b5c292a37c8317a72eb56d18de23298d.ppt
- Количество слайдов: 1
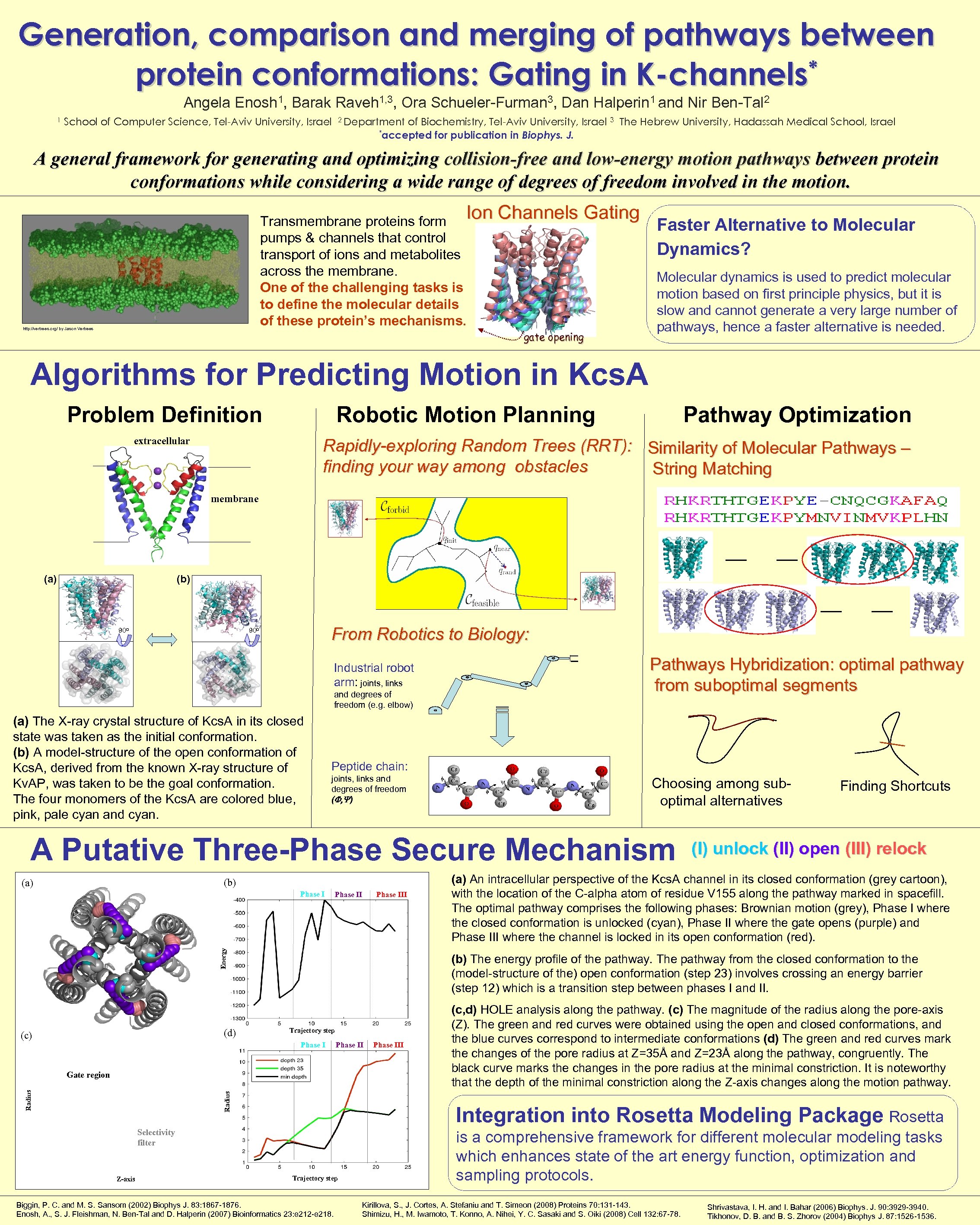
Generation, comparison and merging of pathways between * protein conformations: Gating in K-channels Angela Enosh 1, Barak Raveh 1, 3, Ora Schueler-Furman 3, Dan Halperin 1 and Nir Ben-Tal 2 1 School of Computer Science, Tel-Aviv University, Israel 2 Department of Biochemistry, Tel-Aviv University, Israel *accepted for publication in Biophys. J. 3 The Hebrew University, Hadassah Medical School, Israel A general framework for generating and optimizing collision-free and low-energy motion pathways between protein conformations while considering a wide range of degrees of freedom involved in the motion. Ion Channels Gating Transmembrane proteins form pumps & channels that control transport of ions and metabolites across the membrane. One of the challenging tasks is to define the molecular details of these protein’s mechanisms. http: //vertrees. org/ by Jason Vertrees gate opening Faster Alternative to Molecular Dynamics? Molecular dynamics is used to predict molecular motion based on first principle physics, but it is slow and cannot generate a very large number of pathways, hence a faster alternative is needed. Algorithms for Predicting Motion in Kcs. A Problem Definition Robotic Motion Planning extracellular Pathway Optimization Rapidly-exploring Random Trees (RRT): Similarity of Molecular Pathways – finding your way among obstacles String Matching membrane (a) (b) 90 o From Robotics to Biology: 90 o Industrial robot arm: joints, links and degrees of freedom (e. g. elbow) (a) The X-ray crystal structure of Kcs. A in its closed state was taken as the initial conformation. (b) A model-structure of the open conformation of Kcs. A, derived from the known X-ray structure of Kv. AP, was taken to be the goal conformation. The four monomers of the Kcs. A are colored blue, pink, pale cyan and cyan. Pathways Hybridization: optimal pathway from suboptimal segments Peptide chain: joints, links and degrees of freedom (F, Y) Choosing among suboptimal alternatives A Putative Three-Phase Secure Mechanism (b) (a) Phase III Energy Phase I (d) (c) Trajectory step Phase II Gate region Radius (I) unlock (II) open (III) relock (a) An intracellular perspective of the Kcs. A channel in its closed conformation (grey cartoon), with the location of the C-alpha atom of residue V 155 along the pathway marked in spacefill. The optimal pathway comprises the following phases: Brownian motion (grey), Phase I where the closed conformation is unlocked (cyan), Phase II where the gate opens (purple) and Phase III where the channel is locked in its open conformation (red). (b) The energy profile of the pathway. The pathway from the closed conformation to the (model-structure of the) open conformation (step 23) involves crossing an energy barrier (step 12) which is a transition step between phases I and II. Phase III (c, d) HOLE analysis along the pathway. (c) The magnitude of the radius along the pore-axis (Z). The green and red curves were obtained using the open and closed conformations, and the blue curves correspond to intermediate conformations (d) The green and red curves mark the changes of the pore radius at Z=35Å and Z=23Å along the pathway, congruently. The black curve marks the changes in the pore radius at the minimal constriction. It is noteworthy that the depth of the minimal constriction along the Z-axis changes along the motion pathway. Integration into Rosetta Modeling Package Rosetta Selectivity filter Z-axis Finding Shortcuts Trajectory step Biggin, P. C. and M. S. Sansom (2002) Biophys J. 83: 1867 -1876. Enosh, A. , S. J. Fleishman, N. Ben-Tal and D. Halperin (2007) Bioinformatics 23: e 212 -e 218. is a comprehensive framework for different molecular modeling tasks which enhances state of the art energy function, optimization and sampling protocols. Kirillova, S. , J. Cortes, A. Stefaniu and T. Simeon (2008) Proteins 70: 131 -143. Shimizu, H. , M. Iwamoto, T. Konno, A. Nihei, Y. C. Sasaki and S. Oiki (2008) Cell 132: 67 -78. Shrivastava, I. H. and I. Bahar (2006) Biophys. J. 90: 3929 -3940. Tikhonov, D. B. and B. S. Zhorov (2004) Biophys J. 87: 1526 -1536.
b5c292a37c8317a72eb56d18de23298d.ppt