarity Phenotype similarity (covariance) Heritability – Familial Correlations Distant Relatives 2 nd cousins Cousins Sibs/ DZ twins MZ twins
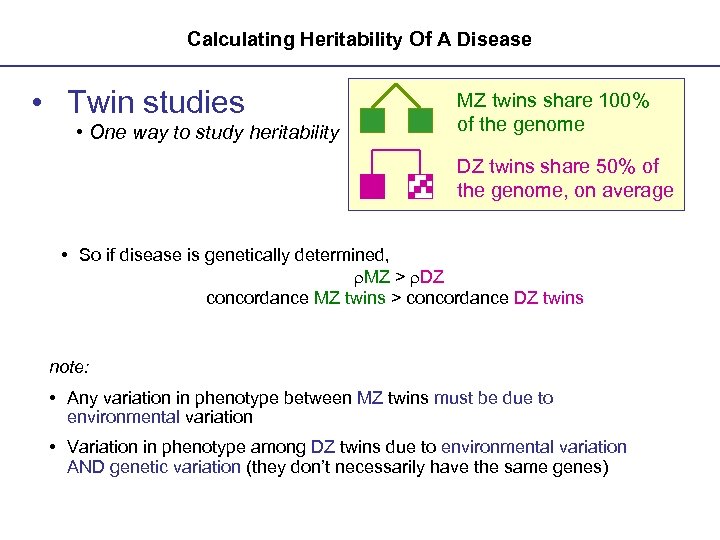
Calculating Heritability Of A Disease • Twin studies • One way to study heritability MZ twins share 100% of the genome DZ twins share 50% of the genome, on average • So if disease is genetically determined, MZ > DZ concordance MZ twins > concordance DZ twins note: • Any variation in phenotype between MZ twins must be due to environmental variation • Variation in phenotype among DZ twins due to environmental variation AND genetic variation (they don’t necessarily have the same genes)
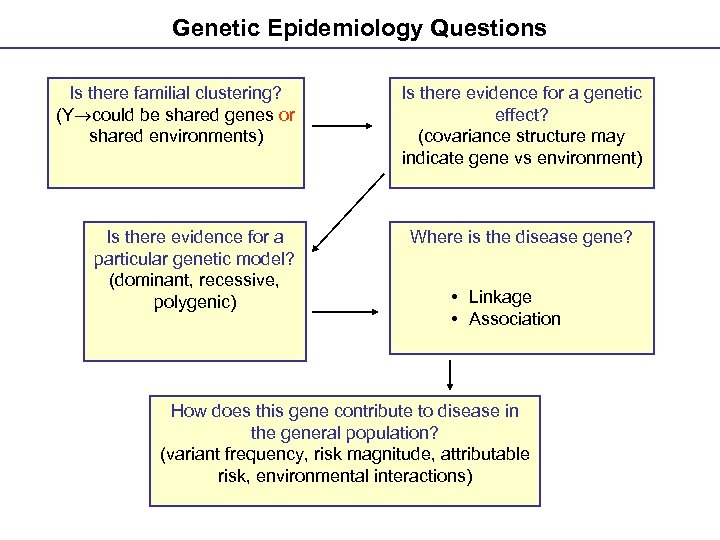
Genetic Epidemiology Questions Is there familial clustering? (Y could be shared genes or shared environments) Is there evidence for a particular genetic model? (dominant, recessive, polygenic) Is there evidence for a genetic effect? (covariance structure may indicate gene vs environment) Where is the disease gene? • Linkage • Association How does this gene contribute to disease in the general population? (variant frequency, risk magnitude, attributable risk, environmental interactions)
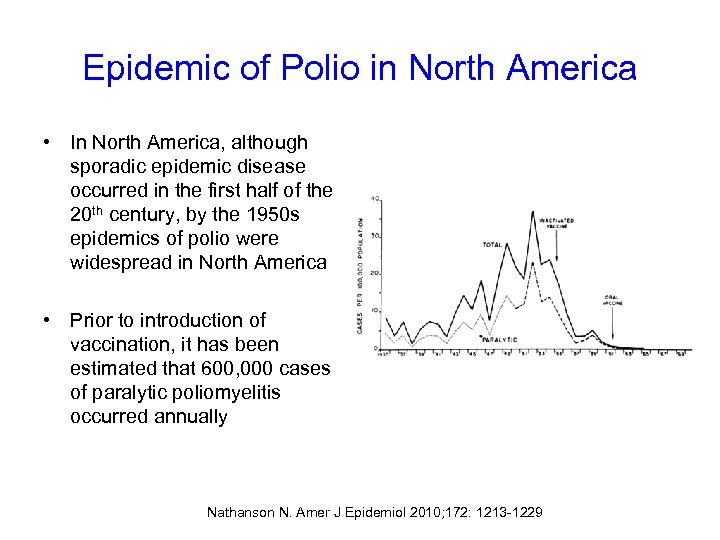
Epidemic of Polio in North America • In North America, although sporadic epidemic disease occurred in the first half of the 20 th century, by the 1950 s epidemics of polio were widespread in North America • Prior to introduction of vaccination, it has been estimated that 600, 000 cases of paralytic poliomyelitis occurred annually Nathanson N. Amer J Epidemiol 2010; 172: 1213 -1229
Host Factors • < 1% of individuals infected with poliovirus developed paralytic polio in pre-vaccine era • In families with a clinical case of poliomyelitis, ratio of inapparent to apparent infection between 3: 1 and 7: 1 versus 100: 1 in the general population
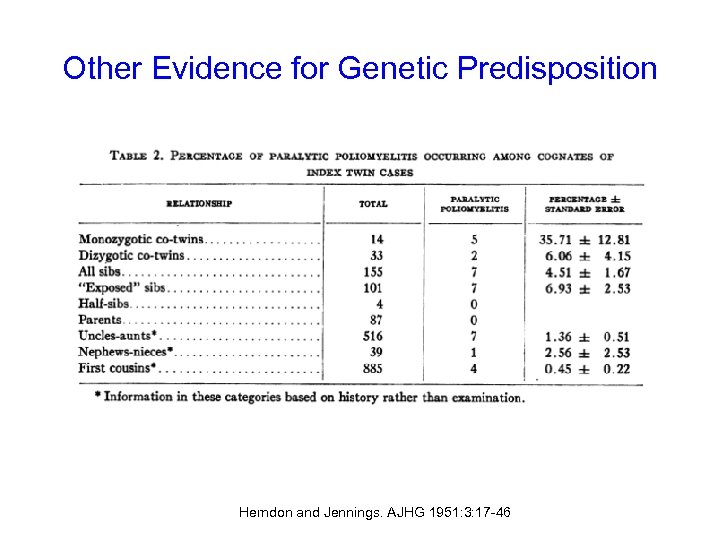
Other Evidence for Genetic Predisposition Herndon and Jennings. AJHG 1951: 3: 17 -46
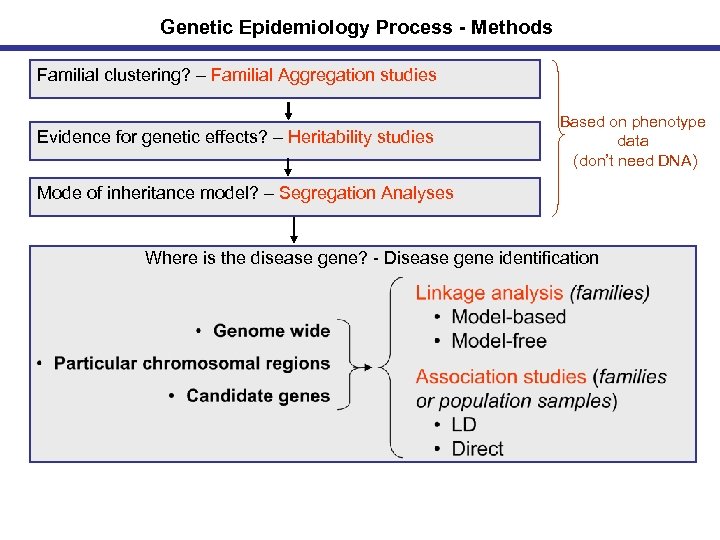
Genetic Epidemiology Process - Methods Familial clustering? – Familial Aggregation studies Evidence for genetic effects? – Heritability studies Based on phenotype data (don’t need DNA) Mode of inheritance model? – Segregation Analyses Where is the disease gene? - Disease gene identification
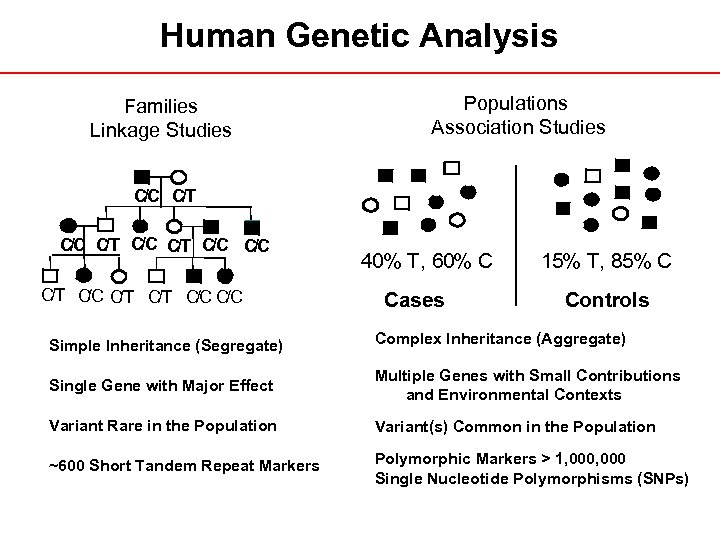
Human Genetic Analysis Families Linkage Studies Populations Association Studies C/C C/T C/C C C/C /T /T C/C C C/T C/C /T 40% T, 60% C Cases 15% T, 85% C Controls Simple Inheritance (Segregate) Complex Inheritance (Aggregate) Single Gene with Major Effect Multiple Genes with Small Contributions and Environmental Contexts Variant Rare in the Population Variant(s) Common in the Population ~600 Short Tandem Repeat Markers Polymorphic Markers > 1, 000 Single Nucleotide Polymorphisms (SNPs)

Q. What is the first step in designing the study?
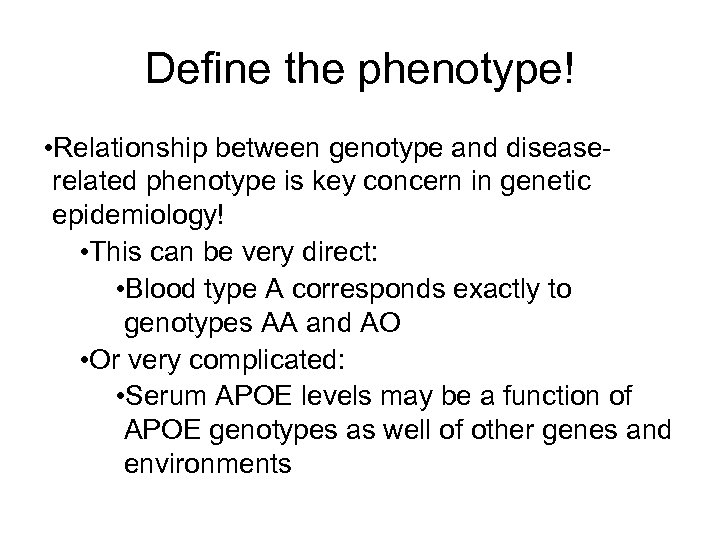
Define the phenotype! • Relationship between genotype and diseaserelated phenotype is key concern in genetic epidemiology! • This can be very direct: • Blood type A corresponds exactly to genotypes AA and AO • Or very complicated: • Serum APOE levels may be a function of APOE genotypes as well of other genes and environments
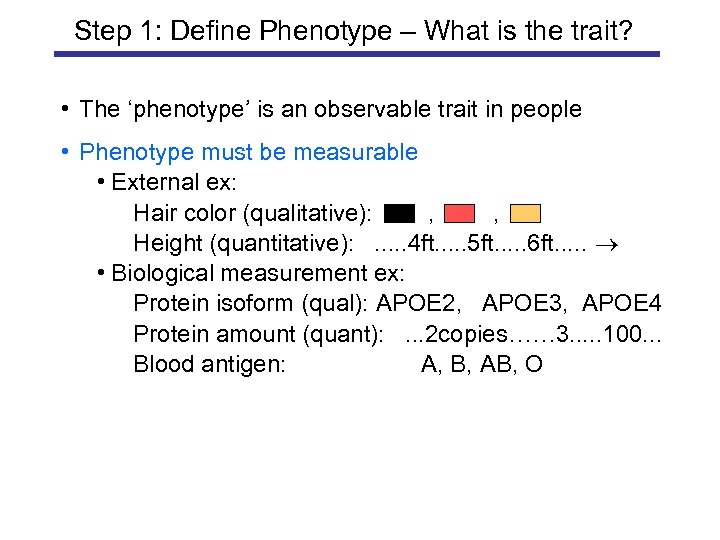
Step 1: Define Phenotype – What is the trait? • The ‘phenotype’ is an observable trait in people • Phenotype must be measurable • External ex: Hair color (qualitative): , , Height (quantitative): . . . 4 ft. . . 5 ft. . . 6 ft. . . • Biological measurement ex: Protein isoform (qual): APOE 2, APOE 3, APOE 4 Protein amount (quant): . . . 2 copies…… 3. . . 100. . . Blood antigen: A, B, AB, O
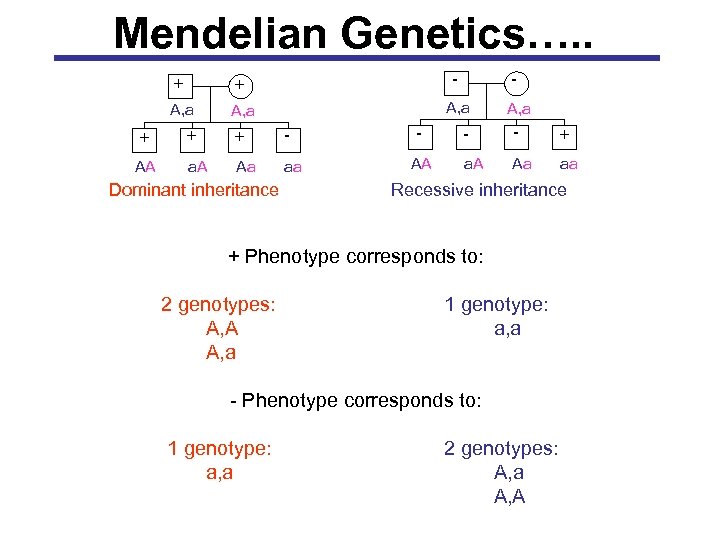
Mendelian Genetics…. . - + + - A, a + + + - AA a. A Aa aa Dominant inheritance AA A, a - - + a. A Aa aa Recessive inheritance + Phenotype corresponds to: 2 genotypes: A, A A, a 1 genotype: a, a - Phenotype corresponds to: 1 genotype: a, a 2 genotypes: A, a A, A
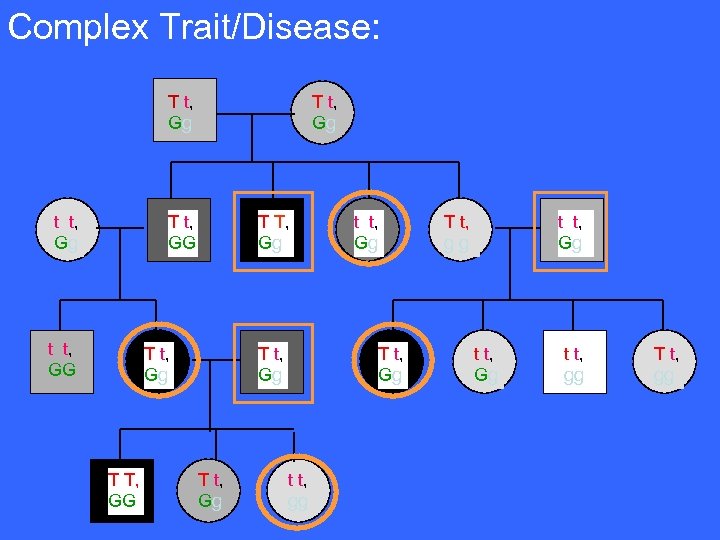
Complex Trait/Disease: gene ‘T’: Family Pedigree following Quantitative Trait/Disease: Recessive Trait/Disease: Dominant Trait/Disease: Tt T t, Gg ttttt, t Gg t T tt, t GG T T t t, GG T T tt, Gg T T, TT TT GG T T t t, Gg Tt. T, TT Gg t. T t t T t, Gg T T t t, Gg Ex: Early-onset Alzheimer’s disease Ex: Cystic Fibrosis t t, tttt gg ttt t, t Gg T T t t, Gg Tt T T t, gg t T tt t t, Gg ttttt, t Gg t T tt t t, gg T T t t, gg
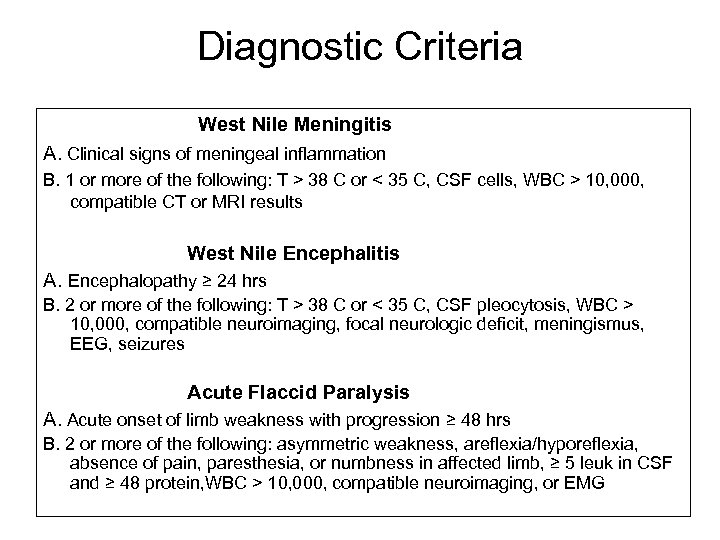
Diagnostic Criteria West Nile Meningitis A. Clinical signs of meningeal inflammation B. 1 or more of the following: T > 38 C or < 35 C, CSF cells, WBC > 10, 000, compatible CT or MRI results West Nile Encephalitis A. Encephalopathy ≥ 24 hrs B. 2 or more of the following: T > 38 C or < 35 C, CSF pleocytosis, WBC > 10, 000, compatible neuroimaging, focal neurologic deficit, meningismus, EEG, seizures Acute Flaccid Paralysis A. Acute onset of limb weakness with progression ≥ 48 hrs B. 2 or more of the following: asymmetric weakness, areflexia/hyporeflexia, absence of pain, paresthesia, or numbness in affected limb, ≥ 5 leuk in CSF and ≥ 48 protein, WBC > 10, 000, compatible neuroimaging, or EMG
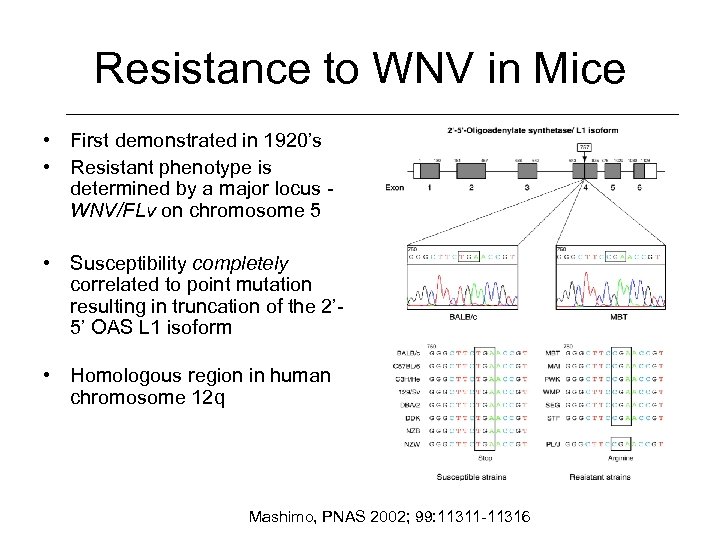
Resistance to WNV in Mice • First demonstrated in 1920’s • Resistant phenotype is determined by a major locus WNV/FLv on chromosome 5 • Susceptibility completely correlated to point mutation resulting in truncation of the 2’ 5’ OAS L 1 isoform • Homologous region in human chromosome 12 q Mashimo, PNAS 2002; 99: 11311 -11316

Clinical Syndromes • 80% asymptomatic • 20% “West Nile Fever” • 1 in 20 of symptomatic patients develop neuroinvasive disease - Meningitis - Encephalitis - Acute Flaccid Paralysis • Apart from increased age, risk factors ill defined

Q. How do you find genes responsible for human disease?
Difficult: • Many risk models (genotype-phenotype correlations do not follow simple patterns – ‘complex disease’) • Many possible genes (~30, 000 human genes)
• Difficult challenge to find a disease gene: like finding a misspelled word in a set of encyclopedias! A 1 Z 24 7 Vol 7 ‘which chromosome? ’ page ‘chromosomal region’ This is a sentence in a paragraph… This it a sentence in a paragraph… ‘gene’ ‘mutation’
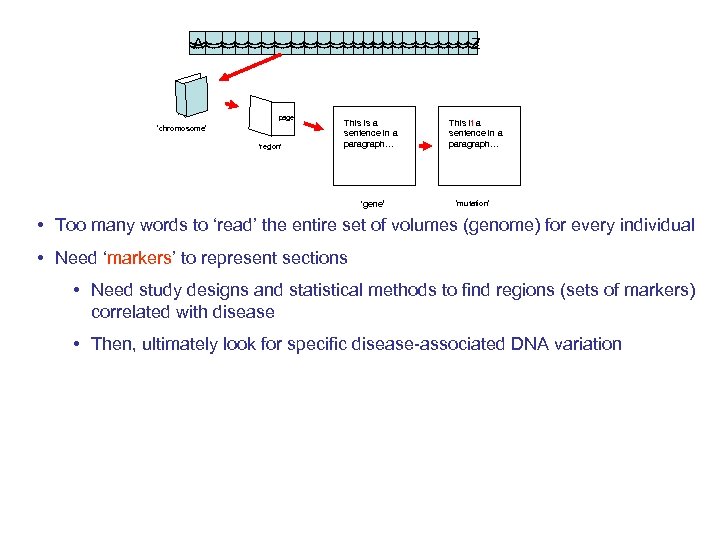
A Z page ‘chromosome’ ‘region’ This is a sentence in a paragraph… ‘gene’ This it a sentence in a paragraph… ‘mutation’ • Too many words to ‘read’ the entire set of volumes (genome) for every individual • Need ‘markers’ to represent sections • Need study designs and statistical methods to find regions (sets of markers) correlated with disease • Then, ultimately look for specific disease-associated DNA variation
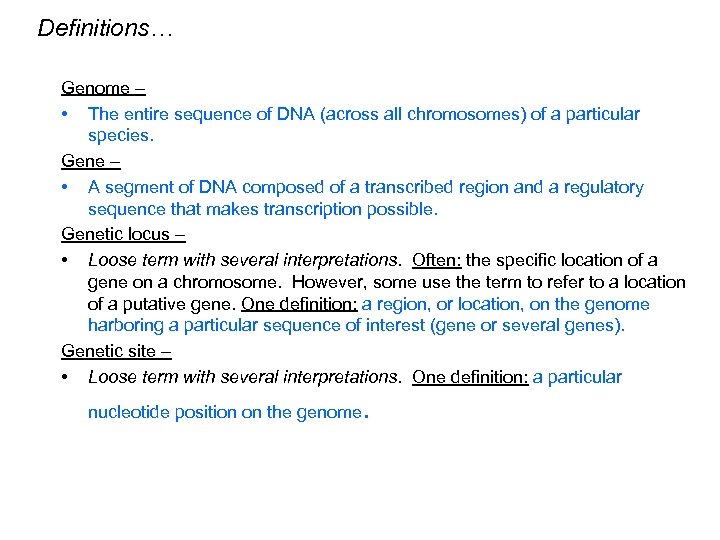
Definitions… Genome – • The entire sequence of DNA (across all chromosomes) of a particular species. Gene – • A segment of DNA composed of a transcribed region and a regulatory sequence that makes transcription possible. Genetic locus – • Loose term with several interpretations. Often: the specific location of a gene on a chromosome. However, some use the term to refer to a location of a putative gene. One definition: a region, or location, on the genome harboring a particular sequence of interest (gene or several genes). Genetic site – • Loose term with several interpretations. One definition: a particular nucleotide position on the genome .
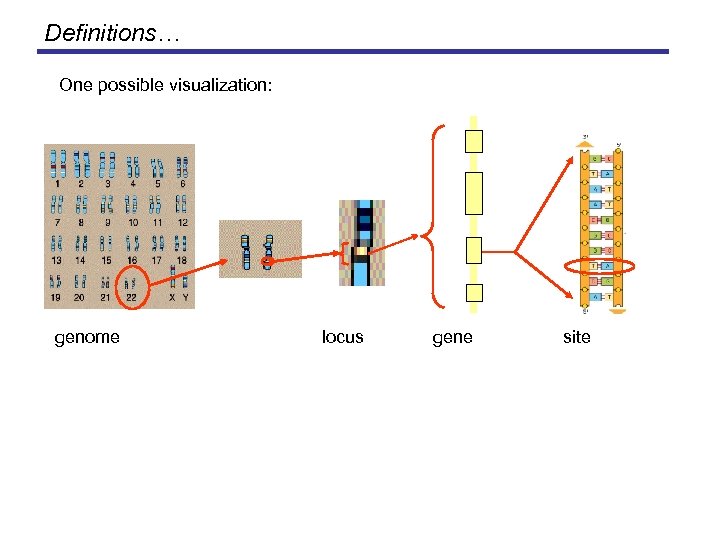
Definitions… One possible visualization: genome locus gene site
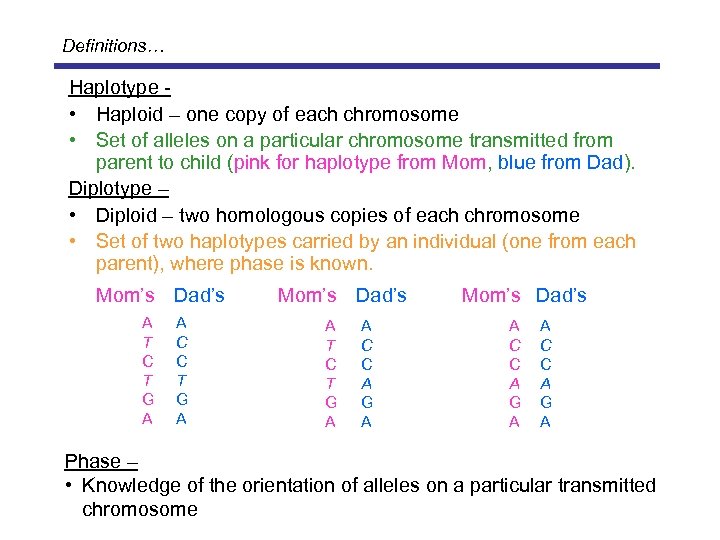
Definitions… Haplotype • Haploid – one copy of each chromosome • Set of alleles on a particular chromosome transmitted from parent to child (pink for haplotype from Mom, blue from Dad). Diplotype – • Diploid – two homologous copies of each chromosome • Set of two haplotypes carried by an individual (one from each parent), where phase is known. Mom’s Dad’s A T C T G A A C C T G A Mom’s Dad’s A T C T G A A C C A G A Mom’s Dad’s A C C A G A Phase – • Knowledge of the orientation of alleles on a particular transmitted chromosome
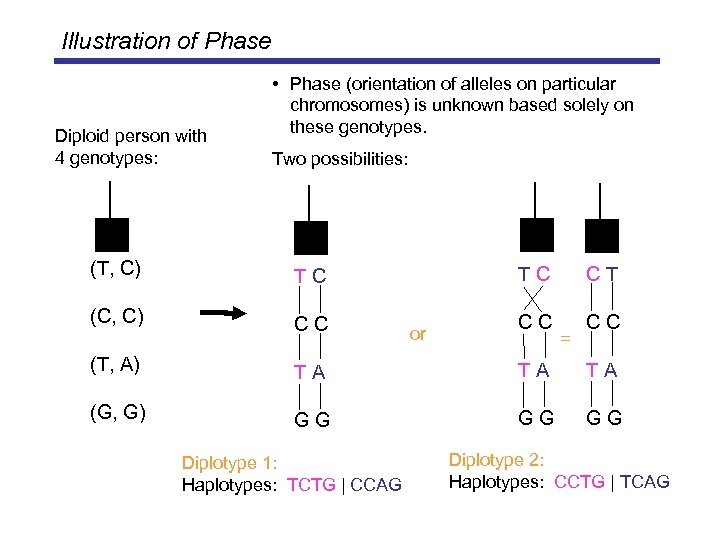
Illustration of Phase Diploid person with 4 genotypes: • Phase (orientation of alleles on particular chromosomes) is unknown based solely on these genotypes. Two possibilities: (T, C) TC (C, C) CC TC or CT CC CC = (T, A) TA TA TA (G, G) GG GG GG Diplotype 1: Haplotypes: TCTG | CCAG Diplotype 2: Haplotypes: CCTG | TCAG
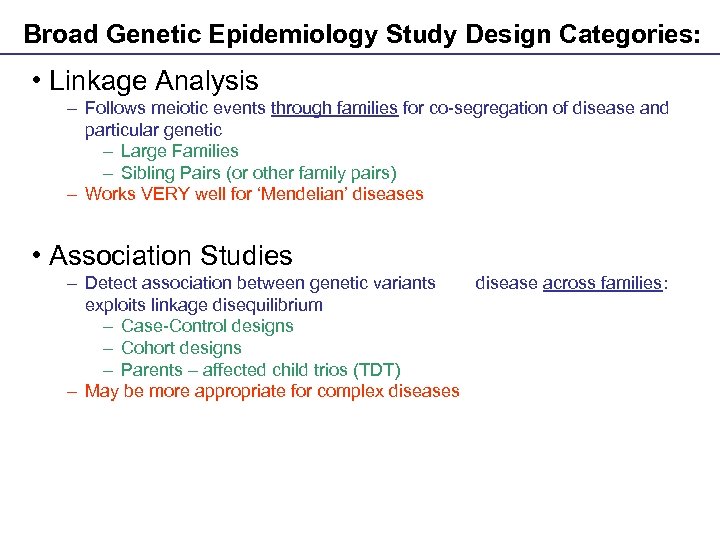
Broad Genetic Epidemiology Study Design Categories: • Linkage Analysis – Follows meiotic events through families for co-segregation of disease and particular genetic variants – Large Families – Sibling Pairs (or other family pairs) – Works VERY well for ‘Mendelian’ diseases • Association Studies – Detect association between genetic variants and disease across families: exploits linkage disequilibrium – Case-Control designs – Cohort designs – Parents – affected child trios (TDT) – May be more appropriate for complex diseases

Q. What approaches exist for association studies ?
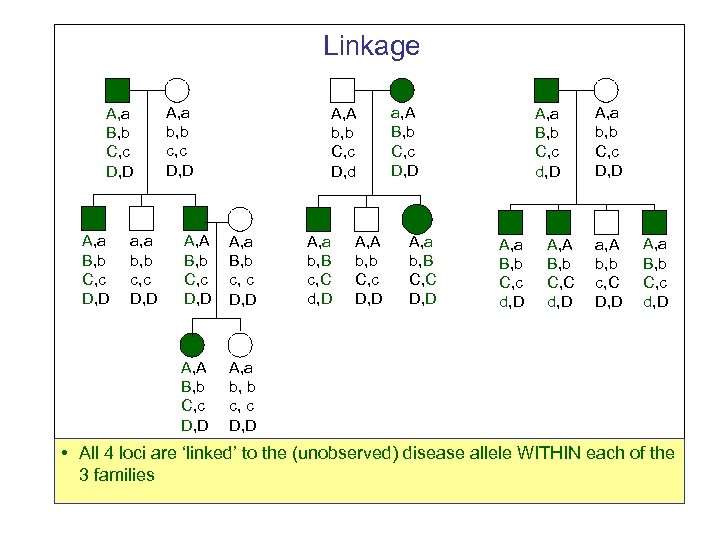
Linkage A, a B, b C, c D, D a, a b, b c, c D, D A, A b, b C, c D, d A, A B, b C, c D, D A, a B, b c, c D, D A, A B, b C, c D, D a, A B, b C, c D, D A, a B, b C, c d, D A, a b, b C, c D, D A, a b, b c, c D, D A, a b, B c, C d, D A, A b, b C, c D, D A, a b, B C, C D, D A, a B, b C, c d, D A, A B, b C, C d, D a, A b, b c, C D, D A, a B, b C, c d, D • All 4 loci are ‘linked’ to the (unobserved) disease allele WITHIN each of the 3 families
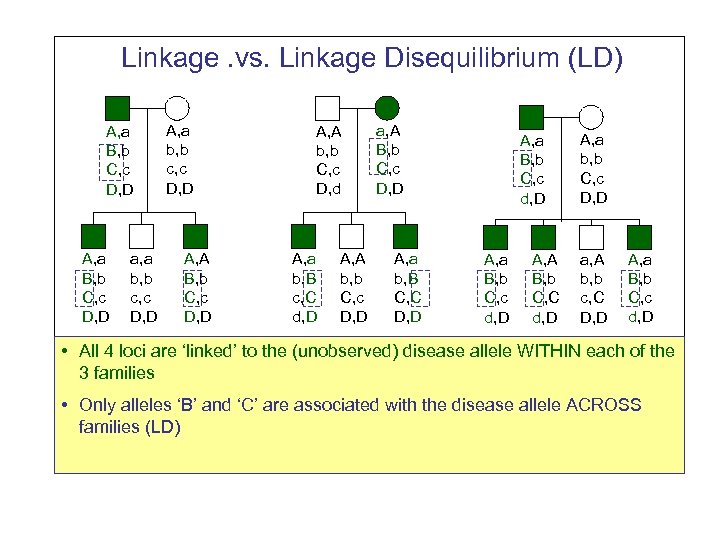
Linkage. vs. Linkage Disequilibrium (LD) A, a B, b C, c D, D a, a b, b c, c D, D A, A B, b C, c D, D A, A b, b C, c D, d A, a b, B c, C d, D A, A b, b C, c D, D a, A B, b C, c D, D A, a b, B C, C D, D A, a B, b C, c d, D A, A B, b C, C d, D A, a b, b C, c D, D a, A b, b c, C D, D A, a B, b C, c d, D • All 4 loci are ‘linked’ to the (unobserved) disease allele WITHIN each of the 3 families • Only alleles ‘B’ and ‘C’ are associated with the disease allele ACROSS families (LD)
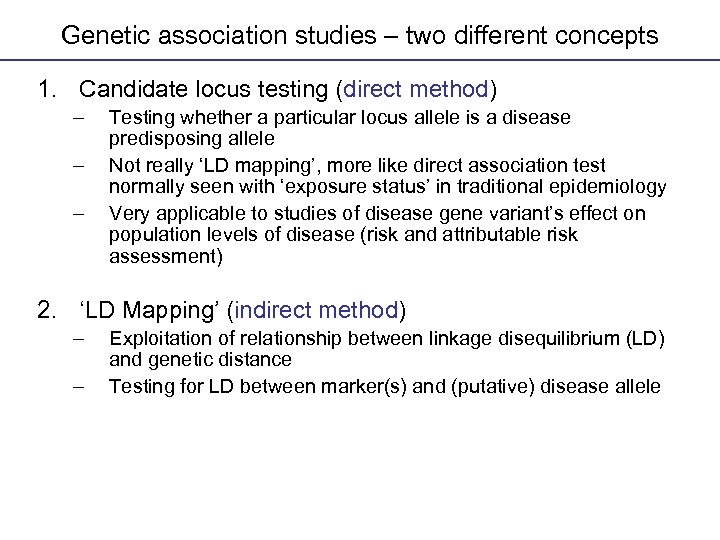
Genetic association studies – two different concepts Genetic Association Studies 1. Candidate locus testing (direct method) – – – Testing whether a particular locus allele is a disease predisposing allele Not really ‘LD mapping’, more like direct association test normally seen with ‘exposure status’ in traditional epidemiology Very applicable to studies of disease gene variant’s effect on population levels of disease (risk and attributable risk assessment) 2. ‘LD Mapping’ (indirect method) – – Exploitation of relationship between linkage disequilibrium (LD) and genetic distance Testing for LD between marker(s) and (putative) disease allele
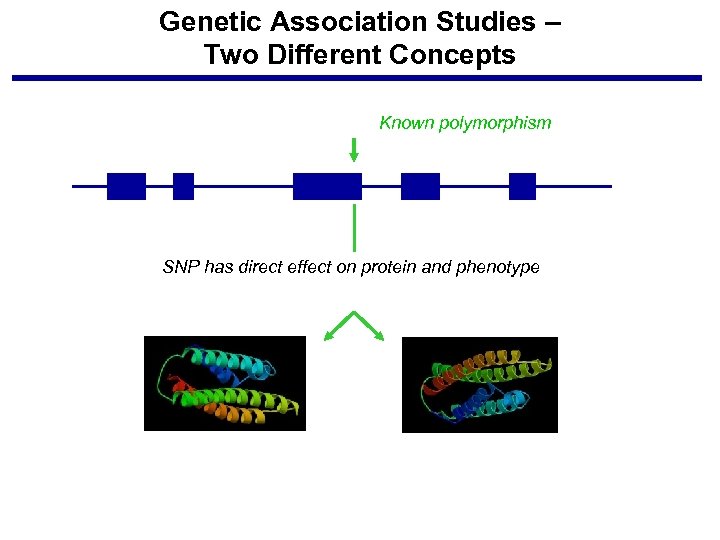
Genetic Association Studies – Two Different Concepts Known polymorphism SNP has direct effect on protein and phenotype
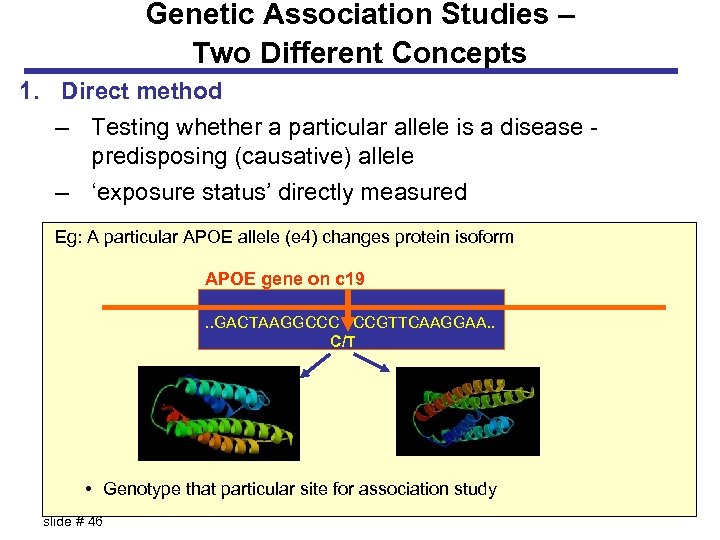
Genetic Association Studies – Two Different Concepts 1. Direct method – Testing whether a particular allele is a disease predisposing (causative) allele – ‘exposure status’ directly measured Eg: A particular APOE allele (e 4) changes protein isoform APOE gene on c 19. . GACTAAGGCCC CCGTTCAAGGAA. . C/T • Genotype that particular site for association study Fallin, L 3 a, 6/21/2005 slide # 46
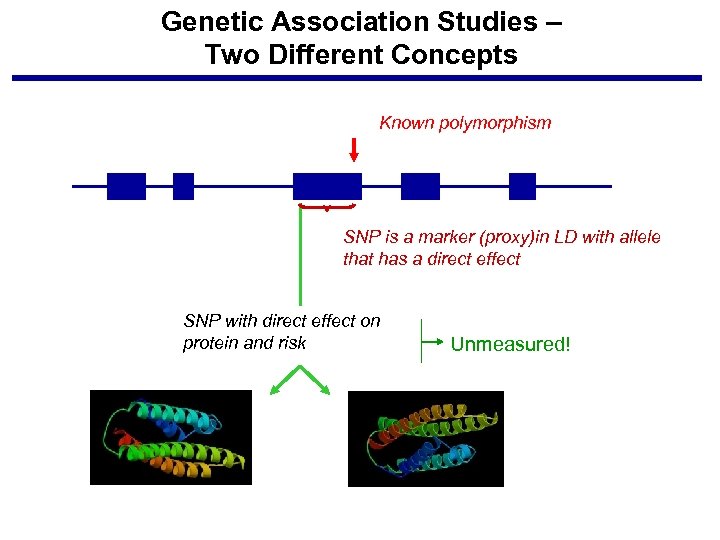
Genetic Association Studies – Two Different Concepts Known polymorphism SNP is a marker (proxy)in LD with allele that has a direct effect SNP with direct effect on protein and risk Unmeasured!
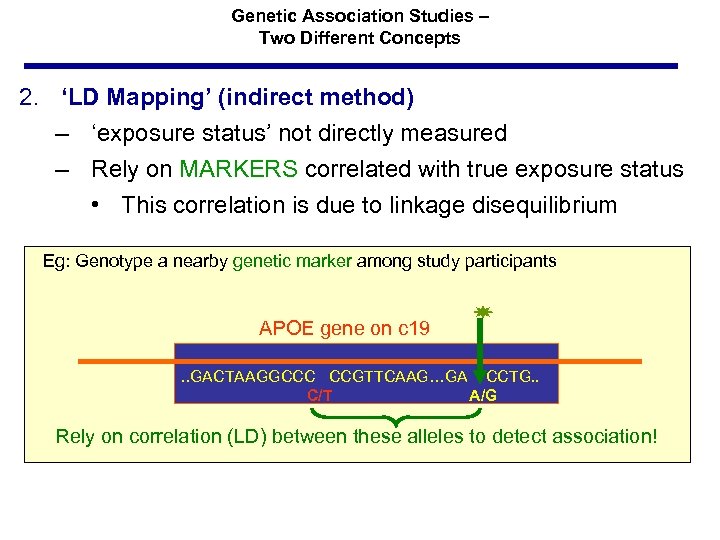
Genetic Association Studies – Two Different Concepts 2. ‘LD Mapping’ (indirect method) – ‘exposure status’ not directly measured – Rely on MARKERS correlated with true exposure status • This correlation is due to linkage disequilibrium Eg: Genotype a nearby genetic marker among study participants APOE gene on c 19. . GACTAAGGCCC CCGTTCAAG…GA CCTG. . C/T A/G Rely on correlation (LD) between these alleles to detect association!
Marker-based Studies • We often do not measure the genetic variant of interest • Instead, we genotype markers at known locations in the genome • Look for markers the may indicate close proximity to a disease-related DNA variant
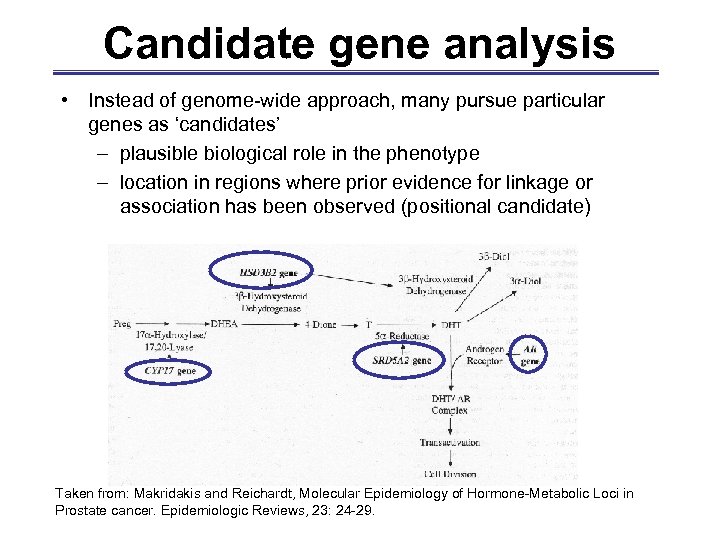
Candidate gene analysis • Instead of genome-wide approach, many pursue particular genes as ‘candidates’ – plausible biological role in the phenotype – location in regions where prior evidence for linkage or association has been observed (positional candidate) Taken from: Makridakis and Reichardt, Molecular Epidemiology of Hormone-Metabolic Loci in Prostate cancer. Epidemiologic Reviews, 23: 24 -29.
Candidate Genes • • • CD 209 (DC-SIGN) VDR Fc γ receptor II TNF- , IL-10 HLA-A, HLA-B TAP 1, TAP 2, and CTLA-4
Case Control Comparison Groups
Genome Wide Association Studies • Large number of individuals with disease and a relevant comparison group • DNA isolation and genotyping • Statistical tests for associations between the SNPs passing quality thresholds and the disease/trait • Replication of identified associations in an independent population sample or examination of functional implications experimentally.

Lessons Learned from initial G WA Studies • • This actually works Size and luck matter! Replication matters Collaboration matters Controls matter, but can be shared sometimes Non-coding SNPs matter Current hypotheses regarding candidate genes and pathways may not matter so much • Several genes influence more than one disease
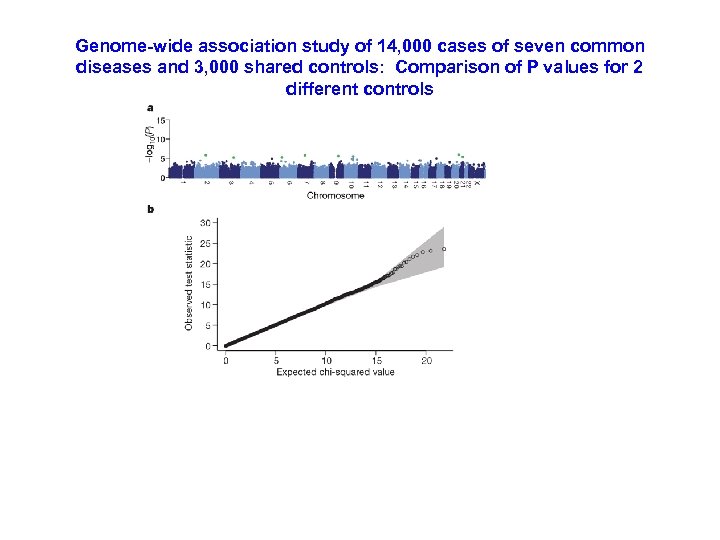
Genome-wide association study of 14, 000 cases of seven common diseases and 3, 000 shared controls: Comparison of P values for 2 different controls

Q. What is the basis for LD?
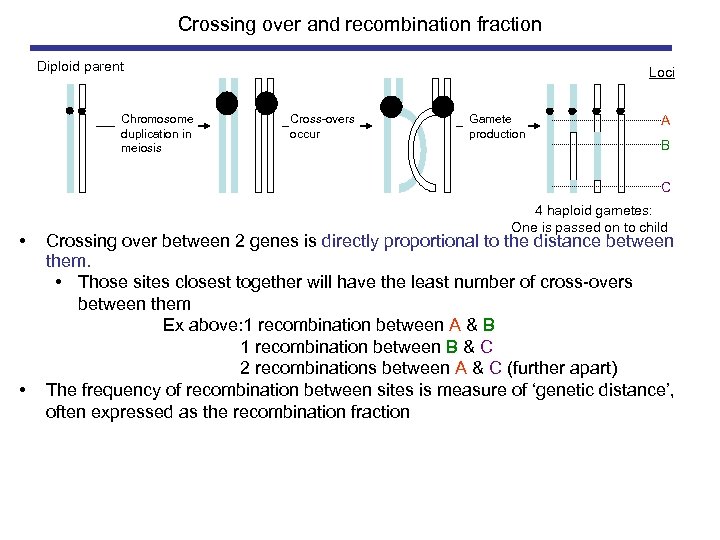
Crossing over and recombination fraction Diploid parent Chromosome duplication in meiosis Loci . Cross-overs occur Gamete production A B C • • 4 haploid gametes: One is passed on to child Crossing over between 2 genes is directly proportional to the distance between them. • Those sites closest together will have the least number of cross-overs between them Ex above: 1 recombination between A & B 1 recombination between B & C 2 recombinations between A & C (further apart) The frequency of recombination between sites is measure of ‘genetic distance’, often expressed as the recombination fraction

LD Mapping Caveats: Other Reasons for Observed Allelic Associations in Populations • Population stratification / subdivision • Recent admixture • Genetic drift • Selection • Assortative mating • Type 1 error

Q. What are key assumptions made in genetic epidemiology studies?
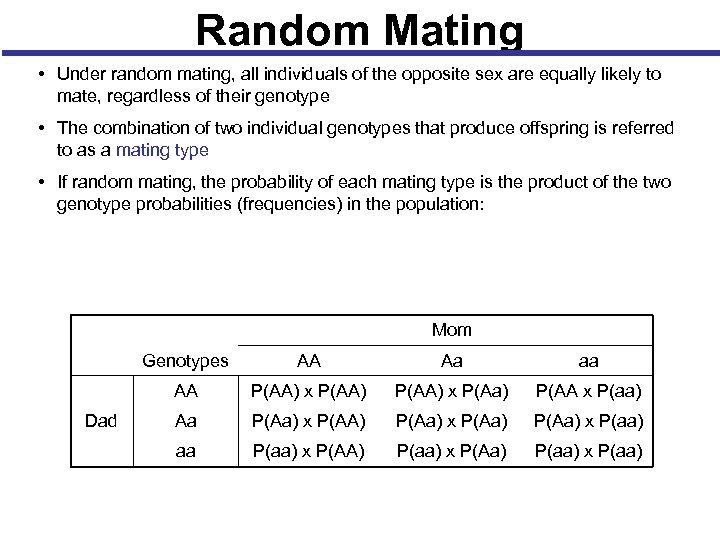
Random Mating • Under random mating, all individuals of the opposite sex are equally likely to mate, regardless of their genotype • The combination of two individual genotypes that produce offspring is referred to as a mating type • If random mating, the probability of each mating type is the product of the two genotype probabilities (frequencies) in the population: Mom Genotypes Aa aa AA Dad AA P(AA) x P(Aa) P(AA x P(aa) Aa P(Aa) x P(AA) P(Aa) x P(aa) aa P(aa) x P(AA) P(aa) x P(Aa) P(aa) x P(aa)
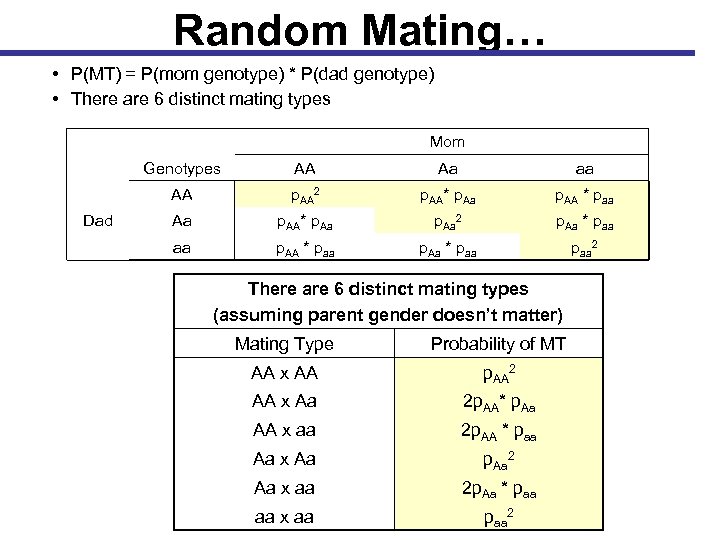
Random Mating… • P(MT) = P(mom genotype) * P(dad genotype) • There are 6 distinct mating types Mom Genotypes Aa aa AA Dad AA p. AA 2 p. AA* p. Aa p. AA * paa Aa p. AA* p. Aa 2 p. Aa * paa aa p. AA * paa p. Aa * paa 2 There are 6 distinct mating types (assuming parent gender doesn’t matter) Mating Type Probability of MT AA x AA p. AA 2 AA x Aa 2 p. AA* p. Aa AA x aa 2 p. AA * paa Aa x Aa p. Aa 2 Aa x aa 2 p. Aa * paa aa x aa paa 2
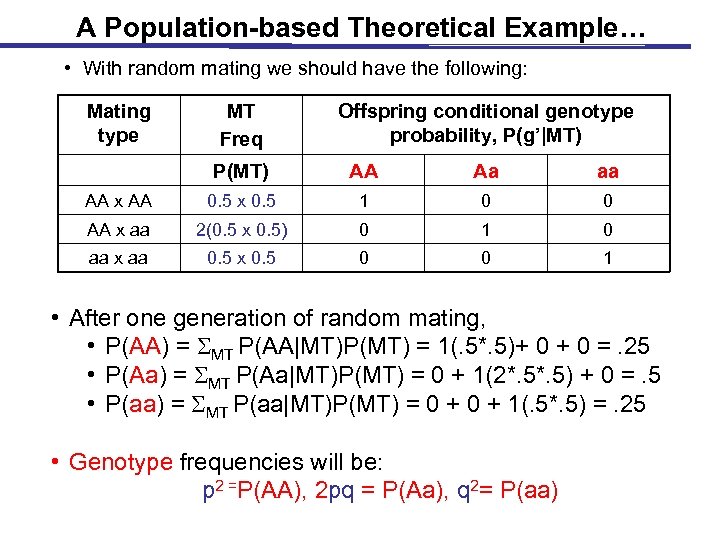
A Population-based Theoretical Example… • With random mating we should have the following: Mating type MT Freq Offspring conditional genotype probability, P(g’|MT) P(MT) AA Aa aa AA x AA 0. 5 x 0. 5 1 0 0 AA x aa 2(0. 5 x 0. 5) 0 1 0 aa x aa 0. 5 x 0. 5 0 0 1 • After one generation of random mating, • P(AA) = SMT P(AA|MT)P(MT) = 1(. 5*. 5)+ 0 =. 25 • P(Aa) = SMT P(Aa|MT)P(MT) = 0 + 1(2*. 5) + 0 =. 5 • P(aa) = SMT P(aa|MT)P(MT) = 0 + 1(. 5*. 5) =. 25 • Genotype frequencies will be: p 2 =P(AA), 2 pq = P(Aa), q 2= P(aa)
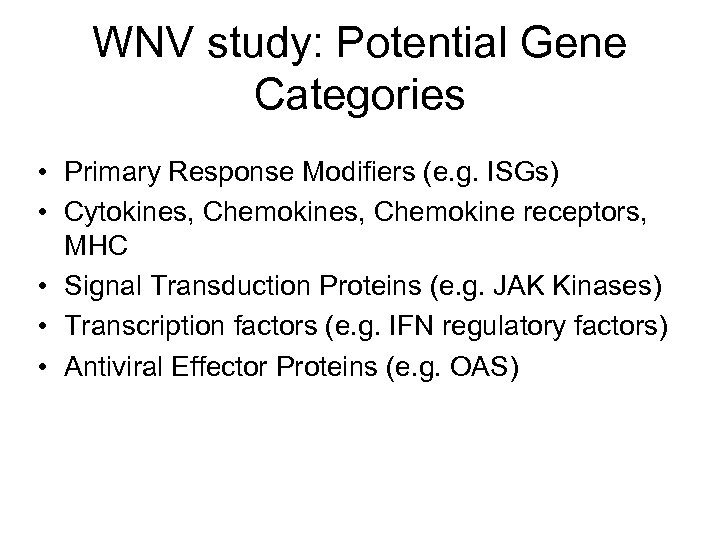
WNV study: Potential Gene Categories • Primary Response Modifiers (e. g. ISGs) • Cytokines, Chemokine receptors, MHC • Signal Transduction Proteins (e. g. JAK Kinases) • Transcription factors (e. g. IFN regulatory factors) • Antiviral Effector Proteins (e. g. OAS)
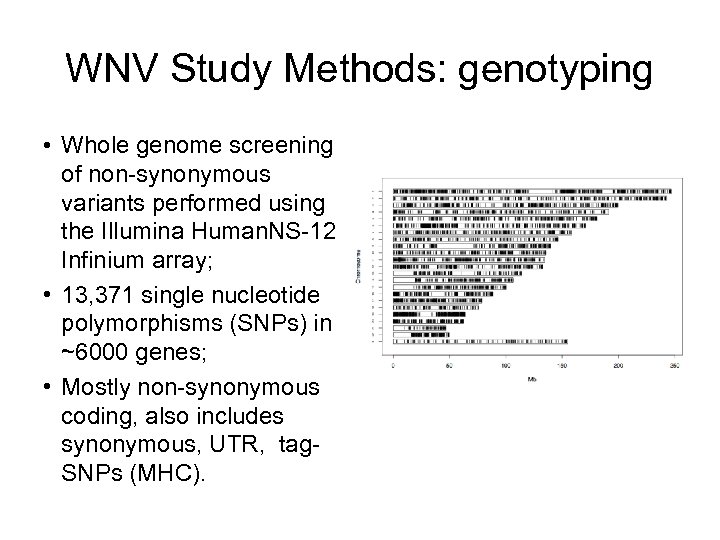
WNV Study Methods: genotyping • Whole genome screening of non-synonymous variants performed using the Illumina Human. NS-12 Infinium array; • 13, 371 single nucleotide polymorphisms (SNPs) in ~6000 genes; • Mostly non-synonymous coding, also includes synonymous, UTR, tag. SNPs (MHC).

Case-Control Study • Cases from states/provinces with highest rates of WNV infection • Meet CDC criteria for WNV infection and have evidence of neuroinvasive diease • Controls are those who meet criteria for infection with WNV but who did not develop neuroinvasive disease
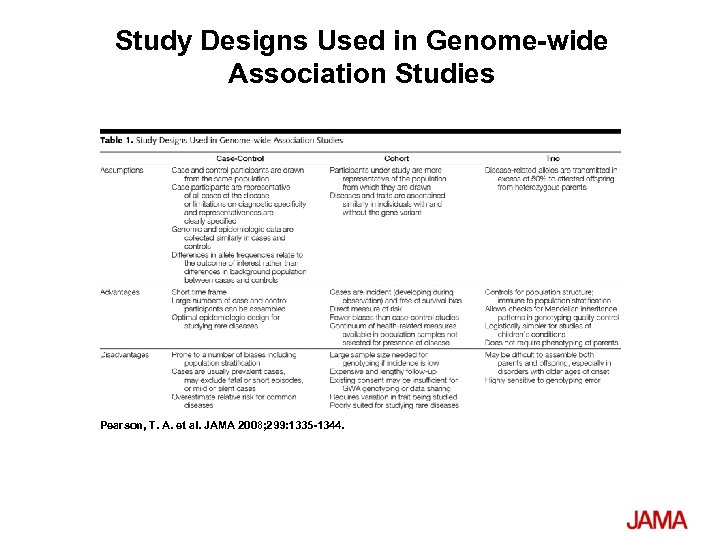
Study Designs Used in Genome-wide Association Studies Pearson, T. A. et al. JAMA 2008; 299: 1335 -1344.

Implementation • State and provincial public health agencies contact all WNV infected individuals in 20022008 • 4 Clinical Centers – Pennsylvania, Texas, Nebraska, Ontario • Whole blood is collected from participants and sent to Mc. Gill Genome Center
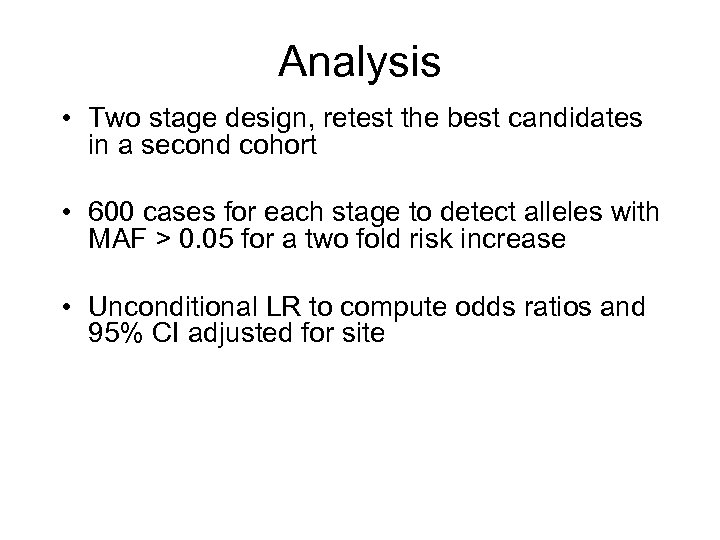
Analysis • Two stage design, retest the best candidates in a second cohort • 600 cases for each stage to detect alleles with MAF > 0. 05 for a two fold risk increase • Unconditional LR to compute odds ratios and 95% CI adjusted for site
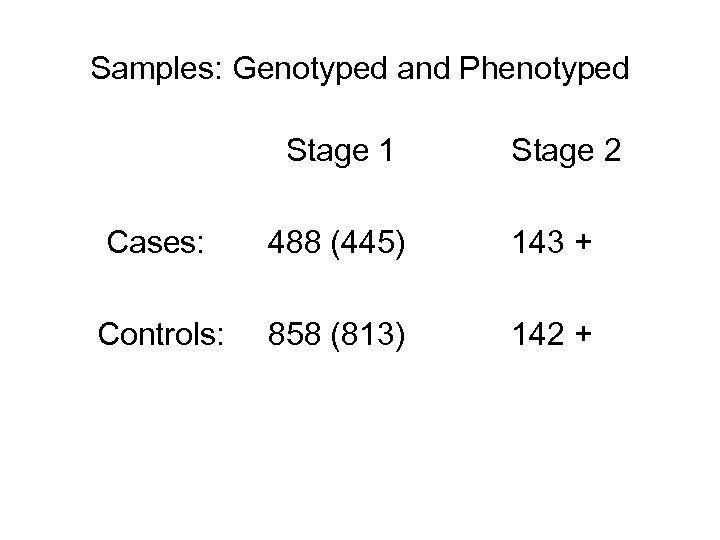
Samples: Genotyped and Phenotyped Stage 1 Stage 2 Cases: 488 (445) 143 + Controls: 858 (813) 142 +
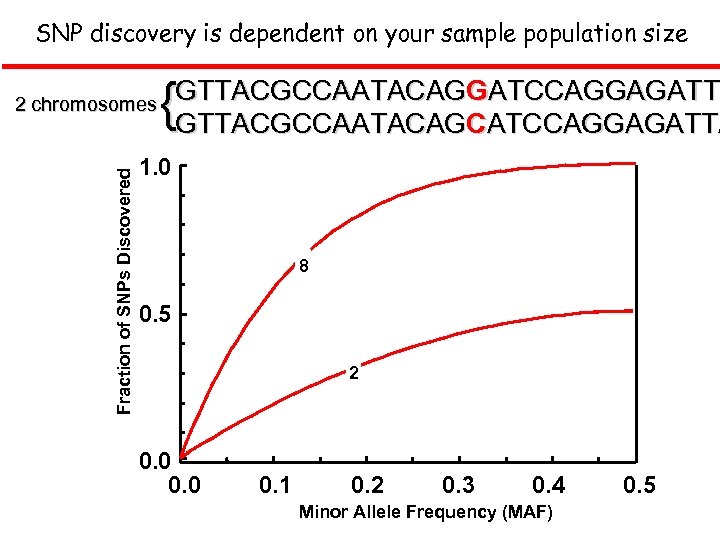
SNP discovery is dependent on your sample population size Fraction of SNPs Discovered 2 chromosomes { GTTACGCCAATACAGGATCCAGGAGATTA ATCCAGGAGATT GTTACGCCAATACAGCATCCAGGAGATTA 1. 0 8 8 0. 5 2 0. 0 0. 1 0. 2 0. 3 0. 4 Minor Allele Frequency (MAF) 0. 5
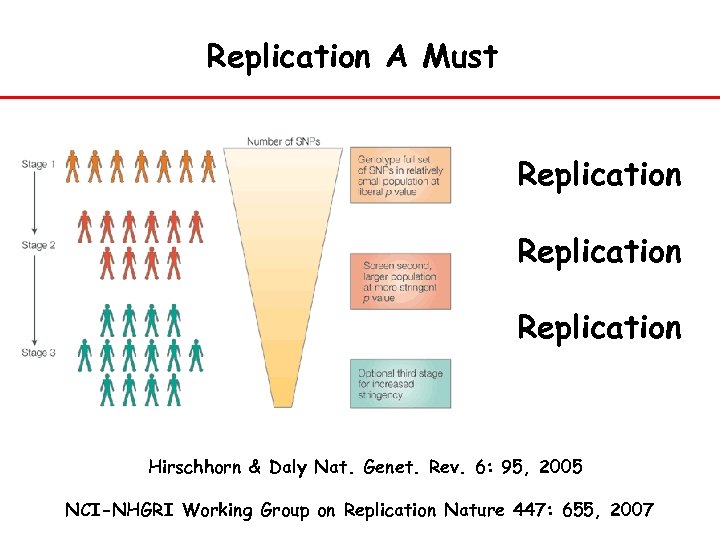
Replication A Must Replication Hirschhorn & Daly Nat. Genet. Rev. 6: 95, 2005 NCI-NHGRI Working Group on Replication Nature 447: 655, 2007
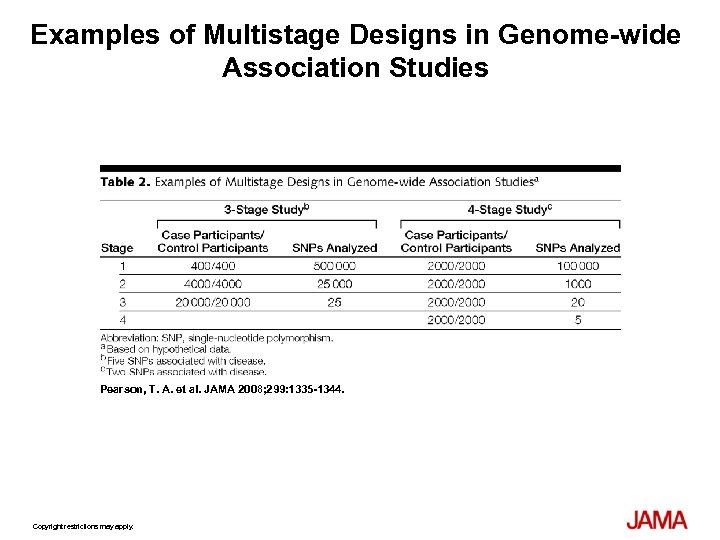
Examples of Multistage Designs in Genome-wide Association Studies Pearson, T. A. et al. JAMA 2008; 299: 1335 -1344. Copyright restrictions may apply.
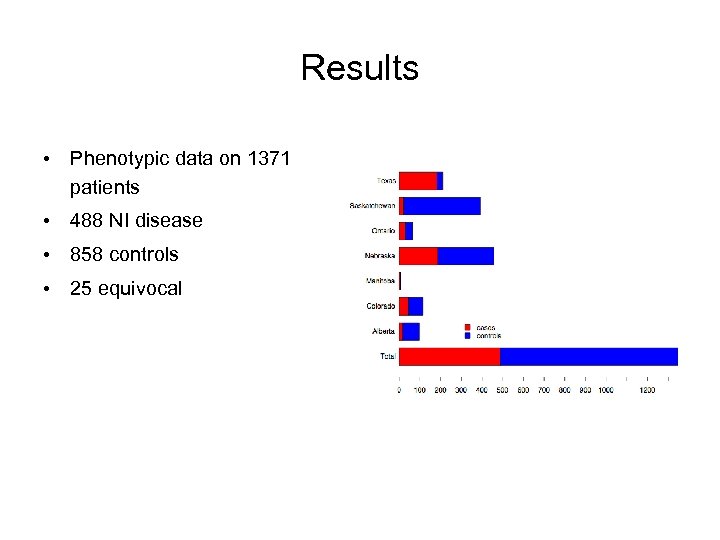
Results • Phenotypic data on 1371 patients • 488 NI disease • 858 controls • 25 equivocal
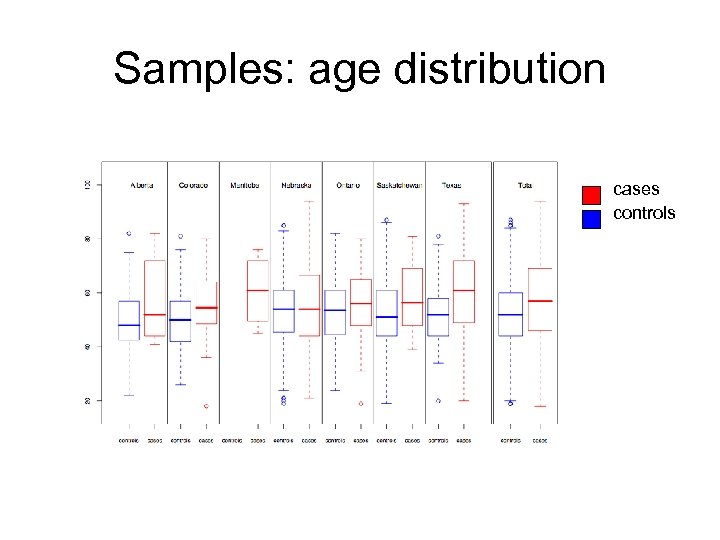
Samples: age distribution cases controls
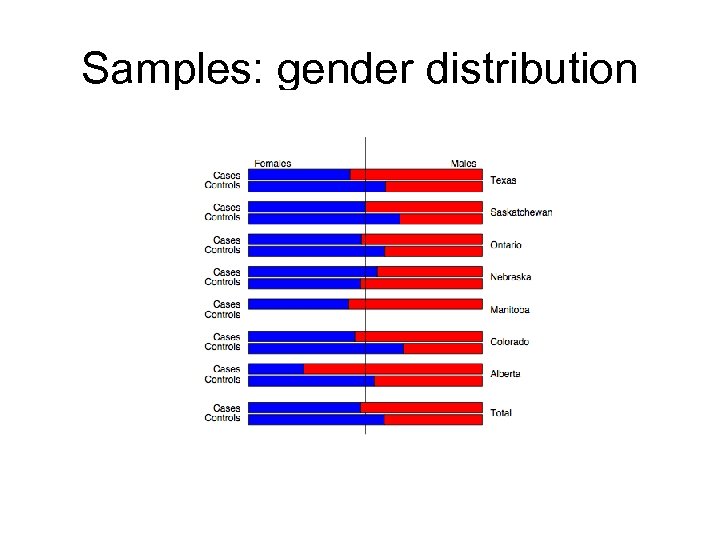
Samples: gender distribution
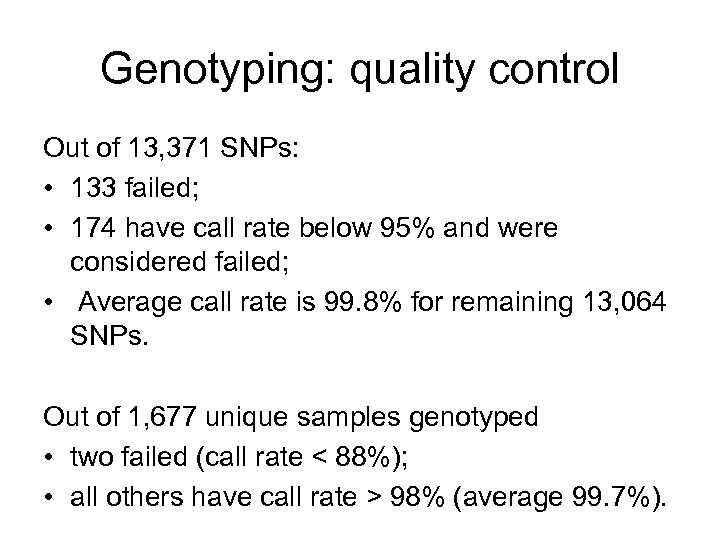
Genotyping: quality control Out of 13, 371 SNPs: • 133 failed; • 174 have call rate below 95% and were considered failed; • Average call rate is 99. 8% for remaining 13, 064 SNPs. Out of 1, 677 unique samples genotyped • two failed (call rate < 88%); • all others have call rate > 98% (average 99. 7%).
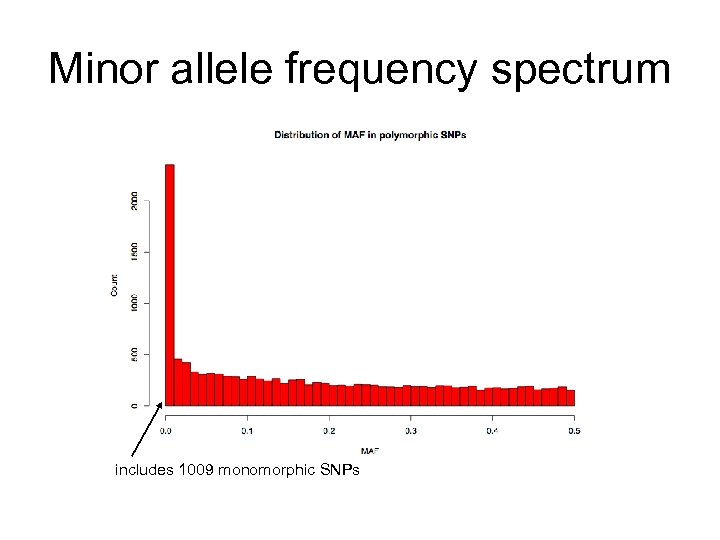
Minor allele frequency spectrum includes 1009 monomorphic SNPs
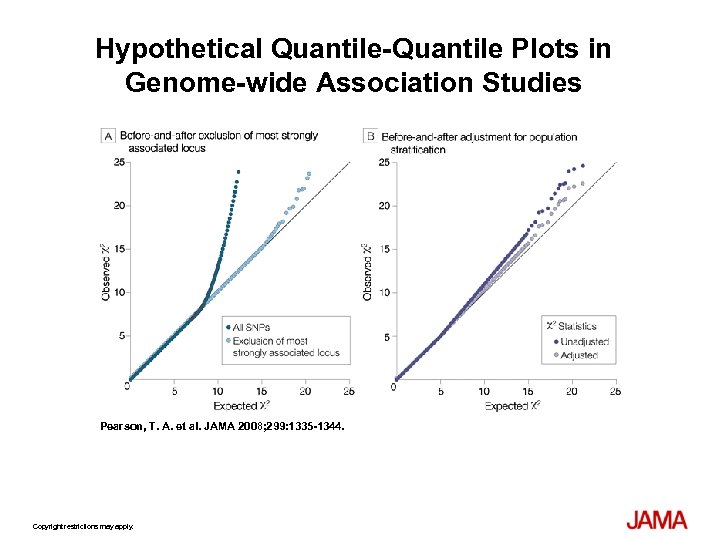
Hypothetical Quantile-Quantile Plots in Genome-wide Association Studies Pearson, T. A. et al. JAMA 2008; 299: 1335 -1344. Copyright restrictions may apply.
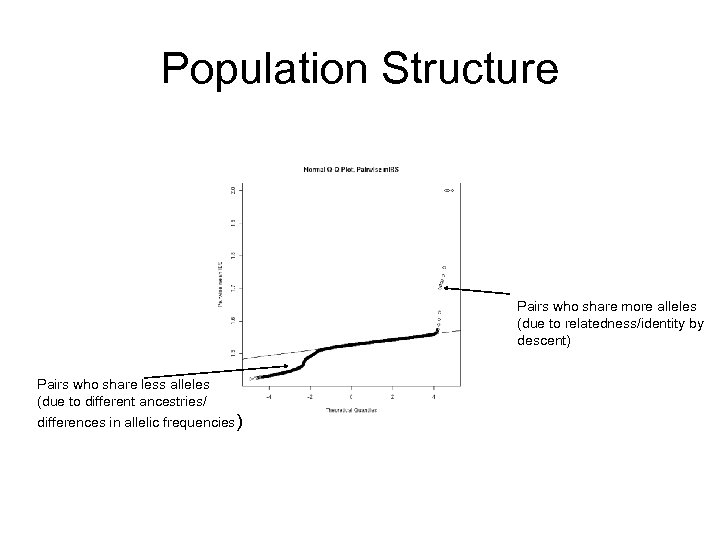
Population Structure Pairs who share more alleles (due to relatedness/identity by descent) Pairs who share less alleles (due to different ancestries/ differences in allelic frequencies)
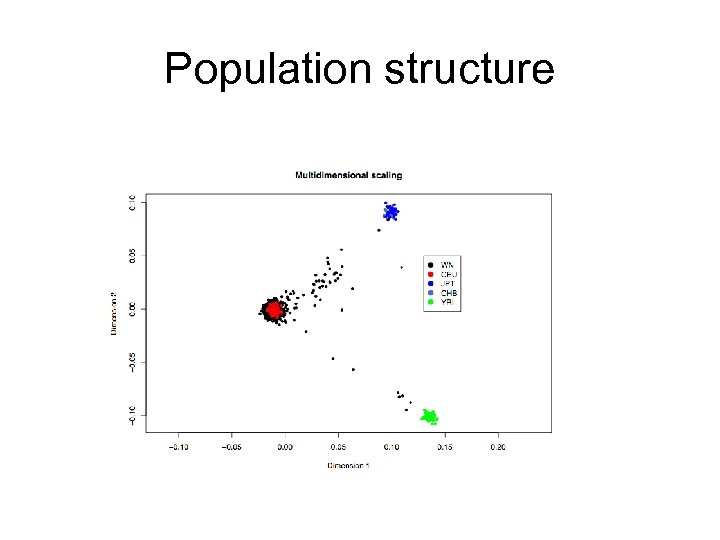
Population structure
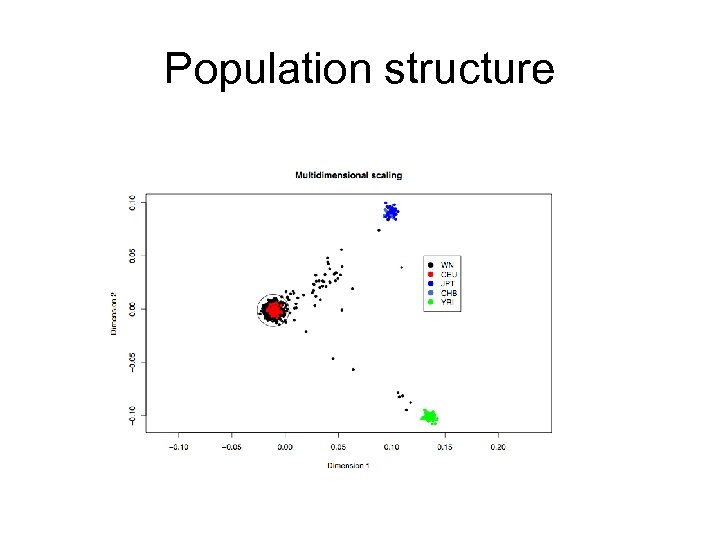
Population structure
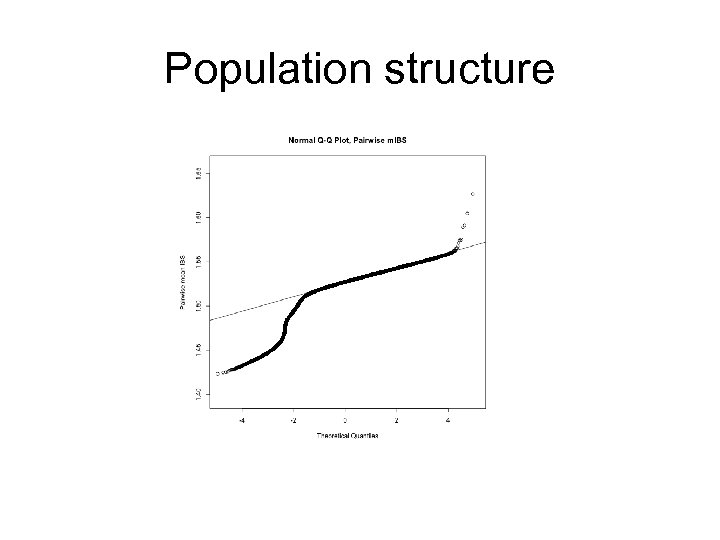
Population structure
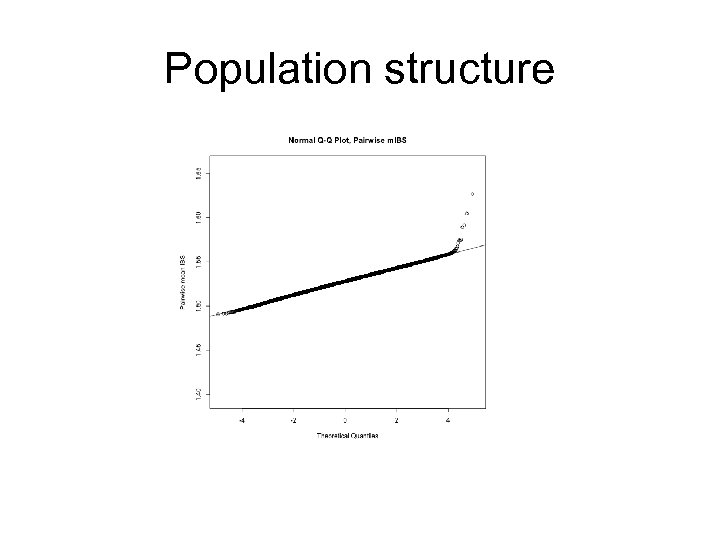
Population structure
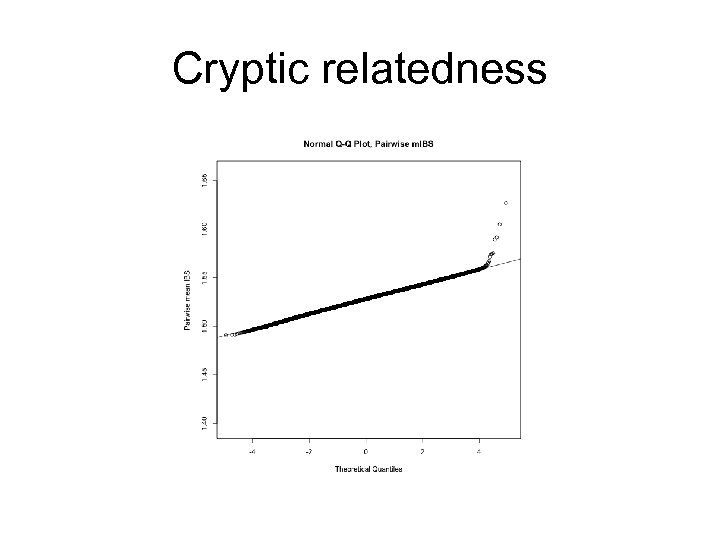
Cryptic relatedness
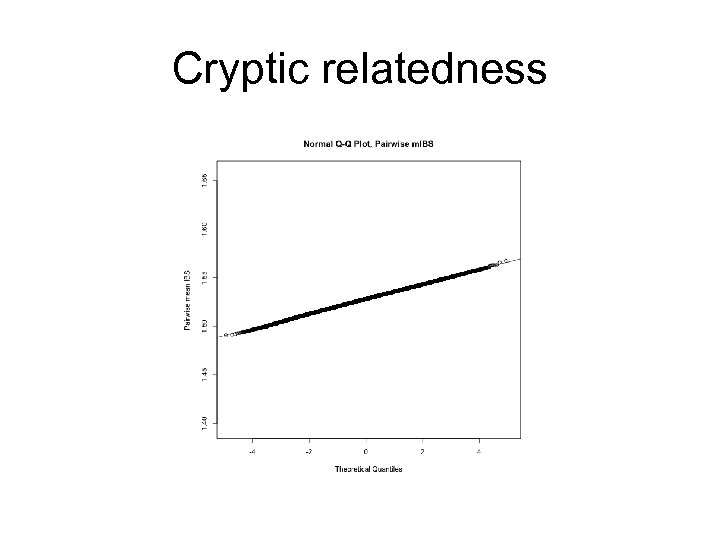
Cryptic relatedness
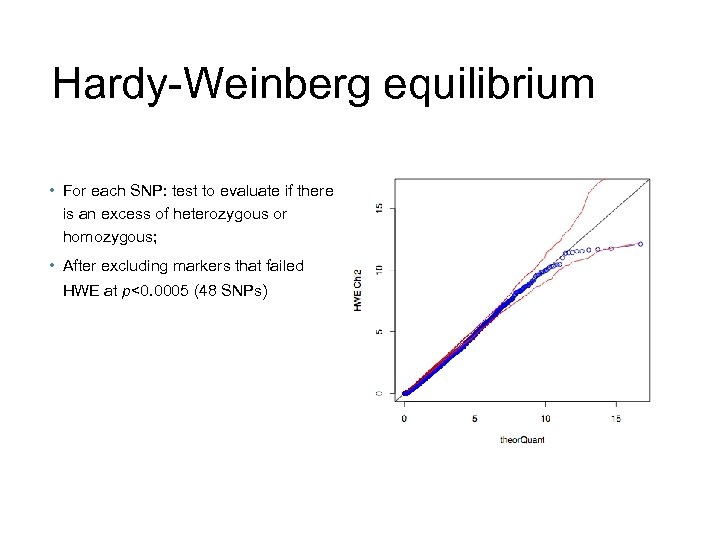
Hardy-Weinberg equilibrium • For each SNP: test to evaluate if there is an excess of heterozygous or homozygous; • After excluding markers that failed HWE at p<0. 0005 (48 SNPs)
Statistical Tests for HWP • Q: Is an observed departure from HWP statistically significant? – Ho: DHW = 0 HA: DHW 0 • Methods: – Chi-square goodness of fit (GOF) • Ho: Do the data fit a model where genotype frequencies equal expected values under HWP? – Likelihood ratio test (LRT) • Ho: Does a model assuming HWE fit the observed data better than a model that does not assume HWE? – I. e. compare likelihood of data, fixing genotype frequencies to HWP (Lo) versus likelihood of the data without fixing genotype frequencies to match HWP (L 1)
Reasons for Departure from HWP • Population allele frequencies can change from generation to generation due to: • • Migration / admixture Chance, in small populations - genetic drift Mutation Selection - depends on fertility of parents and viability of offspring • Survival bias and gender proportions - Allele frequencies can also change with age within a generation, and could be sex dependent. • Abnormal gene segregation (segregation distortion, meiotic drive - all maternal and paternal gametic contributions are not equally probable)
Why is the H-W model useful to Genetic Epidemiology? • Can use HWP assumption to calculate genotype frequencies from observed phenotypes • Can use HWP to obtain haplotype frequencies from observed genotypes – useful for assessing inter-locus equilibrium, later lectures… • Can measure departures from HWP as an indication of population genetic features in a sample: – Inbreeding – Migration / admixture • Can judge potential genotyping errors Ø Important to test for HWE!
Testing for association After applying all QC filters: • 445 neuroinvasive cases, 813 controls; • 10, 591 SNPs with MAF > 1% in controls entered the analysis; • Logistic model, adjusting for collection center; • X chromosome: risk of males = risk of homozygous females; gender as additional covariate.
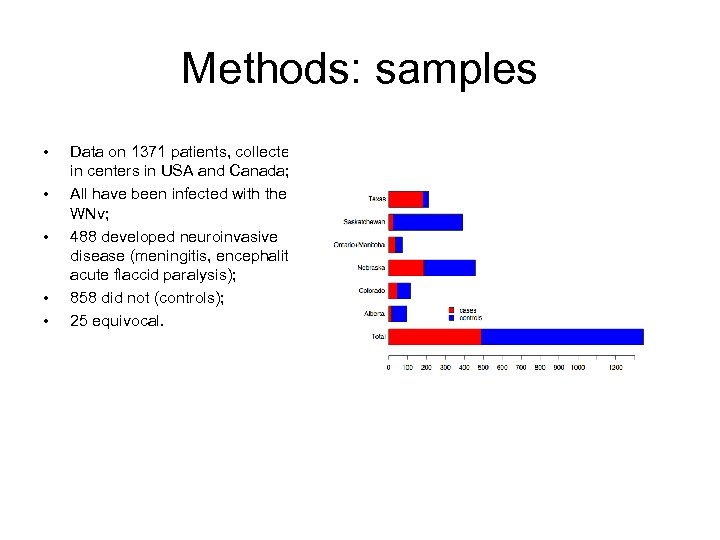
Methods: samples • • • Data on 1371 patients, collected in centers in USA and Canada; All have been infected with the WNv; 488 developed neuroinvasive disease (meningitis, encephalitis, acute flaccid paralysis); 858 did not (controls); 25 equivocal.
Methods: genotyping • hole genome screening of non. W synonymous variants performed using the Illumina Human. NS-12 Infinium array; • 13, 371 single nucleotide polymorphisms (SNPs); • Mostly non-synonymous coding, also includes synonymous, UTR, tag-SNPs (MHC).
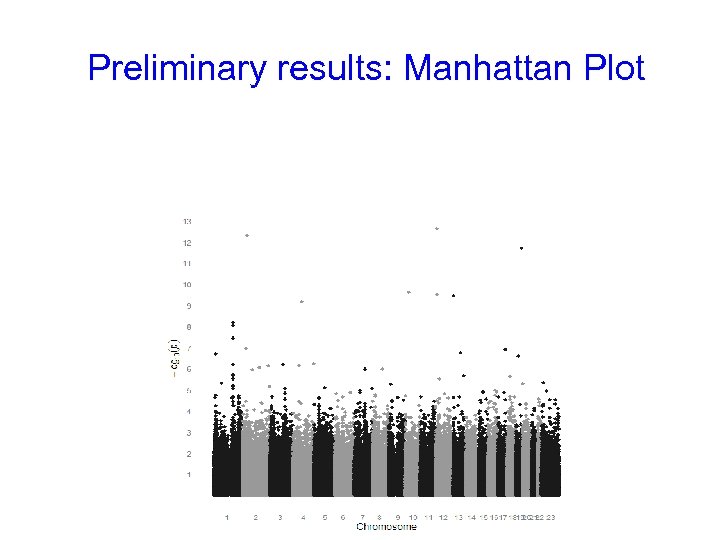
Preliminary results: Manhattan Plot
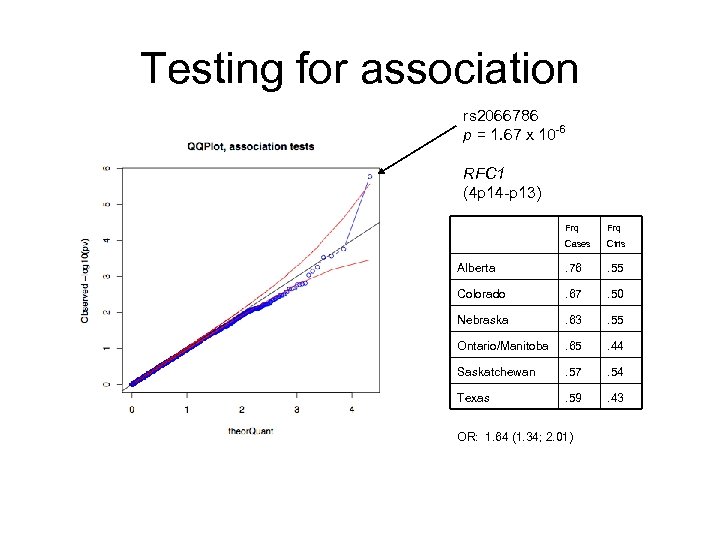
Testing for association rs 2066786 p = 1. 67 x 10 -6 RFC 1 (4 p 14 -p 13) Frq Cases Ctrls Alberta . 76 . 55 Colorado . 67 . 50 Nebraska . 63 . 55 Ontario/Manitoba . 65 . 44 Saskatchewan . 57 . 54 Texas . 59 . 43 OR: 1. 64 (1. 34; 2. 01)
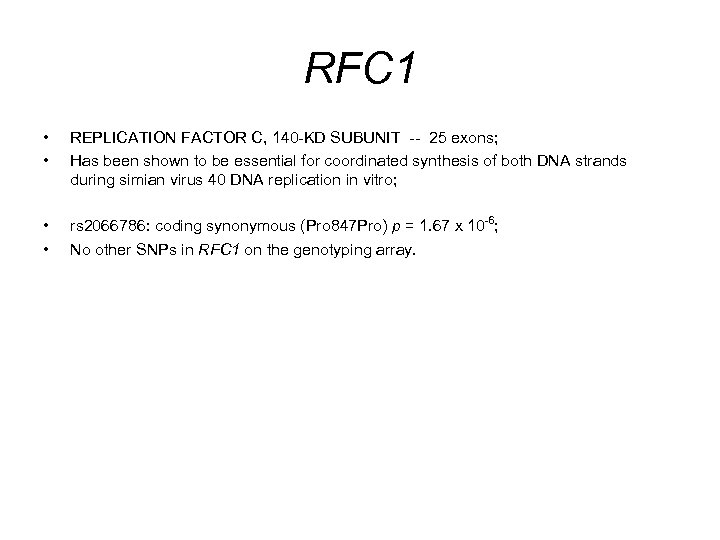
RFC 1 • • REPLICATION FACTOR C, 140 -KD SUBUNIT -- 25 exons; Has been shown to be essential for coordinated synthesis of both DNA strands during simian virus 40 DNA replication in vitro; • • rs 2066786: coding synonymous (Pro 847 Pro) p = 1. 67 x 10 -6; No other SNPs in RFC 1 on the genotyping array.
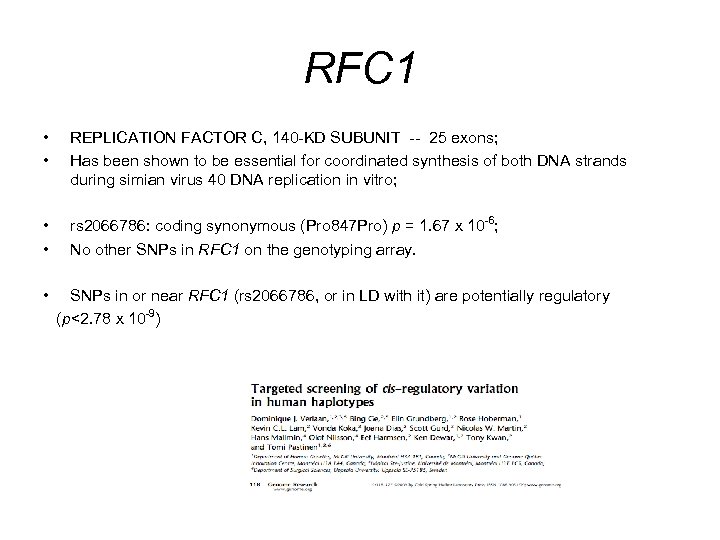
RFC 1 • • REPLICATION FACTOR C, 140 -KD SUBUNIT -- 25 exons; Has been shown to be essential for coordinated synthesis of both DNA strands during simian virus 40 DNA replication in vitro; • • rs 2066786: coding synonymous (Pro 847 Pro) p = 1. 67 x 10 -6; No other SNPs in RFC 1 on the genotyping array. • SNPs in or near RFC 1 (rs 2066786, or in LD with it) are potentially regulatory (p<2. 78 x 10 -9)
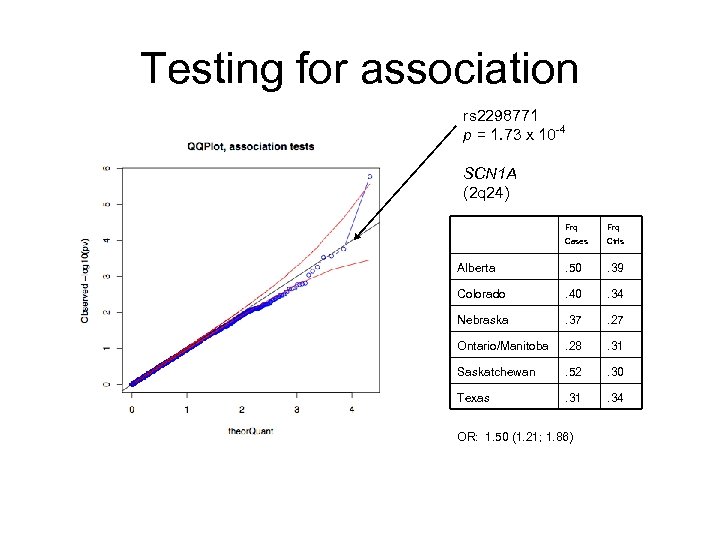
Testing for association rs 2298771 p = 1. 73 x 10 -4 SCN 1 A (2 q 24) Frq Cases Ctrls Alberta . 50 . 39 Colorado . 40 . 34 Nebraska . 37 . 27 Ontario/Manitoba . 28 . 31 Saskatchewan . 52 . 30 Texas . 31 . 34 OR: 1. 50 (1. 21; 1. 86)
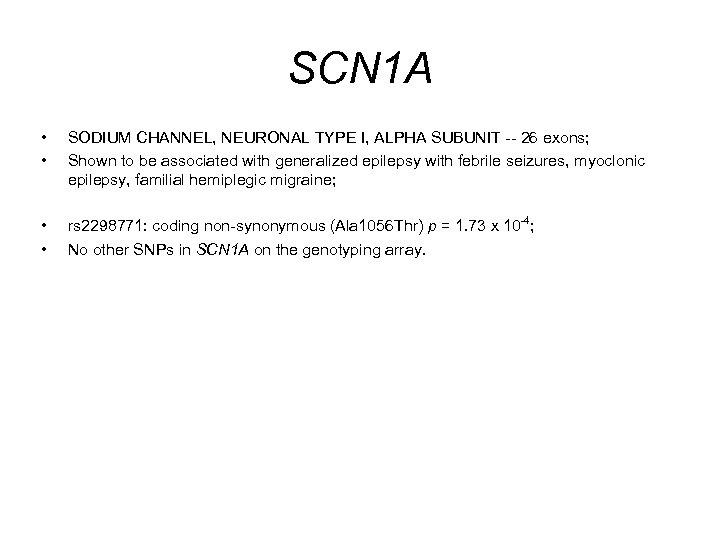
SCN 1 A • • SODIUM CHANNEL, NEURONAL TYPE I, ALPHA SUBUNIT -- 26 exons; Shown to be associated with generalized epilepsy with febrile seizures, myoclonic epilepsy, familial hemiplegic migraine; • • rs 2298771: coding non-synonymous (Ala 1056 Thr) p = 1. 73 x 10 -4; No other SNPs in SCN 1 A on the genotyping array.
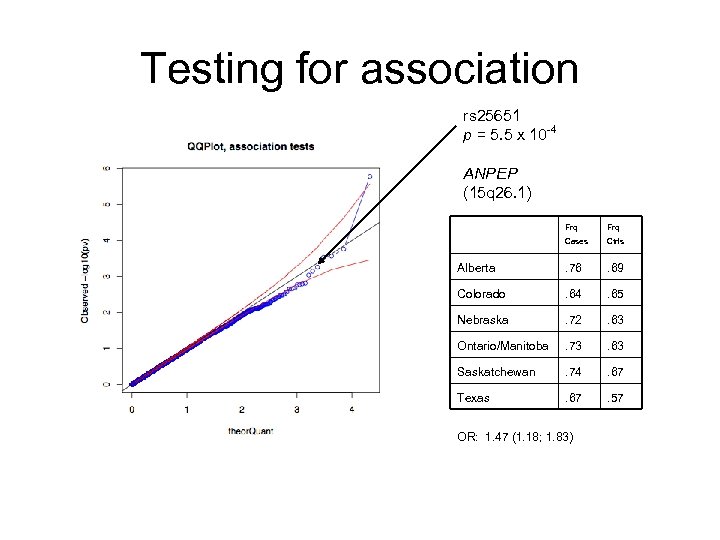
Testing for association rs 25651 p = 5. 5 x 10 -4 ANPEP (15 q 26. 1) Frq Cases Ctrls Alberta . 76 . 69 Colorado . 64 . 65 Nebraska . 72 . 63 Ontario/Manitoba . 73 . 63 Saskatchewan . 74 . 67 Texas . 67 . 57 OR: 1. 47 (1. 18; 1. 83)

ANPEP • • ALANYL AMINOPEPTIDASE -- 20 exons; Serves as receptor for HCo. V-229 E (human coronavirus 229 E); mediates human cytomegalovirus (HCMV) infection; • • rs 25651: coding non-synonymous (Ser 752 Asn) p = 5. 5 x 10 -4; rs 8192297: coding non-synonymous (Ile 603 Met) p = 0. 39.
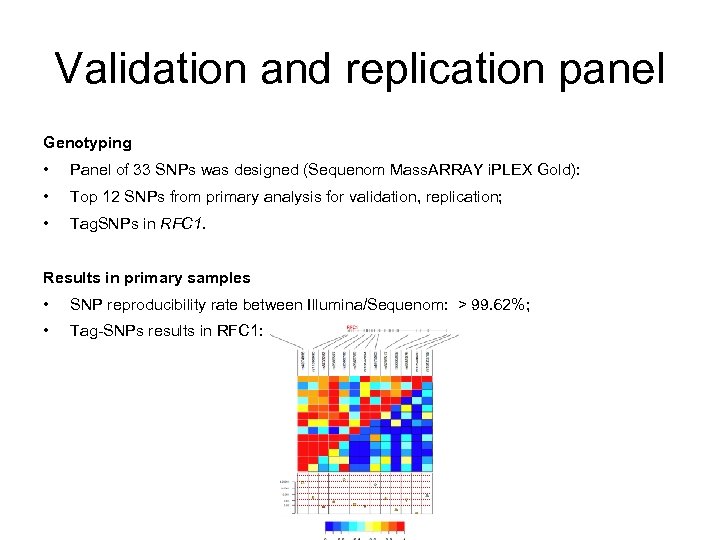
Validation and replication panel Genotyping • Panel of 33 SNPs was designed (Sequenom Mass. ARRAY i. PLEX Gold): • Top 12 SNPs from primary analysis for validation, replication; • Tag. SNPs in RFC 1. Results in primary samples • SNP reproducibility rate between Illumina/Sequenom: > 99. 62%; • Tag-SNPs results in RFC 1:
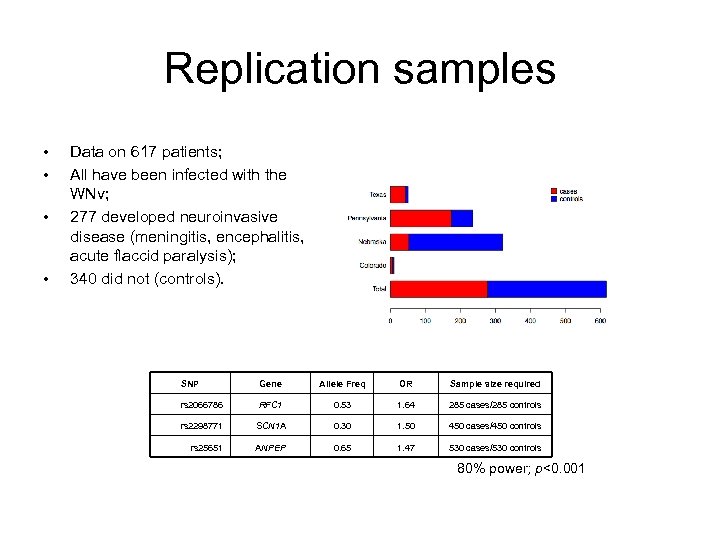
Replication samples • • Data on 617 patients; All have been infected with the WNv; 277 developed neuroinvasive disease (meningitis, encephalitis, acute flaccid paralysis); 340 did not (controls). SNP Gene Allele Freq OR Sample size required rs 2066786 RFC 1 0. 53 1. 64 285 cases/285 controls rs 2298771 SCN 1 A 0. 30 1. 50 450 cases/450 controls rs 25651 ANPEP 0. 65 1. 47 530 cases/530 controls 80% power; p<0. 001
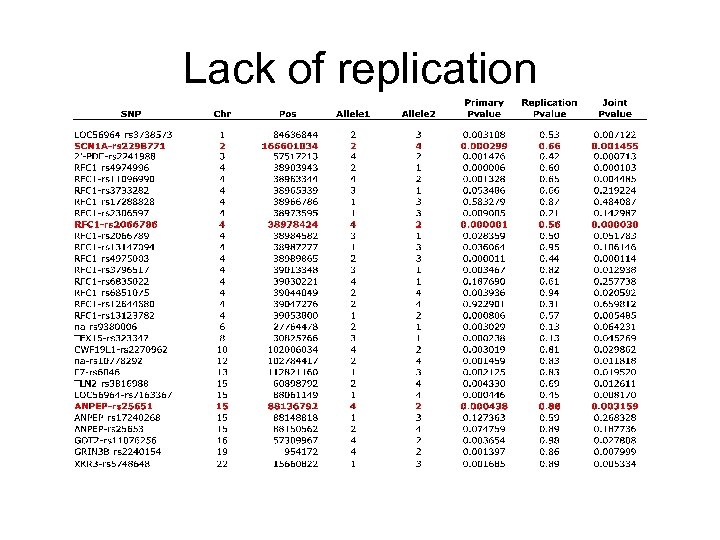
Lack of replication
Age distribution Primary Replication
Gender distribution Primary Replication
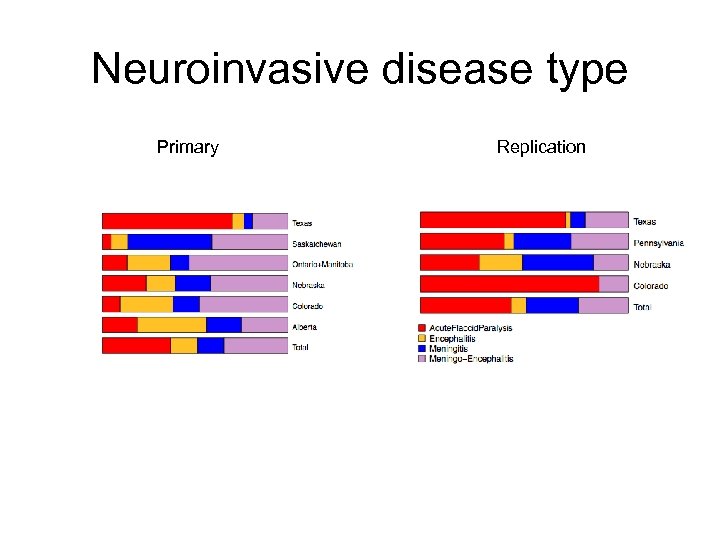
Neuroinvasive disease type Primary Replication
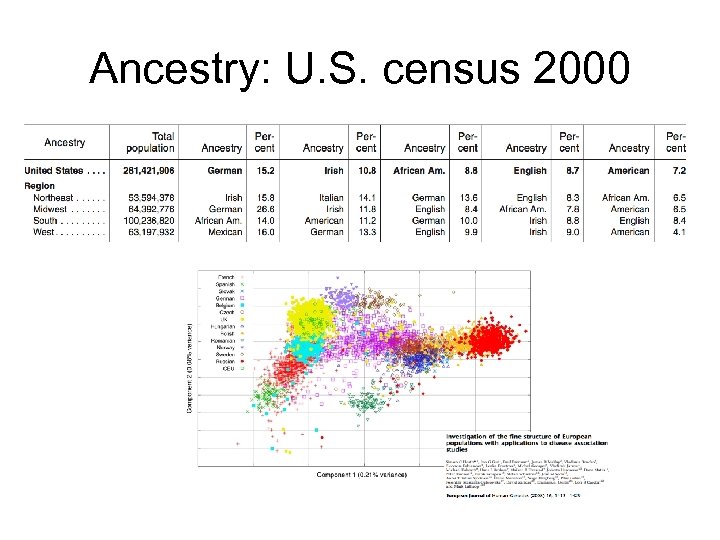
Ancestry: U. S. census 2000
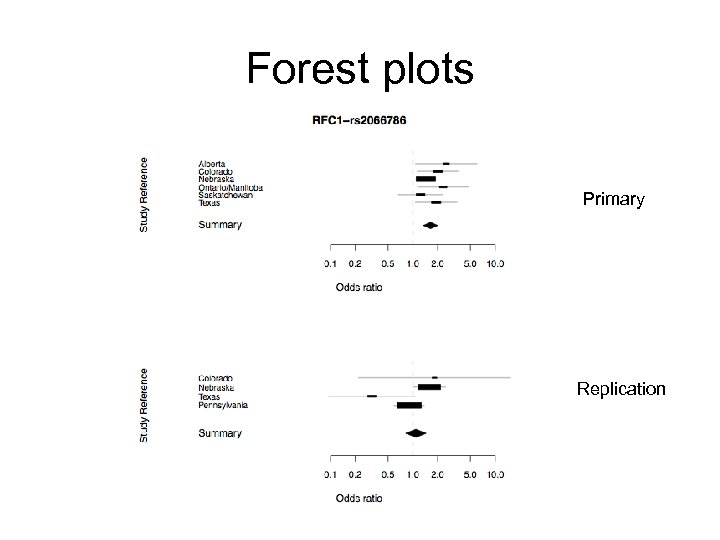
Forest plots Primary Replication
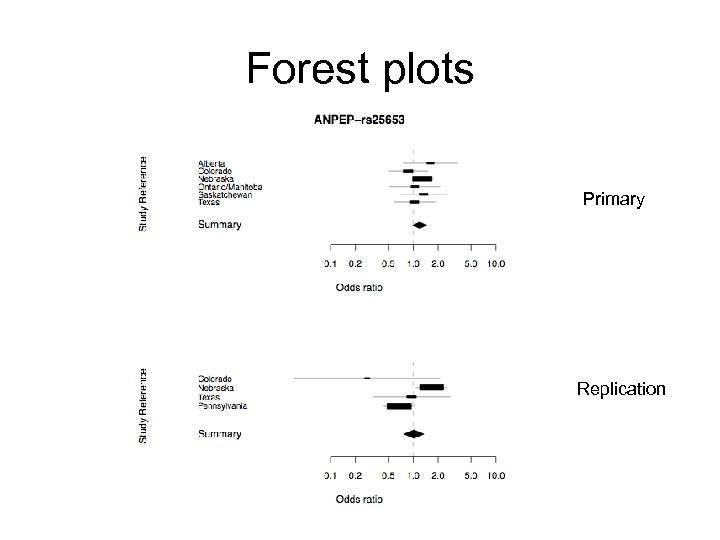
Forest plots Primary Replication
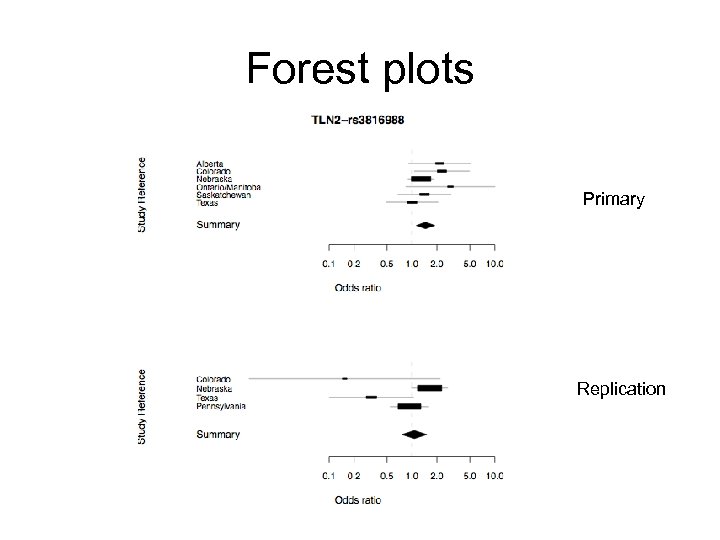
Forest plots Primary Replication
b9c5341ca3e465fd4d0629e4534e6f5a.ppt