37183aa998605faa15a8eaa1323e2731.ppt
- Количество слайдов: 22

Functional Consequences of the SNPs : BRCA 1, Membrane Transporters and More Nebojsa Mirkovic, Carles Ferrer-Costa, Marc A. Marti-Renom, Alvaro N. A. Monteiro, Andrej Sali
Single Nucleotide Polymorphisms (SNPs) • single base pair replacements of appreciable allelic frequency in the population ( >1% ); • predominant form of human genetic variation (90%); • number predicted to range from one to several million per genome; predicted frequency: 1/1000 bp; • predicted number per gene: 4 -12 on average (limited datasets); • classification by genomic location: nc. SNPs, c. SNPs (synonymous, non-synonymous: 24, 000 -40, 000 per genome).
Significance of SNP Analysis • identify gene(s) that underlie numerous genetic disorders and multifactorial traits (eg, pharmacological response); • SNPs are probably the biggest class of pathogenic changes in the human genome (coding and regulatory regions); • several genetic variants connected to genetic disorders (E 4 allele of APOE with Alzheimer disease, FV Leiden allele with -venuos thrombosis, and CCR 532 with resistance to HIV infection); • numerous indirect evidence of functional impact; • markers of choice in genetic studies. deep
Identification of Sequence Variants in Genes of Interest Br Br 42 * Br 32

Project Goal What is the likelihood that a given SNP destroys the function of a protein? Approach • design a classification function that can predict functional impact of a particular SNP and relies on a combination of sequence, structure and genetic properties from a well-characterized set of ns. SNPs; • build Mod. SNP, a structural database of SNPs, containing protein structure models for all known ns. SNPs; • apply the classification function to a number of specific examples and to all the SNPs in Mod. SNP.

BRCA 1 Project
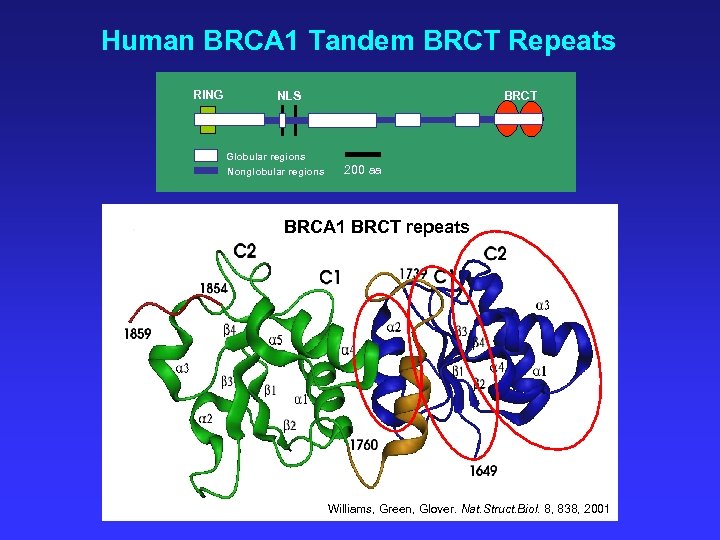
Human BRCA 1 Tandem BRCT Repeats RING BRCT NLS Globular regions Nonglobular regions 200 aa BRCA 1 BRCT repeats Williams, Green, Glover. Nat. Struct. Biol. 8, 838, 2001
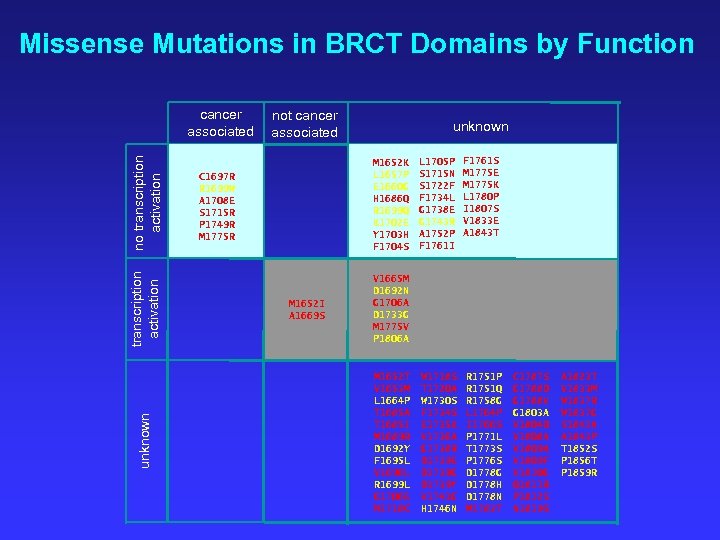
Missense Mutations in BRCT Domains by Function unknown transcription activation no transcription activation cancer associated not cancer associated unknown M 1652 K L 1657 P E 1660 G H 1686 Q R 1699 Q K 1702 E Y 1703 H F 1704 S C 1697 R R 1699 W A 1708 E S 1715 R P 1749 R M 1775 R M 1652 I A 1669 S L 1705 P S 1715 N S 1722 F F 1734 L G 1738 E G 1743 R A 1752 P F 1761 I F 1761 S M 1775 E M 1775 K L 1780 P I 1807 S V 1833 E A 1843 T V 1665 M D 1692 N G 1706 A D 1733 G M 1775 V P 1806 A M 1652 T V 1653 M L 1664 P T 1685 A T 1685 I M 1689 R D 1692 Y F 1695 L V 1696 L R 1699 L G 1706 E W 1718 C W 1718 S T 1720 A W 1730 S F 1734 S E 1735 K V 1736 A G 1738 R D 1739 E D 1739 G D 1739 Y V 1741 G H 1746 N R 1751 P R 1751 Q R 1758 G L 1764 P I 1766 S P 1771 L T 1773 S P 1776 S D 1778 G D 1778 H D 1778 N M 1783 T C 1787 S G 1788 D G 1788 V G 1803 A V 1804 D V 1808 A V 1809 F V 1810 G Q 1811 R P 1812 S N 1819 S A 1823 T V 1833 M W 1837 R W 1837 G S 1841 N A 1843 P T 1852 S P 1856 T P 1859 R
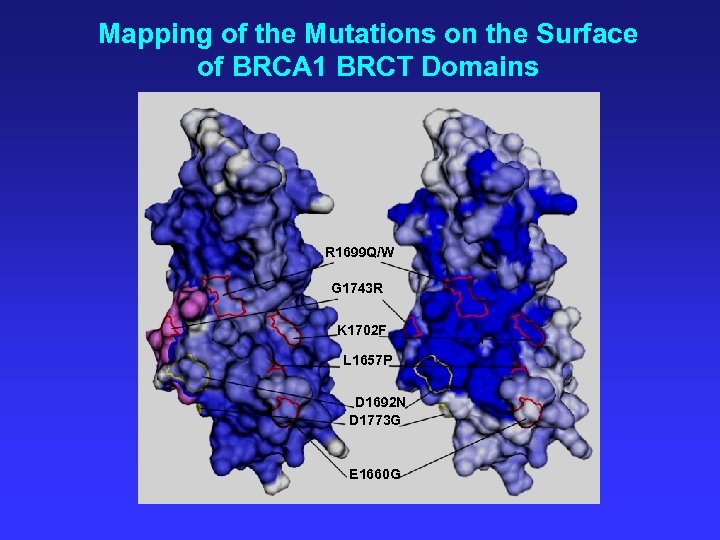
Mapping of the Mutations on the Surface of BRCA 1 BRCT Domains R 1699 Q/W G 1743 R K 1702 F L 1657 P D 1692 N D 1773 G E 1660 G
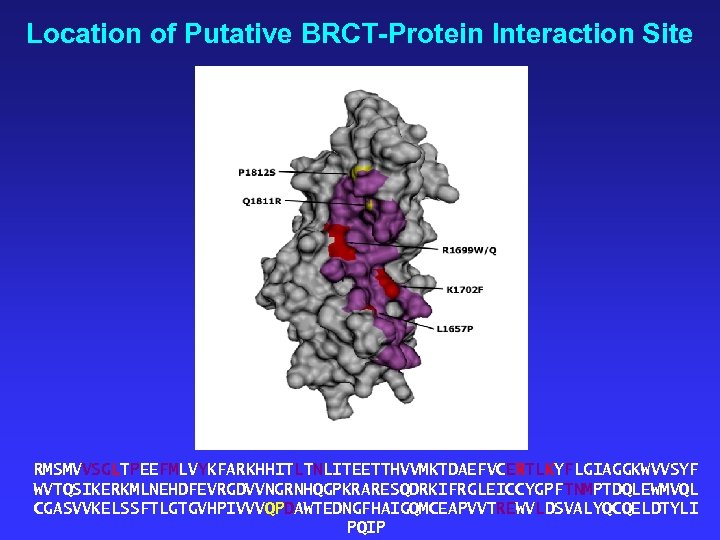
Location of Putative BRCT-Protein Interaction Site RMSMVVSGLTPEEFMLVYKFARKHHITLTNLITEETTHVVMKTDAEFVCERTLKYFLGIAGGKWVVSYF WVTQSIKERKMLNEHDFEVRGDVVNGRNHQGPKRARESQDRKIFRGLEICCYGPFTNMPTDQLEWMVQL CGASVVKELSSFTLGTGVHPIVVVQPDAWTEDNGFHAIGQMCEAPVVTREWVLDSVALYQCQELDTYLI PQIP
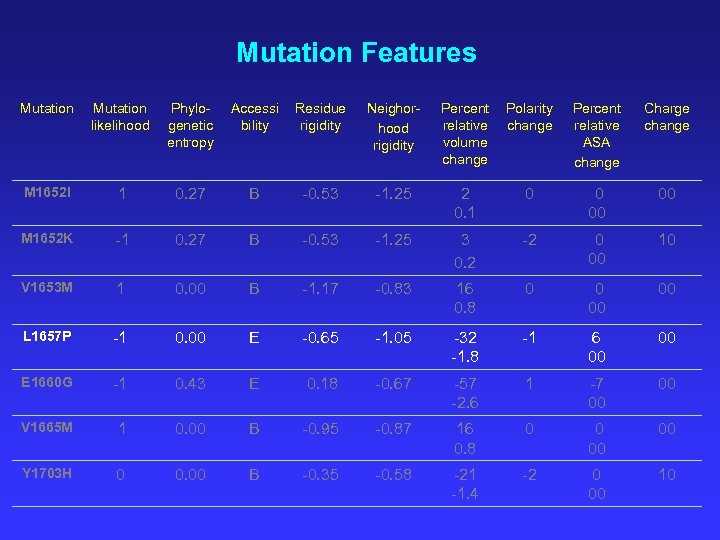
Mutation Features Mutation likelihood Phylogenetic entropy Accessi bility Residue rigidity Neighorhood rigidity Percent relative volume change Polarity change Percent relative ASA change Charge change M 1652 I 1 0. 27 B -0. 53 -1. 25 2 0. 1 0 0 00 00 M 1652 K -1 0. 27 B -0. 53 -1. 25 3 0. 2 -2 0 00 10 V 1653 M 1 0. 00 B -1. 17 -0. 83 16 0. 8 0 0 00 00 L 1657 P -1 0. 00 E -0. 65 -1. 05 -32 -1. 8 -1 6 00 00 E 1660 G -1 0. 43 E 0. 18 -0. 67 -57 -2. 6 1 -7 00 00 V 1665 M 1 0. 00 B -0. 95 -0. 87 16 0. 8 0 0 00 00 Y 1703 H 0 0. 00 B -0. 35 -0. 58 -21 -1. 4 -2 0 00 10
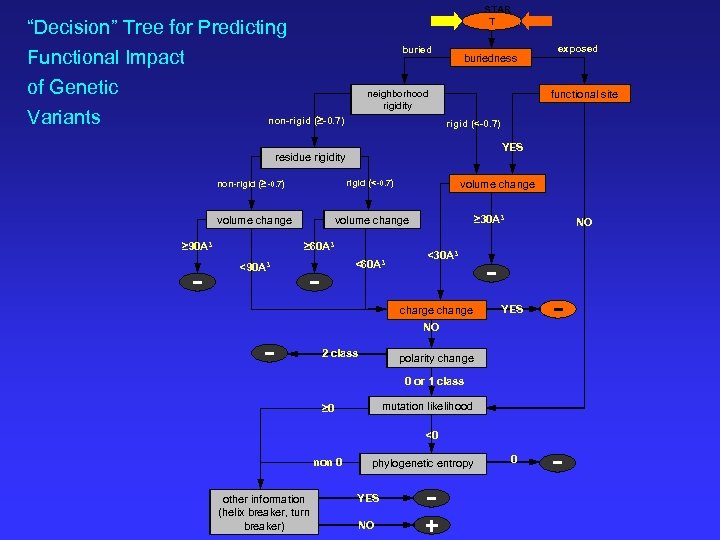
STAR T “Decision” Tree for Predicting Functional Impact of Genetic Variants buriedness neighborhood rigidity non-rigid (≥-0. 7) functional site rigid (<-0. 7) YES residue rigidity non-rigid (≥-0. 7) rigid (<-0. 7) volume change ≥ 90 A 3 - volume change ≥ 30 A 3 volume change ≥ 60 A 3 <90 A 3 <60 A 3 - <30 A 3 charge change - exposed NO YES - 0 - NO 2 class polarity change 0 or 1 class mutation likelihood ≥ 0 <0 non 0 other information (helix breaker, turn breaker) phylogenetic entropy YES NO - +
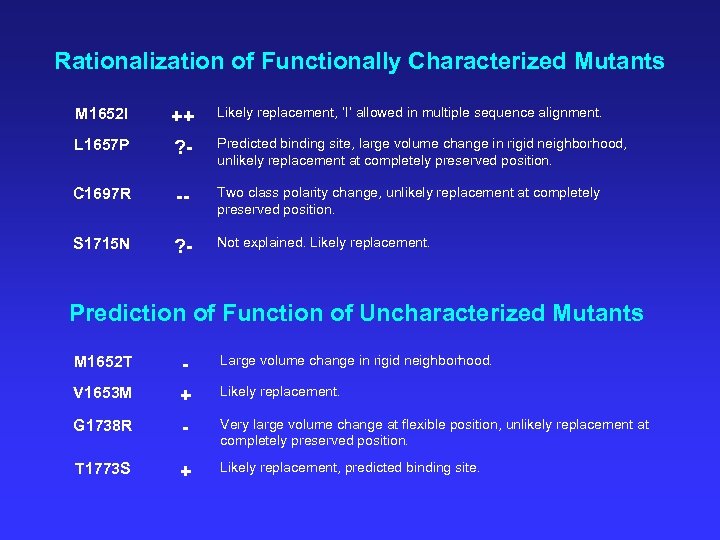
Rationalization of Functionally Characterized Mutants M 1652 I ++ Likely replacement, ‘I’ allowed in multiple sequence alignment. L 1657 P ? - Predicted binding site, large volume change in rigid neighborhood, unlikely replacement at completely preserved position. C 1697 R -- Two class polarity change, unlikely replacement at completely preserved position. S 1715 N ? - Not explained. Likely replacement. Prediction of Function of Uncharacterized Mutants M 1652 T - Large volume change in rigid neighborhood. V 1653 M + Likely replacement. G 1738 R - Very large volume change at flexible position, unlikely replacement at completely preserved position. T 1773 S + Likely replacement, predicted binding site.
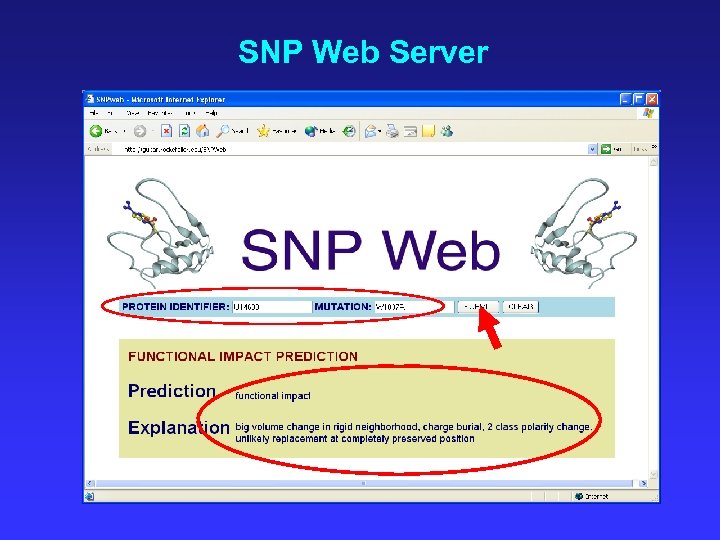
SNP Web Server

Membrane Transporter Project
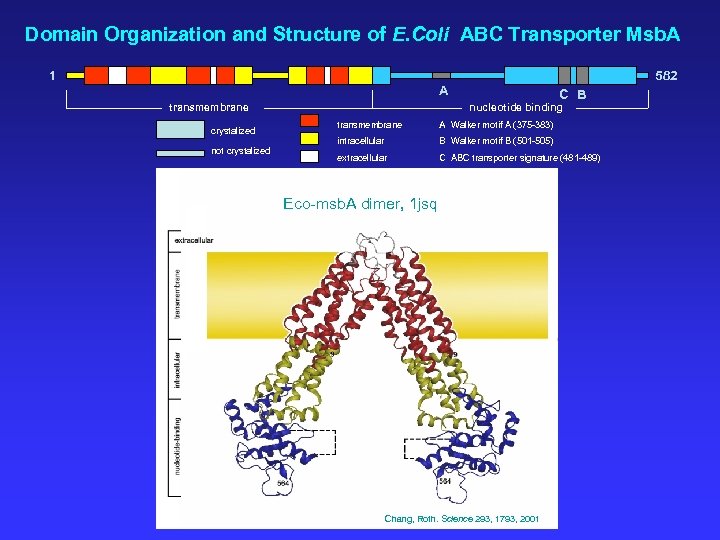
Domain Organization and Structure of E. Coli ABC Transporter Msb. A 1 582 A transmembrane crystalized not crystalized C B nucleotide binding transmembrane A Walker motif A (375 -383) intracellular B Walker motif B (501 -505) extracellular C ABC transporter signature (481 -489) Eco-msb. A dimer, 1 jsq Chang, Roth. Science 293, 1793, 2001
PMT Database • a database of in-site detected and/or verified polymorphisms in various mammalian membrane transporters with ample population genetics data (allelic frequencies, ethnic distribution); • 59 proteins divided in two groups (24 and 25 proteins respectively) depending on the SNP-detection approach; • 10 ABC transporters, 5 with SNPs reported so far (BSEP, MDR 1, MDR 3, MRP 1, MRP 2).
http: //guitar. rockefeller. edu/modbase Pieper et al. , Nucl. Acids Res. 2002.
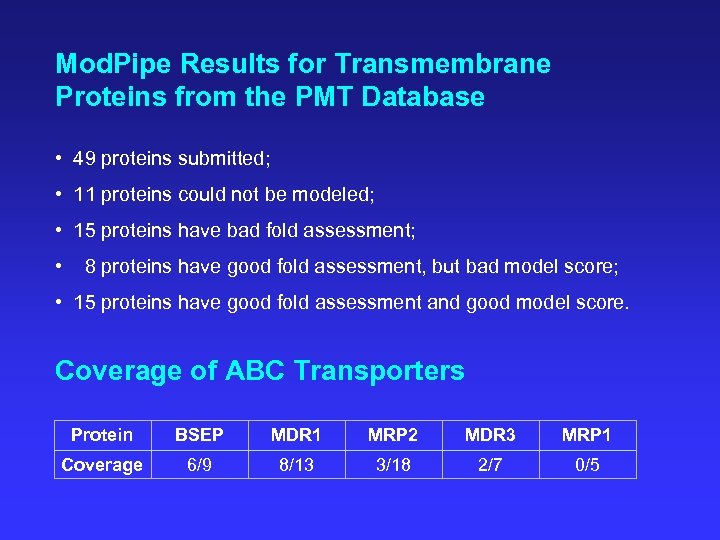
Mod. Pipe Results for Transmembrane Proteins from the PMT Database • 49 proteins submitted; • 11 proteins could not be modeled; • 15 proteins have bad fold assessment; • 8 proteins have good fold assessment, but bad model score; • 15 proteins have good fold assessment and good model score. Coverage of ABC Transporters Protein BSEP MDR 1 MRP 2 MDR 3 MRP 1 Coverage 6/9 8/13 3/18 2/7 0/5
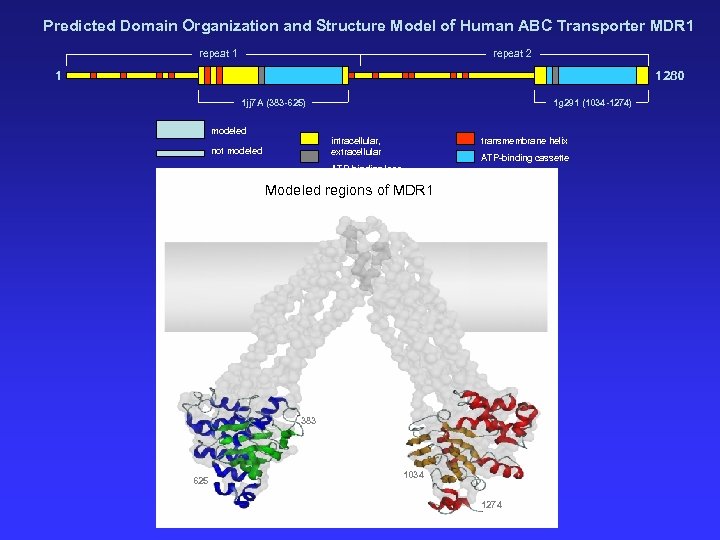
Predicted Domain Organization and Structure Model of Human ABC Transporter MDR 1 repeat 2 1 1280 1 jj 7 A (383 -625) 1 g 291 (1034 -1274) modeled intracellular, extracellular not modeled transmembrane helix ATP-binding cassette ATP-binding loop Modeled regions of MDR 1 383 625 1034 1274
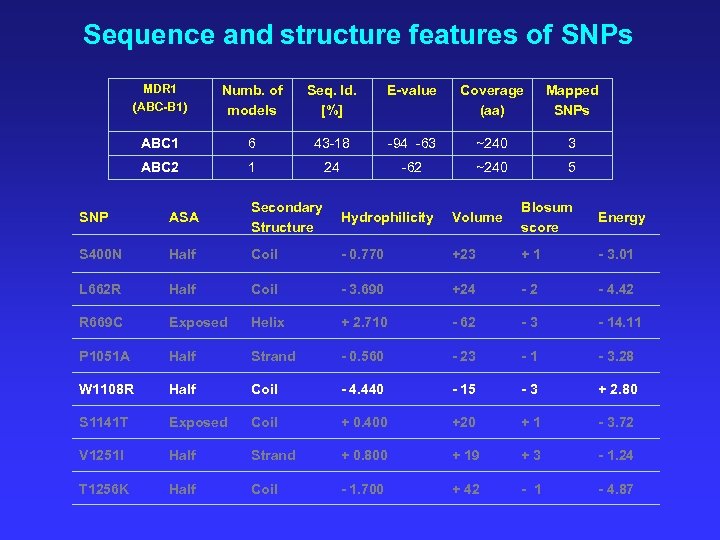
Sequence and structure features of SNPs MDR 1 (ABC-B 1) Numb. of models Seq. Id. [%] E-value Coverage (aa) Mapped SNPs ABC 1 6 43 -18 -94 -63 ~240 3 ABC 2 1 24 -62 ~240 5 SNP ASA Secondary Structure Hydrophilicity Volume Blosum score Energy S 400 N Half Coil - 0. 770 +23 +1 - 3. 01 L 662 R Half Coil - 3. 690 +24 -2 - 4. 42 R 669 C Exposed Helix + 2. 710 - 62 -3 - 14. 11 P 1051 A Half Strand - 0. 560 - 23 -1 - 3. 28 W 1108 R Half Coil - 4. 440 - 15 -3 + 2. 80 S 1141 T Exposed Coil + 0. 400 +20 +1 - 3. 72 V 1251 I Half Strand + 0. 800 + 19 +3 - 1. 24 T 1256 K Half Coil - 1. 700 + 42 - 1 - 4. 87
Significance • a comprehensive structural study of the functional impact of mutations focused on a particular protein family; • a large and informative mutation set used; • rationalization of the characterized mutation set used to develop a predictive tool; • possible contribution to genetic studies (eg, candidate gene approach) and medical practice (with other methods).
37183aa998605faa15a8eaa1323e2731.ppt