0965fe862008f343e05187b373571ed8.ppt
- Количество слайдов: 66
 Functional Genomics: Making mutants and analysing gene transcription Regulation of antibiotic production Engineering lantibiotic production Mervyn Bibb Department of Molecular Microbiology John Innes Centre, Norwich
Functional Genomics: Making mutants and analysing gene transcription Regulation of antibiotic production Engineering lantibiotic production Mervyn Bibb Department of Molecular Microbiology John Innes Centre, Norwich
 Functional genomics: making mutants and analysing gene transcription • Making mutants - Gene disruption, replacement, deletion and point mutation • Homologous recombination • http: //www. jic. bbsrc. ac. uk/SCIENCE/molmicro/ Strepmanual/Manual. htm • PCR-targetting (Redirect) • Analysing gene transcription • Northerns, S 1 nuclease protection, Primer extension, RT-PCR • DNA microarrays – whole genome analysis of gene transcription (QRT-PCR) • By sequencing – e. g. Solexa
Functional genomics: making mutants and analysing gene transcription • Making mutants - Gene disruption, replacement, deletion and point mutation • Homologous recombination • http: //www. jic. bbsrc. ac. uk/SCIENCE/molmicro/ Strepmanual/Manual. htm • PCR-targetting (Redirect) • Analysing gene transcription • Northerns, S 1 nuclease protection, Primer extension, RT-PCR • DNA microarrays – whole genome analysis of gene transcription (QRT-PCR) • By sequencing – e. g. Solexa
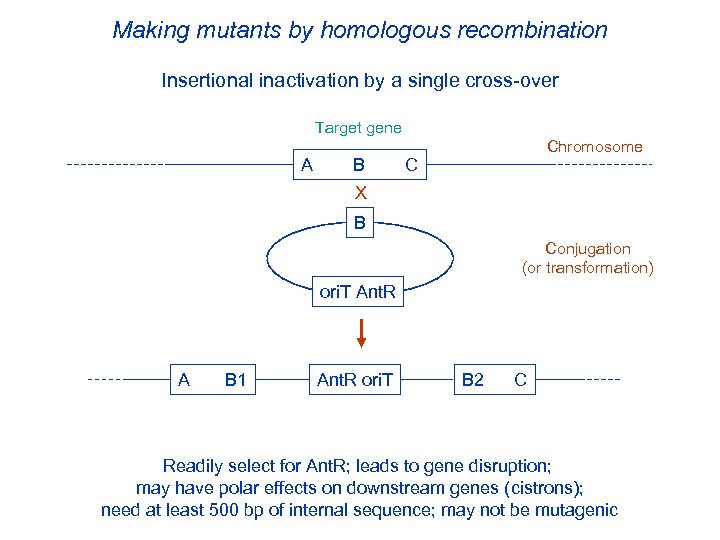 Making mutants by homologous recombination Insertional inactivation by a single cross-over Target gene Chromosome A B C X B Conjugation (or transformation) ori. T Ant. R A B 1 Ant. R ori. T B 2 C Readily select for Ant. R; leads to gene disruption; may have polar effects on downstream genes (cistrons); need at least 500 bp of internal sequence; may not be mutagenic
Making mutants by homologous recombination Insertional inactivation by a single cross-over Target gene Chromosome A B C X B Conjugation (or transformation) ori. T Ant. R A B 1 Ant. R ori. T B 2 C Readily select for Ant. R; leads to gene disruption; may have polar effects on downstream genes (cistrons); need at least 500 bp of internal sequence; may not be mutagenic
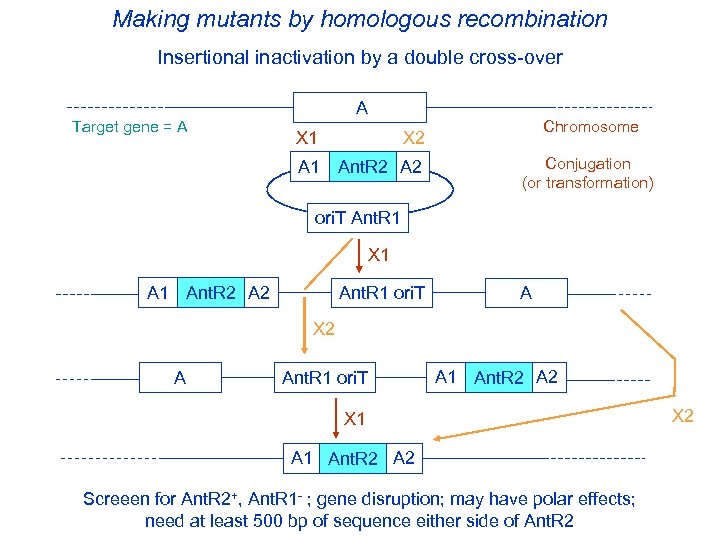 Making mutants by homologous recombination Insertional inactivation by a double cross-over Target gene = A Chromosome X 1 X 2 A 1 Ant. R 2 A 2 Ant. R 2 Conjugation (or transformation) ori. T Ant. R 1 X 1 Ant. R 2 A 2 Ant. R 1 ori. T A X 2 A Ant. R 1 ori. T A 1 Ant. R 2 A 2 Ant. R 2 X 1 Ant. R 2 A 2 Ant. R 2 Screeen for Ant. R 2+, Ant. R 1 - ; gene disruption; may have polar effects; need at least 500 bp of sequence either side of Ant. R 2 X 2
Making mutants by homologous recombination Insertional inactivation by a double cross-over Target gene = A Chromosome X 1 X 2 A 1 Ant. R 2 A 2 Ant. R 2 Conjugation (or transformation) ori. T Ant. R 1 X 1 Ant. R 2 A 2 Ant. R 1 ori. T A X 2 A Ant. R 1 ori. T A 1 Ant. R 2 A 2 Ant. R 2 X 1 Ant. R 2 A 2 Ant. R 2 Screeen for Ant. R 2+, Ant. R 1 - ; gene disruption; may have polar effects; need at least 500 bp of sequence either side of Ant. R 2 X 2
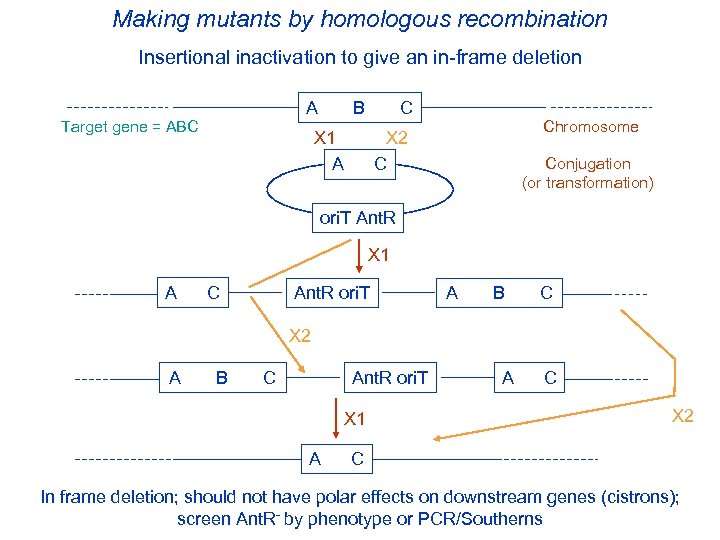 Making mutants by homologous recombination Insertional inactivation to give an in-frame deletion Target gene = ABC A B C X 1 X 2 A C Chromosome Conjugation (or transformation) ori. T Ant. R X 1 A C Ant. R ori. T A B C X 2 A B C Ant. R ori. T X 1 A C X 2 A C In frame deletion; should not have polar effects on downstream genes (cistrons); screen Ant. R- by phenotype or PCR/Southerns
Making mutants by homologous recombination Insertional inactivation to give an in-frame deletion Target gene = ABC A B C X 1 X 2 A C Chromosome Conjugation (or transformation) ori. T Ant. R X 1 A C Ant. R ori. T A B C X 2 A B C Ant. R ori. T X 1 A C X 2 A C In frame deletion; should not have polar effects on downstream genes (cistrons); screen Ant. R- by phenotype or PCR/Southerns
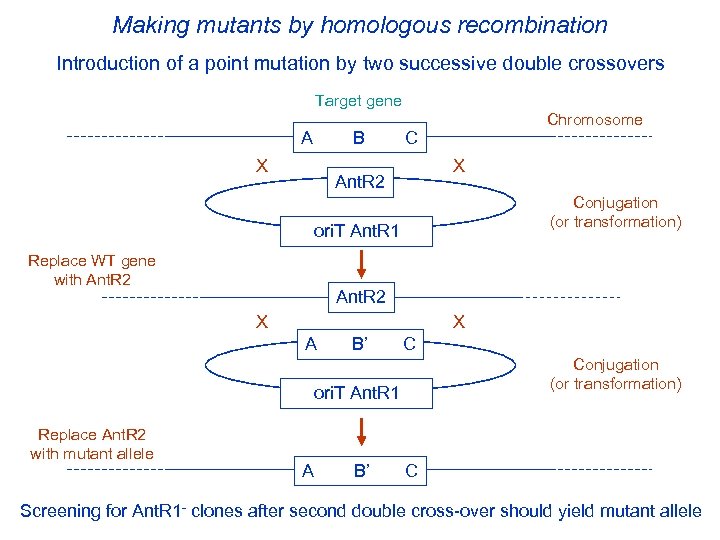 Making mutants by homologous recombination Introduction of a point mutation by two successive double crossovers Target gene A B C Chromosome X X Ant. R 2 ori. T Ant. R 1 Replace WT gene with Ant. R 2 X X A B’ C ori. T Ant. R 1 Replace Ant. R 2 with mutant allele Conjugation (or transformation) A B’ C Screening for Ant. R 1 - clones after second double cross-over should yield mutant allele
Making mutants by homologous recombination Introduction of a point mutation by two successive double crossovers Target gene A B C Chromosome X X Ant. R 2 ori. T Ant. R 1 Replace WT gene with Ant. R 2 X X A B’ C ori. T Ant. R 1 Replace Ant. R 2 with mutant allele Conjugation (or transformation) A B’ C Screening for Ant. R 1 - clones after second double cross-over should yield mutant allele
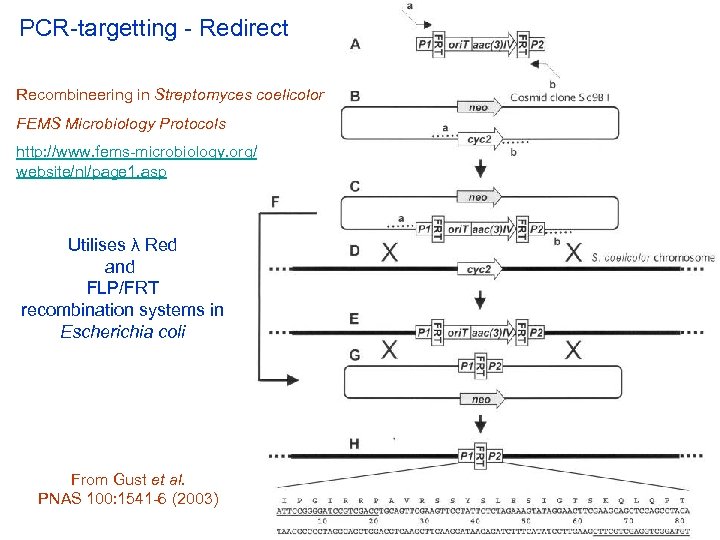 PCR-targetting - Redirect Recombineering in Streptomyces coelicolor FEMS Microbiology Protocols http: //www. fems-microbiology. org/ website/nl/page 1. asp Utilises λ Red and FLP/FRT recombination systems in Escherichia coli From Gust et al. PNAS 100: 1541 -6 (2003)
PCR-targetting - Redirect Recombineering in Streptomyces coelicolor FEMS Microbiology Protocols http: //www. fems-microbiology. org/ website/nl/page 1. asp Utilises λ Red and FLP/FRT recombination systems in Escherichia coli From Gust et al. PNAS 100: 1541 -6 (2003)
 REDIRECT templates (Nick Bird) P 2 p. IJ 779 P 1 FRT aad. A bla ori. T tet att. P int bla p. IJ 789 neo aac(3)IV neo p. IJ 794 P 2 neo ori. T aac(3)IV neo p. IJ 795 neo ori. T vph neo P 2 p. IJ 796 neo ori. T aad. A neo p. IJ 797 P 1 http: //streptomyces. org. uk/redirect/index. html p. IJ 798 bla ori. T hyg FRT P 2 FRT vph P 2 ori. T aac(3)IV p. IJ 788 FRT ori. T P 1 egfp P 2 tip. Ap P 2 aad. A FRT P 1 FRT p. IJ 781 P 1 FRT p. IJ 780 p. IJ 786 aac(3)IV ori. T FRT ori. T P 1 P 2 p. IJ 787 FRT P 1 FRT p. IJ 778 neo bla ori. T aac(3)IV bla tet FRT neo FRT ori. T FRT P 1 FRT p. IJ 777 P 1 FRT p. IJ 776 P 1 ori. T aac(3)IV P 2 p. IJ 784 ori. T FRT p. IJ 775 P 1 p. IJ 785 Swa. I p. IJ 782 FRT Swa. I FRT ori. T aac(3)IV FRT P 2 lox. P P 1 FRT p. IJ 774 ori. T aac(3)IV lox. P P 1 FRT p. IJ 773 bla P 2
REDIRECT templates (Nick Bird) P 2 p. IJ 779 P 1 FRT aad. A bla ori. T tet att. P int bla p. IJ 789 neo aac(3)IV neo p. IJ 794 P 2 neo ori. T aac(3)IV neo p. IJ 795 neo ori. T vph neo P 2 p. IJ 796 neo ori. T aad. A neo p. IJ 797 P 1 http: //streptomyces. org. uk/redirect/index. html p. IJ 798 bla ori. T hyg FRT P 2 FRT vph P 2 ori. T aac(3)IV p. IJ 788 FRT ori. T P 1 egfp P 2 tip. Ap P 2 aad. A FRT P 1 FRT p. IJ 781 P 1 FRT p. IJ 780 p. IJ 786 aac(3)IV ori. T FRT ori. T P 1 P 2 p. IJ 787 FRT P 1 FRT p. IJ 778 neo bla ori. T aac(3)IV bla tet FRT neo FRT ori. T FRT P 1 FRT p. IJ 777 P 1 FRT p. IJ 776 P 1 ori. T aac(3)IV P 2 p. IJ 784 ori. T FRT p. IJ 775 P 1 p. IJ 785 Swa. I p. IJ 782 FRT Swa. I FRT ori. T aac(3)IV FRT P 2 lox. P P 1 FRT p. IJ 774 ori. T aac(3)IV lox. P P 1 FRT p. IJ 773 bla P 2
 Traditional methods for detection and quantitation of specific RNA sequences Northern blotting • A denatured RNA sample is separated on the basis of size by gel electrophoresis and transferred to a membrane • A specific labelled DNA fragment is used as a probe to detect and quantify specific transcripts in the RNA sample S 1 nuclease protection analysis • Partially overlapping 5’ end-labelled DNA-m. RNA hybrid created that covers transcriptional start site • S 1 nuclease treatment removes single-stranded tails revealing transcriptional start site and level of transcription Primer extension mapping (RTase) • Oligonucleotide primer and Reverse Transcriptase used to create c. DNA complementary to 5’ end of m. RNA • Gel electrophoresis used to reveal transcriptional start site and level of transcription
Traditional methods for detection and quantitation of specific RNA sequences Northern blotting • A denatured RNA sample is separated on the basis of size by gel electrophoresis and transferred to a membrane • A specific labelled DNA fragment is used as a probe to detect and quantify specific transcripts in the RNA sample S 1 nuclease protection analysis • Partially overlapping 5’ end-labelled DNA-m. RNA hybrid created that covers transcriptional start site • S 1 nuclease treatment removes single-stranded tails revealing transcriptional start site and level of transcription Primer extension mapping (RTase) • Oligonucleotide primer and Reverse Transcriptase used to create c. DNA complementary to 5’ end of m. RNA • Gel electrophoresis used to reveal transcriptional start site and level of transcription
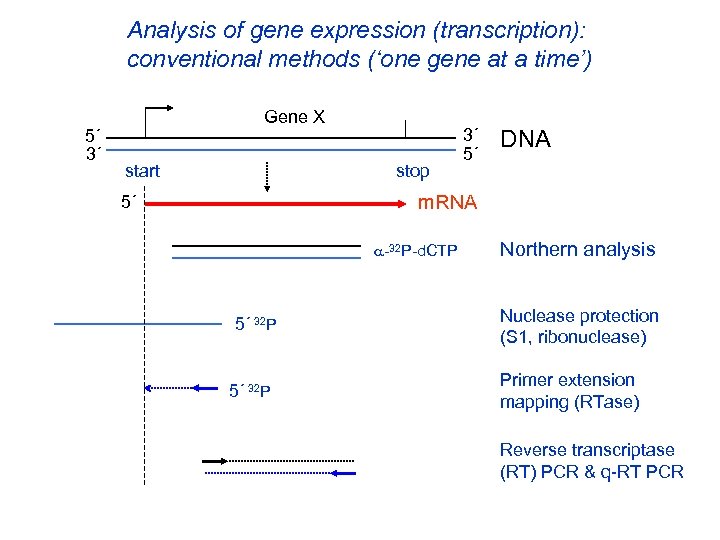 Analysis of gene expression (transcription): conventional methods (‘one gene at a time’) 5´ 3´ Gene X start stop 3´ 5´ DNA m. RNA 5´ -32 P-d. CTP 5´ 32 P Northern analysis Nuclease protection (S 1, ribonuclease) Primer extension mapping (RTase) Reverse transcriptase (RT) PCR & q-RT PCR
Analysis of gene expression (transcription): conventional methods (‘one gene at a time’) 5´ 3´ Gene X start stop 3´ 5´ DNA m. RNA 5´ -32 P-d. CTP 5´ 32 P Northern analysis Nuclease protection (S 1, ribonuclease) Primer extension mapping (RTase) Reverse transcriptase (RT) PCR & q-RT PCR
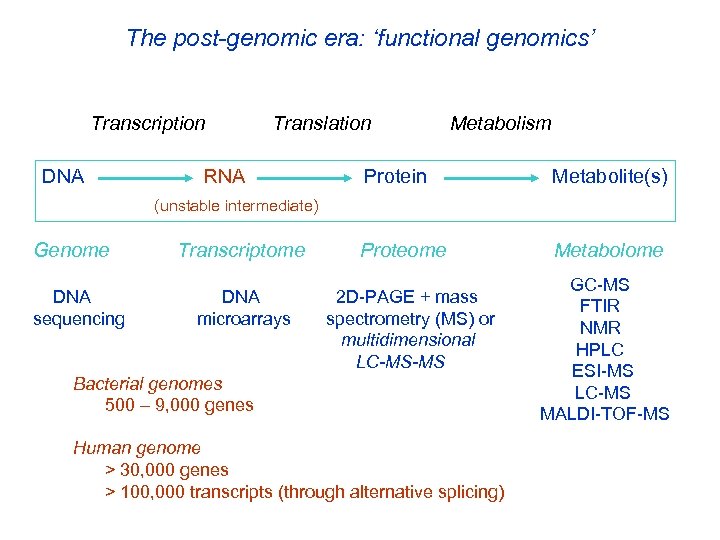 The post-genomic era: ‘functional genomics’ Transcription Translation DNA RNA Metabolism Protein Metabolite(s) (unstable intermediate) Genome Transcriptome Proteome DNA sequencing DNA microarrays 2 D-PAGE + mass spectrometry (MS) or multidimensional LC-MS-MS Bacterial genomes 500 – 9, 000 genes Human genome > 30, 000 genes > 100, 000 transcripts (through alternative splicing) Metabolome GC-MS FTIR NMR HPLC ESI-MS LC-MS MALDI-TOF-MS
The post-genomic era: ‘functional genomics’ Transcription Translation DNA RNA Metabolism Protein Metabolite(s) (unstable intermediate) Genome Transcriptome Proteome DNA sequencing DNA microarrays 2 D-PAGE + mass spectrometry (MS) or multidimensional LC-MS-MS Bacterial genomes 500 – 9, 000 genes Human genome > 30, 000 genes > 100, 000 transcripts (through alternative splicing) Metabolome GC-MS FTIR NMR HPLC ESI-MS LC-MS MALDI-TOF-MS
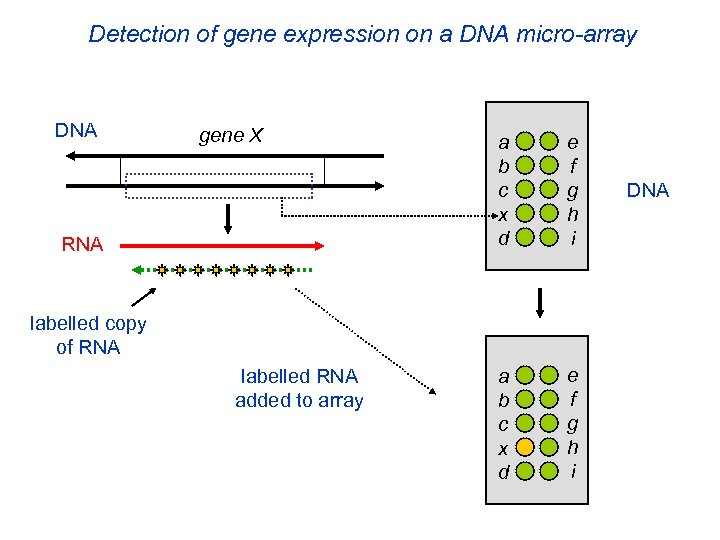 Detection of gene expression on a DNA micro-array DNA gene X RNA a b c x d e f g h i labelled copy of RNA labelled RNA added to array DNA
Detection of gene expression on a DNA micro-array DNA gene X RNA a b c x d e f g h i labelled copy of RNA labelled RNA added to array DNA
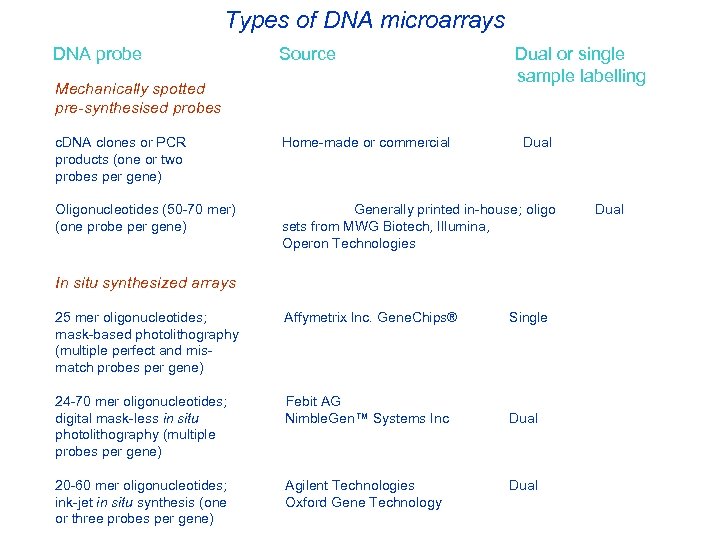 Types of DNA microarrays DNA probe Source Mechanically spotted pre-synthesised probes Dual or single sample labelling c. DNA clones or PCR Home-made or commercial Dual products (one or two probes per gene) Oligonucleotides (50 -70 mer) Generally printed in-house; oligo (one probe per gene) sets from MWG Biotech, Illumina, Operon Technologies In situ synthesized arrays 25 mer oligonucleotides; Affymetrix Inc. Gene. Chips® mask-based photolithography (multiple perfect and mismatch probes per gene) 24 -70 mer oligonucleotides; Febit AG digital mask-less in situ Nimble. Gen™ Systems Inc photolithography (multiple probes per gene) 20 -60 mer oligonucleotides; Agilent Technologies ink-jet in situ synthesis (one Oxford Gene Technology or three probes per gene) Single Dual
Types of DNA microarrays DNA probe Source Mechanically spotted pre-synthesised probes Dual or single sample labelling c. DNA clones or PCR Home-made or commercial Dual products (one or two probes per gene) Oligonucleotides (50 -70 mer) Generally printed in-house; oligo (one probe per gene) sets from MWG Biotech, Illumina, Operon Technologies In situ synthesized arrays 25 mer oligonucleotides; Affymetrix Inc. Gene. Chips® mask-based photolithography (multiple perfect and mismatch probes per gene) 24 -70 mer oligonucleotides; Febit AG digital mask-less in situ Nimble. Gen™ Systems Inc photolithography (multiple probes per gene) 20 -60 mer oligonucleotides; Agilent Technologies ink-jet in situ synthesis (one Oxford Gene Technology or three probes per gene) Single Dual
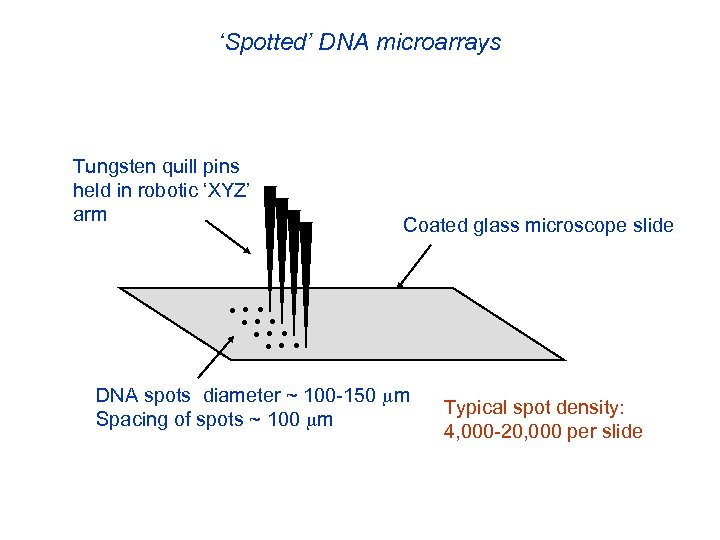 ‘Spotted’ DNA microarrays Tungsten quill pins held in robotic ‘XYZ’ arm Coated glass microscope slide • • • DNA spots diameter ~ 100 -150 m Spacing of spots ~ 100 m Typical spot density: 4, 000 -20, 000 per slide
‘Spotted’ DNA microarrays Tungsten quill pins held in robotic ‘XYZ’ arm Coated glass microscope slide • • • DNA spots diameter ~ 100 -150 m Spacing of spots ~ 100 m Typical spot density: 4, 000 -20, 000 per slide
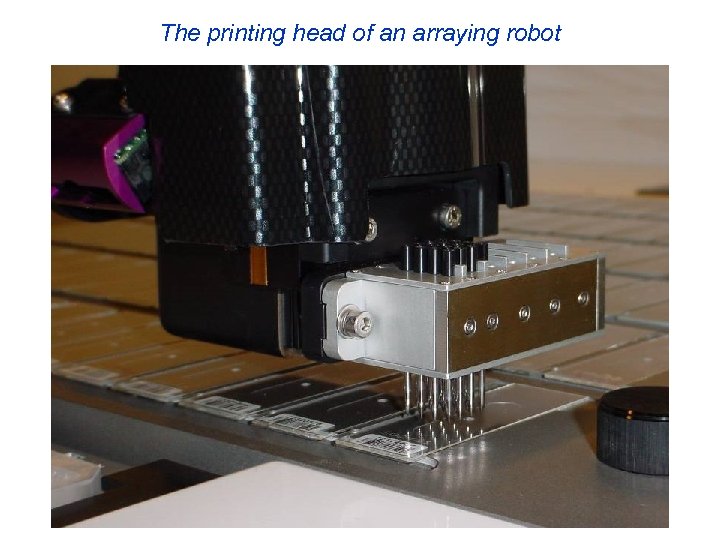 The printing head of an arraying robot
The printing head of an arraying robot
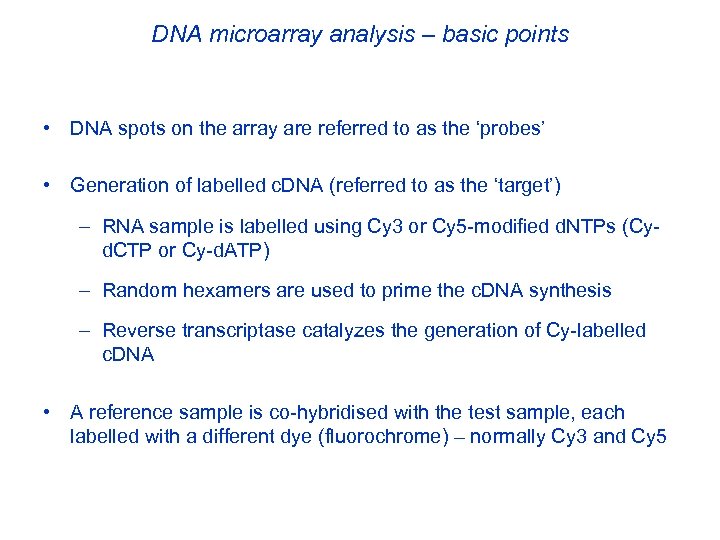 DNA microarray analysis – basic points • DNA spots on the array are referred to as the ‘probes’ • Generation of labelled c. DNA (referred to as the ‘target’) – RNA sample is labelled using Cy 3 or Cy 5 -modified d. NTPs (Cyd. CTP or Cy-d. ATP) – Random hexamers are used to prime the c. DNA synthesis – Reverse transcriptase catalyzes the generation of Cy-labelled c. DNA • A reference sample is co-hybridised with the test sample, each labelled with a different dye (fluorochrome) – normally Cy 3 and Cy 5
DNA microarray analysis – basic points • DNA spots on the array are referred to as the ‘probes’ • Generation of labelled c. DNA (referred to as the ‘target’) – RNA sample is labelled using Cy 3 or Cy 5 -modified d. NTPs (Cyd. CTP or Cy-d. ATP) – Random hexamers are used to prime the c. DNA synthesis – Reverse transcriptase catalyzes the generation of Cy-labelled c. DNA • A reference sample is co-hybridised with the test sample, each labelled with a different dye (fluorochrome) – normally Cy 3 and Cy 5
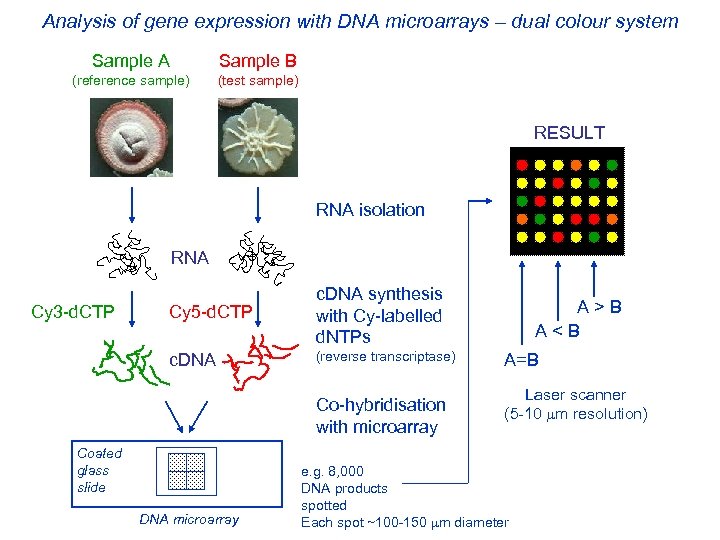 Analysis of gene expression with DNA microarrays – dual colour system Sample A Sample B (reference sample) (test sample) RESULT RNA isolation RNA Cy 5 -d. CTP c. DNA Cy 3 -d. CTP c. DNA synthesis with Cy-labelled d. NTPs (reverse transcriptase) Co-hybridisation with microarray Coated glass slide DNA microarray A > B A < B A=B Laser scanner (5 -10 m resolution) e. g. 8, 000 DNA products spotted Each spot ~100 -150 m diameter
Analysis of gene expression with DNA microarrays – dual colour system Sample A Sample B (reference sample) (test sample) RESULT RNA isolation RNA Cy 5 -d. CTP c. DNA Cy 3 -d. CTP c. DNA synthesis with Cy-labelled d. NTPs (reverse transcriptase) Co-hybridisation with microarray Coated glass slide DNA microarray A > B A < B A=B Laser scanner (5 -10 m resolution) e. g. 8, 000 DNA products spotted Each spot ~100 -150 m diameter
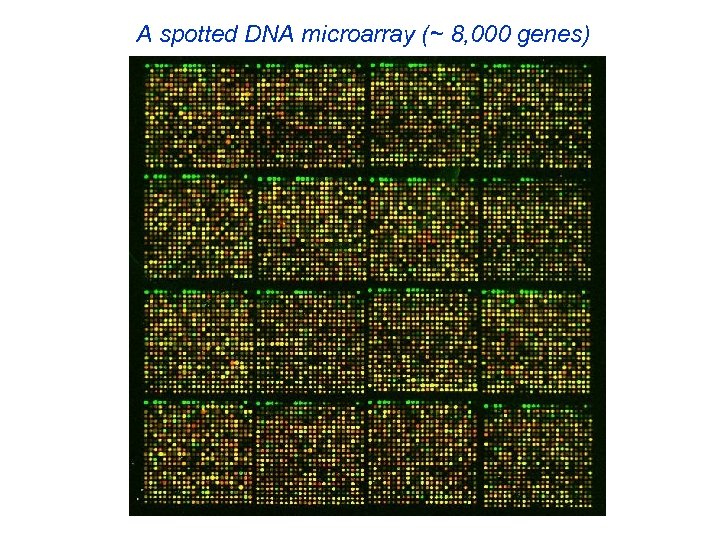 A spotted DNA microarray (~ 8, 000 genes)
A spotted DNA microarray (~ 8, 000 genes)
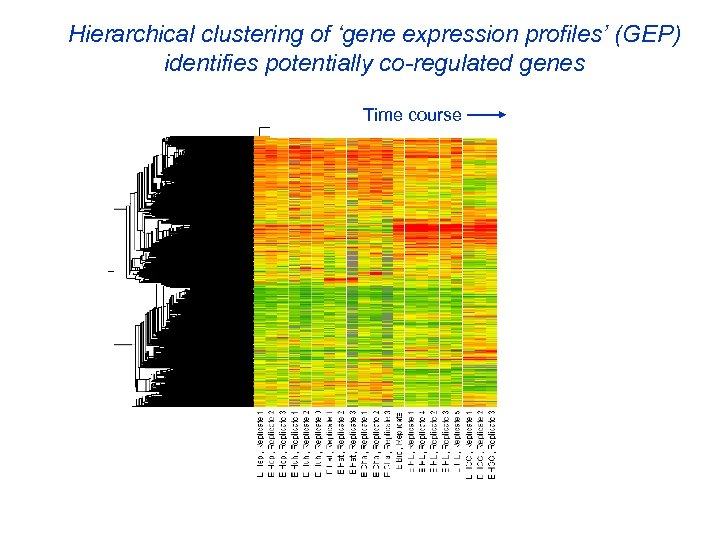 Hierarchical clustering of ‘gene expression profiles’ (GEP) identifies potentially co-regulated genes Time course
Hierarchical clustering of ‘gene expression profiles’ (GEP) identifies potentially co-regulated genes Time course
 Some types of array analysis experiment A. Comparative: Typically RNA vs RNA • wild-type versus mutant • one condition (e. g. induction) versus another • variant versus reference sample (often RNA vs DNA) B. Temporal (time series) analysis C. ‘Ch. IP-on-chip’
Some types of array analysis experiment A. Comparative: Typically RNA vs RNA • wild-type versus mutant • one condition (e. g. induction) versus another • variant versus reference sample (often RNA vs DNA) B. Temporal (time series) analysis C. ‘Ch. IP-on-chip’
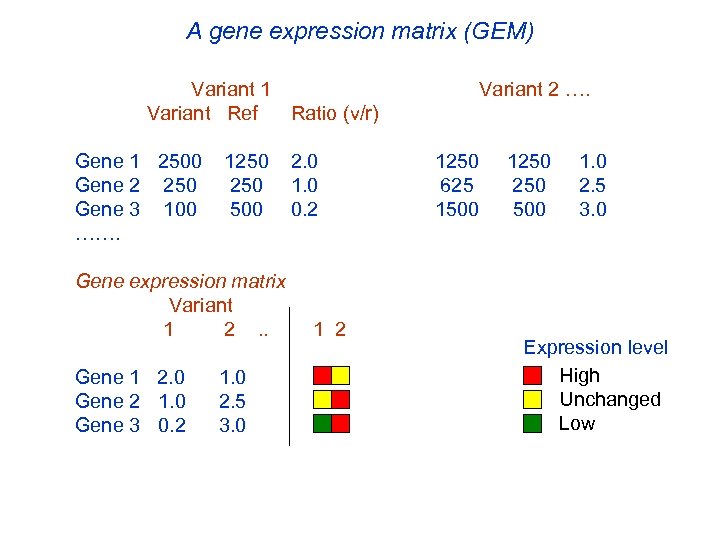 A gene expression matrix (GEM) Variant 1 Variant Ref Ratio (v/r) Gene 1 2500 1250 Gene 2 250 Gene 3 100 500 ……. 2. 0 1. 0 0. 2 Gene expression matrix Variant 1 2 . . 1 2 Gene 1 2. 0 1. 0 Gene 2 1. 0 2. 5 Gene 3 0. 2 3. 0 Variant 2 …. 1250 1. 0 625 250 2. 5 1500 3. 0 Expression level High Unchanged Low
A gene expression matrix (GEM) Variant 1 Variant Ref Ratio (v/r) Gene 1 2500 1250 Gene 2 250 Gene 3 100 500 ……. 2. 0 1. 0 0. 2 Gene expression matrix Variant 1 2 . . 1 2 Gene 1 2. 0 1. 0 Gene 2 1. 0 2. 5 Gene 3 0. 2 3. 0 Variant 2 …. 1250 1. 0 625 250 2. 5 1500 3. 0 Expression level High Unchanged Low
 Factors to take in account in experimental design: standardizing your system-normalization, reciprocal labelling, replicates • Normalization: Compensate for systematic differences not due to the biological system you are studying. Normally per spot and per chip normalization are required • Reciprocal labelling: compensate for labelling bias (cy 3 dye incorporates better than cy 5) • Replicates: accounts for experimental and /or biological variation in the data At least 3 biological replicates are normally required for a micro-array experiment. More replicates allow one to detect more subtle gene expression changes.
Factors to take in account in experimental design: standardizing your system-normalization, reciprocal labelling, replicates • Normalization: Compensate for systematic differences not due to the biological system you are studying. Normally per spot and per chip normalization are required • Reciprocal labelling: compensate for labelling bias (cy 3 dye incorporates better than cy 5) • Replicates: accounts for experimental and /or biological variation in the data At least 3 biological replicates are normally required for a micro-array experiment. More replicates allow one to detect more subtle gene expression changes.
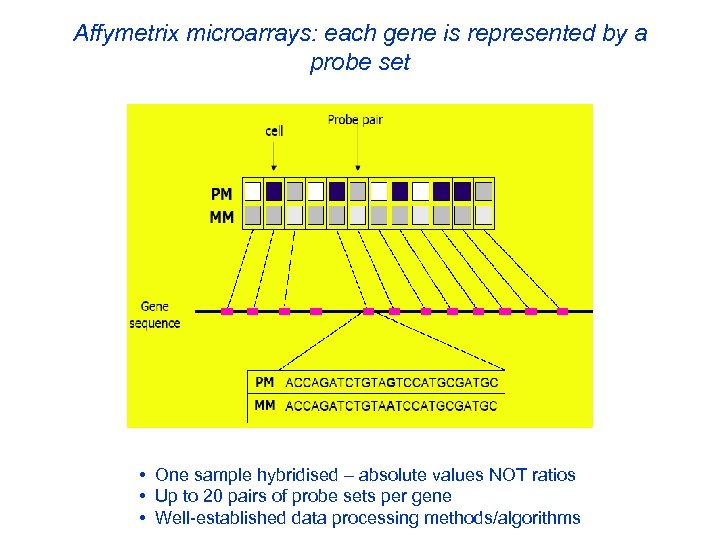 Affymetrix microarrays: each gene is represented by a probe set • One sample hybridised – absolute values NOT ratios • Up to 20 pairs of probe sets per gene • Well-established data processing methods/algorithms
Affymetrix microarrays: each gene is represented by a probe set • One sample hybridised – absolute values NOT ratios • Up to 20 pairs of probe sets per gene • Well-established data processing methods/algorithms
 S. coelicolor and S. venezuelae Microarrays • Affymetrix chips covering both genomes • Chips include a wide range of secondary metabolic gene clusters (ca. 50) • Analyze expression of cloned pathways • With proteome analysis, understand changes in gene expression at the onset of secondary metabolism • Knowledge based strain improvement
S. coelicolor and S. venezuelae Microarrays • Affymetrix chips covering both genomes • Chips include a wide range of secondary metabolic gene clusters (ca. 50) • Analyze expression of cloned pathways • With proteome analysis, understand changes in gene expression at the onset of secondary metabolism • Knowledge based strain improvement
 S. coelicolor and S. venezuelae Microarrays • • Prepare c. DNA from m. RNA Fragment to ~50 mers with DNasel End-label with biotin-dd. UTP using terminal transferase Inject ~10 ug c. DNA fragments onto chip Hybridise at ca. 50 C overnight in 6% DMSO Detect signals by staining with a) Streptavidin b) Anti-streptavidin antibody multiply labelled with biotin c) Streptavidin-phycoerythrin fluorescent conjugate (amplifies the signal). Measure spot intensity with laser scanner Data analysis: • R • Gene. Spring
S. coelicolor and S. venezuelae Microarrays • • Prepare c. DNA from m. RNA Fragment to ~50 mers with DNasel End-label with biotin-dd. UTP using terminal transferase Inject ~10 ug c. DNA fragments onto chip Hybridise at ca. 50 C overnight in 6% DMSO Detect signals by staining with a) Streptavidin b) Anti-streptavidin antibody multiply labelled with biotin c) Streptavidin-phycoerythrin fluorescent conjugate (amplifies the signal). Measure spot intensity with laser scanner Data analysis: • R • Gene. Spring
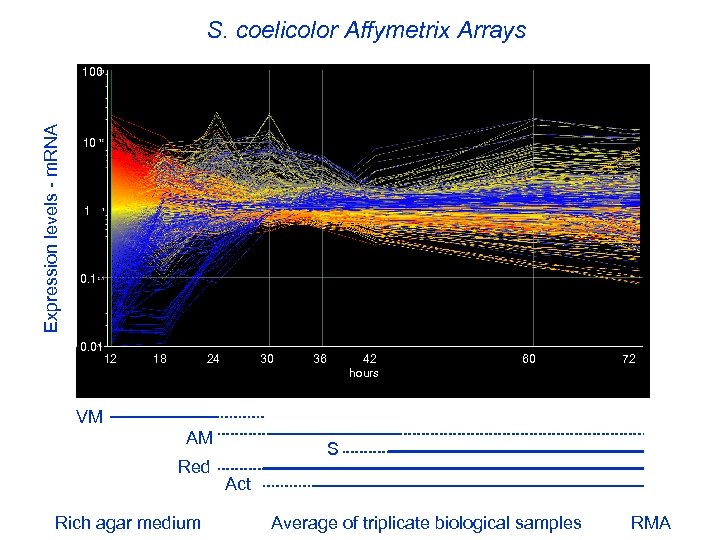 S. coelicolor Affymetrix Arrays Expression levels - m. RNA 100 10 1 0. 01 12 18 24 30 36 42 60 72 hours VM AM Red Rich agar medium S Act Average of triplicate biological samples RMA
S. coelicolor Affymetrix Arrays Expression levels - m. RNA 100 10 1 0. 01 12 18 24 30 36 42 60 72 hours VM AM Red Rich agar medium S Act Average of triplicate biological samples RMA
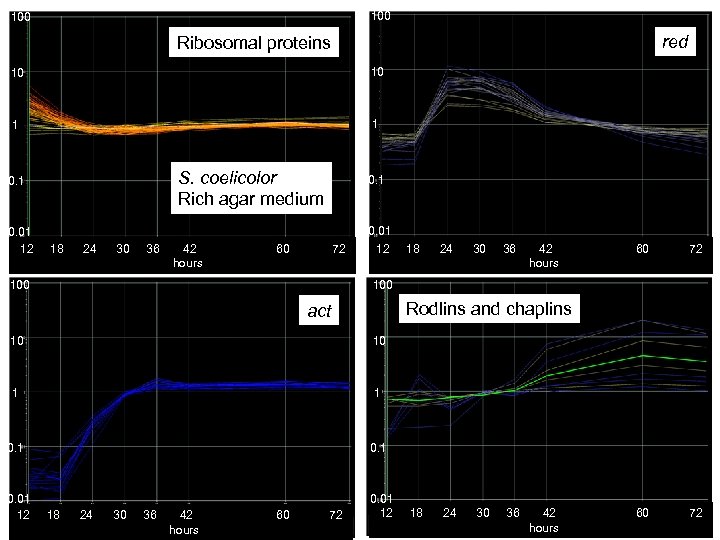 100 red Ribosomal proteins 10 10 1 1 0. 1 S. coelicolor Rich agar medium 0. 01 12 18 24 30 36 42 60 72 hours 100 0. 1 0. 01 12 18 24 30 36 42 60 72 hours 100 Rodlins and chaplins act 10 10 1 1 0. 01 12 18 24 30 36 42 60 72 hours 0. 01 12 18 24 30 36 42 60 72 hours
100 red Ribosomal proteins 10 10 1 1 0. 1 S. coelicolor Rich agar medium 0. 01 12 18 24 30 36 42 60 72 hours 100 0. 1 0. 01 12 18 24 30 36 42 60 72 hours 100 Rodlins and chaplins act 10 10 1 1 0. 01 12 18 24 30 36 42 60 72 hours 0. 01 12 18 24 30 36 42 60 72 hours
 Transcriptome analysis of intracellular signalling by pp. Gpp in Streptomyces coelicolor
Transcriptome analysis of intracellular signalling by pp. Gpp in Streptomyces coelicolor
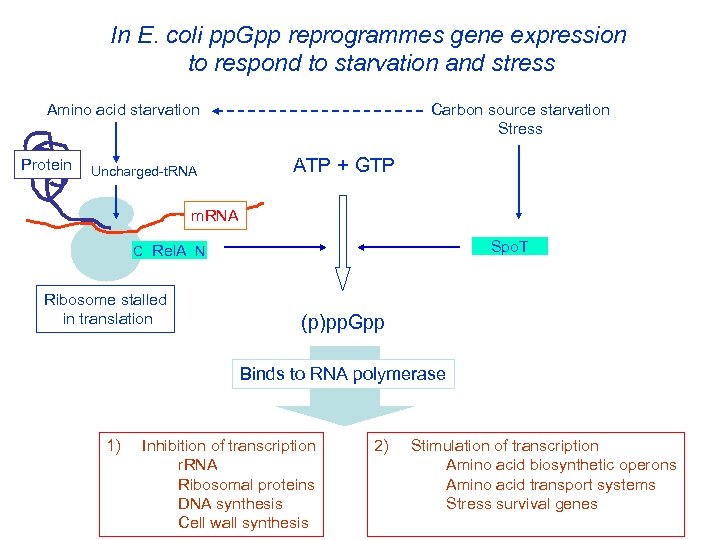 In E. coli pp. Gpp reprogrammes gene expression to respond to starvation and stress Amino acid starvation Protein Uncharged-t. RNA Carbon source starvation Stress ATP + GTP m. RNA Spo. T C Rel. A N Ribosome stalled in translation (p)pp. Gpp Binds to RNA polymerase 1) Inhibition of transcription r. RNA Ribosomal proteins DNA synthesis Cell wall synthesis 2) Stimulation of transcription Amino acid biosynthetic operons Amino acid transport systems Stress survival genes
In E. coli pp. Gpp reprogrammes gene expression to respond to starvation and stress Amino acid starvation Protein Uncharged-t. RNA Carbon source starvation Stress ATP + GTP m. RNA Spo. T C Rel. A N Ribosome stalled in translation (p)pp. Gpp Binds to RNA polymerase 1) Inhibition of transcription r. RNA Ribosomal proteins DNA synthesis Cell wall synthesis 2) Stimulation of transcription Amino acid biosynthetic operons Amino acid transport systems Stress survival genes
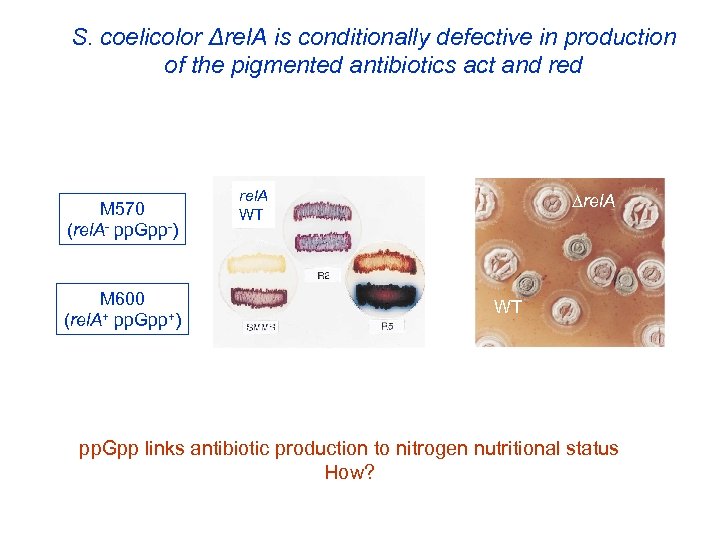 S. coelicolor Δrel. A is conditionally defective in production of the pigmented antibiotics act and red M 570 (rel. A- pp. Gpp-) M 600 (rel. A+ pp. Gpp+) rel. A WT pp. Gpp links antibiotic production to nitrogen nutritional status How?
S. coelicolor Δrel. A is conditionally defective in production of the pigmented antibiotics act and red M 570 (rel. A- pp. Gpp-) M 600 (rel. A+ pp. Gpp+) rel. A WT pp. Gpp links antibiotic production to nitrogen nutritional status How?
 Transcription analysis of the effects of pp. Gpp using Affymetrix microarrays 1. Changes on induction of pp. Gpp synthesis using truncated rel. A 2. Comparison of M 600 (rel. A+, pp. Gpp+) and M 570 (rel. A-, pp. Gpp-) Andy Hesketh • New insights into regulatory network for secondary metabolism • Define the pp. Gpp ‘regulon’
Transcription analysis of the effects of pp. Gpp using Affymetrix microarrays 1. Changes on induction of pp. Gpp synthesis using truncated rel. A 2. Comparison of M 600 (rel. A+, pp. Gpp+) and M 570 (rel. A-, pp. Gpp-) Andy Hesketh • New insights into regulatory network for secondary metabolism • Define the pp. Gpp ‘regulon’
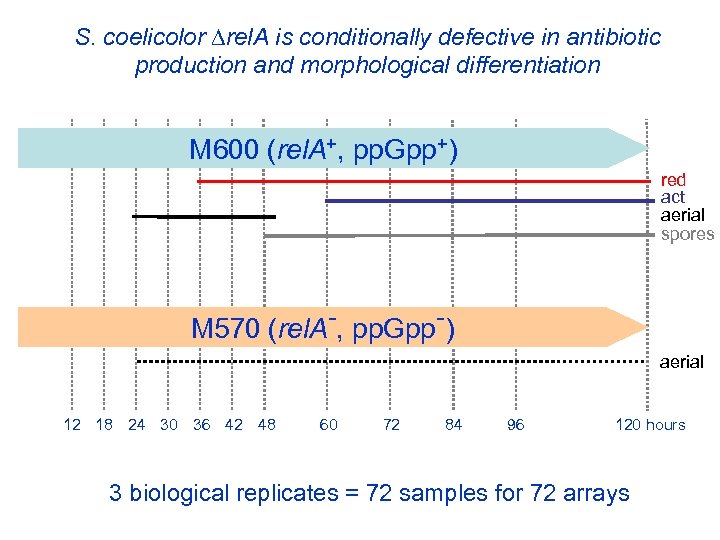 S. coelicolor rel. A is conditionally defective in antibiotic spores production and morphological differentiation red act aerial M 145 (at Diversa) M 600 (rel. A+, pp. Gpp+) red act aerial spores M 570 (rel. A-, pp. Gpp-) aerial 12 18 24 30 36 42 48 60 72 84 96 120 hours 3 biological replicates = 72 samples for 72 arrays
S. coelicolor rel. A is conditionally defective in antibiotic spores production and morphological differentiation red act aerial M 145 (at Diversa) M 600 (rel. A+, pp. Gpp+) red act aerial spores M 570 (rel. A-, pp. Gpp-) aerial 12 18 24 30 36 42 48 60 72 84 96 120 hours 3 biological replicates = 72 samples for 72 arrays
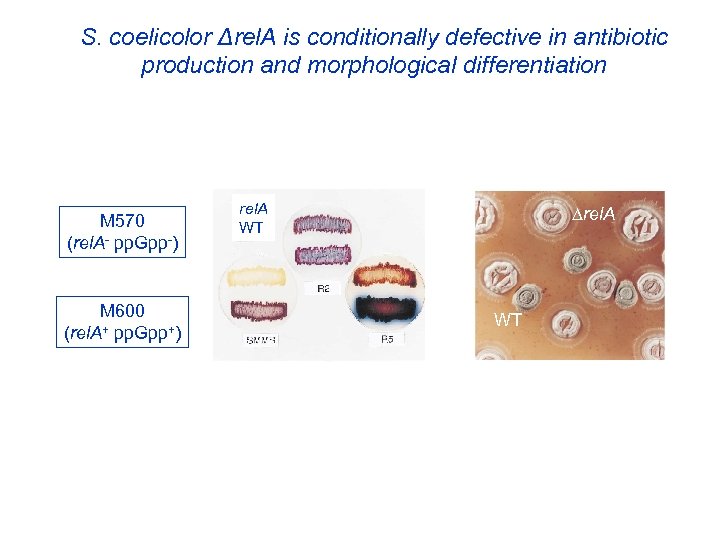 S. coelicolor Δrel. A is conditionally defective in antibiotic production and morphological differentiation M 570 (rel. A- pp. Gpp-) M 600 (rel. A+ pp. Gpp+) rel. A WT
S. coelicolor Δrel. A is conditionally defective in antibiotic production and morphological differentiation M 570 (rel. A- pp. Gpp-) M 600 (rel. A+ pp. Gpp+) rel. A WT
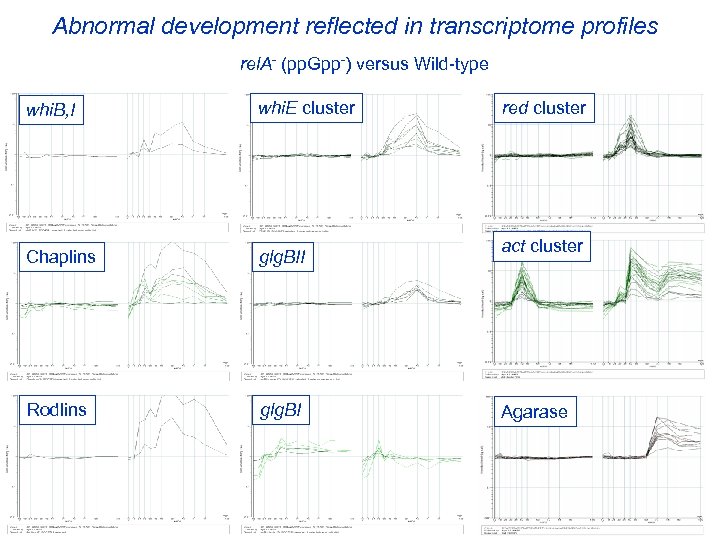 Abnormal development reflected in transcriptome profiles rel. A- (pp. Gpp-) versus Wild-type ram whi. B, I whi. E cluster Chaplins glg. BII Rodlins glg. BI red cluster act cluster Agarase
Abnormal development reflected in transcriptome profiles rel. A- (pp. Gpp-) versus Wild-type ram whi. B, I whi. E cluster Chaplins glg. BII Rodlins glg. BI red cluster act cluster Agarase
 Regulation of secondary metabolism • Secondary metabolites are compounds that are not absolutely required for the survival of an organism under laboratory conditions • While many (most? ) secondary metabolites are produced in stationary phase or at the onset of morphological differentiation, the production of some is growth associated (e. g. chloramphenicol, clavulanic acid) • The production of many antibiotics (just one class of secondary metabolites) is clearly growth phase-dependent and developmentally regulated • Many (but not all) antibiotic biosynthetic gene clusters contain pathway-specific regulatory genes (e. g. SARPs) • Many pathway-specific regulatory genes are controlled by pleiotropic regulatory genes that may also be required for morphological differentiation (e. g. bld genes)
Regulation of secondary metabolism • Secondary metabolites are compounds that are not absolutely required for the survival of an organism under laboratory conditions • While many (most? ) secondary metabolites are produced in stationary phase or at the onset of morphological differentiation, the production of some is growth associated (e. g. chloramphenicol, clavulanic acid) • The production of many antibiotics (just one class of secondary metabolites) is clearly growth phase-dependent and developmentally regulated • Many (but not all) antibiotic biosynthetic gene clusters contain pathway-specific regulatory genes (e. g. SARPs) • Many pathway-specific regulatory genes are controlled by pleiotropic regulatory genes that may also be required for morphological differentiation (e. g. bld genes)
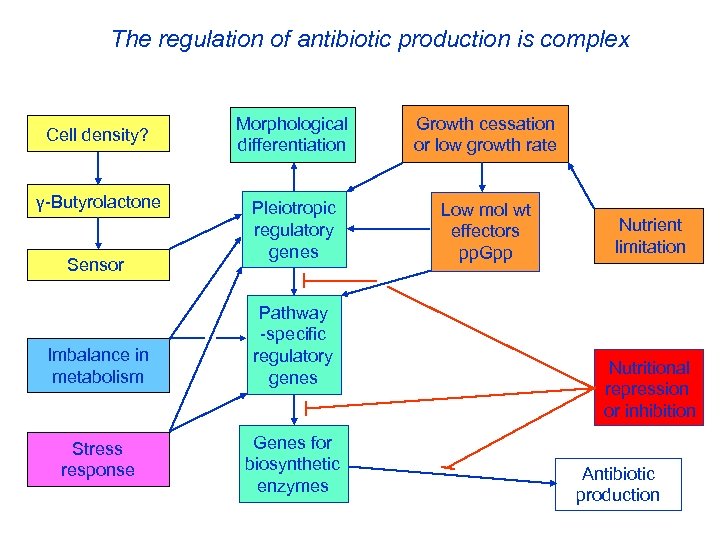 The regulation of antibiotic production is complex Cell density? γ-Butyrolactone Sensor Imbalance in metabolism Stress response Morphological differentiation Growth cessation or low growth rate Pleiotropic regulatory genes Low mol wt effectors pp. Gpp Pathway -specific regulatory genes Genes for biosynthetic enzymes Nutrient limitation Nutritional repression or inhibition Antibiotic production
The regulation of antibiotic production is complex Cell density? γ-Butyrolactone Sensor Imbalance in metabolism Stress response Morphological differentiation Growth cessation or low growth rate Pleiotropic regulatory genes Low mol wt effectors pp. Gpp Pathway -specific regulatory genes Genes for biosynthetic enzymes Nutrient limitation Nutritional repression or inhibition Antibiotic production
 Actinomycete-specific regulators of antibiotic production • The SARP family Winged helix-turn-helix motif towards N-terminus; appear to recognize heptameric repeats: CTCCTGAAAGCGGAGTGAAACCGTAGTGAAAGCGGACGCTCCTAGTGTCGTTCTC Associated with clusters for aromatic polyketides, ribosomally and non-ribosomally synthesized peptides, undecylprodiginines, Type I polyketides, β-lactams and azoxy compounds. Mostly pathway-specific (exceptions: Cca. R, Afs. R). Found only in actinomycetes. • The LAL family Large ATP-binding regulators of the Lux. R family; associated with at least 13 Type I polyketide and two glycopeptide gene clusters. N-terminally located nucleotide triphosphate binding motif and a C-terminal helix-turn-helix motif of the Lux. R family. Homologues with end-to-end similarity confined to the actinomycetes. Bibb, M. J. 2005. Regulation of secondary metabolism in streptomycetes. Current Opinion in Microbiology. 8: 208 -215.
Actinomycete-specific regulators of antibiotic production • The SARP family Winged helix-turn-helix motif towards N-terminus; appear to recognize heptameric repeats: CTCCTGAAAGCGGAGTGAAACCGTAGTGAAAGCGGACGCTCCTAGTGTCGTTCTC Associated with clusters for aromatic polyketides, ribosomally and non-ribosomally synthesized peptides, undecylprodiginines, Type I polyketides, β-lactams and azoxy compounds. Mostly pathway-specific (exceptions: Cca. R, Afs. R). Found only in actinomycetes. • The LAL family Large ATP-binding regulators of the Lux. R family; associated with at least 13 Type I polyketide and two glycopeptide gene clusters. N-terminally located nucleotide triphosphate binding motif and a C-terminal helix-turn-helix motif of the Lux. R family. Homologues with end-to-end similarity confined to the actinomycetes. Bibb, M. J. 2005. Regulation of secondary metabolism in streptomycetes. Current Opinion in Microbiology. 8: 208 -215.
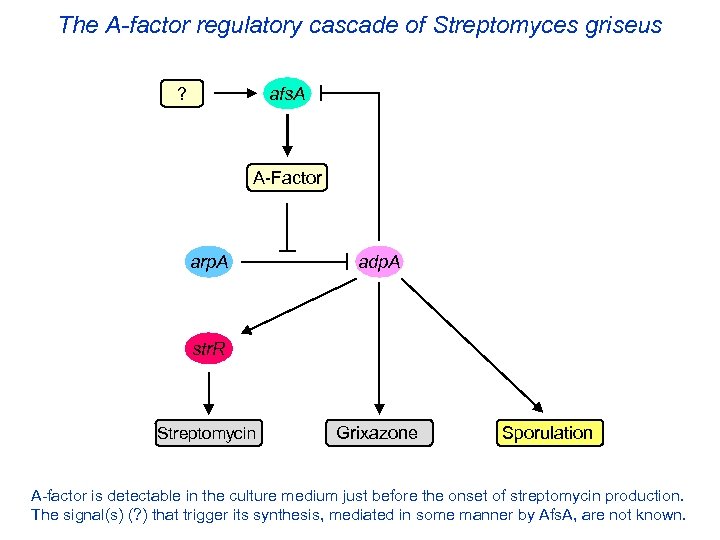 The A-factor regulatory cascade of Streptomyces griseus afs. A ? A-Factor arp. A adp. A str. R Streptomycin Grixazone Sporulation A-factor is detectable in the culture medium just before the onset of streptomycin production. The signal(s) (? ) that trigger its synthesis, mediated in some manner by Afs. A, are not known.
The A-factor regulatory cascade of Streptomyces griseus afs. A ? A-Factor arp. A adp. A str. R Streptomycin Grixazone Sporulation A-factor is detectable in the culture medium just before the onset of streptomycin production. The signal(s) (? ) that trigger its synthesis, mediated in some manner by Afs. A, are not known.
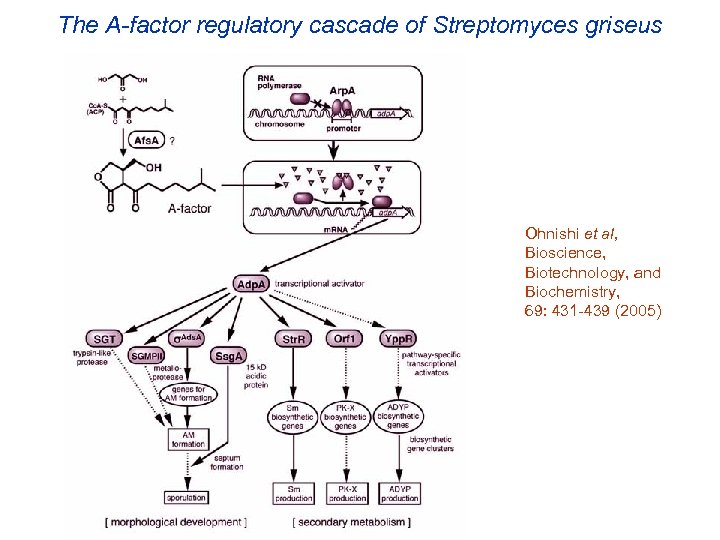 The A-factor regulatory cascade of Streptomyces griseus Ohnishi et al, Bioscience, Biotechnology, and Biochemistry, 69: 431 -439 (2005)
The A-factor regulatory cascade of Streptomyces griseus Ohnishi et al, Bioscience, Biotechnology, and Biochemistry, 69: 431 -439 (2005)
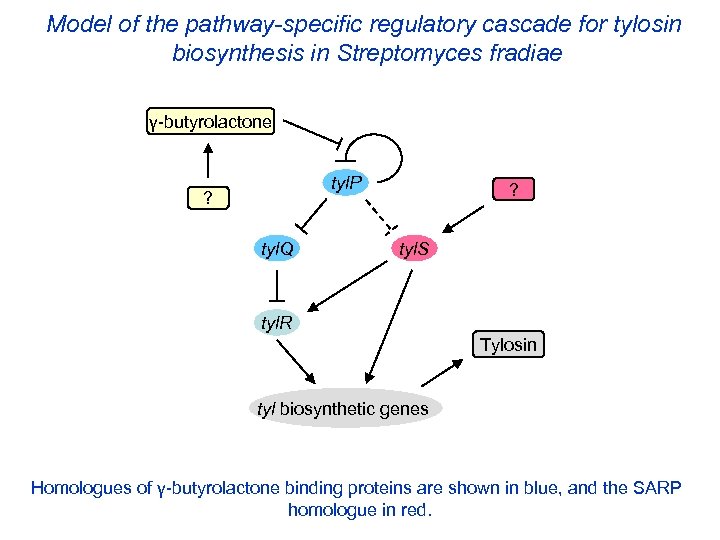 Model of the pathway-specific regulatory cascade for tylosin biosynthesis in Streptomyces fradiae γ-butyrolactone tyl. P ? tyl. Q ? tyl. S tyl. R Tylosin tyl biosynthetic genes Homologues of γ-butyrolactone binding proteins are shown in blue, and the SARP homologue in red.
Model of the pathway-specific regulatory cascade for tylosin biosynthesis in Streptomyces fradiae γ-butyrolactone tyl. P ? tyl. Q ? tyl. S tyl. R Tylosin tyl biosynthetic genes Homologues of γ-butyrolactone binding proteins are shown in blue, and the SARP homologue in red.
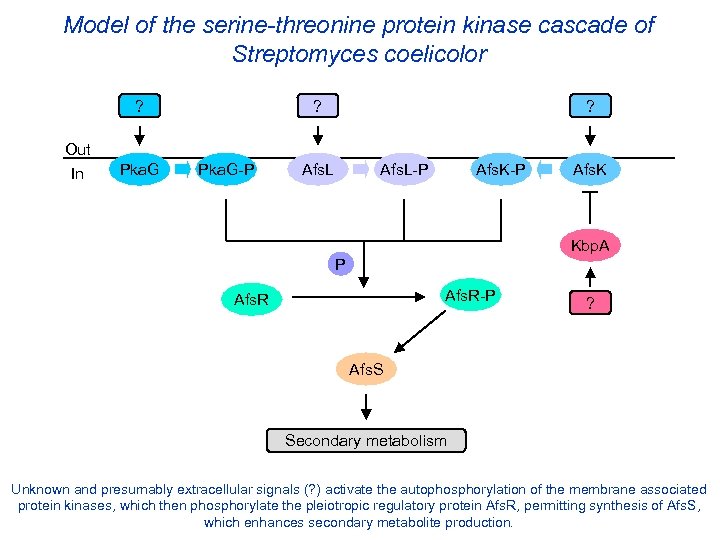 Model of the serine-threonine protein kinase cascade of Streptomyces coelicolor ? Out In Pka. G ? Pka. G-P ? Afs. L-P Afs. K Kbp. A P Afs. R-P Afs. R ? Afs. S Secondary metabolism Unknown and presumably extracellular signals (? ) activate the autophosphorylation of the membrane associated protein kinases, which then phosphorylate the pleiotropic regulatory protein Afs. R, permitting synthesis of Afs. S, which enhances secondary metabolite production.
Model of the serine-threonine protein kinase cascade of Streptomyces coelicolor ? Out In Pka. G ? Pka. G-P ? Afs. L-P Afs. K Kbp. A P Afs. R-P Afs. R ? Afs. S Secondary metabolism Unknown and presumably extracellular signals (? ) activate the autophosphorylation of the membrane associated protein kinases, which then phosphorylate the pleiotropic regulatory protein Afs. R, permitting synthesis of Afs. S, which enhances secondary metabolite production.
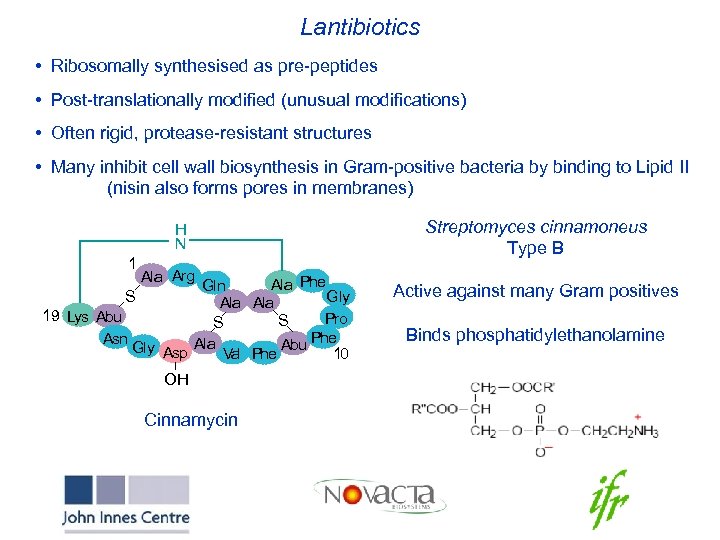 Lantibiotics • Ribosomally synthesised as pre-peptides • Post-translationally modified (unusual modifications) • Often rigid, protease-resistant structures • Many inhibit cell wall biosynthesis in Gram-positive bacteria by binding to Lipid II (nisin also forms pores in membranes) H N 1 Ala Arg Ala Phe Gln S Gly Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 OH Cinnamycin Streptomyces cinnamoneus Type B Active against many Gram positives Binds phosphatidylethanolamine
Lantibiotics • Ribosomally synthesised as pre-peptides • Post-translationally modified (unusual modifications) • Often rigid, protease-resistant structures • Many inhibit cell wall biosynthesis in Gram-positive bacteria by binding to Lipid II (nisin also forms pores in membranes) H N 1 Ala Arg Ala Phe Gln S Gly Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 OH Cinnamycin Streptomyces cinnamoneus Type B Active against many Gram positives Binds phosphatidylethanolamine
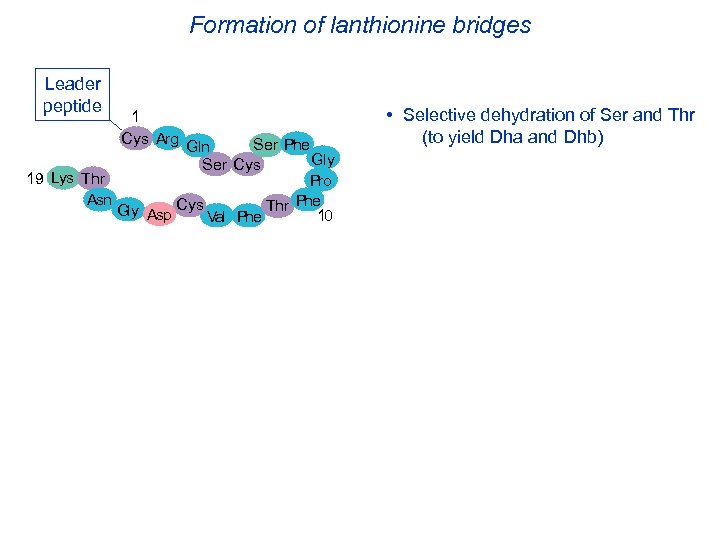 Formation of lanthionine bridges Leader peptide 1 Cys Arg Ser Phe Gln Gly Ser Cys 19 Lys Thr Pro Asn Phe Gly Asp Cys Val Phe Thr 10 • Selective dehydration of Ser and Thr (to yield Dha and Dhb)
Formation of lanthionine bridges Leader peptide 1 Cys Arg Ser Phe Gln Gly Ser Cys 19 Lys Thr Pro Asn Phe Gly Asp Cys Val Phe Thr 10 • Selective dehydration of Ser and Thr (to yield Dha and Dhb)
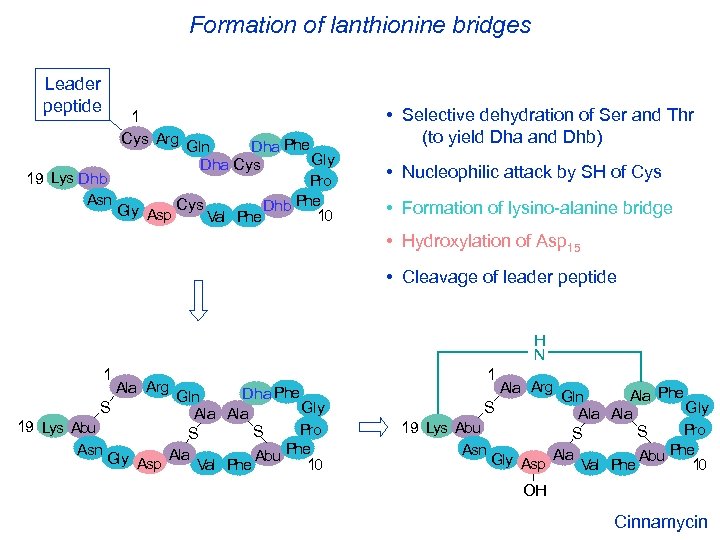 Formation of lanthionine bridges Leader peptide 1 Cys Arg Dha Phe Gln Gly Dha Cys 19 Lys Dhb Pro Asn Phe Gly Asp Cys Val Phe Dhb 10 • Selective dehydration of Ser and Thr (to yield Dha and Dhb) • Nucleophilic attack by SH of Cys • Formation of lysino-alanine bridge • Hydroxylation of Asp 15 • Cleavage of leader peptide H N 1 Ala Arg Dha Phe Gln S Gly Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 1 Ala Arg Ala Phe Gln S Gly Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 OH Cinnamycin
Formation of lanthionine bridges Leader peptide 1 Cys Arg Dha Phe Gln Gly Dha Cys 19 Lys Dhb Pro Asn Phe Gly Asp Cys Val Phe Dhb 10 • Selective dehydration of Ser and Thr (to yield Dha and Dhb) • Nucleophilic attack by SH of Cys • Formation of lysino-alanine bridge • Hydroxylation of Asp 15 • Cleavage of leader peptide H N 1 Ala Arg Dha Phe Gln S Gly Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 1 Ala Arg Ala Phe Gln S Gly Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 OH Cinnamycin
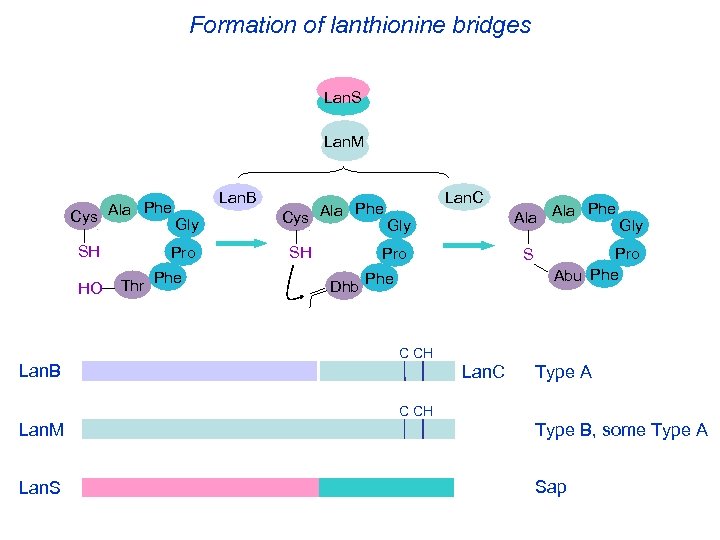 Formation of lanthionine bridges Lan. S Lan. M Phe Cys Ala SH HO Lan. B Gly Pro Thr Phe Cys Ala SH Lan. C Gly Pro Dhb Phe Ala S Phe Gly Pro Abu Phe C CH Lan. B Lan. C Type A C CH Lan. M Type B, some Type A Lan. S Sap
Formation of lanthionine bridges Lan. S Lan. M Phe Cys Ala SH HO Lan. B Gly Pro Thr Phe Cys Ala SH Lan. C Gly Pro Dhb Phe Ala S Phe Gly Pro Abu Phe C CH Lan. B Lan. C Type A C CH Lan. M Type B, some Type A Lan. S Sap
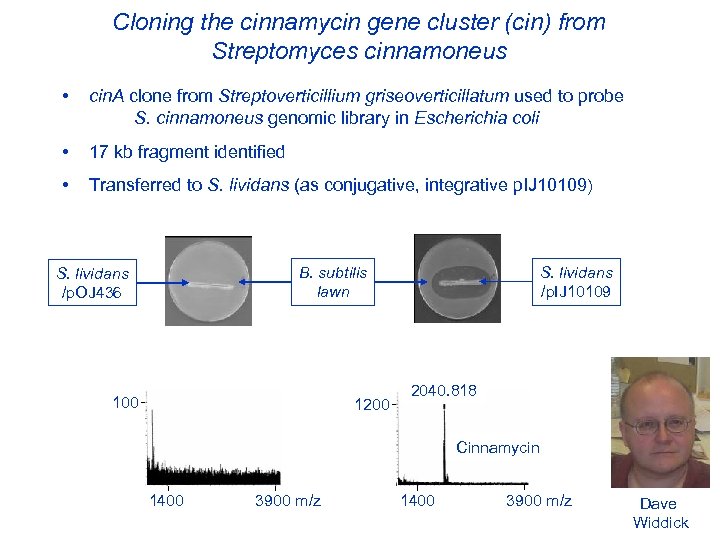 Cloning the cinnamycin gene cluster (cin) from Streptomyces cinnamoneus • cin. A clone from Streptoverticillium griseoverticillatum used to probe S. cinnamoneus genomic library in Escherichia coli • 17 kb fragment identified • Transferred to S. lividans (as conjugative, integrative p. IJ 10109) S. lividans /p. OJ 436 B. subtilis lawn 100 1200 S. lividans /p. IJ 10109 2040. 818 Cinnamycin 1400 3900 m/z Dave Widdick
Cloning the cinnamycin gene cluster (cin) from Streptomyces cinnamoneus • cin. A clone from Streptoverticillium griseoverticillatum used to probe S. cinnamoneus genomic library in Escherichia coli • 17 kb fragment identified • Transferred to S. lividans (as conjugative, integrative p. IJ 10109) S. lividans /p. OJ 436 B. subtilis lawn 100 1200 S. lividans /p. IJ 10109 2040. 818 Cinnamycin 1400 3900 m/z Dave Widdick
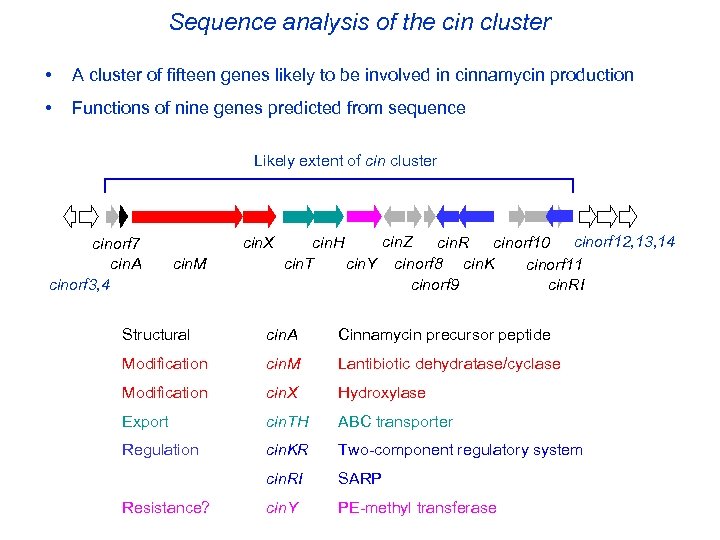 Sequence analysis of the cin cluster • A cluster of fifteen genes likely to be involved in cinnamycin production • Functions of nine genes predicted from sequence Likely extent of cin cluster cinorf 7 cin. A cinorf 3, 4 cin. X cin. M cin. Z cin. H cin. R cinorf 10 cinorf 12, 13, 14 cin. T cin. Y cinorf 8 cin. K cinorf 11 cin. RI cinorf 9 Structural cin. A Cinnamycin precursor peptide Modification cin. M Lantibiotic dehydratase/cyclase Modification cin. X Hydroxylase Export cin. TH ABC transporter Regulation cin. KR Two-component regulatory system cin. RI SARP cin. Y PE-methyl transferase Resistance?
Sequence analysis of the cin cluster • A cluster of fifteen genes likely to be involved in cinnamycin production • Functions of nine genes predicted from sequence Likely extent of cin cluster cinorf 7 cin. A cinorf 3, 4 cin. X cin. M cin. Z cin. H cin. R cinorf 10 cinorf 12, 13, 14 cin. T cin. Y cinorf 8 cin. K cinorf 11 cin. RI cinorf 9 Structural cin. A Cinnamycin precursor peptide Modification cin. M Lantibiotic dehydratase/cyclase Modification cin. X Hydroxylase Export cin. TH ABC transporter Regulation cin. KR Two-component regulatory system cin. RI SARP cin. Y PE-methyl transferase Resistance?
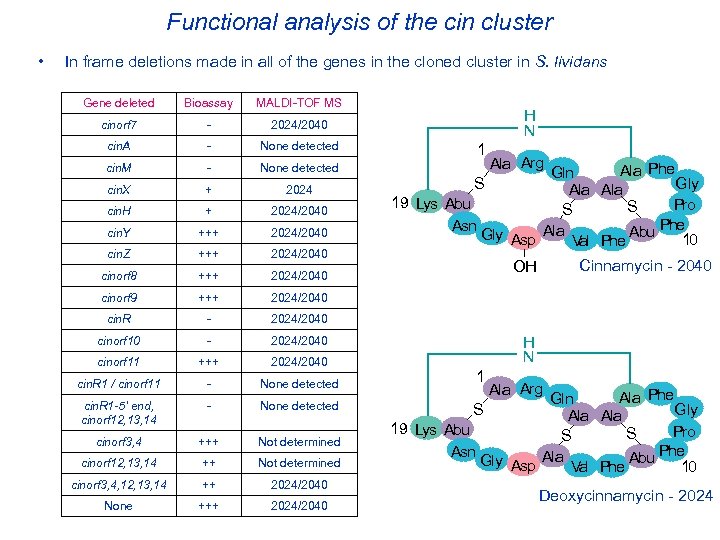 Functional analysis of the cin cluster • In frame deletions made in all of the genes in the cloned cluster in S. lividans Gene deleted Bioassay MALDI-TOF MS cinorf 7 - 2024/2040 cin. A - None detected cin. M - None detected cin. X + 2024 cin. H + 2024/2040 cin. Y +++ 2024/2040 cin. Z +++ 2024/2040 cinorf 8 +++ 2024/2040 cinorf 9 +++ 2024/2040 cin. R - 2024/2040 cinorf 10 - 2024/2040 cinorf 11 +++ 2024/2040 cin. R 1 / cinorf 11 - None detected cin. R 1 -5’ end, cinorf 12, 13, 14 - None detected cinorf 3, 4 +++ Not determined cinorf 12, 13, 14 ++ Not determined cinorf 3, 4, 12, 13, 14 ++ 2024/2040 None +++ 2024/2040 H N 1 Ala Arg Ala Phe Gln S Gly Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 Cinnamycin - 2040 OH H N 1 Ala Arg Ala Phe Gln Gly Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 S Deoxycinnamycin - 2024
Functional analysis of the cin cluster • In frame deletions made in all of the genes in the cloned cluster in S. lividans Gene deleted Bioassay MALDI-TOF MS cinorf 7 - 2024/2040 cin. A - None detected cin. M - None detected cin. X + 2024 cin. H + 2024/2040 cin. Y +++ 2024/2040 cin. Z +++ 2024/2040 cinorf 8 +++ 2024/2040 cinorf 9 +++ 2024/2040 cin. R - 2024/2040 cinorf 10 - 2024/2040 cinorf 11 +++ 2024/2040 cin. R 1 / cinorf 11 - None detected cin. R 1 -5’ end, cinorf 12, 13, 14 - None detected cinorf 3, 4 +++ Not determined cinorf 12, 13, 14 ++ Not determined cinorf 3, 4, 12, 13, 14 ++ 2024/2040 None +++ 2024/2040 H N 1 Ala Arg Ala Phe Gln S Gly Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 Cinnamycin - 2040 OH H N 1 Ala Arg Ala Phe Gln Gly Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 S Deoxycinnamycin - 2024
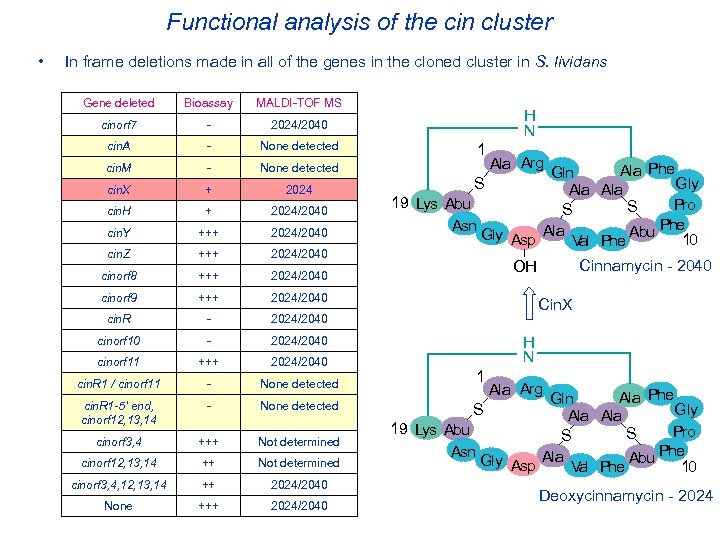 Functional analysis of the cin cluster • In frame deletions made in all of the genes in the cloned cluster in S. lividans Gene deleted Bioassay MALDI-TOF MS cinorf 7 - 2024/2040 cin. A - None detected cin. M - None detected cin. X + 2024 cin. H + 2024/2040 cin. Y +++ 2024/2040 cin. Z +++ 2024/2040 cinorf 8 +++ 2024/2040 cinorf 9 +++ 2024/2040 cin. R - 2024/2040 cinorf 10 - 2024/2040 cinorf 11 +++ 2024/2040 cin. R 1 / cinorf 11 - None detected cin. R 1 -5’ end, cinorf 12, 13, 14 - None detected cinorf 3, 4 +++ Not determined cinorf 12, 13, 14 ++ Not determined cinorf 3, 4, 12, 13, 14 ++ 2024/2040 None +++ 2024/2040 H N 1 Ala Arg Ala Phe Gln S Gly Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 Cinnamycin - 2040 OH Cin. X H N 1 Ala Arg Ala Phe Gln Gly Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 S Deoxycinnamycin - 2024
Functional analysis of the cin cluster • In frame deletions made in all of the genes in the cloned cluster in S. lividans Gene deleted Bioassay MALDI-TOF MS cinorf 7 - 2024/2040 cin. A - None detected cin. M - None detected cin. X + 2024 cin. H + 2024/2040 cin. Y +++ 2024/2040 cin. Z +++ 2024/2040 cinorf 8 +++ 2024/2040 cinorf 9 +++ 2024/2040 cin. R - 2024/2040 cinorf 10 - 2024/2040 cinorf 11 +++ 2024/2040 cin. R 1 / cinorf 11 - None detected cin. R 1 -5’ end, cinorf 12, 13, 14 - None detected cinorf 3, 4 +++ Not determined cinorf 12, 13, 14 ++ Not determined cinorf 3, 4, 12, 13, 14 ++ 2024/2040 None +++ 2024/2040 H N 1 Ala Arg Ala Phe Gln S Gly Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 Cinnamycin - 2040 OH Cin. X H N 1 Ala Arg Ala Phe Gln Gly Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 S Deoxycinnamycin - 2024
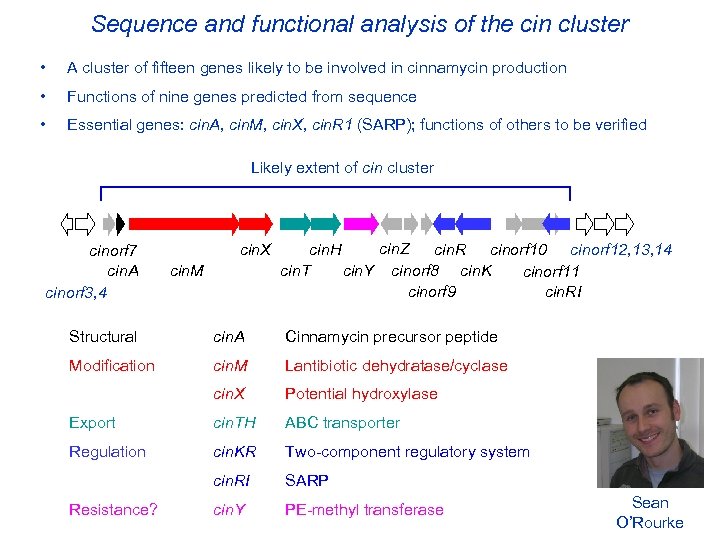 Sequence and functional analysis of the cin cluster • A cluster of fifteen genes likely to be involved in cinnamycin production • Functions of nine genes predicted from sequence • Essential genes: cin. A, cin. M, cin. X, cin. R 1 (SARP); functions of others to be verified Likely extent of cin cluster cinorf 7 cin. A cinorf 3, 4 cin. X cin. M cin. Z cin. H cin. R cinorf 10 cinorf 12, 13, 14 cin. T cin. Y cinorf 8 cin. K cinorf 11 cin. RI cinorf 9 Structural cin. A Cinnamycin precursor peptide Modification cin. M Lantibiotic dehydratase/cyclase cin. X Potential hydroxylase Export cin. TH ABC transporter Regulation cin. KR Two-component regulatory system cin. RI SARP cin. Y PE-methyl transferase Resistance? Sean O’Rourke
Sequence and functional analysis of the cin cluster • A cluster of fifteen genes likely to be involved in cinnamycin production • Functions of nine genes predicted from sequence • Essential genes: cin. A, cin. M, cin. X, cin. R 1 (SARP); functions of others to be verified Likely extent of cin cluster cinorf 7 cin. A cinorf 3, 4 cin. X cin. M cin. Z cin. H cin. R cinorf 10 cinorf 12, 13, 14 cin. T cin. Y cinorf 8 cin. K cinorf 11 cin. RI cinorf 9 Structural cin. A Cinnamycin precursor peptide Modification cin. M Lantibiotic dehydratase/cyclase cin. X Potential hydroxylase Export cin. TH ABC transporter Regulation cin. KR Two-component regulatory system cin. RI SARP cin. Y PE-methyl transferase Resistance? Sean O’Rourke
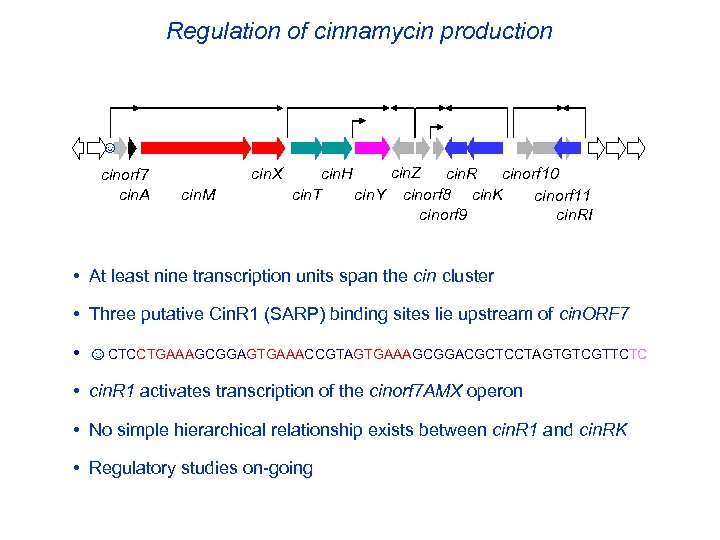 Regulation of cinnamycin production ☺ cinorf 7 cin. A cin. X cin. M cin. Z cin. H cin. R cinorf 10 cin. T cin. Y cinorf 8 cin. K cinorf 11 cin. RI cinorf 9 • At least nine transcription units span the cin cluster • Three putative Cin. R 1 (SARP) binding sites lie upstream of cin. ORF 7 • ☺CTCCTGAAAGCGGAGTGAAACCGTAGTGAAAGCGGACGCTCCTAGTGTCGTTCTC • cin. R 1 activates transcription of the cinorf 7 AMX operon • No simple hierarchical relationship exists between cin. R 1 and cin. RK • Regulatory studies on-going
Regulation of cinnamycin production ☺ cinorf 7 cin. A cin. X cin. M cin. Z cin. H cin. R cinorf 10 cin. T cin. Y cinorf 8 cin. K cinorf 11 cin. RI cinorf 9 • At least nine transcription units span the cin cluster • Three putative Cin. R 1 (SARP) binding sites lie upstream of cin. ORF 7 • ☺CTCCTGAAAGCGGAGTGAAACCGTAGTGAAAGCGGACGCTCCTAGTGTCGTTCTC • cin. R 1 activates transcription of the cinorf 7 AMX operon • No simple hierarchical relationship exists between cin. R 1 and cin. RK • Regulatory studies on-going
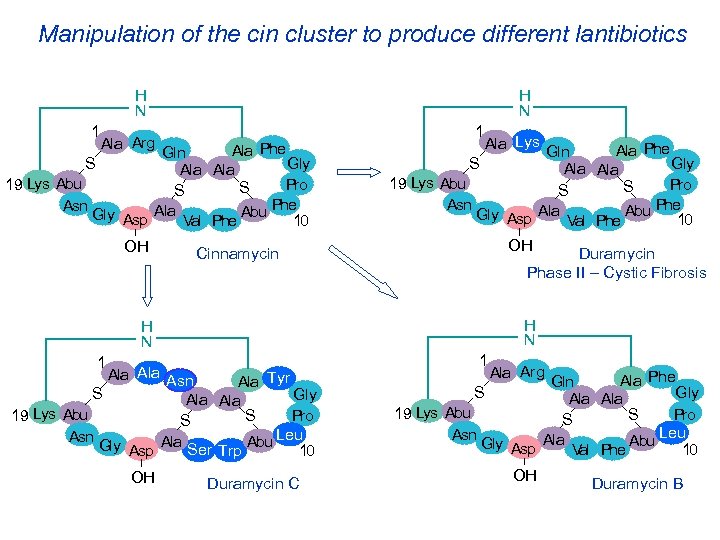 Manipulation of the cin cluster to produce different lantibiotics H N 1 H N Ala Arg Ala Phe Gln Gly S Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 OH 1 Ala Lys Ala Phe Gln Gly S Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 OH Cinnamycin Duramycin Phase II – Cystic Fibrosis H N 1 Ala Asn Ala Tyr S Gly Ala 19 Lys Abu S Pro S Leu Asn Gly Asp Ala Ser Trp Abu 10 OH Duramycin C 1 Ala Arg Ala Phe Gln Gly S Ala 19 Lys Abu Pro S S Leu Asn Gly Asp Ala Val Phe Abu 10 OH Duramycin B
Manipulation of the cin cluster to produce different lantibiotics H N 1 H N Ala Arg Ala Phe Gln Gly S Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 OH 1 Ala Lys Ala Phe Gln Gly S Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 OH Cinnamycin Duramycin Phase II – Cystic Fibrosis H N 1 Ala Asn Ala Tyr S Gly Ala 19 Lys Abu S Pro S Leu Asn Gly Asp Ala Ser Trp Abu 10 OH Duramycin C 1 Ala Arg Ala Phe Gln Gly S Ala 19 Lys Abu Pro S S Leu Asn Gly Asp Ala Val Phe Abu 10 OH Duramycin B
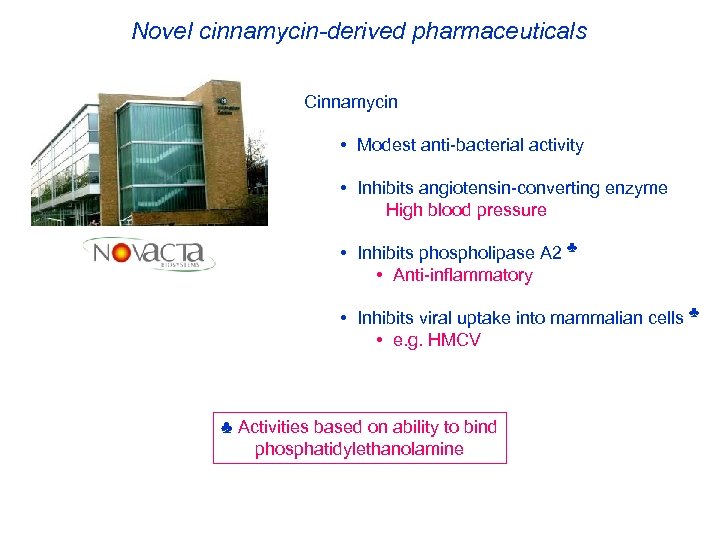 Novel cinnamycin-derived pharmaceuticals Cinnamycin • Modest anti-bacterial activity • Inhibits angiotensin-converting enzyme High blood pressure • Inhibits phospholipase A 2 ♣ • Anti-inflammatory • Inhibits viral uptake into mammalian cells ♣ • e. g. HMCV ♣ Activities based on ability to bind phosphatidylethanolamine
Novel cinnamycin-derived pharmaceuticals Cinnamycin • Modest anti-bacterial activity • Inhibits angiotensin-converting enzyme High blood pressure • Inhibits phospholipase A 2 ♣ • Anti-inflammatory • Inhibits viral uptake into mammalian cells ♣ • e. g. HMCV ♣ Activities based on ability to bind phosphatidylethanolamine
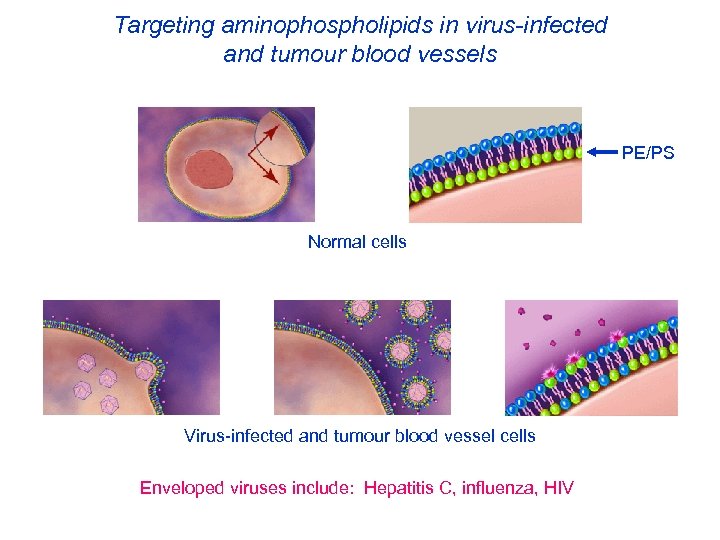 Targeting aminophospholipids in virus-infected and tumour blood vessels PE/PS Normal cells Virus-infected and tumour blood vessel cells Enveloped viruses include: Hepatitis C, influenza, HIV
Targeting aminophospholipids in virus-infected and tumour blood vessels PE/PS Normal cells Virus-infected and tumour blood vessel cells Enveloped viruses include: Hepatitis C, influenza, HIV
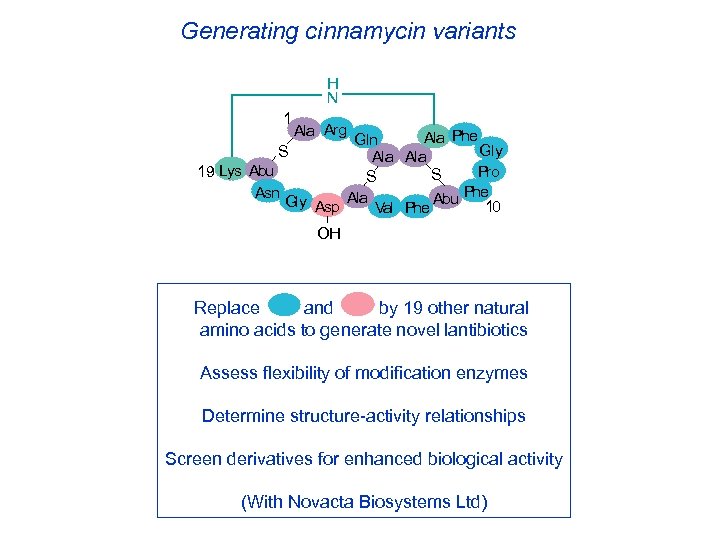 Generating cinnamycin variants H N 1 Ala Arg Ala Phe Gln Gly S Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 OH Replace and by 19 other natural amino acids to generate novel lantibiotics Assess flexibility of modification enzymes Determine structure-activity relationships Screen derivatives for enhanced biological activity (With Novacta Biosystems Ltd)
Generating cinnamycin variants H N 1 Ala Arg Ala Phe Gln Gly S Ala 19 Lys Abu Pro S S Phe Asn Gly Asp Ala Val Phe Abu 10 OH Replace and by 19 other natural amino acids to generate novel lantibiotics Assess flexibility of modification enzymes Determine structure-activity relationships Screen derivatives for enhanced biological activity (With Novacta Biosystems Ltd)
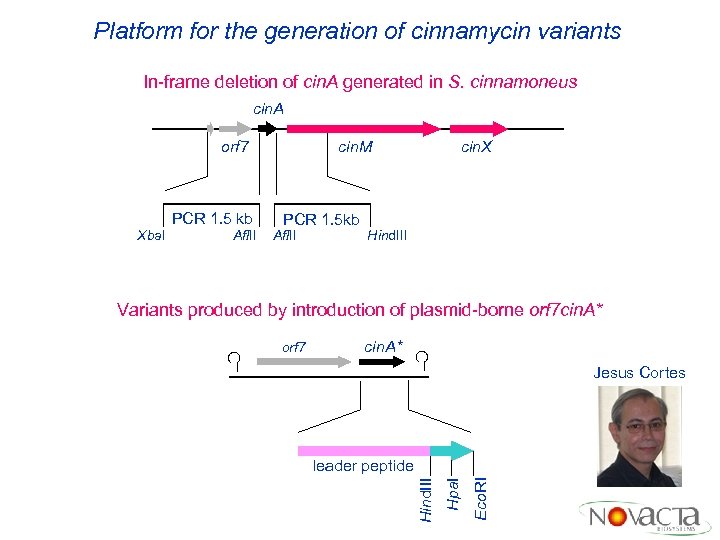 Platform for the generation of cinnamycin variants In-frame deletion of cin. A generated in S. cinnamoneus cin. A orf 7 PCR 1. 5 kb Xba. I Afl. II cin. M PCR 1. 5 kb Afl. II cin. X Hind. III Variants produced by introduction of plasmid-borne orf 7 cin. A* Jesus Cortes Eco. RI Hpa. I leader peptide Hind. III orf 7
Platform for the generation of cinnamycin variants In-frame deletion of cin. A generated in S. cinnamoneus cin. A orf 7 PCR 1. 5 kb Xba. I Afl. II cin. M PCR 1. 5 kb Afl. II cin. X Hind. III Variants produced by introduction of plasmid-borne orf 7 cin. A* Jesus Cortes Eco. RI Hpa. I leader peptide Hind. III orf 7
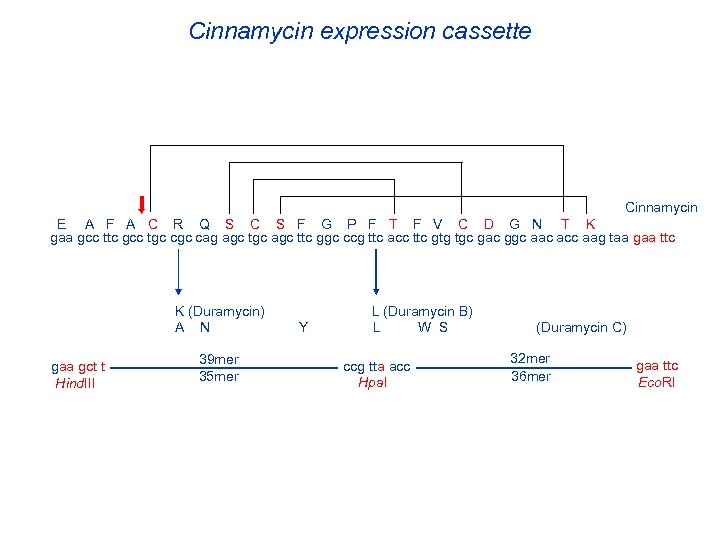 Cinnamycin expression cassette Cinnamycin E A F A C R Q S C S F G P F T F V C D G N T K gaa gcc ttc gcc tgc cag agc tgc agc ttc ggc ccg ttc acc ttc gtg tgc gac ggc aac acc aag taa gaa ttc K (Duramycin) A N gaa gct t Hind. III 39 mer 35 mer Y L (Duramycin B) L W S ccg tta acc Hpa. I (Duramycin C) 32 mer 36 mer gaa ttc Eco. RI
Cinnamycin expression cassette Cinnamycin E A F A C R Q S C S F G P F T F V C D G N T K gaa gcc ttc gcc tgc cag agc tgc agc ttc ggc ccg ttc acc ttc gtg tgc gac ggc aac acc aag taa gaa ttc K (Duramycin) A N gaa gct t Hind. III 39 mer 35 mer Y L (Duramycin B) L W S ccg tta acc Hpa. I (Duramycin C) 32 mer 36 mer gaa ttc Eco. RI
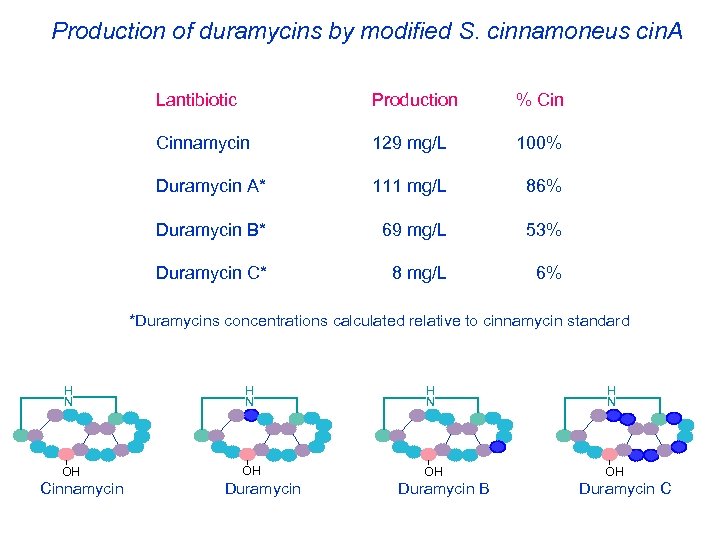 Production of duramycins by modified S. cinnamoneus cin. A Lantibiotic Production % Cinnamycin 129 mg/L 100% Duramycin A* 111 mg/L 86% Duramycin B* 69 mg/L 53% Duramycin C* 8 mg/L 6% *Duramycins concentrations calculated relative to cinnamycin standard H N H N OH OH Cinnamycin Duramycin B Duramycin C
Production of duramycins by modified S. cinnamoneus cin. A Lantibiotic Production % Cinnamycin 129 mg/L 100% Duramycin A* 111 mg/L 86% Duramycin B* 69 mg/L 53% Duramycin C* 8 mg/L 6% *Duramycins concentrations calculated relative to cinnamycin standard H N H N OH OH Cinnamycin Duramycin B Duramycin C
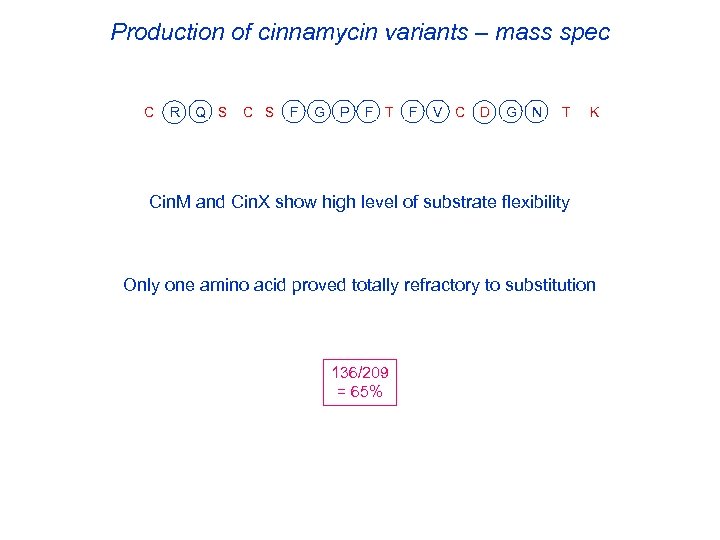 Production of cinnamycin variants – mass spec C R Q S C S F G P F T F V C D G N T K Cin. M and Cin. X show high level of substrate flexibility Only one amino acid proved totally refractory to substitution 136/209 = 65%
Production of cinnamycin variants – mass spec C R Q S C S F G P F T F V C D G N T K Cin. M and Cin. X show high level of substrate flexibility Only one amino acid proved totally refractory to substitution 136/209 = 65%
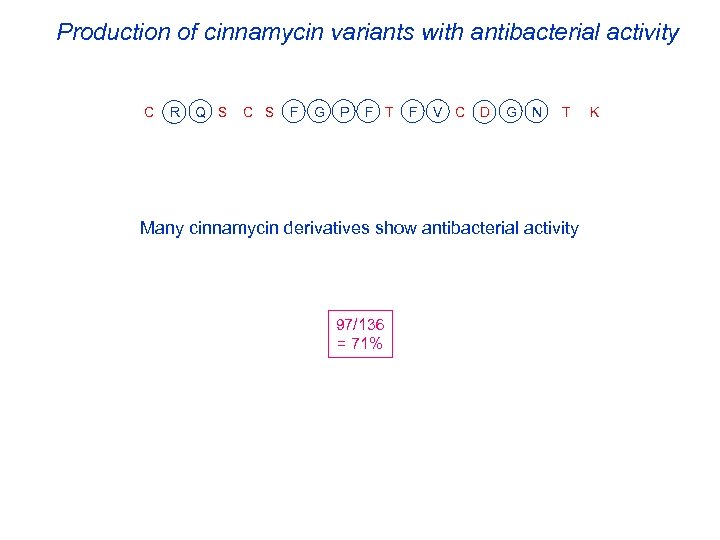 Production of cinnamycin variants with antibacterial activity C R Q S C S F G P F T F V C D G N T K Many cinnamycin derivatives show antibacterial activity 97/136 = 71%
Production of cinnamycin variants with antibacterial activity C R Q S C S F G P F T F V C D G N T K Many cinnamycin derivatives show antibacterial activity 97/136 = 71%
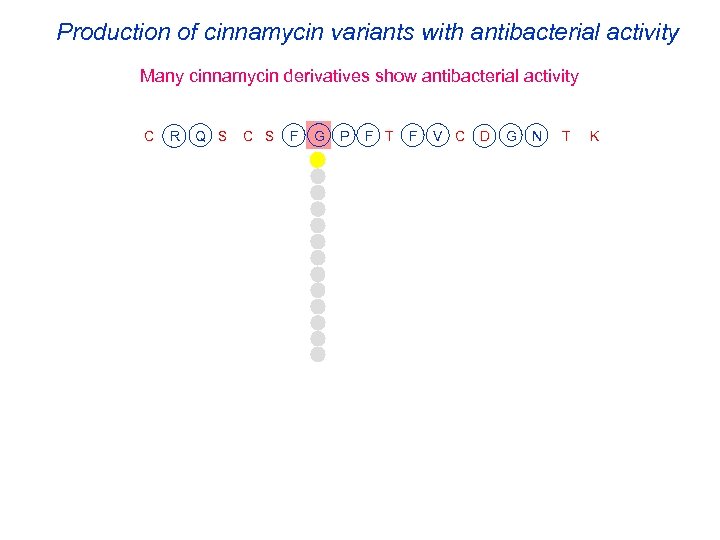 Production of cinnamycin variants with antibacterial activity Many cinnamycin derivatives show antibacterial activity C R Q S C S F G P F T F V C D G N T K
Production of cinnamycin variants with antibacterial activity Many cinnamycin derivatives show antibacterial activity C R Q S C S F G P F T F V C D G N T K
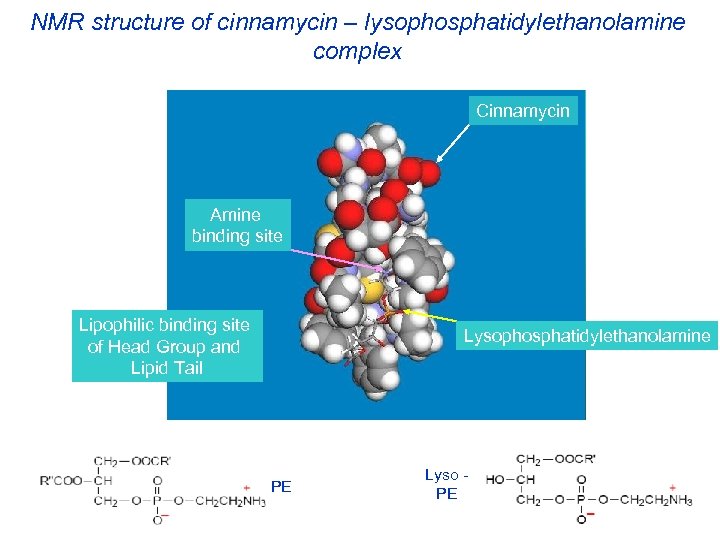 NMR structure of cinnamycin – lysophosphatidylethanolamine complex Cinnamycin Amine binding site Lipophilic binding site of Head Group and Lipid Tail Lysophosphatidylethanolamine PE Lyso PE
NMR structure of cinnamycin – lysophosphatidylethanolamine complex Cinnamycin Amine binding site Lipophilic binding site of Head Group and Lipid Tail Lysophosphatidylethanolamine PE Lyso PE
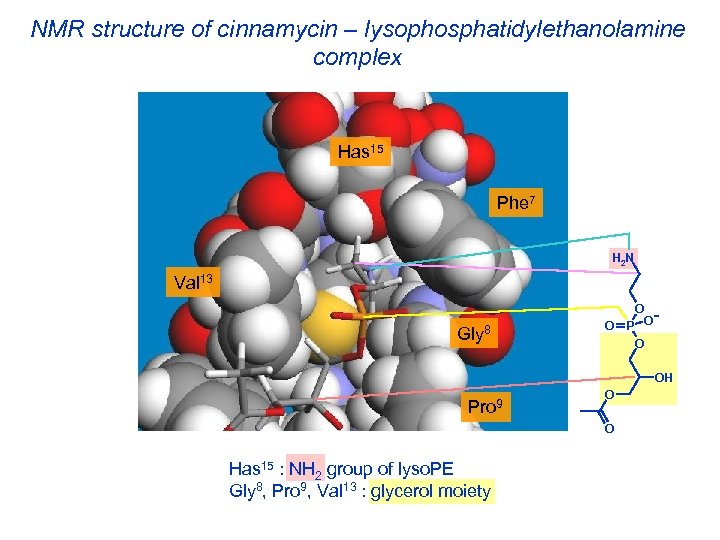 NMR structure of cinnamycin – lysophosphatidylethanolamine complex Has 15 Phe 7 H 2 N Val 13 Gly 8 O O P O O OH Pro 9 O O Has 15 : NH 2 group of lyso. PE Gly 8, Pro 9, Val 13 : glycerol moiety
NMR structure of cinnamycin – lysophosphatidylethanolamine complex Has 15 Phe 7 H 2 N Val 13 Gly 8 O O P O O OH Pro 9 O O Has 15 : NH 2 group of lyso. PE Gly 8, Pro 9, Val 13 : glycerol moiety
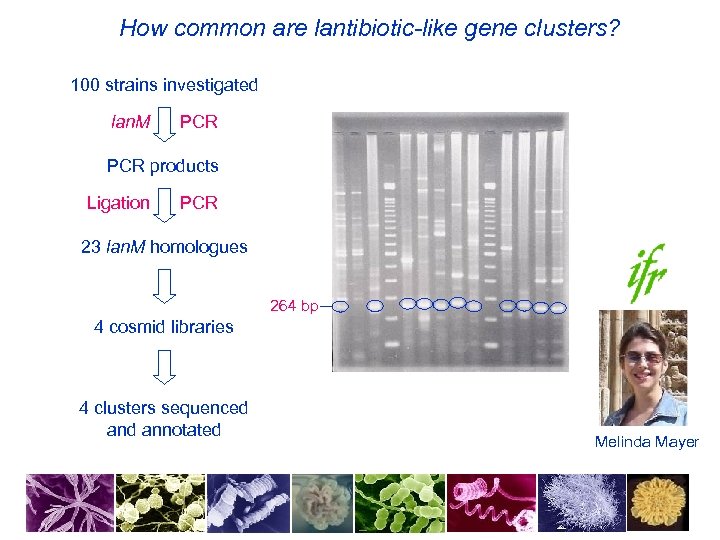 How common are lantibiotic-like gene clusters? 100 strains investigated lan. M PCR products Ligation PCR 23 lan. M homologues 264 bp 4 cosmid libraries 4 clusters sequenced annotated Melinda Mayer
How common are lantibiotic-like gene clusters? 100 strains investigated lan. M PCR products Ligation PCR 23 lan. M homologues 264 bp 4 cosmid libraries 4 clusters sequenced annotated Melinda Mayer
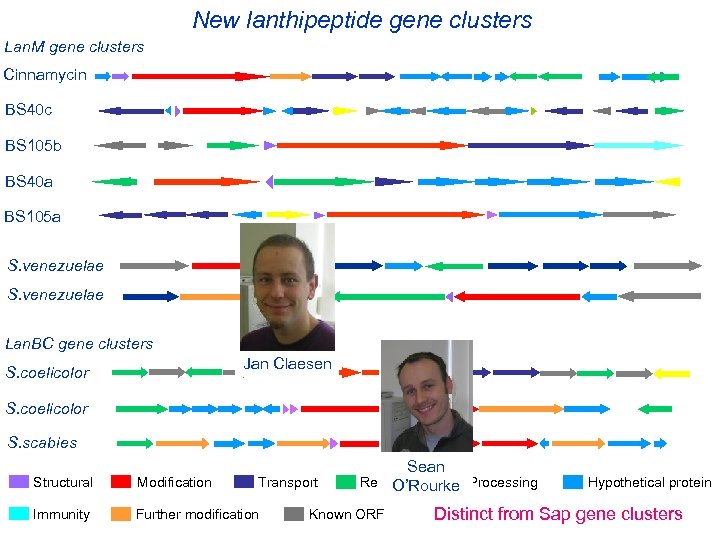 New lanthipeptide gene clusters Lan. M gene clusters Cinnamycin BS 40 c BS 105 b BS 40 a BS 105 a S. venezuelae Lan. BC gene clusters Jan Claesen S. coelicolor S. scabies Structural Modification Transport Immunity Further modification Sean Regulation O’Rourke Processing Known ORF Hypothetical protein Distinct from Sap gene clusters
New lanthipeptide gene clusters Lan. M gene clusters Cinnamycin BS 40 c BS 105 b BS 40 a BS 105 a S. venezuelae Lan. BC gene clusters Jan Claesen S. coelicolor S. scabies Structural Modification Transport Immunity Further modification Sean Regulation O’Rourke Processing Known ORF Hypothetical protein Distinct from Sap gene clusters
 Many thanks to Colin Smith (spotted micro-arrays) and Andy Hesketh (Affymetrix – rel. A analysis) for providing slides
Many thanks to Colin Smith (spotted micro-arrays) and Andy Hesketh (Affymetrix – rel. A analysis) for providing slides


