c873c724f86e4c2925cdbb02e9b10ce1.ppt
- Количество слайдов: 50

general: Activators protein-DNA interaction
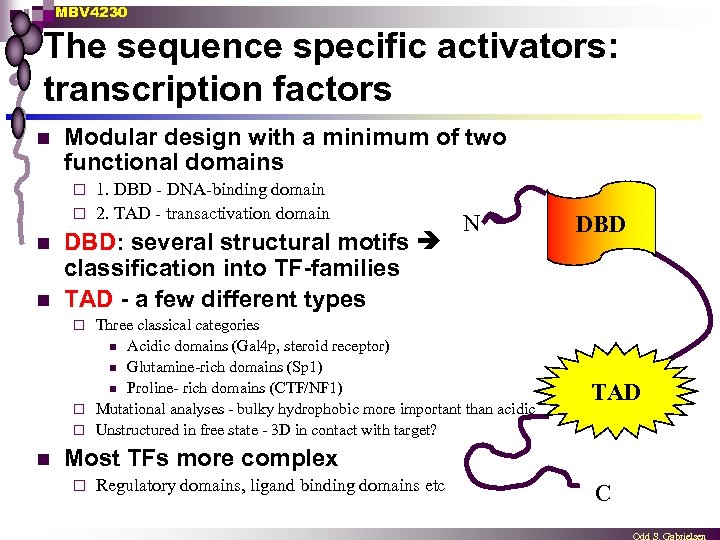
MBV 4230 The sequence specific activators: transcription factors n Modular design with a minimum of two functional domains 1. DBD - DNA-binding domain ¨ 2. TAD - transactivation domain ¨ n n DBD: several structural motifs classification into TF-families TAD - a few different types N Three classical categories n Acidic domains (Gal 4 p, steroid receptor) n Glutamine-rich domains (Sp 1) n Proline- rich domains (CTF/NF 1) ¨ Mutational analyses - bulky hydrophobic more important than acidic ¨ Unstructured in free state - 3 D in contact with target? DBD ¨ n TAD Most TFs more complex ¨ Regulatory domains, ligand binding domains etc C
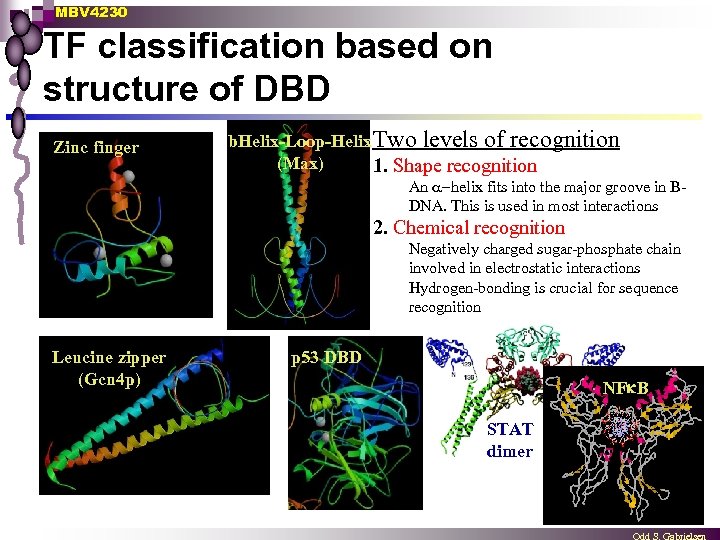
MBV 4230 TF classification based on structure of DBD Zinc finger b. Helix-Loop-Helix. Two levels of recognition (Max) 1. Shape recognition An helix fits into the major groove in BDNA. This is used in most interactions 2. Chemical recognition Negatively charged sugar-phosphate chain involved in electrostatic interactions Hydrogen-bonding is crucial for sequence recognition Leucine zipper (Gcn 4 p) p 53 DBD NFk. B STAT dimer
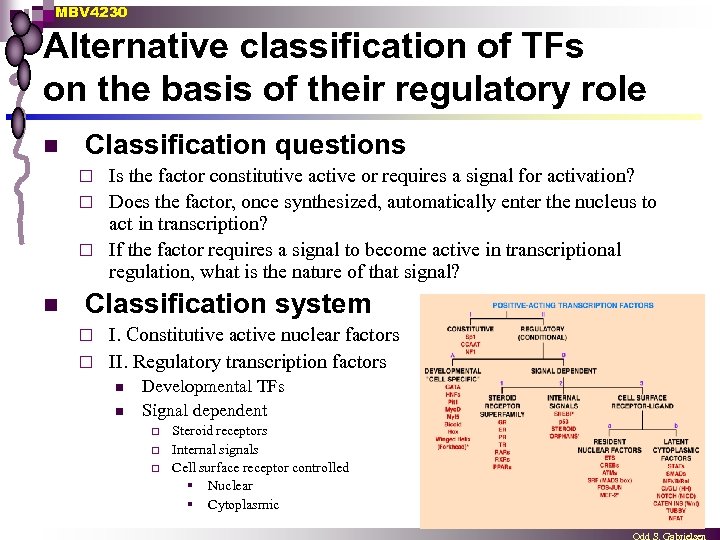
MBV 4230 Alternative classification of TFs on the basis of their regulatory role n Classification questions Is the factor constitutive active or requires a signal for activation? ¨ Does the factor, once synthesized, automatically enter the nucleus to act in transcription? ¨ If the factor requires a signal to become active in transcriptional regulation, what is the nature of that signal? ¨ n Classification system I. Constitutive active nuclear factors ¨ II. Regulatory transcription factors ¨ n n Developmental TFs Signal dependent ¨ ¨ ¨ Steroid receptors Internal signals Cell surface receptor controlled § Nuclear § Cytoplasmic
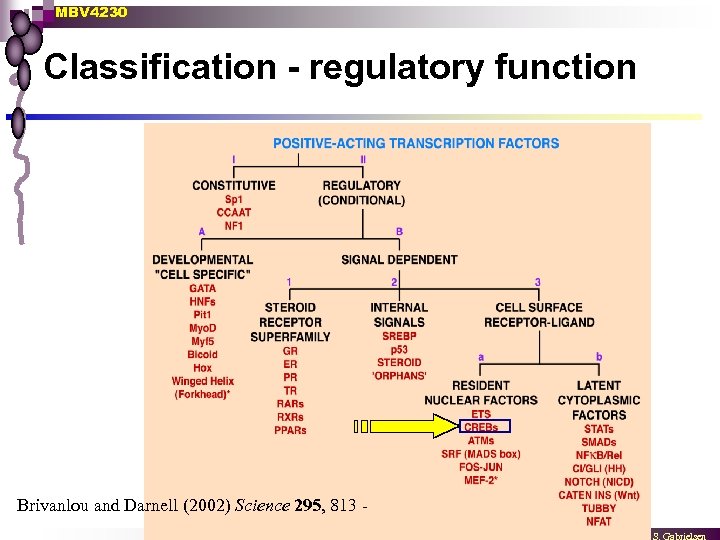
MBV 4230 Classification - regulatory function Brivanlou and Darnell (2002) Science 295, 813 -
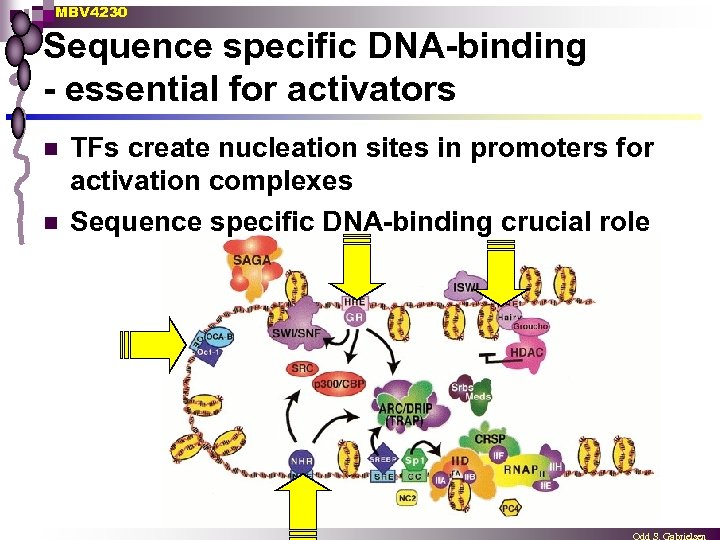
MBV 4230 Sequence specific DNA-binding - essential for activators n n TFs create nucleation sites in promoters for activation complexes Sequence specific DNA-binding crucial role

Principles of sequence specific DNA-binding
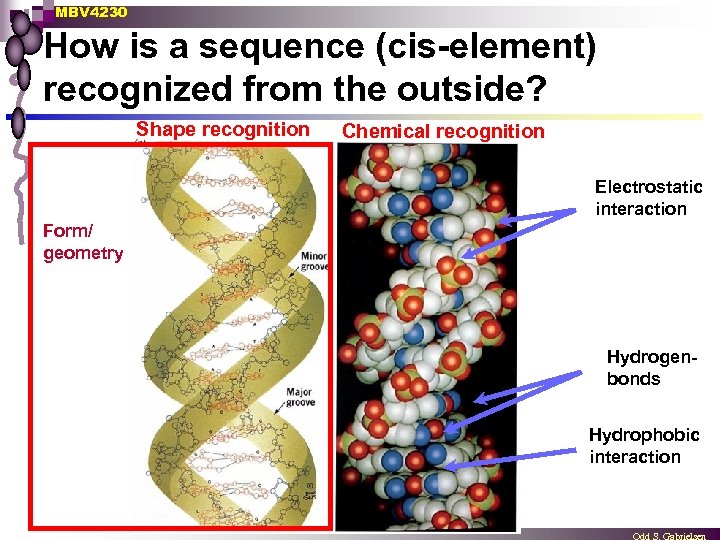
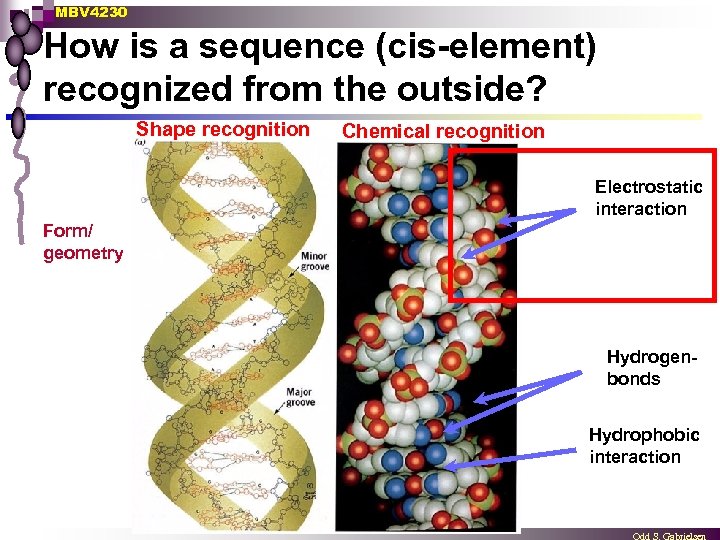
MBV 4230 How is a sequence (cis-element) recognized from the outside? Shape recognition Chemical recognition Electrostatic interaction Form/ geometry Hydrogenbonds Hydrophobic interaction
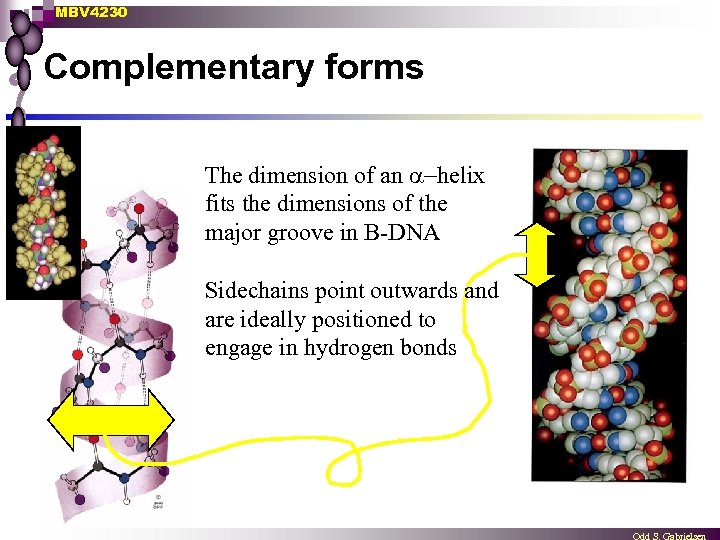
MBV 4230 Complementary forms The dimension of an helix fits the dimensions of the major groove in B-DNA Sidechains point outwards and are ideally positioned to engage in hydrogen bonds
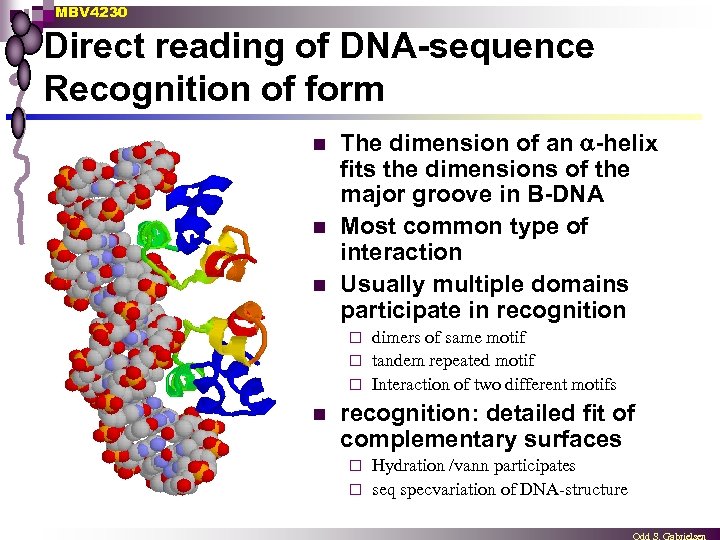
MBV 4230 Direct reading of DNA-sequence Recognition of form n n n The dimension of an -helix fits the dimensions of the major groove in B-DNA Most common type of interaction Usually multiple domains participate in recognition dimers of same motif ¨ tandem repeated motif ¨ Interaction of two different motifs ¨ n recognition: detailed fit of complementary surfaces Hydration /vann participates ¨ seq specvariation of DNA-structure ¨
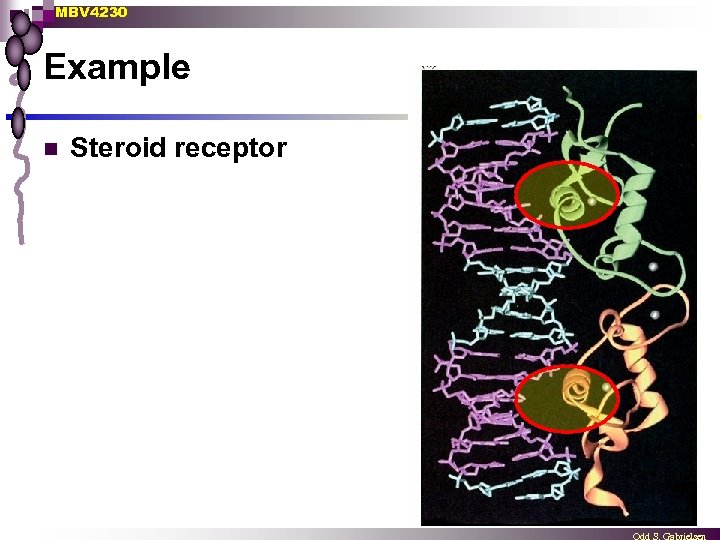
MBV 4230 Example n Steroid receptor
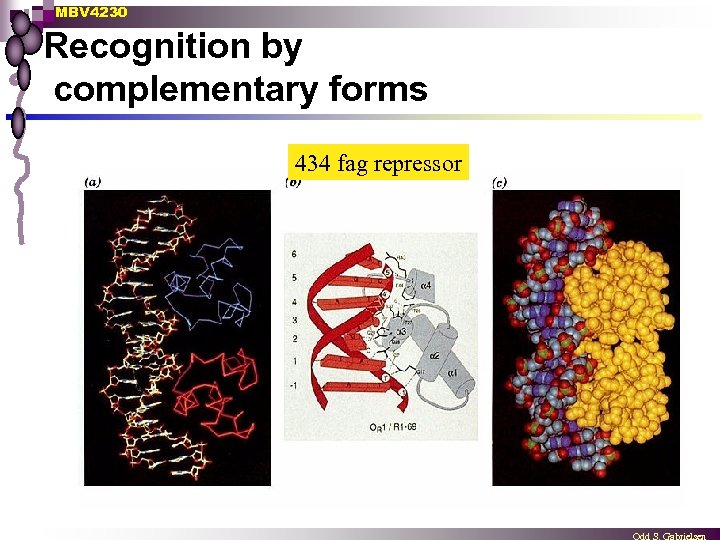
MBV 4230 Recognition by complementary forms 434 fag repressor
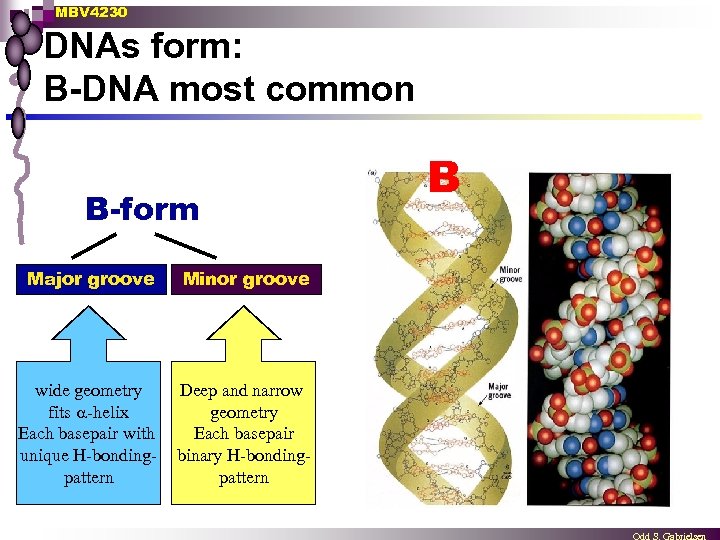
MBV 4230 DNAs form: B-DNA most common B-form Major groove Minor groove wide geometry fits -helix Each basepair with unique H-bondingpattern Deep and narrow geometry Each basepair binary H-bondingpattern B
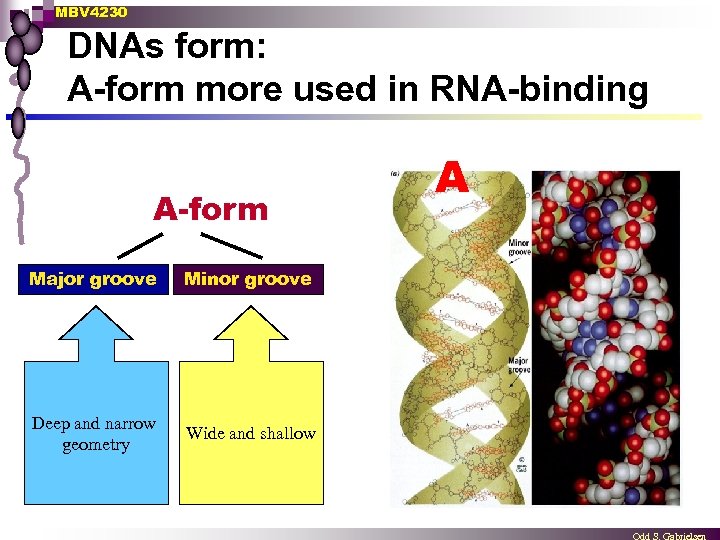
MBV 4230 DNAs form: A-form more used in RNA-binding A-form Major groove Minor groove Deep and narrow geometry Wide and shallow A
MBV 4230 How is a sequence (cis-element) recognized from the outside? Shape recognition Chemical recognition Electrostatic interaction Form/ geometry Hydrogenbonds Hydrophobic interaction
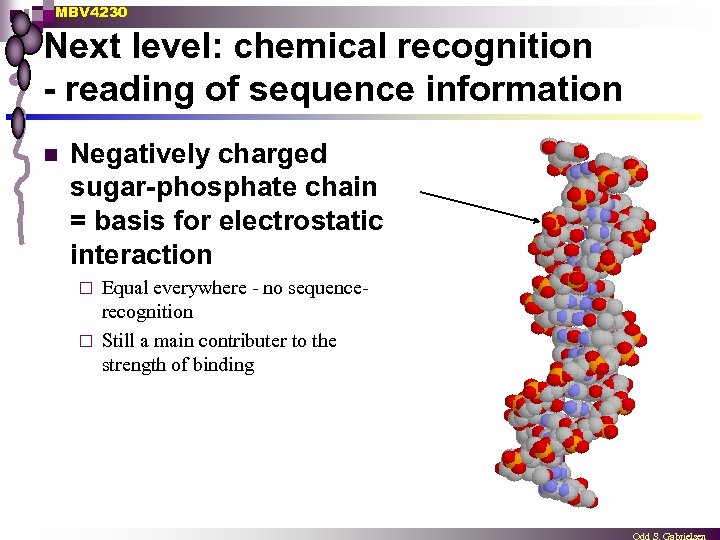
MBV 4230 Next level: chemical recognition - reading of sequence information n Negatively charged sugar-phosphate chain = basis for electrostatic interaction Equal everywhere - no sequencerecognition ¨ Still a main contributer to the strength of binding ¨
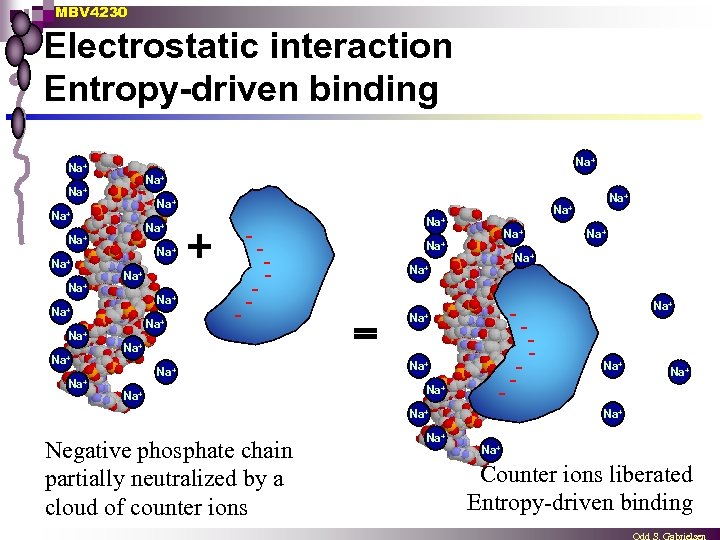
MBV 4230 Electrostatic interaction Entropy-driven binding Na+ Na+ Na+ Na+ - Na+ - - Na+ Na+ Negative phosphate chain partially neutralized by a cloud of counter ions Na+ Na+ - Na+ Na+ Na+ Counter ions liberated Entropy-driven binding
MBV 4230 How is a sequence (cis-element) recognized from the outside? Shape recognition Chemical recognition Electrostatic interaction Form/ geometry Hydrogenbonds Hydrophobic interaction
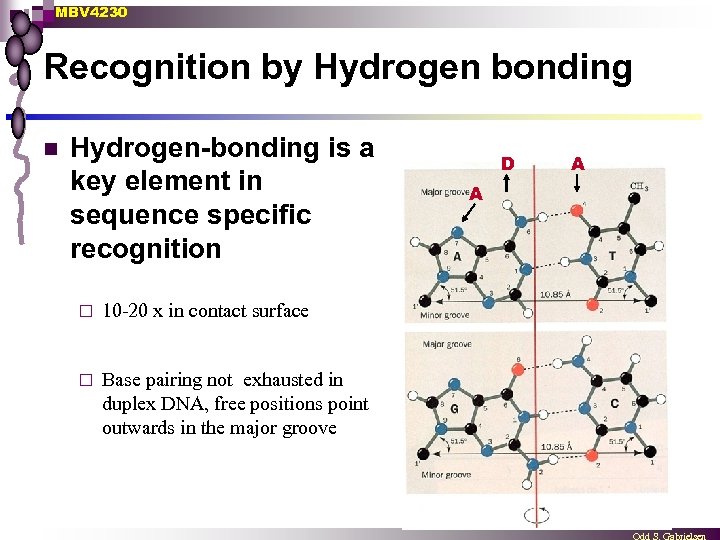
MBV 4230 Recognition by Hydrogen bonding n Hydrogen-bonding is a key element in sequence specific recognition ¨ 10 -20 x in contact surface ¨ Base pairing not exhausted in duplex DNA, free positions point outwards in the major groove D A A
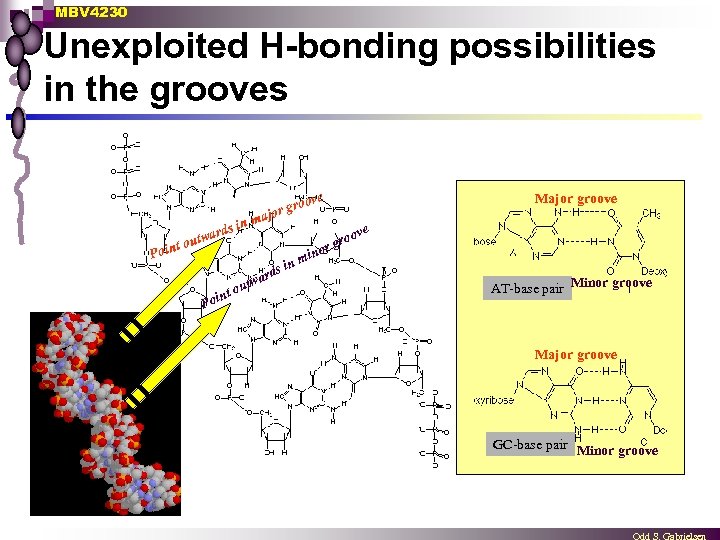
MBV 4230 Unexploited H-bonding possibilities in the grooves ove ds in t Poin ar outw gro ajor m ar nm ds i Poi n tw t ou Major groove gro or in AT-base pair Minor groove Major groove GC-base pair Minor groove
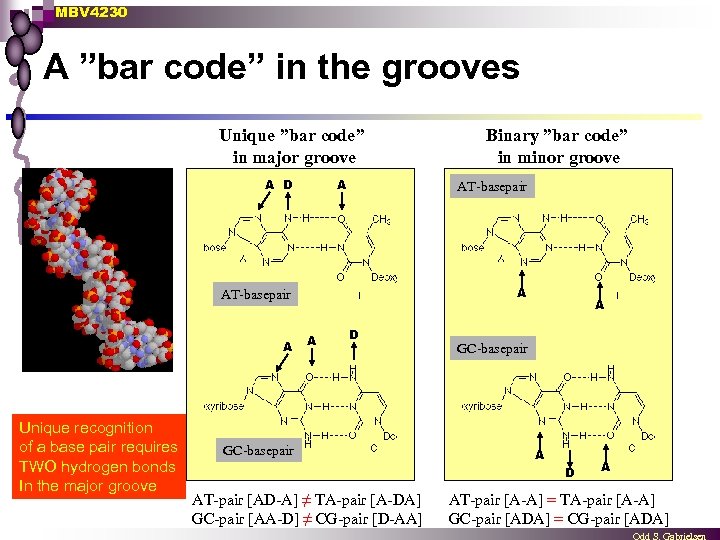
MBV 4230 A ”bar code” in the grooves Unique ”bar code” in major groove A D A AT-basepair A Unique recognition of a base pair requires TWO hydrogen bonds In the major groove Binary ”bar code” in minor groove A D GC-basepair A D AT-pair [AD-A] ≠ TA-pair [A-DA] GC-pair [AA-D] ≠ CG-pair [D-AA] A AT-pair [A-A] = TA-pair [A-A] GC-pair [ADA] = CG-pair [ADA]
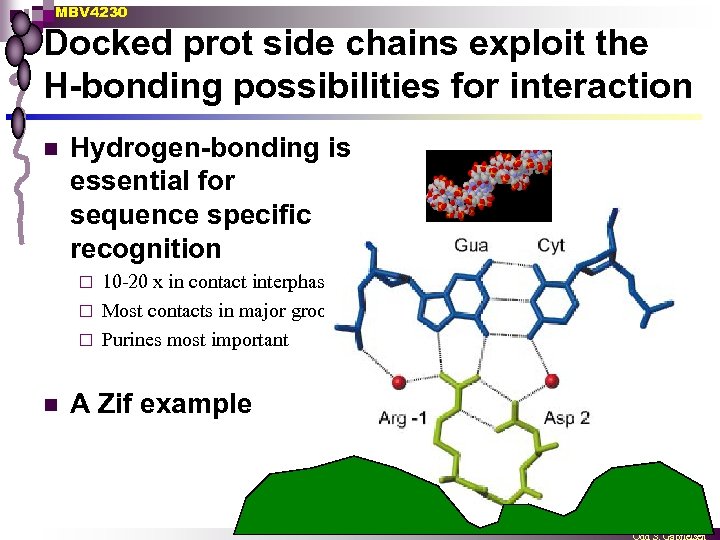
MBV 4230 Docked prot side chains exploit the H-bonding possibilities for interaction n Hydrogen-bonding is essential for sequence specific recognition 10 -20 x in contact interphase ¨ Most contacts in major groove ¨ Purines most important ¨ n A Zif example
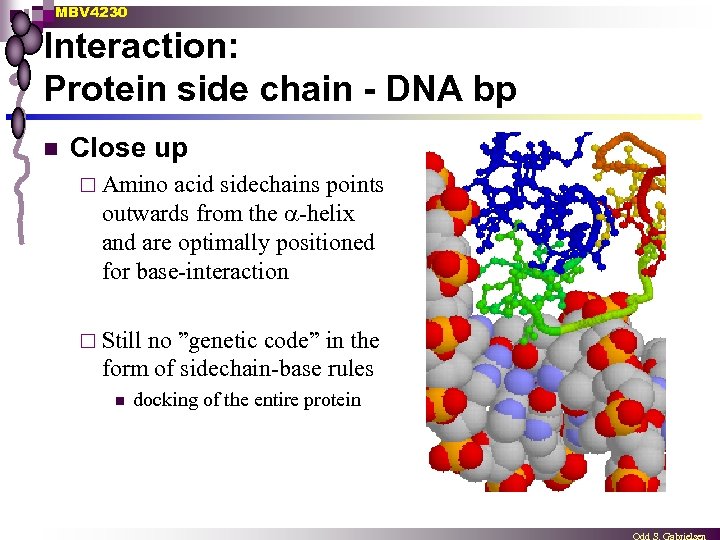
MBV 4230 Interaction: Protein side chain - DNA bp n Close up ¨ Amino acid sidechains points outwards from the -helix and are optimally positioned for base-interaction ¨ Still no ”genetic code” in the form of sidechain-base rules n docking of the entire protein
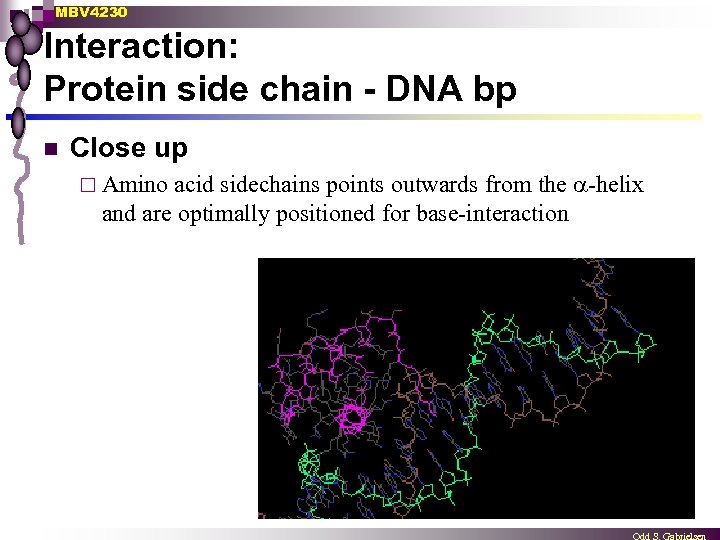
MBV 4230 Interaction: Protein side chain - DNA bp n Close up acid sidechains points outwards from the -helix and are optimally positioned for base-interaction ¨ Amino
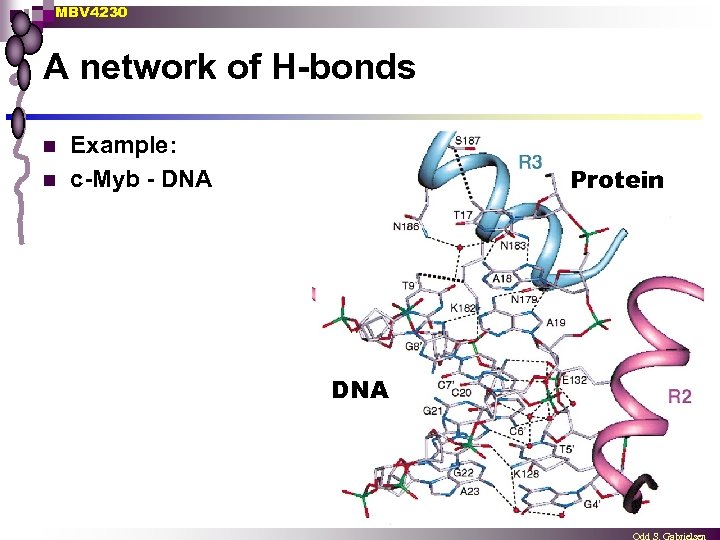
MBV 4230 A network of H-bonds n n Example: c-Myb - DNA Protein DNA
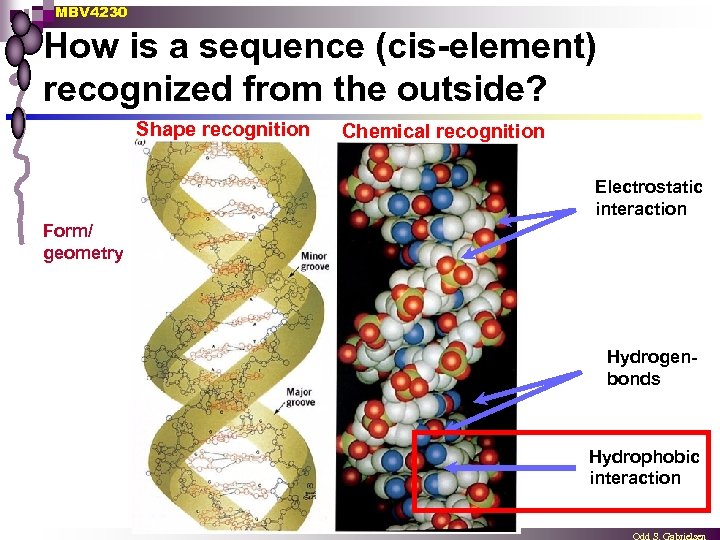
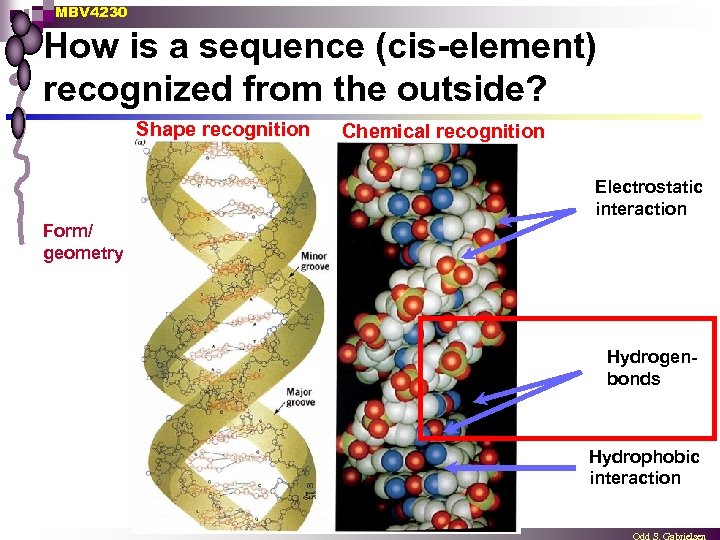
MBV 4230 How is a sequence (cis-element) recognized from the outside? Shape recognition Chemical recognition Electrostatic interaction Form/ geometry Hydrogenbonds Hydrophobic interaction
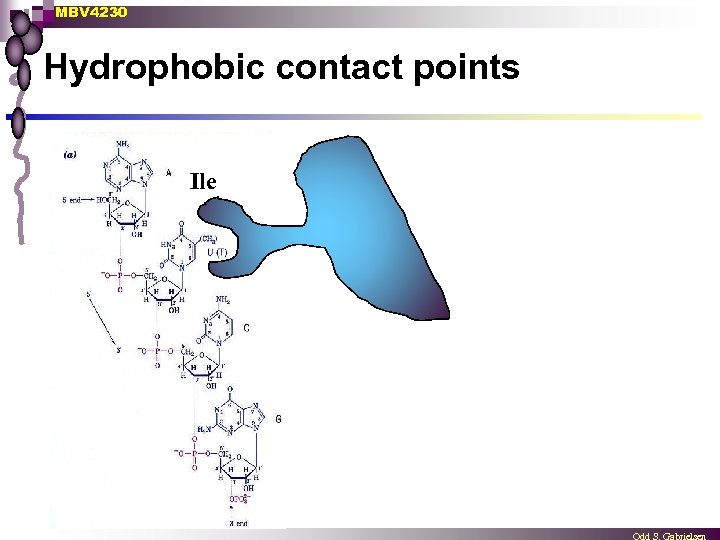
MBV 4230 Hydrophobic contact points Ile

Homeodomains
MBV 4230 The Homeodomain-family: common DBD-structure n Homeotic genes - biology Regulation of Drosophila development ¨ Striking phenotypes of mutants - bodyparts move ¨ Control genetic developmental program ¨ n Homeobox / homeodomain Conservered DNA-sequence “homeobox” in a large number of genes ¨ Encode a 60 aa “homeodomain” ¨ A stably folded structure that binds DNA ¨ Similarity with prokaryotic helix-turn-helix ¨ n 3 D-structure determined for several HDs Drosophila Antennapedia HD (NMR) ¨ Drosophila Engrailed HD-DNA kompleks (crystal) ¨ Yeast MAT 2 ¨
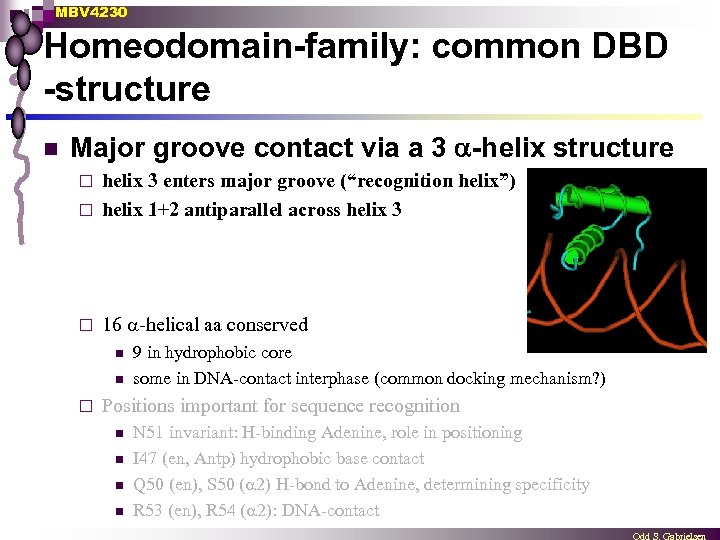
MBV 4230 Homeodomain-family: common DBD -structure n Major groove contact via a 3 -helix structure helix 3 enters major groove (“recognition helix”) ¨ helix 1+2 antiparallel across helix 3 ¨ ¨ 16 -helical aa conserved n n ¨ 9 in hydrophobic core some in DNA-contact interphase (common docking mechanism? ) Positions important for sequence recognition n n N 51 invariant: H-binding Adenine, role in positioning I 47 (en, Antp) hydrophobic base contact Q 50 (en), S 50 ( 2) H-bond to Adenine, determining specificity R 53 (en), R 54 ( 2): DNA-contact
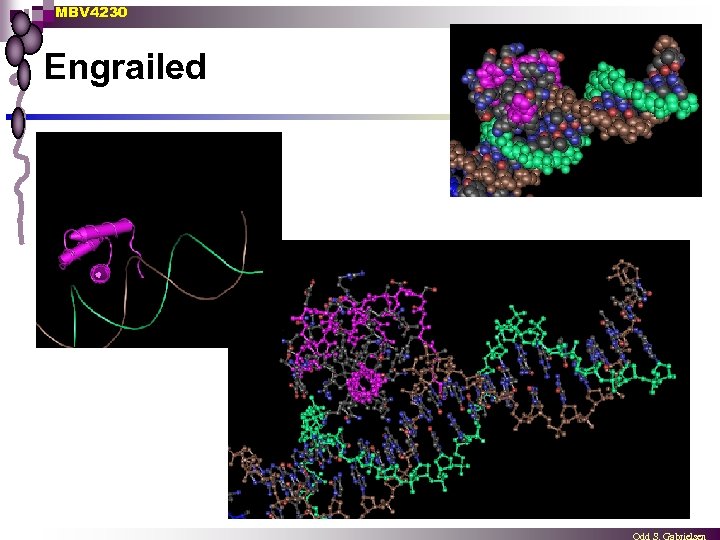
MBV 4230 Engrailed
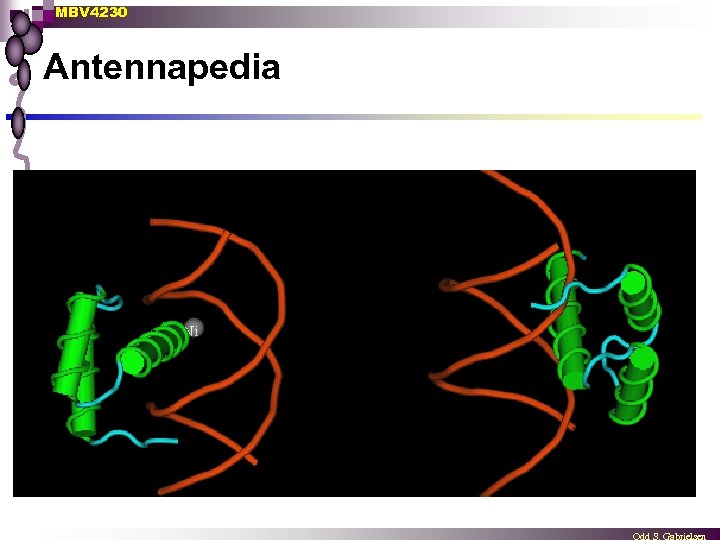
MBV 4230 Antennapedia
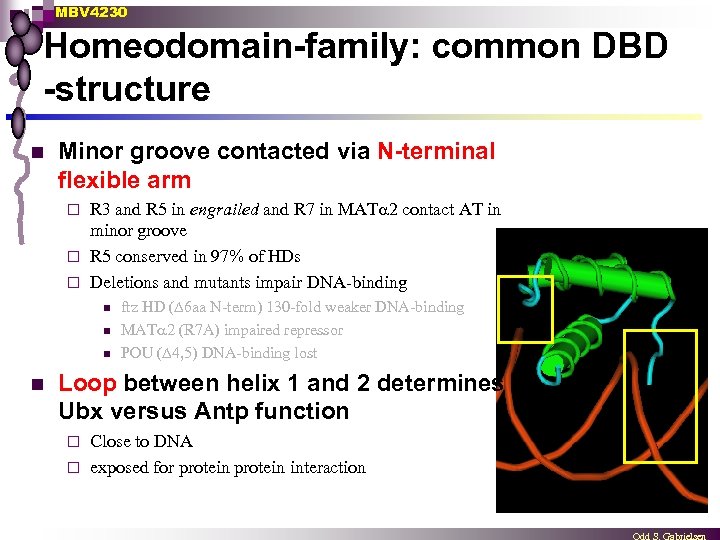
MBV 4230 Homeodomain-family: common DBD -structure n Minor groove contacted via N-terminal flexible arm R 3 and R 5 in engrailed and R 7 in MAT 2 contact AT in minor groove ¨ R 5 conserved in 97% of HDs ¨ Deletions and mutants impair DNA-binding ¨ n n ftz HD (∆6 aa N-term) 130 -fold weaker DNA-binding MAT 2 (R 7 A) impaired repressor POU (∆4, 5) DNA-binding lost Loop between helix 1 and 2 determines Ubx versus Antp function Close to DNA ¨ exposed for protein interaction ¨
MBV 4230 HD-paradox: what determines sequence specificity? n n Drosophila Ultrabithorax (Ubx), Antennapedia (Antp), Deformed (Dfd) and Sex combs reduced (Scr): closely similar HD, biological rolle very different Minor differences in DNA-binding in vitro TAAT-motif bound by most HD-factors ¨ contrast between promiscuity in vitro and specific effects in vivo ¨ n Swaps reveal that surprisingly much of the specificity is determined by the N-terminal arm which contacts the minor groove Swaps: Antp with Scr-type N-term arm shows Scr-type specificity in vivo ¨ Swaps: Dfd with Ubx-type N-term arm shows Ubx-type specificity in vivo ¨ n N-terminal arm more divergent than the rest of HD R 5 and R 7 (contacting DNA) are present in both Ubx, Antp, Dfd, and Scr ¨ Other tail aa diverge much more ¨
MBV 4230 Solutions of the paradox n Conformational effects mediated by N-term arm ¨ n Even if the -helical HDs are very similar, a much larger diversity is found in the N-terminal arms that contact the minor groove Protein-protein interaction with other TFs through the N-terminal arm - enhanced affinity/specificity - the basis of combinatorial control MAT 2 interaction with MCM 1 - cooperative interactions ¨ Ultrabithorax- Extradenticle in Drosophila ¨ Hox-Pbx 1 in mammals ¨
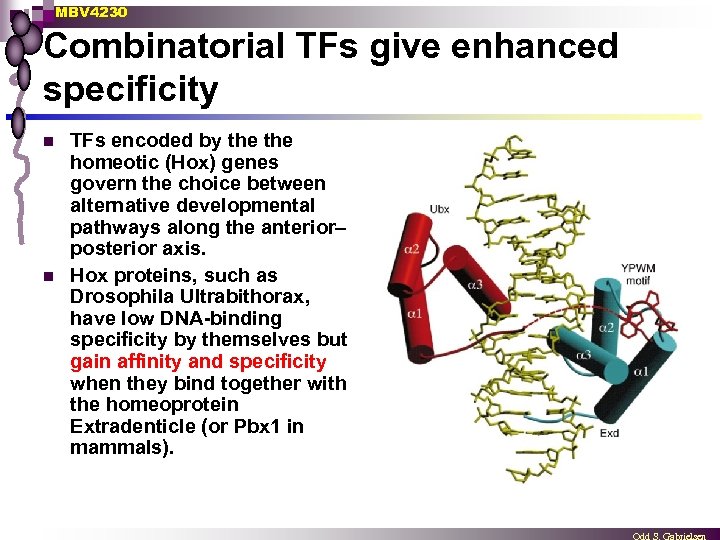
MBV 4230 Combinatorial TFs give enhanced specificity n n TFs encoded by the homeotic (Hox) genes govern the choice between alternative developmental pathways along the anterior– posterior axis. Hox proteins, such as Drosophila Ultrabithorax, have low DNA-binding specificity by themselves but gain affinity and specificity when they bind together with the homeoprotein Extradenticle (or Pbx 1 in mammals).
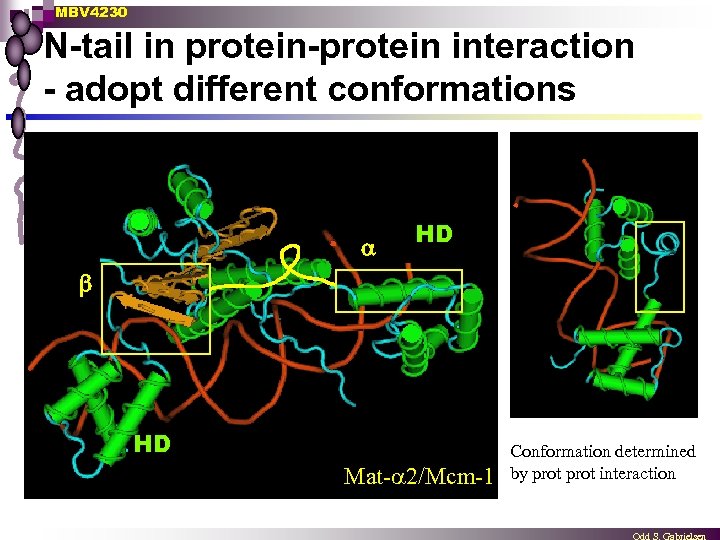
MBV 4230 N-tail in protein-protein interaction - adopt different conformations HD b HD Mat- 2/Mcm-1 Conformation determined by prot interaction
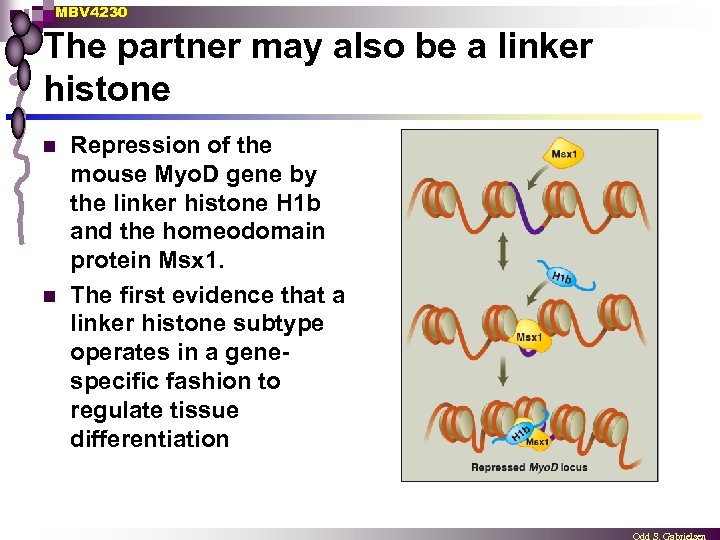
MBV 4230 The partner may also be a linker histone n n Repression of the mouse Myo. D gene by the linker histone H 1 b and the homeodomain protein Msx 1. The first evidence that a linker histone subtype operates in a genespecific fashion to regulate tissue differentiation
MBV 4230 It works impressively well n Hox genes

POU family
MBV 4230 POU-family: common DBD-structure n The POU-name : Pit-1 pituitary specific TF ¨ Oct-1 and Oct-2 lymphoide TFs ¨ Unc 86 TF that regulates neuronal development in C. elegans ¨ n A bipartite 160 aa homeodomain-related DBD a POU-type HD subdomain (C-terminally located) ¨ et POU-specific subdomain (N-terminally located) ¨ Coupled by a variabel linker (15 -30 aa) ¨ n POU is a structurally bipartite motif that arose by the fusion of genes encoding two different types of DNA-binding domain.
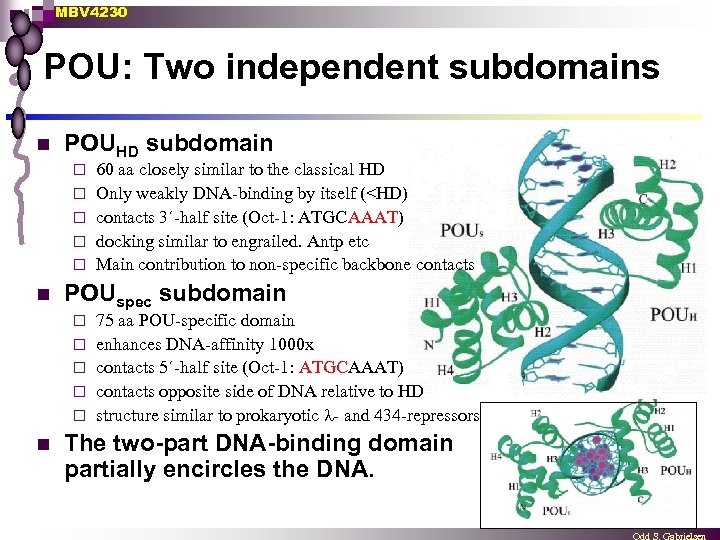
MBV 4230 POU: Two independent subdomains n POUHD subdomain ¨ ¨ ¨ n POUspec subdomain ¨ ¨ ¨ n 60 aa closely similar to the classical HD Only weakly DNA-binding by itself (<HD) contacts 3´-half site (Oct-1: ATGCAAAT) docking similar to engrailed. Antp etc Main contribution to non-specific backbone contacts 75 aa POU-specific domain enhances DNA-affinity 1000 x contacts 5´-half site (Oct-1: ATGCAAAT) contacts opposite side of DNA relative to HD structure similar to prokaryotic - and 434 -repressors The two-part DNA-binding domain partially encircles the DNA.
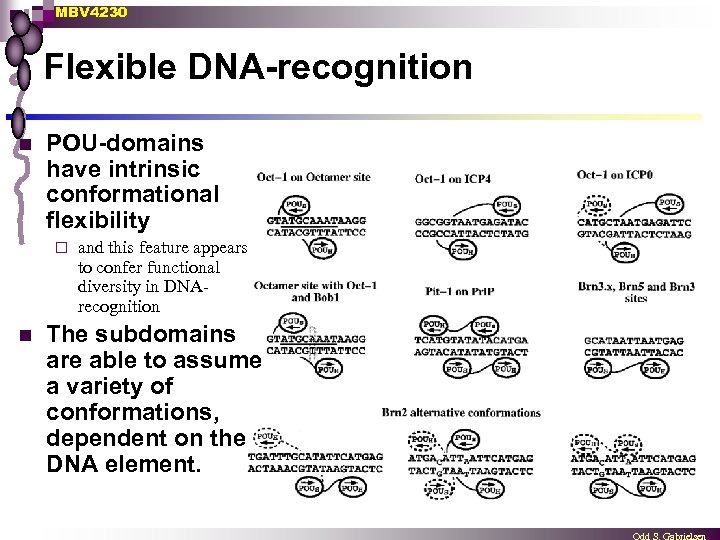
MBV 4230 Flexible DNA-recognition n POU-domains have intrinsic conformational flexibility ¨ n and this feature appears to confer functional diversity in DNArecognition The subdomains are able to assume a variety of conformations, dependent on the DNA element.
MBV 4230 A POU prototype: Oct-1 n n Ubiquitously expressed Oct-1 (≠ cell type specific Oct-2) Oct-1 performs many divergent roles in cellular trx regulation ¨ n partly owing to its flexibility in DNA binding and ability to associate with multiple and varied co-regulators Oct-1 activates transcription of genes that are involved in basic cellular processes Oct-1 activates small nuclear RNA (sn. RNA) and ¨ S-phase histone H 2 B gene transcription ¨ cell-specific promoters, particularly in the immune and nervous systems ¨ immunoglobulin (Ig) heavy- and lightchains ¨ n Activate target genes by bidning to the “octamer” ciselement ATGCAAAT ¨ Hence the name “Octamer-motif binding protein”
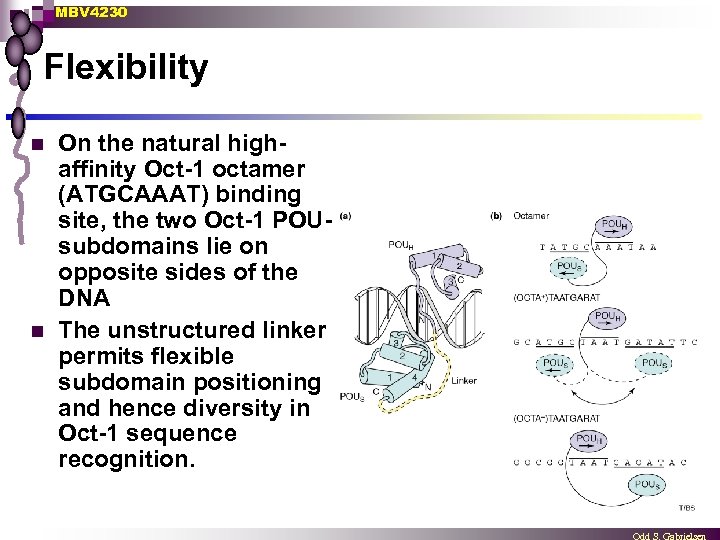
MBV 4230 Flexibility n n On the natural highaffinity Oct-1 octamer (ATGCAAAT) binding site, the two Oct-1 POUsubdomains lie on opposite sides of the DNA The unstructured linker permits flexible subdomain positioning and hence diversity in Oct-1 sequence recognition.
MBV 4230 Oct-1: associates with multiple and varied co-regulators n Oct-1 associates with a B-cell specific coregulator OCA-B (OBF-1). OCA-B stabilizes Oct-1 on DNA and provides a transcriptional activation domain. ¨ ¨ ¨ n B-cell specific activation of immunoglobulin genes - for long a paradox Depended on octamer cis-elements B-cell express both ubiquitous Oct-1 and the cell type specific Oct-2 Hypothesis: Oct-2 aktivates Ig. Gs (Wrong!) oct-2 deficient mouse normal development of early B-cells and cell lines without Oct-2 produce abundant amounts of Ig A B-cell specific coactivator mediates Oct-1 transactivation VP 16 - a virus strategy to exploit a host TF
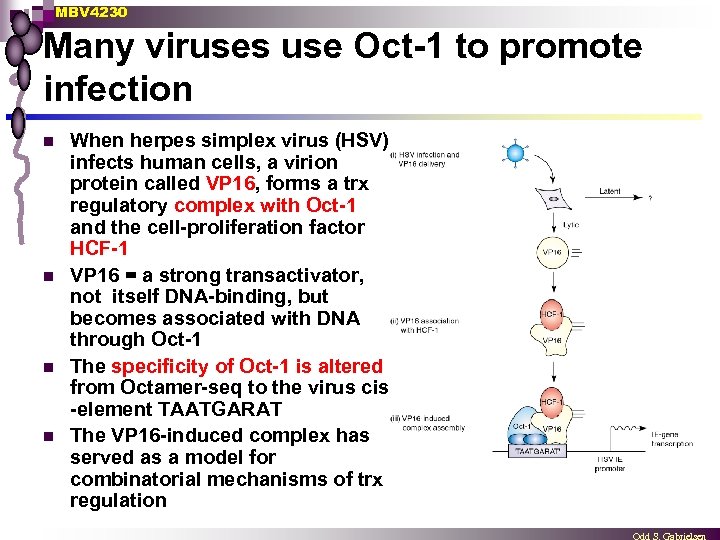
MBV 4230 Many viruses use Oct-1 to promote infection n n When herpes simplex virus (HSV) infects human cells, a virion protein called VP 16, forms a trx regulatory complex with Oct-1 and the cell-proliferation factor HCF-1 VP 16 = a strong transactivator, not itself DNA-binding, but becomes associated with DNA through Oct-1 The specificity of Oct-1 is altered from Octamer-seq to the virus cis -element TAATGARAT The VP 16 -induced complex has served as a model for combinatorial mechanisms of trx regulation

Pax family
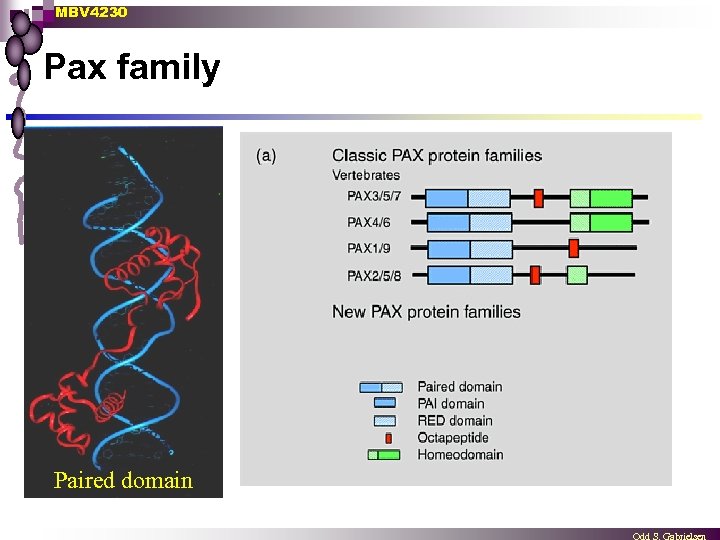
MBV 4230 Pax family Paired domain
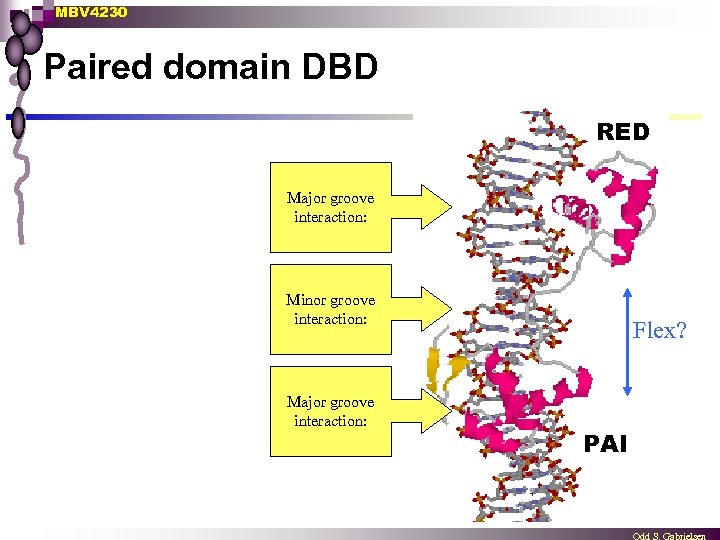
MBV 4230 Paired domain DBD RED Major groove interaction: Minor groove interaction: Major groove interaction: Flex? PAI
c873c724f86e4c2925cdbb02e9b10ce1.ppt