3716840ce12b7d29858ab1423c489fb9.ppt
- Количество слайдов: 47
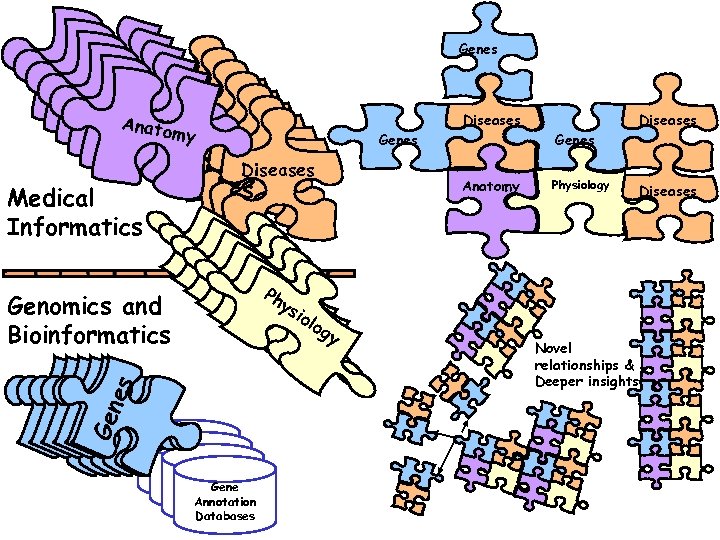 Genes Anato An m y Aatom n y Aatom Diseas n n tom y Aaatoes. Diseas my Diseas y es Genes es. Diseases es Medical Informatics Genes Gene es s Genomics and Bioinformatics Ph hs y Py. Pyio y hsilo hs o y Py. Pyilg y hshg log y Pyisilso y g ilo o Gene Annotation Databases Annotation Databases Diseases log g Anatomy y Genes Physiology Novel relationships & Deeper insights Diseases
Genes Anato An m y Aatom n y Aatom Diseas n n tom y Aaatoes. Diseas my Diseas y es Genes es. Diseases es Medical Informatics Genes Gene es s Genomics and Bioinformatics Ph hs y Py. Pyio y hsilo hs o y Py. Pyilg y hshg log y Pyisilso y g ilo o Gene Annotation Databases Annotation Databases Diseases log g Anatomy y Genes Physiology Novel relationships & Deeper insights Diseases
 Identification and Prioritization of Novel Disease Candidate Genes Systems Biology Based Integrative Approaches Bioinformatics to Systems Biology November 16, 2007 Anil Jegga Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center (CCHMC) Department of Pediatrics, University of Cincinnati, Ohio - 45229 Anil. Jegga@cchmc. org http: //anil. cchmc. org
Identification and Prioritization of Novel Disease Candidate Genes Systems Biology Based Integrative Approaches Bioinformatics to Systems Biology November 16, 2007 Anil Jegga Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center (CCHMC) Department of Pediatrics, University of Cincinnati, Ohio - 45229 Anil. Jegga@cchmc. org http: //anil. cchmc. org
 Acknowledgements • Jing Chen • Eric Bardes • Bruce Aronow Support • All the publicly available gene annotation resources especially NCBI, MGI and UCSC Cincinnati Children’s Hospital Medical Center Computational Medical Center, Cincinnati Mouse Models of Human Cancers Consortium University of Cincinnati College of Medicine
Acknowledgements • Jing Chen • Eric Bardes • Bruce Aronow Support • All the publicly available gene annotation resources especially NCBI, MGI and UCSC Cincinnati Children’s Hospital Medical Center Computational Medical Center, Cincinnati Mouse Models of Human Cancers Consortium University of Cincinnati College of Medicine
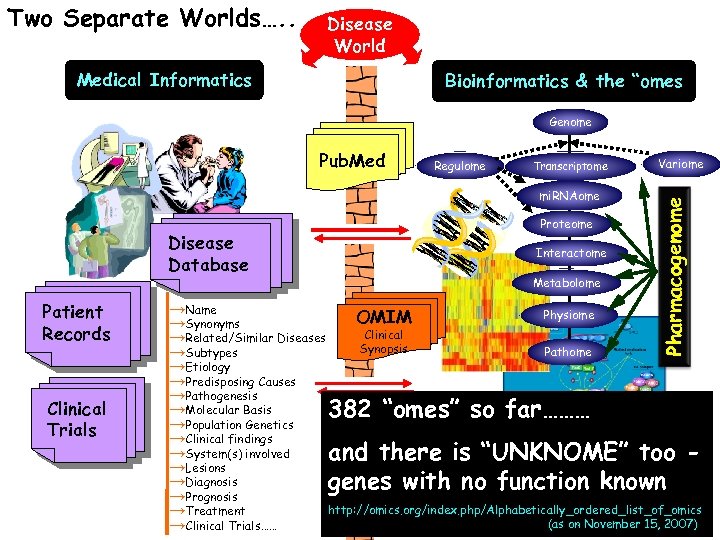 Two Separate Worlds…. . Disease World Medical Informatics Bioinformatics & the “omes Genome Regulome Transcriptome mi. RNAome Disease Database Patient Records Clinical Trials Proteome Interactome Metabolome Variome Pharmacogenome Pub. Med →Name Physiome OMIM →Synonyms Clinical →Related/Similar Diseases Synopsis Pathome →Subtypes →Etiology →Predisposing Causes →Pathogenesis →Molecular Basis 382 “omes” so far……… →Population Genetics →Clinical findings →System(s) involved and there is “UNKNOME” too →Lesions →Diagnosis genes with no function known →Prognosis http: //omics. org/index. php/Alphabetically_ordered_list_of_omics →Treatment (as on Exchange… →Clinical Trials…… With Some Data November 15, 2007)
Two Separate Worlds…. . Disease World Medical Informatics Bioinformatics & the “omes Genome Regulome Transcriptome mi. RNAome Disease Database Patient Records Clinical Trials Proteome Interactome Metabolome Variome Pharmacogenome Pub. Med →Name Physiome OMIM →Synonyms Clinical →Related/Similar Diseases Synopsis Pathome →Subtypes →Etiology →Predisposing Causes →Pathogenesis →Molecular Basis 382 “omes” so far……… →Population Genetics →Clinical findings →System(s) involved and there is “UNKNOME” too →Lesions →Diagnosis genes with no function known →Prognosis http: //omics. org/index. php/Alphabetically_ordered_list_of_omics →Treatment (as on Exchange… →Clinical Trials…… With Some Data November 15, 2007)
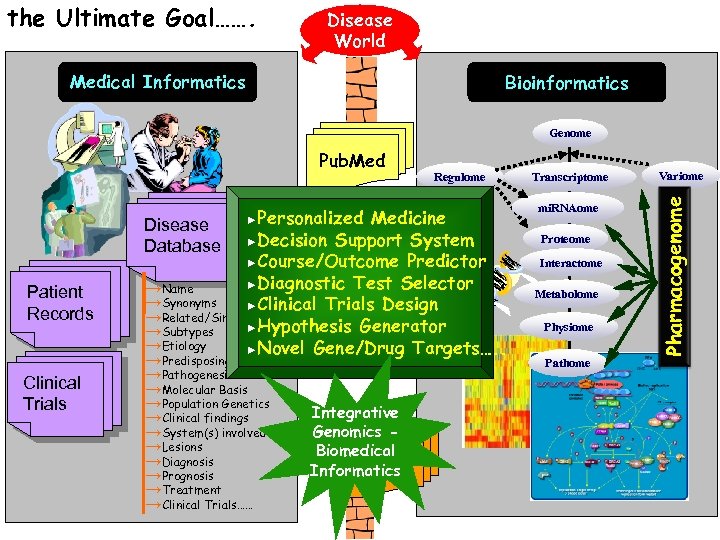 the Ultimate Goal……. Disease World Medical Informatics Bioinformatics Genome ►Personalized Patient Records Clinical Trials Regulome Medicine Disease Database ►Decision Support System ►Course/Outcome Predictor ►Diagnostic Test Selector →Name →Synonyms ►Clinical Trials Design →Related/Similar Diseases ►Hypothesis Generator →Subtypes →Etiology ►Novel Gene/Drug Targets…. . →Predisposing Causes →Pathogenesis →Molecular Basis →Population Genetics →Clinical findings →System(s) involved →Lesions →Diagnosis →Prognosis →Treatment →Clinical Trials…… Integrative Genomics Biomedical OMIM Informatics Transcriptome mi. RNAome Proteome Interactome Metabolome Physiome Pathome Variome Pharmacogenome Pub. Med
the Ultimate Goal……. Disease World Medical Informatics Bioinformatics Genome ►Personalized Patient Records Clinical Trials Regulome Medicine Disease Database ►Decision Support System ►Course/Outcome Predictor ►Diagnostic Test Selector →Name →Synonyms ►Clinical Trials Design →Related/Similar Diseases ►Hypothesis Generator →Subtypes →Etiology ►Novel Gene/Drug Targets…. . →Predisposing Causes →Pathogenesis →Molecular Basis →Population Genetics →Clinical findings →System(s) involved →Lesions →Diagnosis →Prognosis →Treatment →Clinical Trials…… Integrative Genomics Biomedical OMIM Informatics Transcriptome mi. RNAome Proteome Interactome Metabolome Physiome Pathome Variome Pharmacogenome Pub. Med
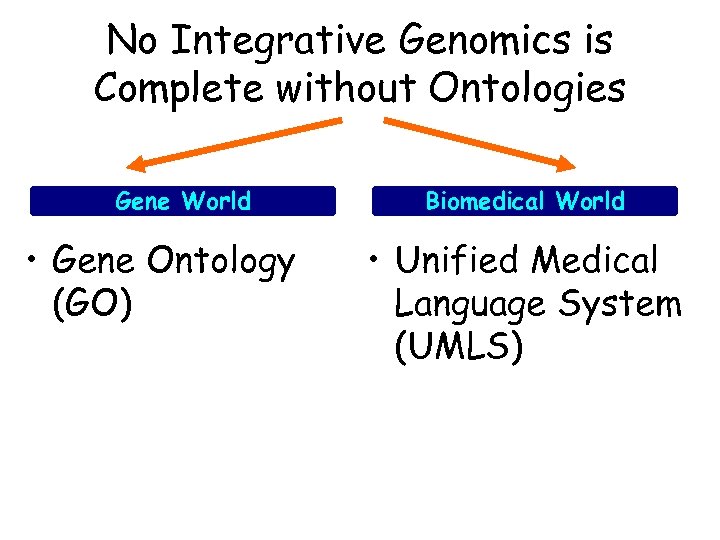 No Integrative Genomics is Complete without Ontologies Gene World • Gene Ontology (GO) Biomedical World • Unified Medical Language System (UMLS)
No Integrative Genomics is Complete without Ontologies Gene World • Gene Ontology (GO) Biomedical World • Unified Medical Language System (UMLS)
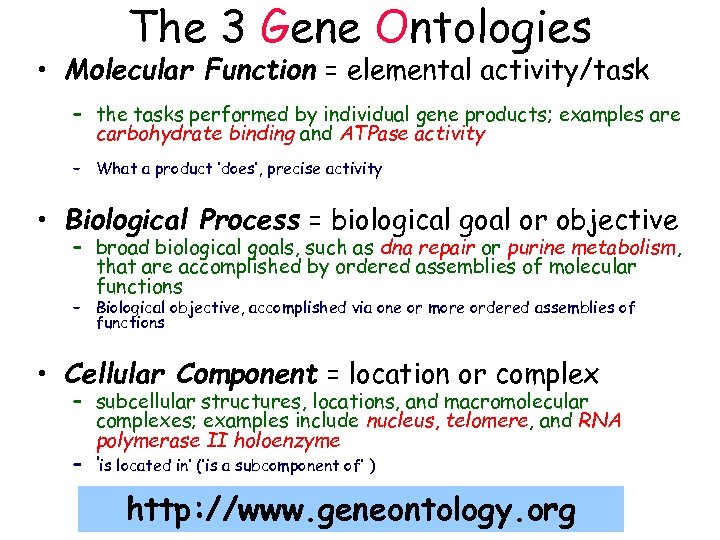 The 3 Gene Ontologies • Molecular Function = elemental activity/task – the tasks performed by individual gene products; examples are carbohydrate binding and ATPase activity – What a product ‘does’, precise activity • Biological Process = biological goal or objective – broad biological goals, such as dna repair or purine metabolism, that are accomplished by ordered assemblies of molecular functions – Biological objective, accomplished via one or more ordered assemblies of functions • Cellular Component = location or complex – subcellular structures, locations, and macromolecular complexes; examples include nucleus, telomere, and RNA polymerase II holoenzyme – ‘is located in’ (‘is a subcomponent of’ ) http: //www. geneontology. org
The 3 Gene Ontologies • Molecular Function = elemental activity/task – the tasks performed by individual gene products; examples are carbohydrate binding and ATPase activity – What a product ‘does’, precise activity • Biological Process = biological goal or objective – broad biological goals, such as dna repair or purine metabolism, that are accomplished by ordered assemblies of molecular functions – Biological objective, accomplished via one or more ordered assemblies of functions • Cellular Component = location or complex – subcellular structures, locations, and macromolecular complexes; examples include nucleus, telomere, and RNA polymerase II holoenzyme – ‘is located in’ (‘is a subcomponent of’ ) http: //www. geneontology. org
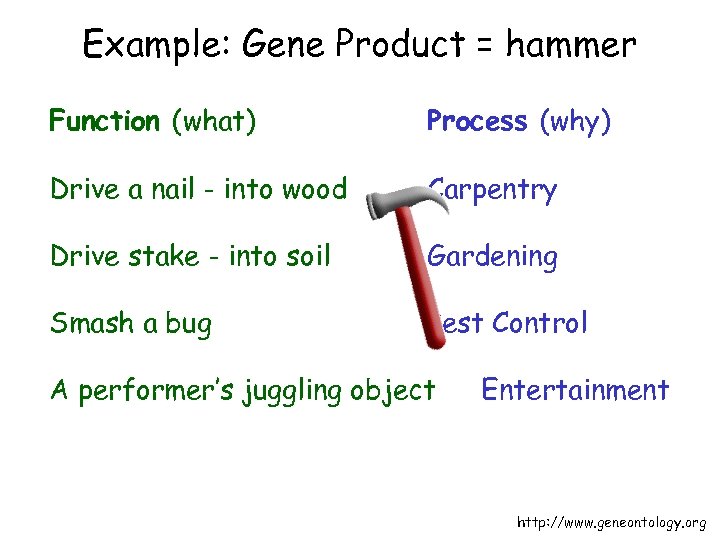 Example: Gene Product = hammer Function (what) Process (why) Drive a nail - into wood Carpentry Drive stake - into soil Gardening Smash a bug Pest Control A performer’s juggling object Entertainment http: //www. geneontology. org
Example: Gene Product = hammer Function (what) Process (why) Drive a nail - into wood Carpentry Drive stake - into soil Gardening Smash a bug Pest Control A performer’s juggling object Entertainment http: //www. geneontology. org
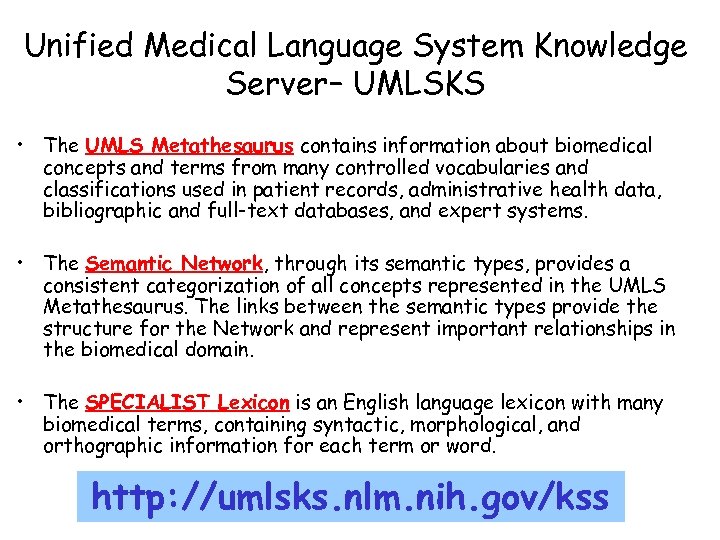 Unified Medical Language System Knowledge Server– UMLSKS • The UMLS Metathesaurus contains information about biomedical concepts and terms from many controlled vocabularies and classifications used in patient records, administrative health data, bibliographic and full-text databases, and expert systems. • The Semantic Network, through its semantic types, provides a consistent categorization of all concepts represented in the UMLS Metathesaurus. The links between the semantic types provide the structure for the Network and represent important relationships in the biomedical domain. • The SPECIALIST Lexicon is an English language lexicon with many biomedical terms, containing syntactic, morphological, and orthographic information for each term or word. http: //umlsks. nlm. nih. gov/kss
Unified Medical Language System Knowledge Server– UMLSKS • The UMLS Metathesaurus contains information about biomedical concepts and terms from many controlled vocabularies and classifications used in patient records, administrative health data, bibliographic and full-text databases, and expert systems. • The Semantic Network, through its semantic types, provides a consistent categorization of all concepts represented in the UMLS Metathesaurus. The links between the semantic types provide the structure for the Network and represent important relationships in the biomedical domain. • The SPECIALIST Lexicon is an English language lexicon with many biomedical terms, containing syntactic, morphological, and orthographic information for each term or word. http: //umlsks. nlm. nih. gov/kss
 • • • Unified Medical Language System Metathesaurus about over 1 million biomedical concepts About 5 million concept names from more than 100 controlled vocabularies and classifications (some in multiple languages) used in patient records, administrative health data, bibliographic and full-text databases and expert systems. The Metathesaurus is organized by concept or meaning. Alternate names for the same concept (synonyms, lexical variants, and translations) are linked together. Each Metathesaurus concept has attributes that help to define its meaning, e. g. , the semantic type(s) or categories to which it belongs, its position in the hierarchical contexts from various source vocabularies, and, for many concepts, a definition. Customizable: Users can exclude vocabularies that are not relevant for specific purposes or not licensed for use in their institutions. Metamorpho. Sys, the multi-platform Java install and customization program distributed with the UMLS resources, helps users to generate pre-defined or custom subsets of the Metathesaurus. • Uses: – linking between different clinical or biomedical vocabularies – information retrieval from databases with human assigned subject index terms and from free-text information sources – linking patient records to related information in bibliographic, full-text, or factual databases – natural language processing and automated indexing research
• • • Unified Medical Language System Metathesaurus about over 1 million biomedical concepts About 5 million concept names from more than 100 controlled vocabularies and classifications (some in multiple languages) used in patient records, administrative health data, bibliographic and full-text databases and expert systems. The Metathesaurus is organized by concept or meaning. Alternate names for the same concept (synonyms, lexical variants, and translations) are linked together. Each Metathesaurus concept has attributes that help to define its meaning, e. g. , the semantic type(s) or categories to which it belongs, its position in the hierarchical contexts from various source vocabularies, and, for many concepts, a definition. Customizable: Users can exclude vocabularies that are not relevant for specific purposes or not licensed for use in their institutions. Metamorpho. Sys, the multi-platform Java install and customization program distributed with the UMLS resources, helps users to generate pre-defined or custom subsets of the Metathesaurus. • Uses: – linking between different clinical or biomedical vocabularies – information retrieval from databases with human assigned subject index terms and from free-text information sources – linking patient records to related information in bibliographic, full-text, or factual databases – natural language processing and automated indexing research
 Open biomedical ontologies http: //obo. sourceforge. net/
Open biomedical ontologies http: //obo. sourceforge. net/
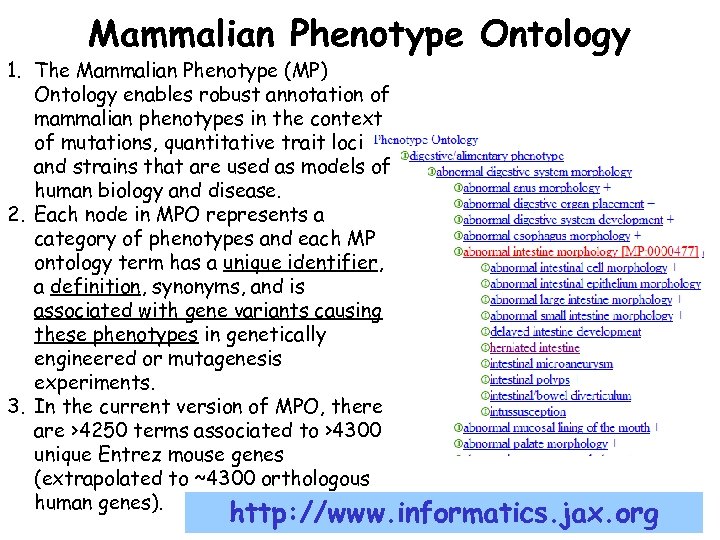 Mammalian Phenotype Ontology 1. The Mammalian Phenotype (MP) Ontology enables robust annotation of mammalian phenotypes in the context of mutations, quantitative trait loci and strains that are used as models of human biology and disease. 2. Each node in MPO represents a category of phenotypes and each MP ontology term has a unique identifier, a definition, synonyms, and is associated with gene variants causing these phenotypes in genetically engineered or mutagenesis experiments. 3. In the current version of MPO, there are >4250 terms associated to >4300 unique Entrez mouse genes (extrapolated to ~4300 orthologous human genes). http: //www. informatics. jax. org
Mammalian Phenotype Ontology 1. The Mammalian Phenotype (MP) Ontology enables robust annotation of mammalian phenotypes in the context of mutations, quantitative trait loci and strains that are used as models of human biology and disease. 2. Each node in MPO represents a category of phenotypes and each MP ontology term has a unique identifier, a definition, synonyms, and is associated with gene variants causing these phenotypes in genetically engineered or mutagenesis experiments. 3. In the current version of MPO, there are >4250 terms associated to >4300 unique Entrez mouse genes (extrapolated to ~4300 orthologous human genes). http: //www. informatics. jax. org
 Disease Gene Identification and Prioritization Hypothesis: Majority of genes that impact or cause disease share membership in any of several functional relationships OR Functionally similar or related genes cause similar phenotype. Functional Similarity – Common/shared • Gene Ontology term • Pathway • Phenotype • Chromosomal location • Expression • Cis regulatory elements (Transcription factor binding sites) • mi. RNA regulators • Interactions • Other features…. .
Disease Gene Identification and Prioritization Hypothesis: Majority of genes that impact or cause disease share membership in any of several functional relationships OR Functionally similar or related genes cause similar phenotype. Functional Similarity – Common/shared • Gene Ontology term • Pathway • Phenotype • Chromosomal location • Expression • Cis regulatory elements (Transcription factor binding sites) • mi. RNA regulators • Interactions • Other features…. .
 Background, Problems & Issues 1. Most of the common diseases are multifactorial and modified by genetically and mechanistically complex polygenic interactions and environmental factors. 2. High-throughput genome-wide studies like linkage analysis and gene expression profiling, tend to be most useful for classification and characterization but do not provide sufficient information to identify or prioritize specific disease causal genes.
Background, Problems & Issues 1. Most of the common diseases are multifactorial and modified by genetically and mechanistically complex polygenic interactions and environmental factors. 2. High-throughput genome-wide studies like linkage analysis and gene expression profiling, tend to be most useful for classification and characterization but do not provide sufficient information to identify or prioritize specific disease causal genes.
 Background, Problems & Issues 3. Since multiple genes are associated with same or similar disease phenotypes, it is reasonable to expect the underlying genes to be functionally related. 4. Such functional relatedness (common pathway, interaction, biological process, etc. ) can be exploited to aid in the finding of novel disease genes. For e. g. , genetically heterogeneous hereditary diseases such as Hermansky-Pudlak syndrome and Fanconi anaemia have been shown to be caused by mutations in different interacting proteins.
Background, Problems & Issues 3. Since multiple genes are associated with same or similar disease phenotypes, it is reasonable to expect the underlying genes to be functionally related. 4. Such functional relatedness (common pathway, interaction, biological process, etc. ) can be exploited to aid in the finding of novel disease genes. For e. g. , genetically heterogeneous hereditary diseases such as Hermansky-Pudlak syndrome and Fanconi anaemia have been shown to be caused by mutations in different interacting proteins.
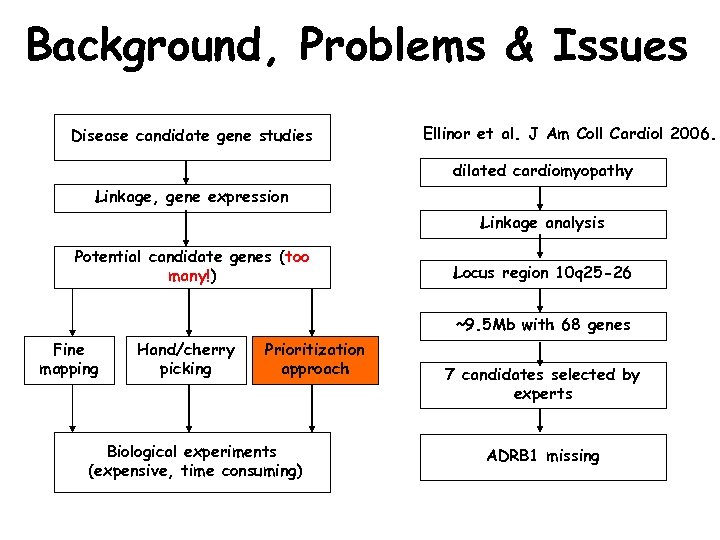 Background, Problems & Issues Disease candidate gene studies Ellinor et al. J Am Coll Cardiol 2006. dilated cardiomyopathy Linkage, gene expression Linkage analysis Potential candidate genes (too many!) Locus region 10 q 25 -26 ~9. 5 Mb with 68 genes Fine mapping Hand/cherry picking Prioritization approach Biological experiments (expensive, time consuming) 7 candidates selected by experts ADRB 1 missing
Background, Problems & Issues Disease candidate gene studies Ellinor et al. J Am Coll Cardiol 2006. dilated cardiomyopathy Linkage, gene expression Linkage analysis Potential candidate genes (too many!) Locus region 10 q 25 -26 ~9. 5 Mb with 68 genes Fine mapping Hand/cherry picking Prioritization approach Biological experiments (expensive, time consuming) 7 candidates selected by experts ADRB 1 missing
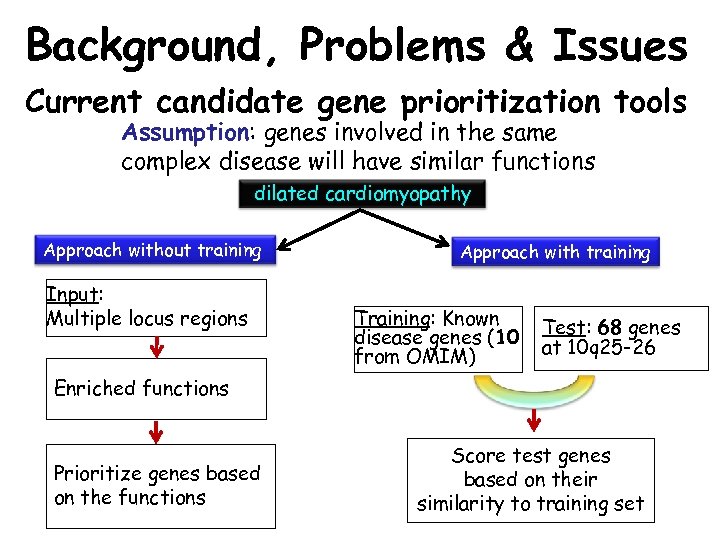 Background, Problems & Issues Current candidate gene prioritization tools Assumption: genes involved in the same complex disease will have similar functions dilated cardiomyopathy Approach without training Input: Multiple locus regions Approach with training Training: Known disease genes (10 from OMIM) Test: 68 genes at 10 q 25 -26 Enriched functions Prioritize genes based on the functions Score test genes based on their similarity to training set
Background, Problems & Issues Current candidate gene prioritization tools Assumption: genes involved in the same complex disease will have similar functions dilated cardiomyopathy Approach without training Input: Multiple locus regions Approach with training Training: Known disease genes (10 from OMIM) Test: 68 genes at 10 q 25 -26 Enriched functions Prioritize genes based on the functions Score test genes based on their similarity to training set
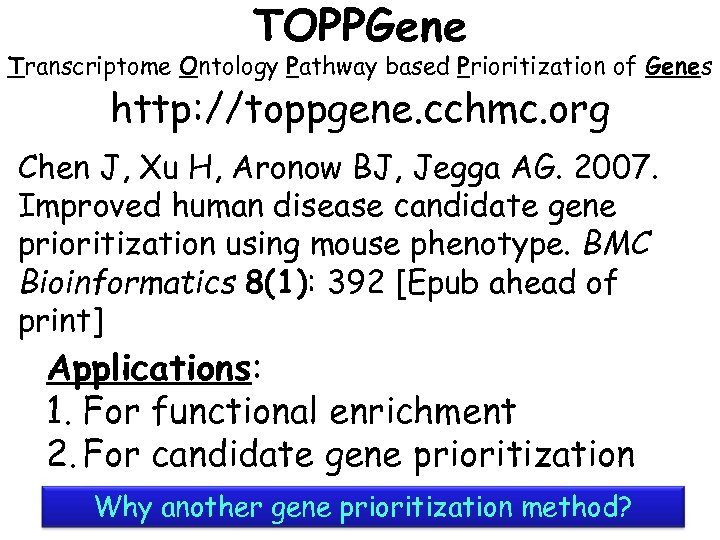 TOPPGene Transcriptome Ontology Pathway based Prioritization of Genes http: //toppgene. cchmc. org Chen J, Xu H, Aronow BJ, Jegga AG. 2007. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics 8(1): 392 [Epub ahead of print] Applications: 1. For functional enrichment 2. For candidate gene prioritization Why another gene prioritization method?
TOPPGene Transcriptome Ontology Pathway based Prioritization of Genes http: //toppgene. cchmc. org Chen J, Xu H, Aronow BJ, Jegga AG. 2007. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics 8(1): 392 [Epub ahead of print] Applications: 1. For functional enrichment 2. For candidate gene prioritization Why another gene prioritization method?
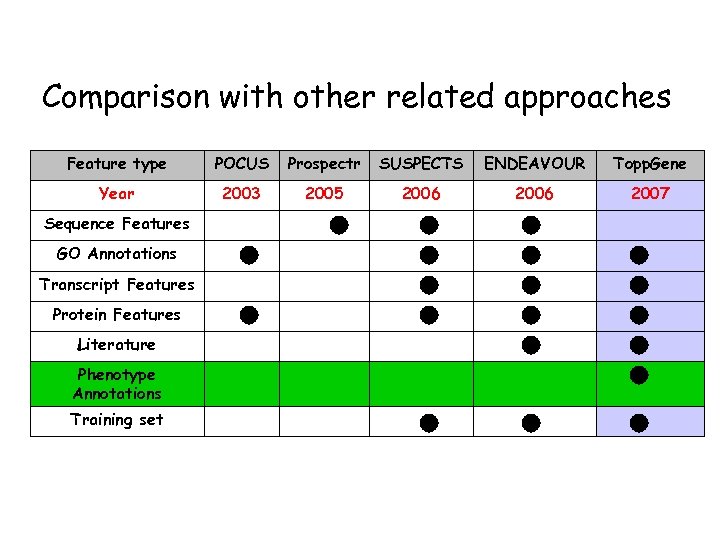 Comparison with other related approaches Feature type POCUS Prospectr SUSPECTS ENDEAVOUR Topp. Gene Year 2003 2005 2006 2007 Sequence Features GO Annotations Transcript Features Protein Features Literature Phenotype Annotations Training set
Comparison with other related approaches Feature type POCUS Prospectr SUSPECTS ENDEAVOUR Topp. Gene Year 2003 2005 2006 2007 Sequence Features GO Annotations Transcript Features Protein Features Literature Phenotype Annotations Training set
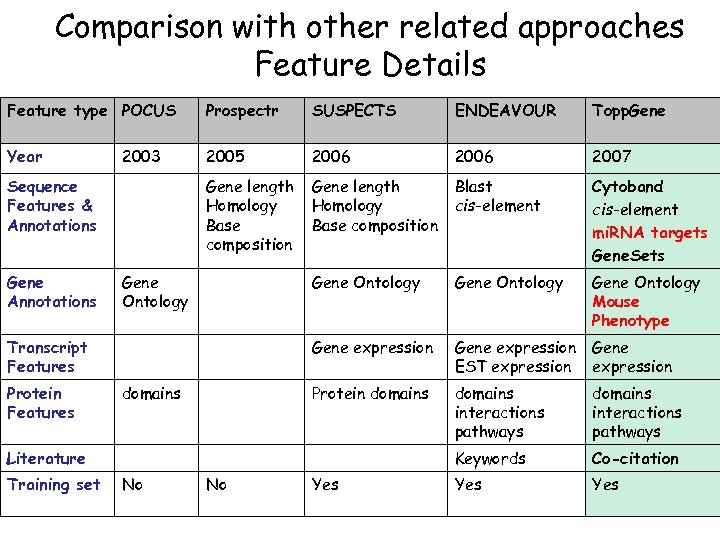 Comparison with other related approaches Feature Details Feature type POCUS Prospectr SUSPECTS ENDEAVOUR Topp. Gene Year 2005 2006 2007 Gene length Homology Base composition Blast cis-element Cytoband cis-element mi. RNA targets Gene. Sets Gene Ontology Mouse Phenotype Gene expression EST expression Gene expression Protein domains interactions pathways Keywords Co-citation Yes 2003 Sequence Features & Annotations Gene Ontology Transcript Features Protein Features domains Literature Training set No No Yes
Comparison with other related approaches Feature Details Feature type POCUS Prospectr SUSPECTS ENDEAVOUR Topp. Gene Year 2005 2006 2007 Gene length Homology Base composition Blast cis-element Cytoband cis-element mi. RNA targets Gene. Sets Gene Ontology Mouse Phenotype Gene expression EST expression Gene expression Protein domains interactions pathways Keywords Co-citation Yes 2003 Sequence Features & Annotations Gene Ontology Transcript Features Protein Features domains Literature Training set No No Yes
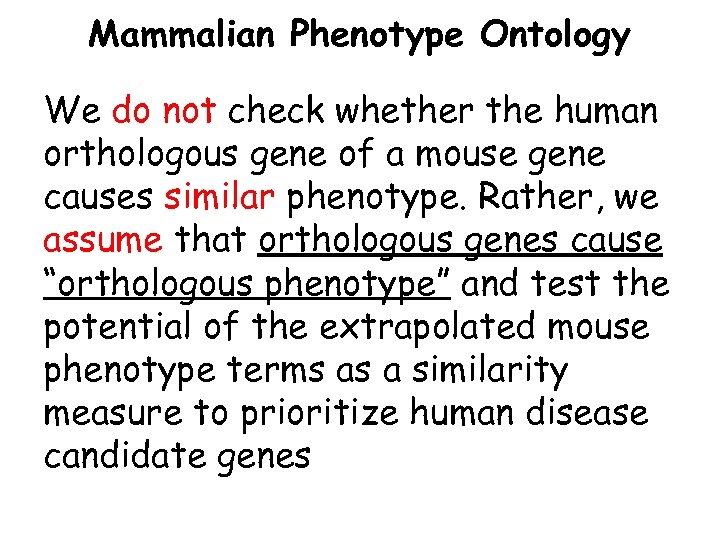 Mammalian Phenotype Ontology We do not check whether the human orthologous gene of a mouse gene causes similar phenotype. Rather, we assume that orthologous genes cause “orthologous phenotype” and test the potential of the extrapolated mouse phenotype terms as a similarity measure to prioritize human disease candidate genes
Mammalian Phenotype Ontology We do not check whether the human orthologous gene of a mouse gene causes similar phenotype. Rather, we assume that orthologous genes cause “orthologous phenotype” and test the potential of the extrapolated mouse phenotype terms as a similarity measure to prioritize human disease candidate genes
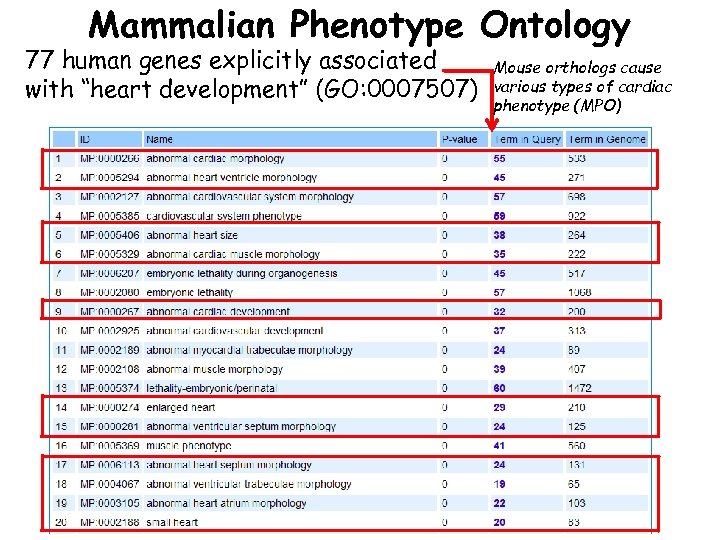 Mammalian Phenotype Ontology 77 human genes explicitly associated with “heart development” (GO: 0007507) Mouse orthologs cause various types of cardiac phenotype (MPO)
Mammalian Phenotype Ontology 77 human genes explicitly associated with “heart development” (GO: 0007507) Mouse orthologs cause various types of cardiac phenotype (MPO)
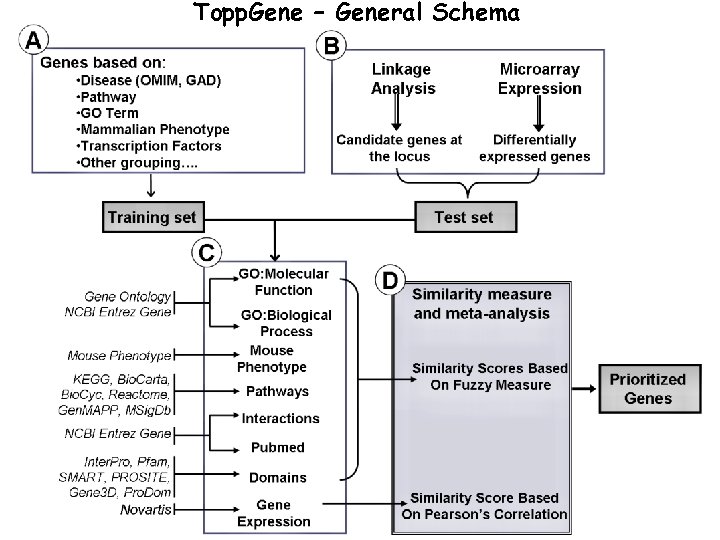 Topp. Gene – General Schema
Topp. Gene – General Schema
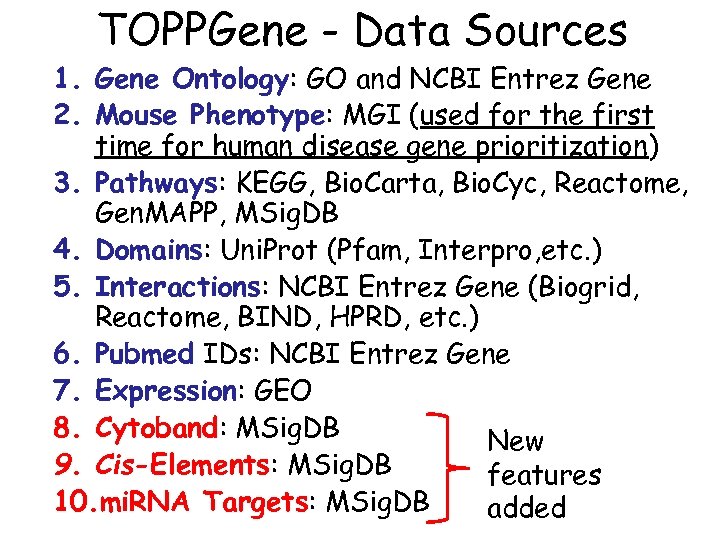 TOPPGene - Data Sources 1. Gene Ontology: GO and NCBI Entrez Gene 2. Mouse Phenotype: MGI (used for the first time for human disease gene prioritization) 3. Pathways: KEGG, Bio. Carta, Bio. Cyc, Reactome, Gen. MAPP, MSig. DB 4. Domains: Uni. Prot (Pfam, Interpro, etc. ) 5. Interactions: NCBI Entrez Gene (Biogrid, Reactome, BIND, HPRD, etc. ) 6. Pubmed IDs: NCBI Entrez Gene 7. Expression: GEO 8. Cytoband: MSig. DB New 9. Cis-Elements: MSig. DB features 10. mi. RNA Targets: MSig. DB added
TOPPGene - Data Sources 1. Gene Ontology: GO and NCBI Entrez Gene 2. Mouse Phenotype: MGI (used for the first time for human disease gene prioritization) 3. Pathways: KEGG, Bio. Carta, Bio. Cyc, Reactome, Gen. MAPP, MSig. DB 4. Domains: Uni. Prot (Pfam, Interpro, etc. ) 5. Interactions: NCBI Entrez Gene (Biogrid, Reactome, BIND, HPRD, etc. ) 6. Pubmed IDs: NCBI Entrez Gene 7. Expression: GEO 8. Cytoband: MSig. DB New 9. Cis-Elements: MSig. DB features 10. mi. RNA Targets: MSig. DB added
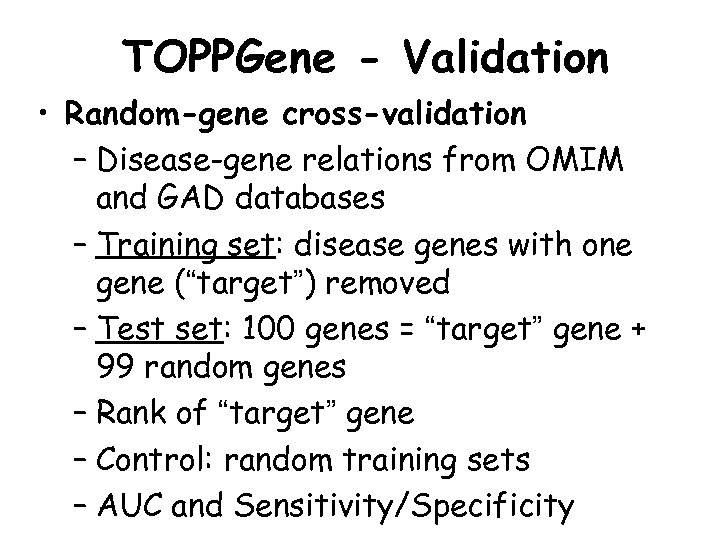 TOPPGene - Validation • Random-gene cross-validation – Disease-gene relations from OMIM and GAD databases – Training set: disease genes with one gene (“target”) removed – Test set: 100 genes = “target” gene + 99 random genes – Rank of “target” gene – Control: random training sets – AUC and Sensitivity/Specificity
TOPPGene - Validation • Random-gene cross-validation – Disease-gene relations from OMIM and GAD databases – Training set: disease genes with one gene (“target”) removed – Test set: 100 genes = “target” gene + 99 random genes – Rank of “target” gene – Control: random training sets – AUC and Sensitivity/Specificity
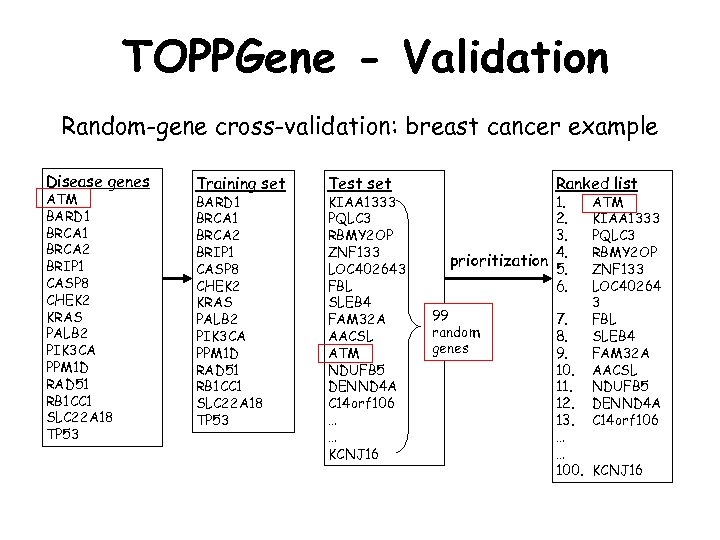 TOPPGene - Validation Random-gene cross-validation: breast cancer example Disease genes ATM BARD 1 BRCA 2 BRIP 1 CASP 8 CHEK 2 KRAS PALB 2 PIK 3 CA PPM 1 D RAD 51 RB 1 CC 1 SLC 22 A 18 TP 53 Training set BARD 1 BRCA 2 BRIP 1 CASP 8 CHEK 2 KRAS PALB 2 PIK 3 CA PPM 1 D RAD 51 RB 1 CC 1 SLC 22 A 18 TP 53 Test set KIAA 1333 PQLC 3 RBMY 2 OP ZNF 133 LOC 402643 FBL SLEB 4 FAM 32 A AACSL ATM NDUFB 5 DENND 4 A C 14 orf 106 … … KCNJ 16 Ranked list 1. 2. 3. 4. prioritization 5. 6. 99 random genes ATM KIAA 1333 PQLC 3 RBMY 2 OP ZNF 133 LOC 40264 3 FBL SLEB 4 FAM 32 A AACSL NDUFB 5 DENND 4 A C 14 orf 106 7. 8. 9. 10. 11. 12. 13. … … 100. KCNJ 16
TOPPGene - Validation Random-gene cross-validation: breast cancer example Disease genes ATM BARD 1 BRCA 2 BRIP 1 CASP 8 CHEK 2 KRAS PALB 2 PIK 3 CA PPM 1 D RAD 51 RB 1 CC 1 SLC 22 A 18 TP 53 Training set BARD 1 BRCA 2 BRIP 1 CASP 8 CHEK 2 KRAS PALB 2 PIK 3 CA PPM 1 D RAD 51 RB 1 CC 1 SLC 22 A 18 TP 53 Test set KIAA 1333 PQLC 3 RBMY 2 OP ZNF 133 LOC 402643 FBL SLEB 4 FAM 32 A AACSL ATM NDUFB 5 DENND 4 A C 14 orf 106 … … KCNJ 16 Ranked list 1. 2. 3. 4. prioritization 5. 6. 99 random genes ATM KIAA 1333 PQLC 3 RBMY 2 OP ZNF 133 LOC 40264 3 FBL SLEB 4 FAM 32 A AACSL NDUFB 5 DENND 4 A C 14 orf 106 7. 8. 9. 10. 11. 12. 13. … … 100. KCNJ 16
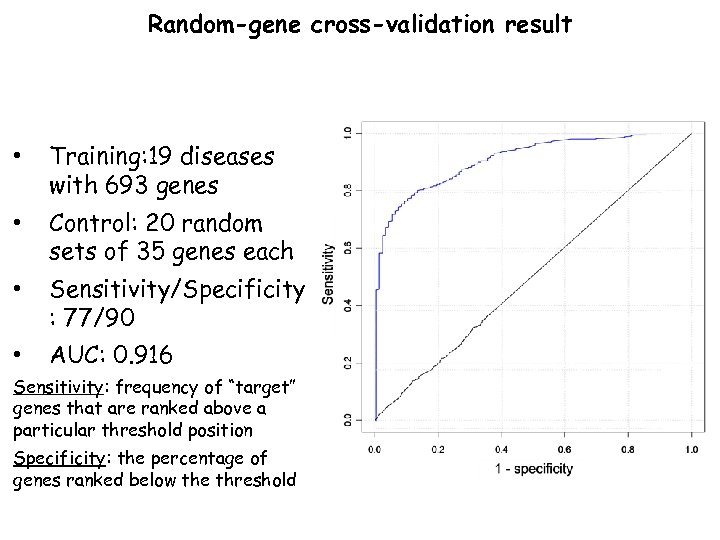 Random-gene cross-validation result • Training: 19 diseases with 693 genes • Control: 20 random sets of 35 genes each • Sensitivity/Specificity : 77/90 • AUC: 0. 916 Sensitivity: frequency of “target” genes that are ranked above a particular threshold position Specificity: the percentage of genes ranked below the threshold
Random-gene cross-validation result • Training: 19 diseases with 693 genes • Control: 20 random sets of 35 genes each • Sensitivity/Specificity : 77/90 • AUC: 0. 916 Sensitivity: frequency of “target” genes that are ranked above a particular threshold position Specificity: the percentage of genes ranked below the threshold
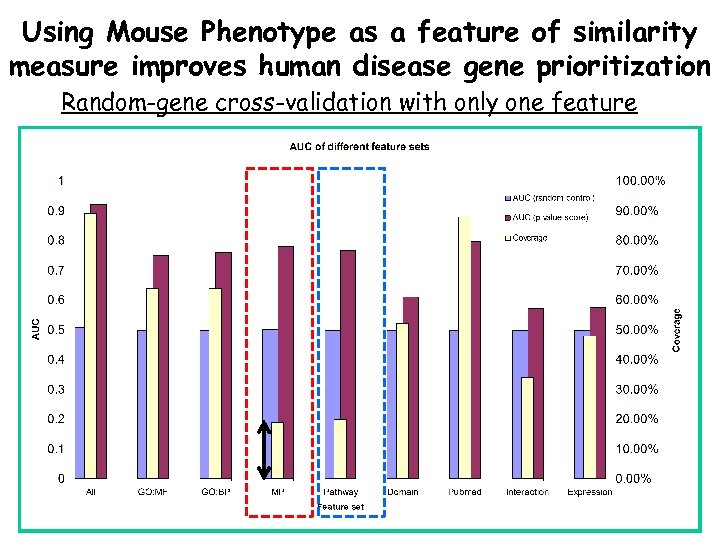 Using Mouse Phenotype as a feature of similarity measure improves human disease gene prioritization Random-gene cross-validation with only one feature
Using Mouse Phenotype as a feature of similarity measure improves human disease gene prioritization Random-gene cross-validation with only one feature
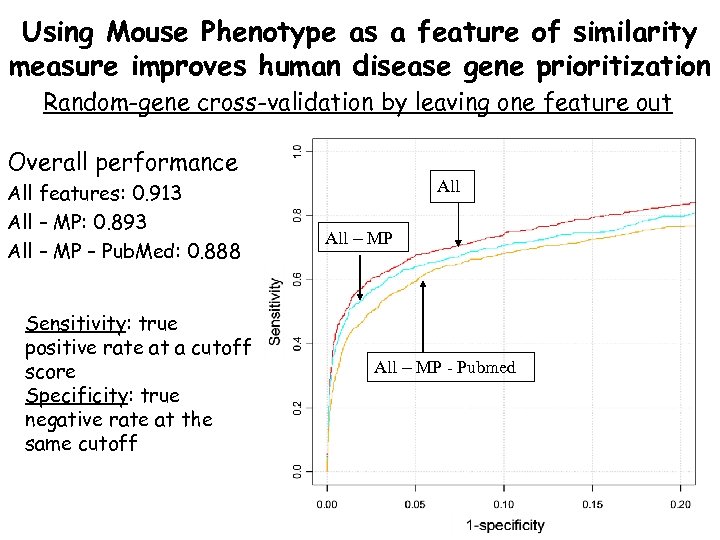 Using Mouse Phenotype as a feature of similarity measure improves human disease gene prioritization Random-gene cross-validation by leaving one feature out Overall performance All features: 0. 913 All – MP: 0. 893 All – MP – Pub. Med: 0. 888 Sensitivity: true positive rate at a cutoff score Specificity: true negative rate at the same cutoff All – MP - Pubmed
Using Mouse Phenotype as a feature of similarity measure improves human disease gene prioritization Random-gene cross-validation by leaving one feature out Overall performance All features: 0. 913 All – MP: 0. 893 All – MP – Pub. Med: 0. 888 Sensitivity: true positive rate at a cutoff score Specificity: true negative rate at the same cutoff All – MP - Pubmed
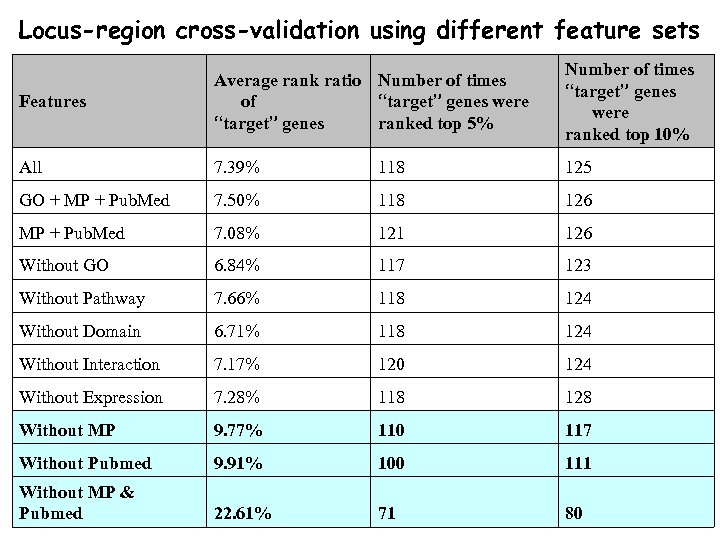 Locus-region cross-validation using different feature sets Features Average rank ratio Number of times of “target” genes were “target” genes ranked top 5% Number of times “target” genes were ranked top 10% All 7. 39% 118 125 GO + MP + Pub. Med 7. 50% 118 126 MP + Pub. Med 7. 08% 121 126 Without GO 6. 84% 117 123 Without Pathway 7. 66% 118 124 Without Domain 6. 71% 118 124 Without Interaction 7. 17% 120 124 Without Expression 7. 28% 118 128 Without MP 9. 77% 110 117 Without Pubmed 9. 91% 100 111 Without MP & Pubmed 22. 61% 71 80
Locus-region cross-validation using different feature sets Features Average rank ratio Number of times of “target” genes were “target” genes ranked top 5% Number of times “target” genes were ranked top 10% All 7. 39% 118 125 GO + MP + Pub. Med 7. 50% 118 126 MP + Pub. Med 7. 08% 121 126 Without GO 6. 84% 117 123 Without Pathway 7. 66% 118 124 Without Domain 6. 71% 118 124 Without Interaction 7. 17% 120 124 Without Expression 7. 28% 118 128 Without MP 9. 77% 110 117 Without Pubmed 9. 91% 100 111 Without MP & Pubmed 22. 61% 71 80
 Topp. Gene web server (http: //toppgene. cchmc. org) For functional enrichment analysis
Topp. Gene web server (http: //toppgene. cchmc. org) For functional enrichment analysis
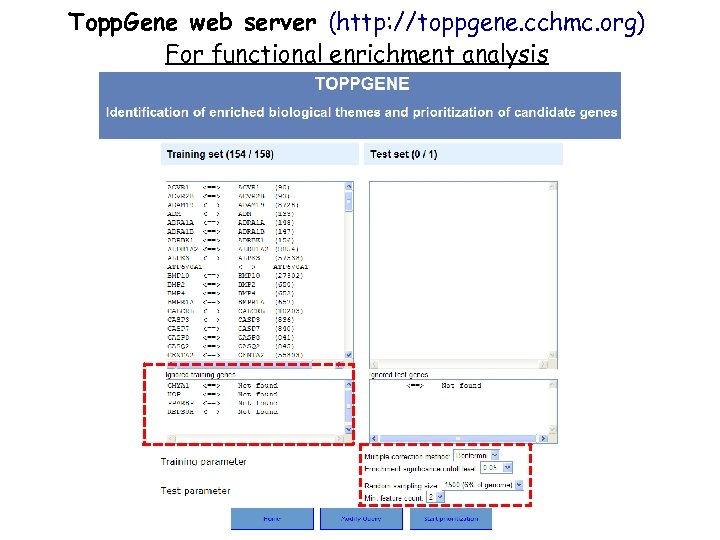 Topp. Gene web server (http: //toppgene. cchmc. org) For functional enrichment analysis
Topp. Gene web server (http: //toppgene. cchmc. org) For functional enrichment analysis
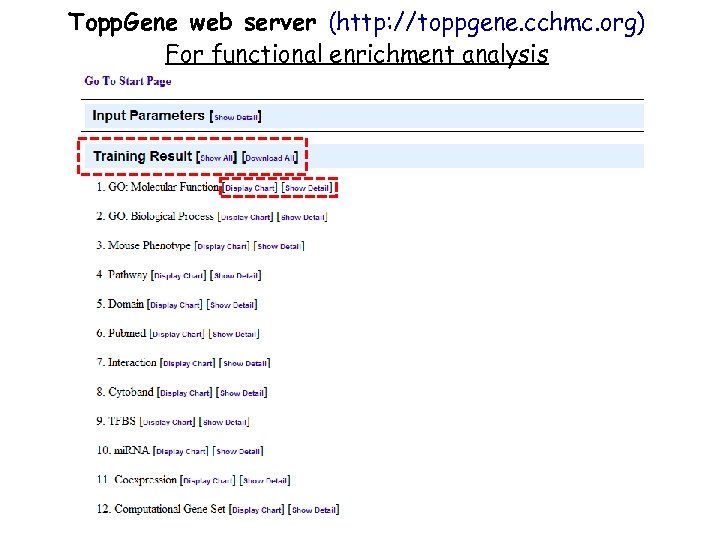 Topp. Gene web server (http: //toppgene. cchmc. org) For functional enrichment analysis
Topp. Gene web server (http: //toppgene. cchmc. org) For functional enrichment analysis
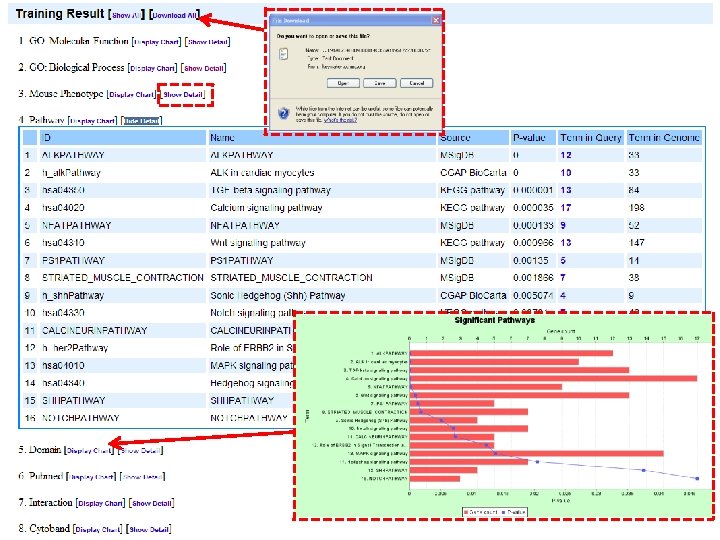 Topp. Gene web server (http: //toppgene. cchmc. org) For functional enrichment analysis
Topp. Gene web server (http: //toppgene. cchmc. org) For functional enrichment analysis
 PPI - Predicting Disease Genes 1. Direct protein–protein interactions (PPI) are one of the strongest manifestations of a functional relation between genes. 2. Hypothesis: Interacting proteins lead to same or similar disease phenotypes when mutated. 3. Several genetically heterogeneous hereditary diseases are shown to be caused by mutations in different interacting proteins. For e. g. Hermansky-Pudlak syndrome and Fanconi anaemia. Hence, protein–protein interactions might in principle be used to identify potentially interesting disease gene candidates.
PPI - Predicting Disease Genes 1. Direct protein–protein interactions (PPI) are one of the strongest manifestations of a functional relation between genes. 2. Hypothesis: Interacting proteins lead to same or similar disease phenotypes when mutated. 3. Several genetically heterogeneous hereditary diseases are shown to be caused by mutations in different interacting proteins. For e. g. Hermansky-Pudlak syndrome and Fanconi anaemia. Hence, protein–protein interactions might in principle be used to identify potentially interesting disease gene candidates.
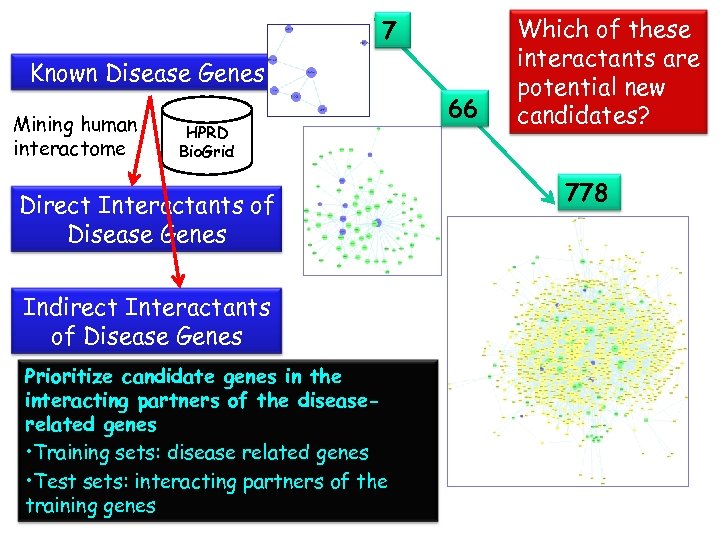 7 Known Disease Genes Mining human interactome HPRD Bio. Grid Direct Interactants of Disease Genes Indirect Interactants of Disease Genes Prioritize candidate genes in the interacting partners of the diseaserelated genes • Training sets: disease related genes • Test sets: interacting partners of the training genes 66 Which of these interactants are potential new candidates? 778
7 Known Disease Genes Mining human interactome HPRD Bio. Grid Direct Interactants of Disease Genes Indirect Interactants of Disease Genes Prioritize candidate genes in the interacting partners of the diseaserelated genes • Training sets: disease related genes • Test sets: interacting partners of the training genes 66 Which of these interactants are potential new candidates? 778
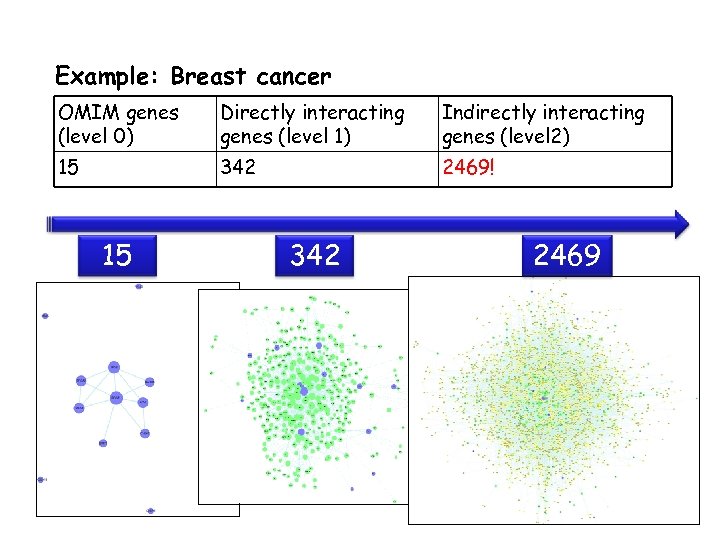 Example: Breast cancer OMIM genes (level 0) Directly interacting genes (level 1) Indirectly interacting genes (level 2) 15 342 2469! 15 342 2469
Example: Breast cancer OMIM genes (level 0) Directly interacting genes (level 1) Indirectly interacting genes (level 2) 15 342 2469! 15 342 2469
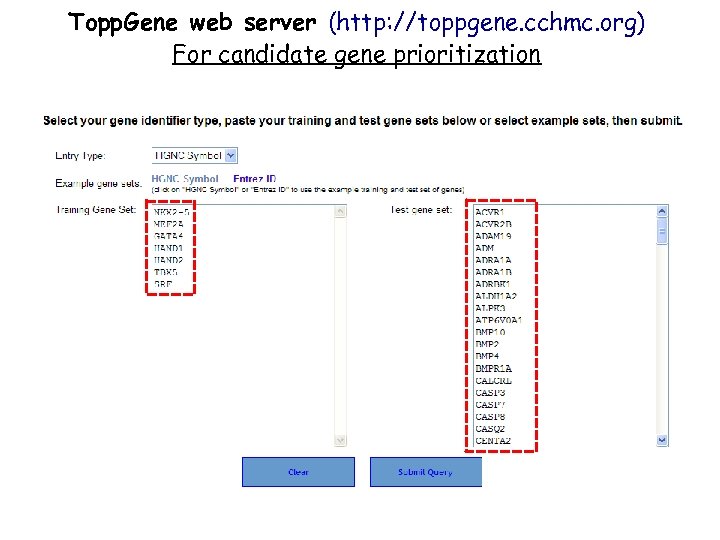 Topp. Gene web server (http: //toppgene. cchmc. org) For candidate gene prioritization
Topp. Gene web server (http: //toppgene. cchmc. org) For candidate gene prioritization
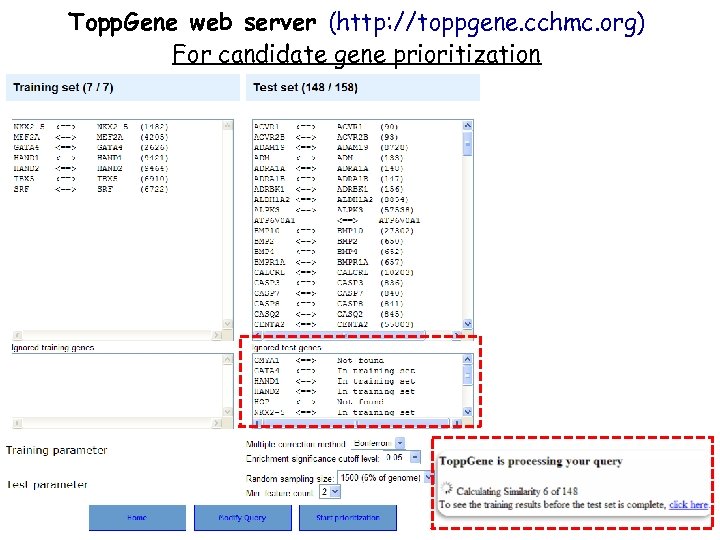 Topp. Gene web server (http: //toppgene. cchmc. org) For candidate gene prioritization
Topp. Gene web server (http: //toppgene. cchmc. org) For candidate gene prioritization
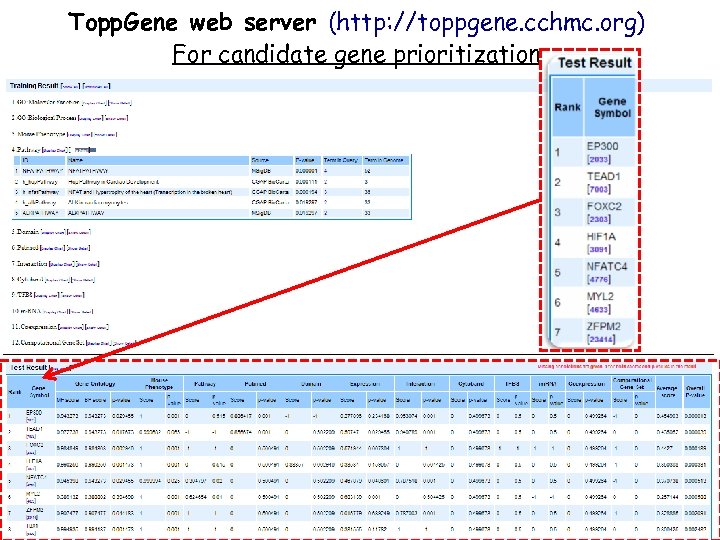 Topp. Gene web server (http: //toppgene. cchmc. org) For candidate gene prioritization
Topp. Gene web server (http: //toppgene. cchmc. org) For candidate gene prioritization
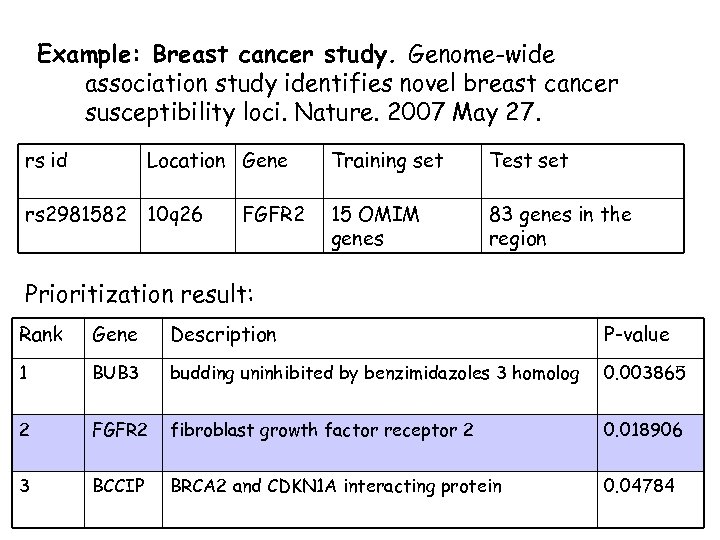 Example: Breast cancer study. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007 May 27. rs id Location Gene Training set Test set rs 2981582 10 q 26 15 OMIM genes 83 genes in the region FGFR 2 Prioritization result: Rank Gene Description P-value 1 BUB 3 budding uninhibited by benzimidazoles 3 homolog 0. 003865 2 FGFR 2 fibroblast growth factor receptor 2 0. 018906 3 BCCIP BRCA 2 and CDKN 1 A interacting protein 0. 04784
Example: Breast cancer study. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007 May 27. rs id Location Gene Training set Test set rs 2981582 10 q 26 15 OMIM genes 83 genes in the region FGFR 2 Prioritization result: Rank Gene Description P-value 1 BUB 3 budding uninhibited by benzimidazoles 3 homolog 0. 003865 2 FGFR 2 fibroblast growth factor receptor 2 0. 018906 3 BCCIP BRCA 2 and CDKN 1 A interacting protein 0. 04784
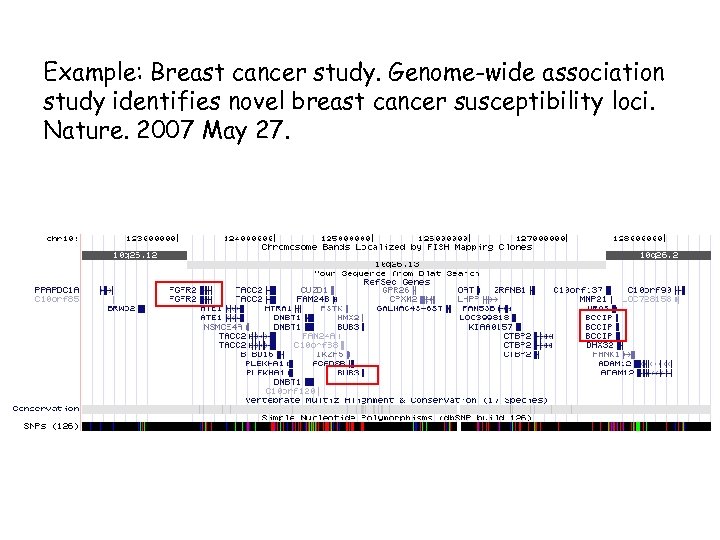 Example: Breast cancer study. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007 May 27.
Example: Breast cancer study. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007 May 27.
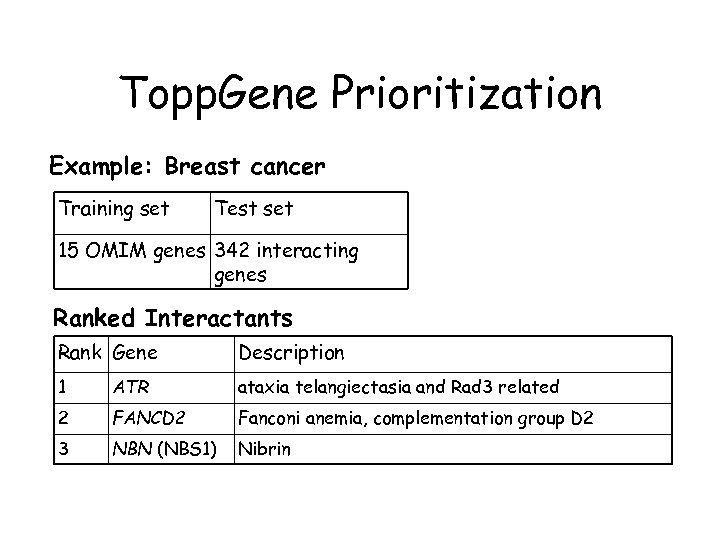 Topp. Gene Prioritization Example: Breast cancer Training set Test set 15 OMIM genes 342 interacting genes Ranked Interactants Rank Gene Description 1 ATR ataxia telangiectasia and Rad 3 related 2 FANCD 2 Fanconi anemia, complementation group D 2 3 NBN (NBS 1) Nibrin
Topp. Gene Prioritization Example: Breast cancer Training set Test set 15 OMIM genes 342 interacting genes Ranked Interactants Rank Gene Description 1 ATR ataxia telangiectasia and Rad 3 related 2 FANCD 2 Fanconi anemia, complementation group D 2 3 NBN (NBS 1) Nibrin
 Limitations General limitations of any training-test strategy: • Prior knowledge of disease-gene associations. • Assumption that the disease genes yet to discover will be consistent with what is already known about a disease. • Depend on the accuracy and completeness of the functional annotations. – Only one-fifth of the known human genes have pathway or phenotype annotations and there are still more than 40% genes whose functions are not defined! Chen et al. , 2007; BMC Bioinformatics
Limitations General limitations of any training-test strategy: • Prior knowledge of disease-gene associations. • Assumption that the disease genes yet to discover will be consistent with what is already known about a disease. • Depend on the accuracy and completeness of the functional annotations. – Only one-fifth of the known human genes have pathway or phenotype annotations and there are still more than 40% genes whose functions are not defined! Chen et al. , 2007; BMC Bioinformatics
 Mouse Phenotype - Limitations 1. MP is not a disease-centric ontology and the phenotype of a same gene mutation can vary depending on specific mouse strains or their genetic backgrounds. 2. Orthologous genes need not necessarily result in orthologous phenotypes. Possible Solutions - Future Directions More efficient cross-species phenome extrapolation where in the mouse phenotype terms are mapped to human phenotype concepts (from UMLS) semantically (“orthologous phenotype”) and the resultant orthologous genes associated with an orthologous phenotype are identified. Chen et al. , 2007; BMC Bioinformatics
Mouse Phenotype - Limitations 1. MP is not a disease-centric ontology and the phenotype of a same gene mutation can vary depending on specific mouse strains or their genetic backgrounds. 2. Orthologous genes need not necessarily result in orthologous phenotypes. Possible Solutions - Future Directions More efficient cross-species phenome extrapolation where in the mouse phenotype terms are mapped to human phenotype concepts (from UMLS) semantically (“orthologous phenotype”) and the resultant orthologous genes associated with an orthologous phenotype are identified. Chen et al. , 2007; BMC Bioinformatics
 PPIs for disease gene identification Limitations 1. Noisy interactome data • In vitro Vs in vivo (for e. g. only 5. 8% of yeast twohybrid predicted interactions were confirmed by HPRD) • Extrapolation of interactions from one species to another • Bias towards “well-studied” genes/proteins 2. Too many interactants! Hub proteins 3. Two interacting proteins need not lead to similar phenotype when mutated 4. Disease proteins may lie at different points in a pathway and need not interact directly 5. Lastly, disease mutations need not always involve proteins Oti et al. , 2006; J Med Gen
PPIs for disease gene identification Limitations 1. Noisy interactome data • In vitro Vs in vivo (for e. g. only 5. 8% of yeast twohybrid predicted interactions were confirmed by HPRD) • Extrapolation of interactions from one species to another • Bias towards “well-studied” genes/proteins 2. Too many interactants! Hub proteins 3. Two interacting proteins need not lead to similar phenotype when mutated 4. Disease proteins may lie at different points in a pathway and need not interact directly 5. Lastly, disease mutations need not always involve proteins Oti et al. , 2006; J Med Gen
 http: //anil. cchmc. org (under presentations) And PRIORITIZATION too! Thank You! http: //sbw. kgi. edu/
http: //anil. cchmc. org (under presentations) And PRIORITIZATION too! Thank You! http: //sbw. kgi. edu/


