4700a3fae36da74a59d4835b29cdeb9f.ppt
- Количество слайдов: 39
 Gene Structure and Function
Gene Structure and Function
 Genetic Code All genomes, from virus to humans, are designed around linear sequences of nucleotides, share a universal code. An m. RNA specify amino acid sequence through the genetic code. We know one amino acid only could specify one nucleotide. Two nucleotide combinations could only specify 16 amino acids. Three nucleotides (64 possibilities), called a codon, is enough to specify each amino acid. Each 3 nucleotide code for one amino acid. • The first codon is the start codon, and usually coincides with the Amino Acid Methionine. (M which has codon code ‘ATG’) • The last codon is the stop codon and does NOT code for an amino acid. It is sometimes represented by ‘*’ to indicate the ‘STOP’ codon. • A coding region (abbreviation CDS) starts at the START codon and ends at the STOP codon.
Genetic Code All genomes, from virus to humans, are designed around linear sequences of nucleotides, share a universal code. An m. RNA specify amino acid sequence through the genetic code. We know one amino acid only could specify one nucleotide. Two nucleotide combinations could only specify 16 amino acids. Three nucleotides (64 possibilities), called a codon, is enough to specify each amino acid. Each 3 nucleotide code for one amino acid. • The first codon is the start codon, and usually coincides with the Amino Acid Methionine. (M which has codon code ‘ATG’) • The last codon is the stop codon and does NOT code for an amino acid. It is sometimes represented by ‘*’ to indicate the ‘STOP’ codon. • A coding region (abbreviation CDS) starts at the START codon and ends at the STOP codon.
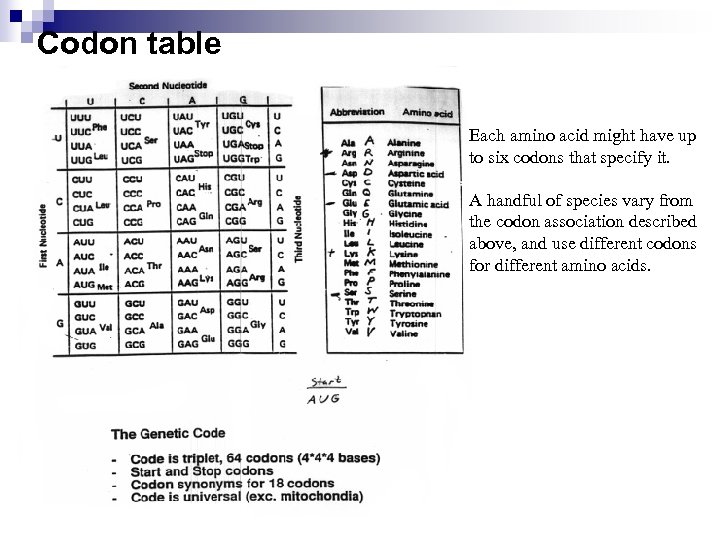 Codon table Each amino acid might have up to six codons that specify it. A handful of species vary from the codon association described above, and use different codons for different amino acids.
Codon table Each amino acid might have up to six codons that specify it. A handful of species vary from the codon association described above, and use different codons for different amino acids.
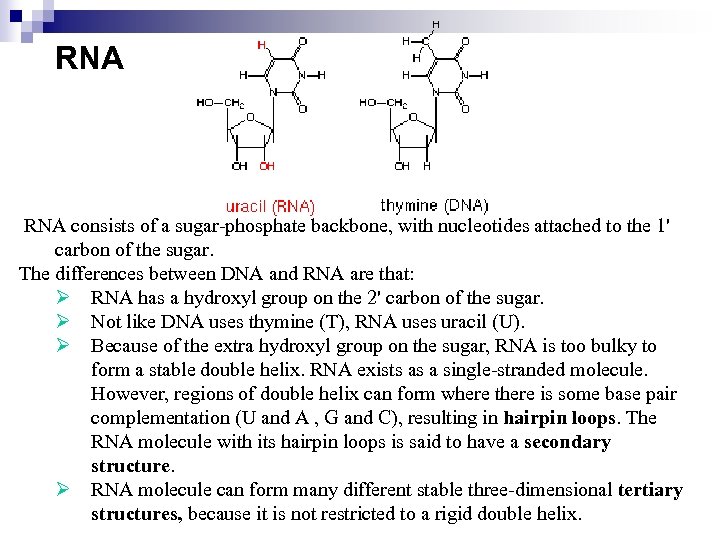 RNA consists of a sugar-phosphate backbone, with nucleotides attached to the 1' carbon of the sugar. The differences between DNA and RNA are that: Ø RNA has a hydroxyl group on the 2' carbon of the sugar. Ø Not like DNA uses thymine (T), RNA uses uracil (U). Ø Because of the extra hydroxyl group on the sugar, RNA is too bulky to form a stable double helix. RNA exists as a single-stranded molecule. However, regions of double helix can form where there is some base pair complementation (U and A , G and C), resulting in hairpin loops. The RNA molecule with its hairpin loops is said to have a secondary structure. Ø RNA molecule can form many different stable three-dimensional tertiary structures, because it is not restricted to a rigid double helix.
RNA consists of a sugar-phosphate backbone, with nucleotides attached to the 1' carbon of the sugar. The differences between DNA and RNA are that: Ø RNA has a hydroxyl group on the 2' carbon of the sugar. Ø Not like DNA uses thymine (T), RNA uses uracil (U). Ø Because of the extra hydroxyl group on the sugar, RNA is too bulky to form a stable double helix. RNA exists as a single-stranded molecule. However, regions of double helix can form where there is some base pair complementation (U and A , G and C), resulting in hairpin loops. The RNA molecule with its hairpin loops is said to have a secondary structure. Ø RNA molecule can form many different stable three-dimensional tertiary structures, because it is not restricted to a rigid double helix.
 Open Reading Frames (ORF) On a given piece of DNA, there can be 6 possible frames. The ORF can be either on the + or - strand on any of 3 possible frames Frame 1: 1 st base of start codon can either start at base 1, 4, 7, 10, . . . Frame 2: 1 st base of start codon can either start at base 2, 5, 8, 11, . . . Frame 3: 1 st base of start codon can either start at base 3, 6, 9, 12, . . . (frame – 1, -2, -3 are on minus strand) An open reading frames starts with ATG in most species, and ends with a stop codon (TAA, TAG or TGA) A program called SIXFRAME, you can visit the site directly http: //searchlauncher. bcm. tmc. edu/seq-util/Options/sixframe. html
Open Reading Frames (ORF) On a given piece of DNA, there can be 6 possible frames. The ORF can be either on the + or - strand on any of 3 possible frames Frame 1: 1 st base of start codon can either start at base 1, 4, 7, 10, . . . Frame 2: 1 st base of start codon can either start at base 2, 5, 8, 11, . . . Frame 3: 1 st base of start codon can either start at base 3, 6, 9, 12, . . . (frame – 1, -2, -3 are on minus strand) An open reading frames starts with ATG in most species, and ends with a stop codon (TAA, TAG or TGA) A program called SIXFRAME, you can visit the site directly http: //searchlauncher. bcm. tmc. edu/seq-util/Options/sixframe. html
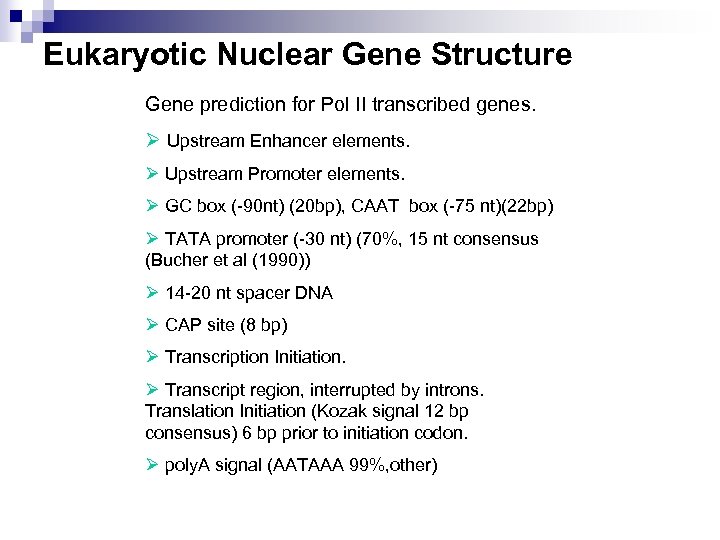 Eukaryotic Nuclear Gene Structure Gene prediction for Pol II transcribed genes. Ø Upstream Enhancer elements. Ø Upstream Promoter elements. Ø GC box (-90 nt) (20 bp), CAAT box (-75 nt)(22 bp) Ø TATA promoter (-30 nt) (70%, 15 nt consensus (Bucher et al (1990)) Ø 14 -20 nt spacer DNA Ø CAP site (8 bp) Ø Transcription Initiation. Ø Transcript region, interrupted by introns. Translation Initiation (Kozak signal 12 bp consensus) 6 bp prior to initiation codon. Ø poly. A signal (AATAAA 99%, other)
Eukaryotic Nuclear Gene Structure Gene prediction for Pol II transcribed genes. Ø Upstream Enhancer elements. Ø Upstream Promoter elements. Ø GC box (-90 nt) (20 bp), CAAT box (-75 nt)(22 bp) Ø TATA promoter (-30 nt) (70%, 15 nt consensus (Bucher et al (1990)) Ø 14 -20 nt spacer DNA Ø CAP site (8 bp) Ø Transcription Initiation. Ø Transcript region, interrupted by introns. Translation Initiation (Kozak signal 12 bp consensus) 6 bp prior to initiation codon. Ø poly. A signal (AATAAA 99%, other)
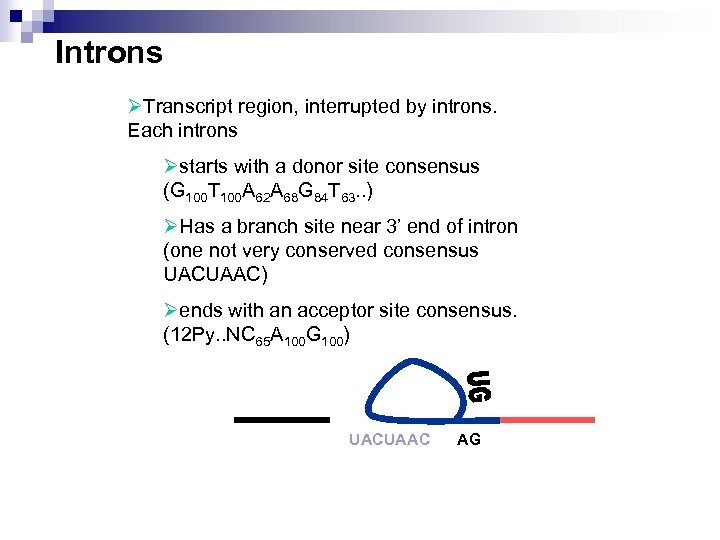 Introns ØTranscript region, interrupted by introns. Each introns Østarts with a donor site consensus (G 100 T 100 A 62 A 68 G 84 T 63. . ) ØHas a branch site near 3’ end of intron (one not very conserved consensus UACUAAC) Øends with an acceptor site consensus. (12 Py. . NC 65 A 100 G 100) UACUAAC AG
Introns ØTranscript region, interrupted by introns. Each introns Østarts with a donor site consensus (G 100 T 100 A 62 A 68 G 84 T 63. . ) ØHas a branch site near 3’ end of intron (one not very conserved consensus UACUAAC) Øends with an acceptor site consensus. (12 Py. . NC 65 A 100 G 100) UACUAAC AG
 Exons ØThe exons of the transcript region are composed of: Ø 5’UTR (mean length of 769 bp) with a specific base composition, that depends on local G+C content of genome) ØAUG (or other start codon) ØRemainder of coding region ØStop Codon Ø 3’ UTR (mean length of 457, with a specific base composition that depends on local G+C content of genome)
Exons ØThe exons of the transcript region are composed of: Ø 5’UTR (mean length of 769 bp) with a specific base composition, that depends on local G+C content of genome) ØAUG (or other start codon) ØRemainder of coding region ØStop Codon Ø 3’ UTR (mean length of 457, with a specific base composition that depends on local G+C content of genome)
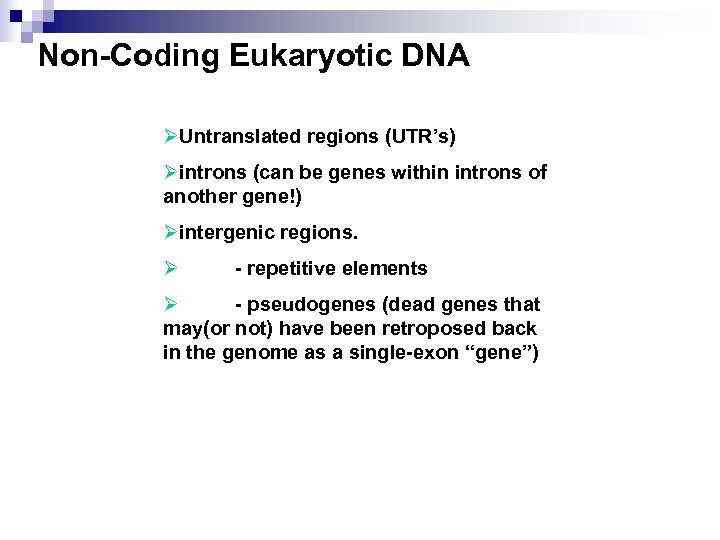 Non-Coding Eukaryotic DNA ØUntranslated regions (UTR’s) Øintrons (can be genes within introns of another gene!) Øintergenic regions. Ø - repetitive elements Ø - pseudogenes (dead genes that may(or not) have been retroposed back in the genome as a single-exon “gene”)
Non-Coding Eukaryotic DNA ØUntranslated regions (UTR’s) Øintrons (can be genes within introns of another gene!) Øintergenic regions. Ø - repetitive elements Ø - pseudogenes (dead genes that may(or not) have been retroposed back in the genome as a single-exon “gene”)
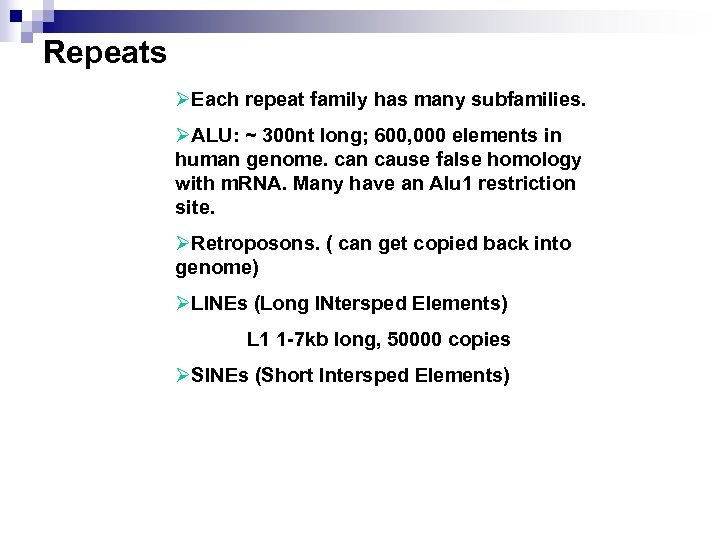 Repeats ØEach repeat family has many subfamilies. ØALU: ~ 300 nt long; 600, 000 elements in human genome. can cause false homology with m. RNA. Many have an Alu 1 restriction site. ØRetroposons. ( can get copied back into genome) ØLINEs (Long INtersped Elements) L 1 1 -7 kb long, 50000 copies ØSINEs (Short Intersped Elements)
Repeats ØEach repeat family has many subfamilies. ØALU: ~ 300 nt long; 600, 000 elements in human genome. can cause false homology with m. RNA. Many have an Alu 1 restriction site. ØRetroposons. ( can get copied back into genome) ØLINEs (Long INtersped Elements) L 1 1 -7 kb long, 50000 copies ØSINEs (Short Intersped Elements)
 Low-Complexity Elements Ø Ø When analyzing sequences, one often rely on the fact that two stretches are similar to infer that they are homologous (and therefore related). . But sequences with repeated patterns will match without there being any philogenetic relation! Sequences like ATATATACTTATATA which are mostly two letters are called low-complexity. Triplet repeats (particularly CAG) have a tendency to make the replication machinery stutter. . So they are amplified. The low-complexity sequence can also be hidden at the translated protein level.
Low-Complexity Elements Ø Ø When analyzing sequences, one often rely on the fact that two stretches are similar to infer that they are homologous (and therefore related). . But sequences with repeated patterns will match without there being any philogenetic relation! Sequences like ATATATACTTATATA which are mostly two letters are called low-complexity. Triplet repeats (particularly CAG) have a tendency to make the replication machinery stutter. . So they are amplified. The low-complexity sequence can also be hidden at the translated protein level.
 Structure of the Eukaryotic Genome Ø~6 -12% of human DNA encodes proteins. Ø~10% of human DNA codes for UTR Ø~90% of human DNA is non-coding.
Structure of the Eukaryotic Genome Ø~6 -12% of human DNA encodes proteins. Ø~10% of human DNA codes for UTR Ø~90% of human DNA is non-coding.
 Masking ØTo avoid finding spurious matches in alignment programs, you should always mask out the query sequence. ØBefore predicting genes it is a good idea to mask out repeats (at least those containing ORFs). ØBefore running blastn against a genomic record, you must mask out the repeats. ØMost used Programs: Gen. Scan: http: //genes. mit. edu/GENSCAN. html Repeat Masker: http: //ftp. genome. washington. edu/cgi-bin/Repeat. Masker
Masking ØTo avoid finding spurious matches in alignment programs, you should always mask out the query sequence. ØBefore predicting genes it is a good idea to mask out repeats (at least those containing ORFs). ØBefore running blastn against a genomic record, you must mask out the repeats. ØMost used Programs: Gen. Scan: http: //genes. mit. edu/GENSCAN. html Repeat Masker: http: //ftp. genome. washington. edu/cgi-bin/Repeat. Masker
 Chromosomal structure • Located in the nucleus • Each chromosome consists of a single molecule of DNA and its associated proteins ØThe DNA and protein complex found in eukaryotic chromosomes is called chromatin Ø 1/3 DNA and 2/3 protein • Complex interactions between proteins and nucleic acids in the chromosomes regulate gene and chromosomal function
Chromosomal structure • Located in the nucleus • Each chromosome consists of a single molecule of DNA and its associated proteins ØThe DNA and protein complex found in eukaryotic chromosomes is called chromatin Ø 1/3 DNA and 2/3 protein • Complex interactions between proteins and nucleic acids in the chromosomes regulate gene and chromosomal function
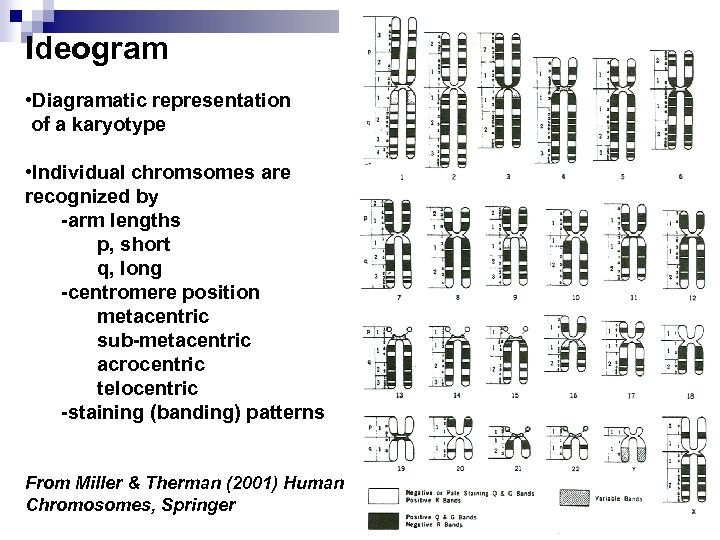 Ideogram • Diagramatic representation of a karyotype • Individual chromsomes are recognized by -arm lengths p, short q, long -centromere position metacentric sub-metacentric acrocentric telocentric -staining (banding) patterns From Miller & Therman (2001) Human Chromosomes, Springer
Ideogram • Diagramatic representation of a karyotype • Individual chromsomes are recognized by -arm lengths p, short q, long -centromere position metacentric sub-metacentric acrocentric telocentric -staining (banding) patterns From Miller & Therman (2001) Human Chromosomes, Springer
 Chromsome banding n n n Q (quinicrine) & G (Giemsa) banding preferentially stain AT rich regions R (reverse banding) preferentially stains GCrich regions C-banding (denaturation & staining) preferentially stains constitutive heterochromatin, found in the centromere regions and distal Yq
Chromsome banding n n n Q (quinicrine) & G (Giemsa) banding preferentially stain AT rich regions R (reverse banding) preferentially stains GCrich regions C-banding (denaturation & staining) preferentially stains constitutive heterochromatin, found in the centromere regions and distal Yq
 June 26, 2000 at the Whitehouse
June 26, 2000 at the Whitehouse
 Initial Analysis of the Human Genome
Initial Analysis of the Human Genome
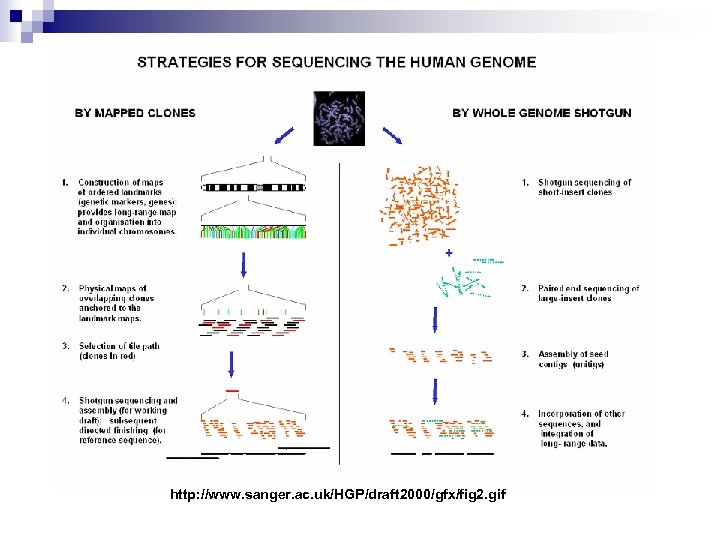 http: //www. sanger. ac. uk/HGP/draft 2000/gfx/fig 2. gif
http: //www. sanger. ac. uk/HGP/draft 2000/gfx/fig 2. gif
 Genome Mapping STS – sequence-tagged sites (short segments of unique DNA on every chromosome – defined by a pair of PCR primers that amplified only one segment of the genome) BAC – Bacterial artificial chromosome, 100 -400 kb YAC – Yeast artificial chromosome, 150 kb-1. 5 Mb Contig – assembled contiguous overlapping segments of DNA from BACs and YACs ESTs – Expressed Sequence Tags Uni. Gene Database – a database for ESTs
Genome Mapping STS – sequence-tagged sites (short segments of unique DNA on every chromosome – defined by a pair of PCR primers that amplified only one segment of the genome) BAC – Bacterial artificial chromosome, 100 -400 kb YAC – Yeast artificial chromosome, 150 kb-1. 5 Mb Contig – assembled contiguous overlapping segments of DNA from BACs and YACs ESTs – Expressed Sequence Tags Uni. Gene Database – a database for ESTs
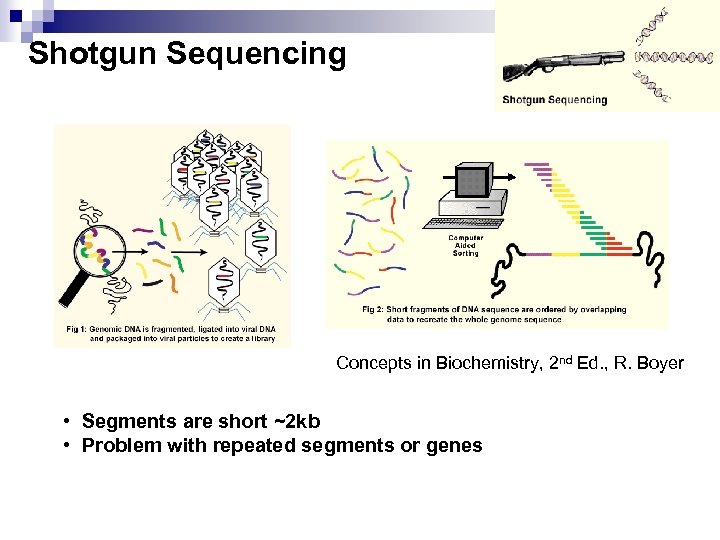 Shotgun Sequencing Concepts in Biochemistry, 2 nd Ed. , R. Boyer • Segments are short ~2 kb • Problem with repeated segments or genes
Shotgun Sequencing Concepts in Biochemistry, 2 nd Ed. , R. Boyer • Segments are short ~2 kb • Problem with repeated segments or genes
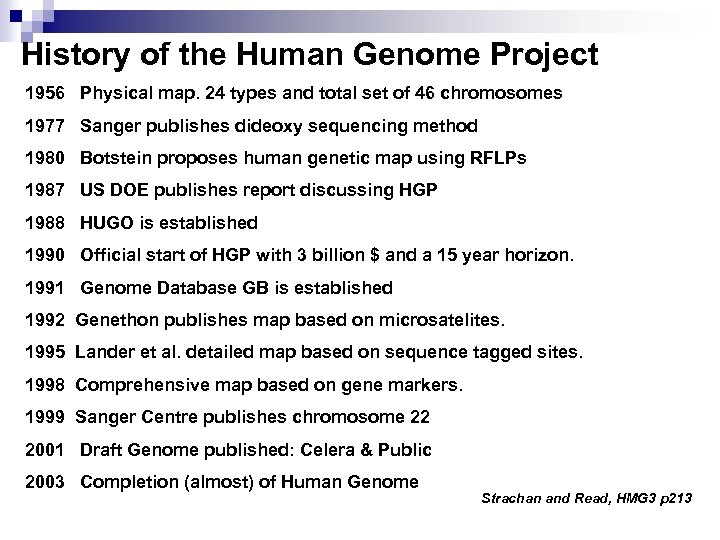 History of the Human Genome Project 1956 Physical map. 24 types and total set of 46 chromosomes 1977 Sanger publishes dideoxy sequencing method 1980 Botstein proposes human genetic map using RFLPs 1987 US DOE publishes report discussing HGP 1988 HUGO is established 1990 Official start of HGP with 3 billion $ and a 15 year horizon. 1991 Genome Database GB is established 1992 Genethon publishes map based on microsatelites. 1995 Lander et al. detailed map based on sequence tagged sites. 1998 Comprehensive map based on gene markers. 1999 Sanger Centre publishes chromosome 22 2001 Draft Genome published: Celera & Public 2003 Completion (almost) of Human Genome Strachan and Read, HMG 3 p 213
History of the Human Genome Project 1956 Physical map. 24 types and total set of 46 chromosomes 1977 Sanger publishes dideoxy sequencing method 1980 Botstein proposes human genetic map using RFLPs 1987 US DOE publishes report discussing HGP 1988 HUGO is established 1990 Official start of HGP with 3 billion $ and a 15 year horizon. 1991 Genome Database GB is established 1992 Genethon publishes map based on microsatelites. 1995 Lander et al. detailed map based on sequence tagged sites. 1998 Comprehensive map based on gene markers. 1999 Sanger Centre publishes chromosome 22 2001 Draft Genome published: Celera & Public 2003 Completion (almost) of Human Genome Strachan and Read, HMG 3 p 213
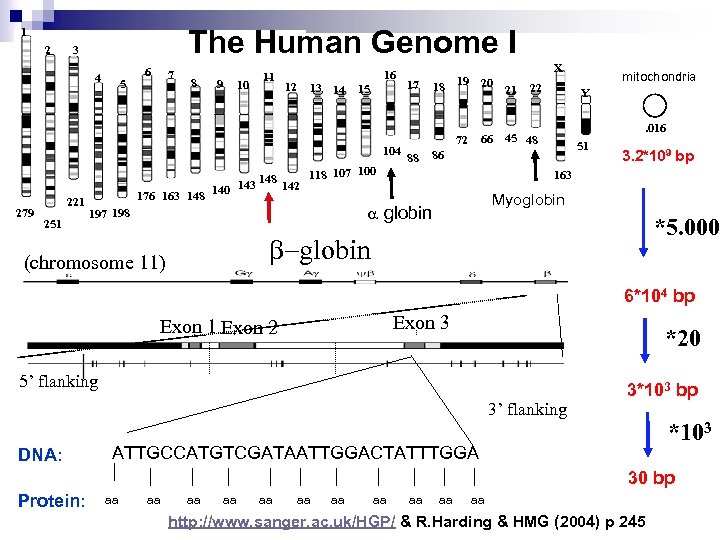 The Human Genome I 1 2 3 4 5 6 7 8 9 10 11 12 13 14 16 15 104 279 221 251 17 18 19 20 66 72 88 21 22 mitochondria Y. 016 45 48 51 86 118 107 100 148 143 142 176 163 148 140 3. 2*109 bp 163 Myoglobin a globin 197 198 X *5. 000 b-globin (chromosome 11) 6*104 bp Exon 3 Exon 1 Exon 2 *20 5’ flanking 3’ flanking DNA: 3*103 bp *103 ATTGCCATGTCGATAATTGGACTATTTGGA 30 bp Protein: aa aa aa http: //www. sanger. ac. uk/HGP/ & R. Harding & HMG (2004) p 245
The Human Genome I 1 2 3 4 5 6 7 8 9 10 11 12 13 14 16 15 104 279 221 251 17 18 19 20 66 72 88 21 22 mitochondria Y. 016 45 48 51 86 118 107 100 148 143 142 176 163 148 140 3. 2*109 bp 163 Myoglobin a globin 197 198 X *5. 000 b-globin (chromosome 11) 6*104 bp Exon 3 Exon 1 Exon 2 *20 5’ flanking 3’ flanking DNA: 3*103 bp *103 ATTGCCATGTCGATAATTGGACTATTTGGA 30 bp Protein: aa aa aa http: //www. sanger. ac. uk/HGP/ & R. Harding & HMG (2004) p 245
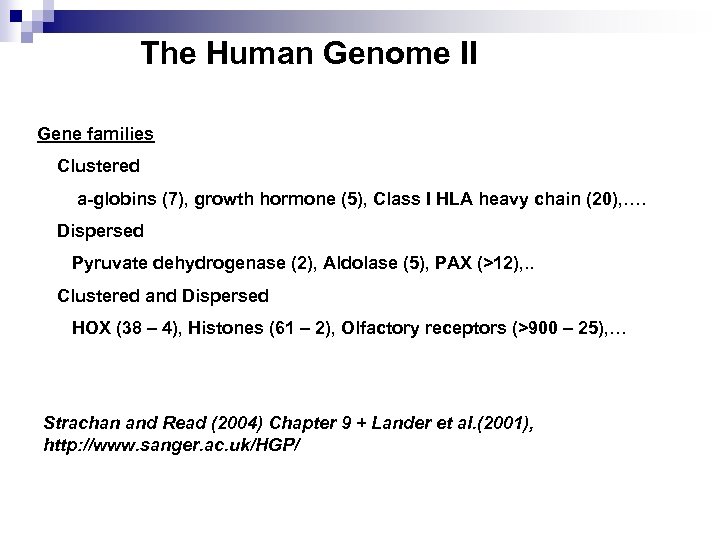 The Human Genome II Gene families Clustered a-globins (7), growth hormone (5), Class I HLA heavy chain (20), …. Dispersed Pyruvate dehydrogenase (2), Aldolase (5), PAX (>12), . . Clustered and Dispersed HOX (38 – 4), Histones (61 – 2), Olfactory receptors (>900 – 25), … Strachan and Read (2004) Chapter 9 + Lander et al. (2001), http: //www. sanger. ac. uk/HGP/
The Human Genome II Gene families Clustered a-globins (7), growth hormone (5), Class I HLA heavy chain (20), …. Dispersed Pyruvate dehydrogenase (2), Aldolase (5), PAX (>12), . . Clustered and Dispersed HOX (38 – 4), Histones (61 – 2), Olfactory receptors (>900 – 25), … Strachan and Read (2004) Chapter 9 + Lander et al. (2001), http: //www. sanger. ac. uk/HGP/
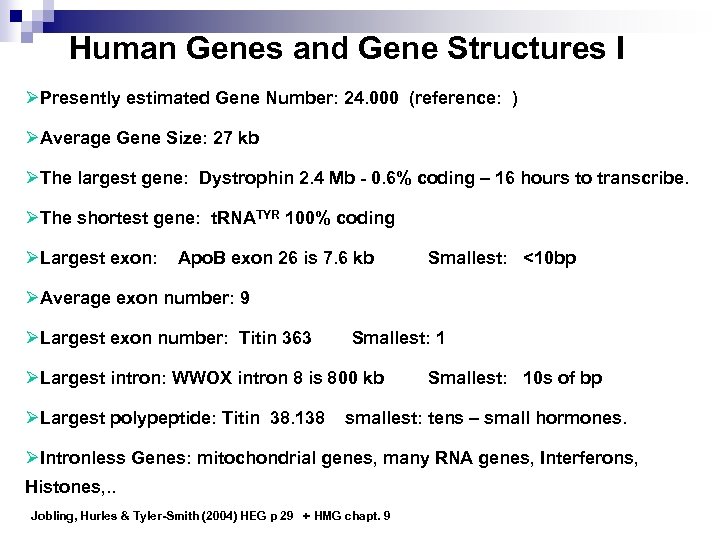 Human Genes and Gene Structures I ØPresently estimated Gene Number: 24. 000 (reference: ) ØAverage Gene Size: 27 kb ØThe largest gene: Dystrophin 2. 4 Mb - 0. 6% coding – 16 hours to transcribe. ØThe shortest gene: t. RNATYR 100% coding ØLargest exon: Apo. B exon 26 is 7. 6 kb Smallest: <10 bp ØAverage exon number: 9 ØLargest exon number: Titin 363 Smallest: 1 ØLargest intron: WWOX intron 8 is 800 kb ØLargest polypeptide: Titin 38. 138 Smallest: 10 s of bp smallest: tens – small hormones. ØIntronless Genes: mitochondrial genes, many RNA genes, Interferons, Histones, . . Jobling, Hurles & Tyler-Smith (2004) HEG p 29 + HMG chapt. 9
Human Genes and Gene Structures I ØPresently estimated Gene Number: 24. 000 (reference: ) ØAverage Gene Size: 27 kb ØThe largest gene: Dystrophin 2. 4 Mb - 0. 6% coding – 16 hours to transcribe. ØThe shortest gene: t. RNATYR 100% coding ØLargest exon: Apo. B exon 26 is 7. 6 kb Smallest: <10 bp ØAverage exon number: 9 ØLargest exon number: Titin 363 Smallest: 1 ØLargest intron: WWOX intron 8 is 800 kb ØLargest polypeptide: Titin 38. 138 Smallest: 10 s of bp smallest: tens – small hormones. ØIntronless Genes: mitochondrial genes, many RNA genes, Interferons, Histones, . . Jobling, Hurles & Tyler-Smith (2004) HEG p 29 + HMG chapt. 9
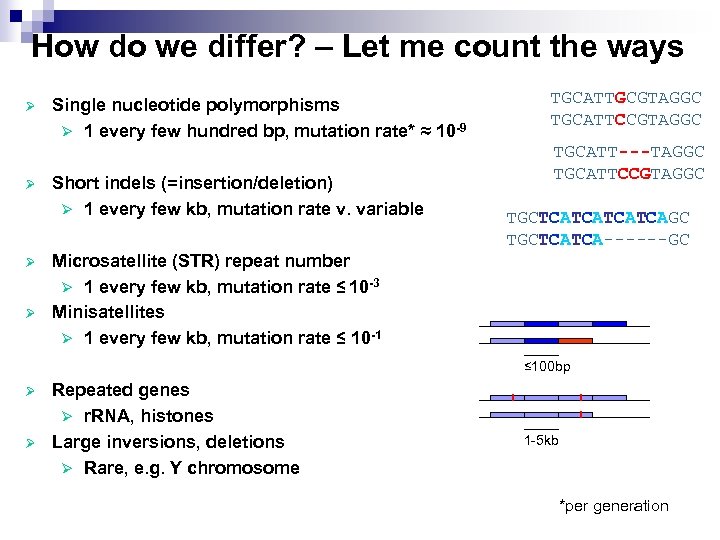 How do we differ? – Let me count the ways Ø Ø Single nucleotide polymorphisms Ø 1 every few hundred bp, mutation rate* ≈ 10 -9 Short indels (=insertion/deletion) Ø 1 every few kb, mutation rate v. variable Microsatellite (STR) repeat number Ø 1 every few kb, mutation rate ≤ 10 -3 Minisatellites Ø 1 every few kb, mutation rate ≤ 10 -1 TGCATTGCGTAGGC TGCATTCCGTAGGC TGCATT---TAGGC TGCATTCCGTAGGC TGCTCATCAGC TGCTCATCA------GC ≤ 100 bp Ø Ø Repeated genes Ø r. RNA, histones Large inversions, deletions Ø Rare, e. g. Y chromosome 1 -5 kb *per generation
How do we differ? – Let me count the ways Ø Ø Single nucleotide polymorphisms Ø 1 every few hundred bp, mutation rate* ≈ 10 -9 Short indels (=insertion/deletion) Ø 1 every few kb, mutation rate v. variable Microsatellite (STR) repeat number Ø 1 every few kb, mutation rate ≤ 10 -3 Minisatellites Ø 1 every few kb, mutation rate ≤ 10 -1 TGCATTGCGTAGGC TGCATTCCGTAGGC TGCATT---TAGGC TGCATTCCGTAGGC TGCTCATCAGC TGCTCATCA------GC ≤ 100 bp Ø Ø Repeated genes Ø r. RNA, histones Large inversions, deletions Ø Rare, e. g. Y chromosome 1 -5 kb *per generation
![Gene Number Ø Ø Ø Ø Ø Walter Gilbert [1980 s] 100 k Antequera Gene Number Ø Ø Ø Ø Ø Walter Gilbert [1980 s] 100 k Antequera](https://present5.com/presentation/4700a3fae36da74a59d4835b29cdeb9f/image-27.jpg) Gene Number Ø Ø Ø Ø Ø Walter Gilbert [1980 s] 100 k Antequera & Bird [1993] 70 -80 k John Quackenbush et al. (TIGR) [2000] 120 k Ewing & Green [2000] 30 k Tetraodon analysis [2001] 35 k Human Genome Project (public) [2001] ~ 31 k Human Genome Project (Celera) [2001] 24 -40 k Mouse Genome Project (public) [2002] 25 k -30 k Lee Rowen [2003] 25, 947
Gene Number Ø Ø Ø Ø Ø Walter Gilbert [1980 s] 100 k Antequera & Bird [1993] 70 -80 k John Quackenbush et al. (TIGR) [2000] 120 k Ewing & Green [2000] 30 k Tetraodon analysis [2001] 35 k Human Genome Project (public) [2001] ~ 31 k Human Genome Project (Celera) [2001] 24 -40 k Mouse Genome Project (public) [2002] 25 k -30 k Lee Rowen [2003] 25, 947
 Gene finding Ø Rules Ø Ø Compositional features Ø Ø Ø ATG TAA, TGA, TAG GT…. . AG Exon lengths Intron lengths Codon bias General genomic properties Homology ? ?
Gene finding Ø Rules Ø Ø Compositional features Ø Ø Ø ATG TAA, TGA, TAG GT…. . AG Exon lengths Intron lengths Codon bias General genomic properties Homology ? ?
 Visualization • Ensembl (http: //www. ensembl. org/index. html) • Genome Browser (http: //genome. ucsc. edu/) • Map Viewer (http: //www. ncbi. nlm. nih. gov/mapview/) • VEGA – VErtebrate Genome Annotation database (http: //vega. sanger. ac. uk/index. html)
Visualization • Ensembl (http: //www. ensembl. org/index. html) • Genome Browser (http: //genome. ucsc. edu/) • Map Viewer (http: //www. ncbi. nlm. nih. gov/mapview/) • VEGA – VErtebrate Genome Annotation database (http: //vega. sanger. ac. uk/index. html)
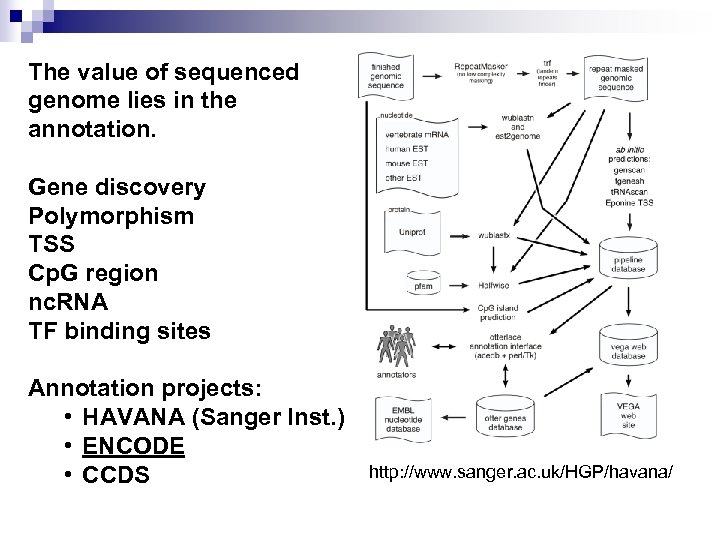 The value of sequenced genome lies in the annotation. Gene discovery Polymorphism TSS Cp. G region nc. RNA TF binding sites Annotation projects: • HAVANA (Sanger Inst. ) • ENCODE • CCDS http: //www. sanger. ac. uk/HGP/havana/
The value of sequenced genome lies in the annotation. Gene discovery Polymorphism TSS Cp. G region nc. RNA TF binding sites Annotation projects: • HAVANA (Sanger Inst. ) • ENCODE • CCDS http: //www. sanger. ac. uk/HGP/havana/
 ENCODE Project n n n ENCODE: Encyclopedia Of DNA Elements The project started in September 2003 funded by National Human Genome Research Institute (NHGRI) Task: identify all functional elements in the human genome sequence. Initial Phases: a pilot phase and a technology development phase. Production phase: A large scale.
ENCODE Project n n n ENCODE: Encyclopedia Of DNA Elements The project started in September 2003 funded by National Human Genome Research Institute (NHGRI) Task: identify all functional elements in the human genome sequence. Initial Phases: a pilot phase and a technology development phase. Production phase: A large scale.
 Initial Phases n n The pilot phase: analyze 1% human genome, including 44 genome regions with 30 million DNA base pairs. The conclusions from this pilot project were published in June 2007 in Nature and Genome Research [genome. org]. The findings highlighted the success of the project to identify and characterize functional elements in the human genome. The technology development phase also has been a success with the promotion of several new technologies to generate high throughput data on functional elements.
Initial Phases n n The pilot phase: analyze 1% human genome, including 44 genome regions with 30 million DNA base pairs. The conclusions from this pilot project were published in June 2007 in Nature and Genome Research [genome. org]. The findings highlighted the success of the project to identify and characterize functional elements in the human genome. The technology development phase also has been a success with the promotion of several new technologies to generate high throughput data on functional elements.
 Major findings n n The ENCODE consortium generated more than 200 datasets and analyzed more than 600 million data points. The ENCODE consortium's major findings include : 1) the majority of DNA in the human genome is transcribed into functional molecules RNA, and that these transcripts extensively overlap one another. This broad pattern of transcription challenges the long-standing view that the human genome consists of a relatively small set of discrete genes, along with a vast amount of so-called junk DNA that is not biologically active. 2) The genome contains very little unused sequences and, is a complex, interwoven network. In this network, genes are just one of many types of DNA sequences that have a functional impact. 3) Half of functional elements in the human genome do not appear to have been constrained during evolution. This may indicate that many species' genomes contain a pool of functional elements that provide no specific benefits in terms of survival or reproduction. As this pool turns over during evolutionary time, it may serve as a "warehouse for natural selection" by acting as a source of functional elements unique to each species and of elements that perform the similar functions among species despite having sequences that appear dissimilar. 4) Identification of numerous previously unrecognized start sites for DNA transcription. 5) Evidence that, contrary to traditional views, regulatory sequences are just as likely to be located downstream of a transcription start site on a DNA strand as upstream. 6) Identification of specific signatures of change in histones, and correlation of these signatures with different genomic functions. 7) Deeper understanding of how DNA replication is coordinated by modifications in histones.
Major findings n n The ENCODE consortium generated more than 200 datasets and analyzed more than 600 million data points. The ENCODE consortium's major findings include : 1) the majority of DNA in the human genome is transcribed into functional molecules RNA, and that these transcripts extensively overlap one another. This broad pattern of transcription challenges the long-standing view that the human genome consists of a relatively small set of discrete genes, along with a vast amount of so-called junk DNA that is not biologically active. 2) The genome contains very little unused sequences and, is a complex, interwoven network. In this network, genes are just one of many types of DNA sequences that have a functional impact. 3) Half of functional elements in the human genome do not appear to have been constrained during evolution. This may indicate that many species' genomes contain a pool of functional elements that provide no specific benefits in terms of survival or reproduction. As this pool turns over during evolutionary time, it may serve as a "warehouse for natural selection" by acting as a source of functional elements unique to each species and of elements that perform the similar functions among species despite having sequences that appear dissimilar. 4) Identification of numerous previously unrecognized start sites for DNA transcription. 5) Evidence that, contrary to traditional views, regulatory sequences are just as likely to be located downstream of a transcription start site on a DNA strand as upstream. 6) Identification of specific signatures of change in histones, and correlation of these signatures with different genomic functions. 7) Deeper understanding of how DNA replication is coordinated by modifications in histones.
 Production Phase n n NHGRI funded new awards in September 2007 to scale the ENCODE Project to a production phase on the entire genome. The ENCODE team: is an open consortium and includes investigators with diverse backgrounds and expertise in the production and analysis of data.
Production Phase n n NHGRI funded new awards in September 2007 to scale the ENCODE Project to a production phase on the entire genome. The ENCODE team: is an open consortium and includes investigators with diverse backgrounds and expertise in the production and analysis of data.
 Tools http: //www. geneontology. org/GO. tools. shtml Examples: • DAVID - http: //david. abcc. ncifcrf. gov • Bi. NGO
Tools http: //www. geneontology. org/GO. tools. shtml Examples: • DAVID - http: //david. abcc. ncifcrf. gov • Bi. NGO
 Tools http: //www. geneontology. org/GO. tools. shtml Examples: • DAVID - http: //david. abcc. ncifcrf. gov • Bi. NGO
Tools http: //www. geneontology. org/GO. tools. shtml Examples: • DAVID - http: //david. abcc. ncifcrf. gov • Bi. NGO


